In mammals, the histone H1 family includes five somatic replication-dependent (H1.1–H1.5) and two replication-independent (H1.10 and H1.0) subtypes. Izzo et al. analyze the contributions of all somatic H1 subtypes to the chromatin landscape during reprogramming in preimplantation embryo and primordial germ cell development.
Abstract
In mammals, histone H1 consists of a family of related proteins, including five replication-dependent (H1.1–H1.5) and two replication-independent (H1.10 and H1.0) subtypes, all expressed in somatic cells. To systematically study the expression and function of H1 subtypes, we generated knockin mouse lines in which endogenous H1 subtypes are tagged. We focused on key developmental periods when epigenetic reprogramming occurs: early mouse embryos and primordial germ cell development. We found that dynamic changes in H1 subtype expression and localization are tightly linked with chromatin remodeling and might be crucial for transitions in chromatin structure during reprogramming. Although all somatic H1 subtypes are present in the blastocyst, each stage of preimplantation development is characterized by a different combination of H1 subtypes. Similarly, the relative abundance of somatic H1 subtypes can distinguish male and female chromatin upon sex differentiation in developing germ cells. Overall, our data provide new insights into the chromatin changes underlying epigenetic reprogramming. We suggest that distinct H1 subtypes may mediate the extensive chromatin remodeling occurring during epigenetic reprogramming and that they may be key players in the acquisition of cellular totipotency and the establishment of specific cellular states.
Introduction
Linker histone H1 is a key regulator of chromatin organization and function. Higher-order chromatin structures are formed through the binding of histone H1 to the nucleosomal core particle and to the linker DNA entering and exiting the nucleosome core (Allan et al., 1980; Syed et al., 2010). Higher eukaryotes contain a variable number of H1 proteins, often referred to as subtypes or variants. In the mouse, 11 H1 subtypes have been identified, of which 7 (H1.1/H1a, H1.2/H1c, H1.3/H1d, H1.4/H1e, H1.5/H1b, H1.0, and H1.10/H1.x) have been classified as being primarily expressed in somatic cells, and the remaining four subtypes are thought to be mainly present in specific differentiated cell types. However, a systematic analysis of the expression of all mouse H1 subtypes in different cell types or tissues is still missing.
The mouse H1 subtypes H1.1, H1.2, H1.3, H1.4, and H1.5 are preferentially transcribed and synthesized in S-phase, whereas H1.0 and H1.10 are expressed throughout the cell cycle (Kamakaka and Biggins, 2005; Izzo et al., 2008). The amino acid sequence of individual H1 subtypes is conserved between species but is more divergent between individual subtypes, suggesting that H1 subtypes have acquired specific functions during evolution (Ponte et al., 1998). However, knockout studies of individual H1 subtypes in mice have failed to reveal any obvious phenotype, which might be a result of compensatory mechanisms, such as up-regulation of other H1 subtypes (Fan et al., 2001). A careful analysis of H1 depletion in several organisms and cell lines showed that specific H1 subtypes are indeed involved in the up- and down-regulation of specific genes (Shen and Gorovsky, 1996; Alami et al., 2003). Moreover, H1 subtypes are subject to a wide variety of posttranslational modifications, which can confer additional specific functions to individual subtypes (Garcia et al., 2004; Izzo and Schneider, 2015). Additionally, H1 subtypes differ in their ability to condense nucleosomes in vitro as well as in their affinity for chromatin in vivo (Liao and Cole, 1981; Th’ng et al., 2005). In agreement with this, H1 subtypes display differences in their localization between active and inactive chromatin and might have a role in nuclear architecture (Cao et al., 2013; Izzo et al., 2013).
Changes in chromatin organization occur during the development of multicellular organisms. The transitions in cellular identity are accompanied by distinctive structural and functional alterations of chromatin architecture. In particular, epigenetic reprogramming refers to a genome-wide removal of chromatin modifications that resets a differentiated state into a more plastic state (Hemberger et al., 2009). In mammals, epigenetic reprogramming occurs twice during the life cycle: first, upon fertilization of the oocyte by the sperm, when both the maternal and paternal genomes undergo extensive chromatin reorganization processes (Hajkova, 2010; Burton and Torres-Padilla, 2014), and second, during the development of the embryonic germ line, in primordial germ cells (PGCs; Seki et al., 2007; Hajkova et al., 2008). Nascent PGCs are derived from pluripotent postimplantation epiblast cells. To enable the generation of gametes, the epigenome of PGCs needs to be reset (Surani et al., 2007).
Although in recent years our mechanistic understanding of epigenetic reprogramming and germ line formation has improved, major aspects remain unresolved. In particular, the contribution of histone H1 and its somatic subtypes to reprogramming and subsequent differentiation has not been addressed. Here we provide the first systematic study of all somatic H1 subtypes and analyze their contribution to the chromatin landscape during the two major reprogramming events in the mammalian life cycle, the preimplantation embryo and primordial germ cell development. Our results show that the expression of H1 subtypes is highly dynamic during epigenetic reprogramming and that specific changes in H1 subtype localization are temporally linked with reorganization of chromatin architecture. By focusing on the fifth histone, the linker histone H1, the data presented here advance our understanding of the chromatin changes underlying epigenetic reprogramming and suggest a possible contribution of H1 subtypes to the molecular mechanisms responsible for resetting the epigenome. The knockin mouse strains we have generated here represent an important tool for understanding the specific role of H1 subtypes in chromatin dynamics in vivo and can be used, for example, for future mapping and developmental studies.
Results and discussion
Generation of somatic Flag-HA-H1 knockin mice
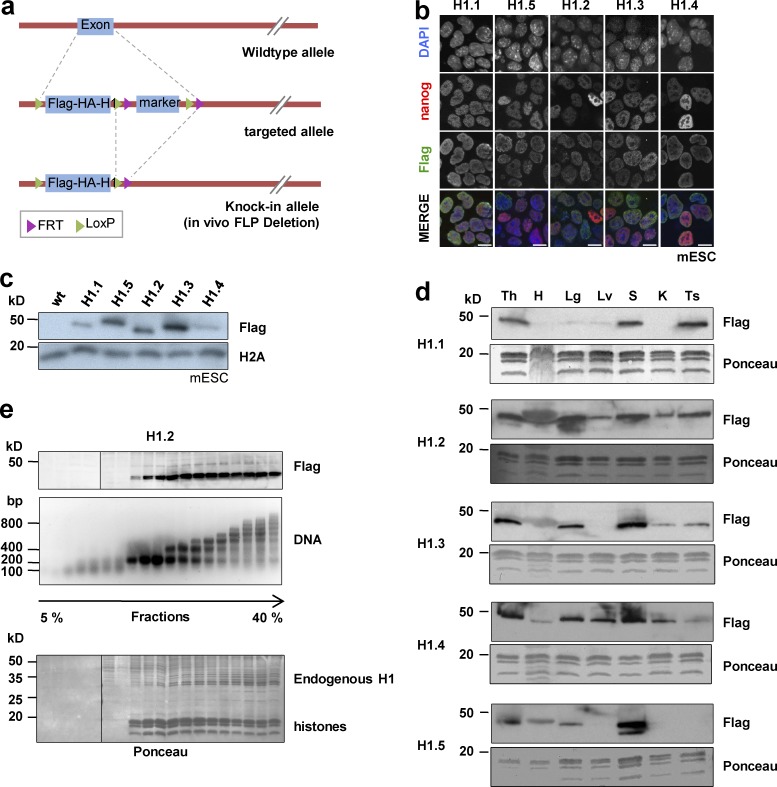
To overcome the lack of reliable H1 subtype–specific antibodies as well as issues with epitope exclusion for the detection of specific H1 subtypes caused by, for example, covalent modifications of H1, we generated knockin mice in which individual somatic H1 subtypes were replaced by the corresponding N-terminally Flag-2xHA–tagged versions. We implemented targeting strategies for all the replication-dependent somatic H1 subtypes (H1.1, H1.2, H1.3, H1.4, and H1.5) at their endogenous loci through homologous recombination in embryonic stem cells (ESCs; Fig. 1 a). Importantly, in this approach, H1 subtypes remain expressed under the control of their endogenous promoters, eliminating the possibility of adverse effects on chromatin structure associated with histone overexpression or compensation between subtypes (Gunjan et al., 1999; Groth et al., 2007). We used N-terminal tagging of H1 because, in contrast to C-terminal tagging, it has been shown to have only minimal effects on chromatin binding of H1 (Th’ng et al., 2005). Homozygous Flag-HA-H1 mice had no overt morphological or physiological phenotype. Initially, we performed immunostaining and immunoblot analyses in knockin ESC lines. Analyses showed that all five H1 somatic subtypes are present in ESCs and correctly localize into the nucleus (Fig. 1, b and c). We confirmed that the tagged H1 subtypes are expressed in a panel of mouse organs, in agreement with previous analyses at both mRNA and protein levels (Fan et al., 2001, 2003; Wisniewski et al., 2007; Medrzycki et al., 2012; Fig. 1 d). We found tissue-specific enrichments of H1 subtypes: for example, whereas H1.2 and H1.4 are present with slightly different abundance in all tissues analyzed, H1.1 is detectable mainly in thymus, spleen, and testis. H1.3 is highly abundant in thymus, lung, and spleen, whereas H1.5 is highly expressed in thymus and spleen. Furthermore, we verified by micrococcal nuclease (Mnase) digestion and sedimentation analysis that each tagged H1 subtype is incorporated into nucleosomes (Fig. 1 e).
Figure 1.
Generation and characterization of Flag-HA-H1 knockin mouse lines. (a) Schematic of the strategy used to generate the Flag-HA-H1 knockin mice. (b) Immunostaining of the ESC lines used to generate the corresponding Flag-HA-H1 knockin mice with anti-Flag antibodies (green). NANOG staining (red) was used as a positive control. DNA was counterstained with DAPI (blue). Bars, 10 µm. (c) Immunoblot analysis of the expression of the indicated Flag-HA-H1 subtypes (H1.1–H1.5) in total extracts from the corresponding knockin ESC lines using anti-Flag antibody (Flag). H2A was used as loading control. (d) Expression of somatic Flag-HA-H1 subtypes (H1.1–H1.5) in several organs (Th, thymus; H, heart; Lg, lung; Lv, liver; S, spleen; K, kidney; and Ts, testis) from the corresponding Flag-HA-H1 knockin mice detected using anti-Flag antibody (Flag). Ponceau is shown as loading control. (e) Flag-HA–tagged H1 subtypes are incorporated into nucleosomes. Nuclei purified from the Flag-HA-H1.2 knockin cells were partially digested with Mnase followed by sedimentation of the digested chromatin over a 5–40% sucrose gradient. The gradient fractions were analyzed by immunoblot with anti-Flag antibody (Flag) and by agarose gel electrophoresis followed by ethidium bromide staining (DNA). The Ponceau staining of the membrane is shown, and the running positions of endogenous H1 and histones are indicated. Note that the same results were obtained for all H1 subtypes.
Somatic H1 subtypes are present in gonadal PGCs before germline reprogramming
To study the dynamics of H1 expression during germline reprogramming, we first assessed the presence, abundance, and localization of H1 subtypes in gonadal PGCs before the epigenetic reprogramming window, which occurs at approximately embryonic day 11.5 (E11.5; Hajkova et al., 2008). For this, we stained sections of genital ridges from embryos of each Flag-HA-H1 mouse line at E10.5 (Figs. S1 and S2) with an anti-Flag antibody and identified germ cells with an anti-OCT4 specific antibody. To analyze H1.0 and H1.10 distribution, we used H1.0 and H1.10 specific antibodies (Fig. S2 d). By comparing the signals and their intensity in OCT4-positive and -negative cells, we found that the distribution and levels of H1.1–H1.5 and H1.10 subtypes in PGCs were similar to those of surrounding somatic cells at E10.5, before gonadal reprogramming (Figs. S1 and S2). Note that H1.0 expression was undetectable in germ cells at all the developmental stages examined (Fig. S2 c).
Somatic H1 subtypes are transiently lost from PGC chromatin during epigenetic reprogramming with different temporal dynamics
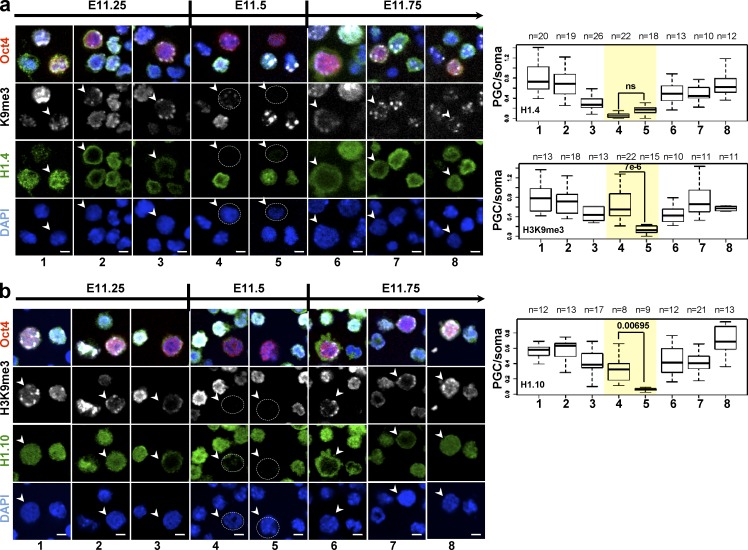
To obtain specific information on the expression of specific H1 subtypes during epigenetic reprogramming, we next investigated the H1 subtype presence and localization in PGCs at ∼E11.5, when chromatin undergoes major epigenetic reprogramming (Hajkova et al., 2008). It has been previously shown that the gonadal reprogramming is associated with changes in numerous histone modifications. In this context, we were interested in the dynamics of H1 subtypes in comparison to the previously reported changes in H3K9me3 (Hajkova et al., 2008). We found that all somatic H1 subtypes expressed before reprogramming (H1.1–H1.5 and H1.10) are transiently not detected on chromatin in PGCs at approximately the same time that H3K9me3 becomes undetectable (Figs. 2 and S3). However, the temporal and spatial dynamics of each of the H1 subtypes was different. In early E11.5 PGCs, we observed localization of H1.1–H1.5 and H1.10 at the nuclear periphery, where they formed a ring-like structure close to the nuclear membrane (Figs. 2, S2 e, and S3). Subsequently, the intensity of H1.1–H1.5 and H1.10 signals started to decrease progressively. Interestingly, loss of H1 signal from the chromatin of PGCs seems to precede the loss of H3K9me3 staining for all subtypes (Figs. 2 a and S3) with the exception of H1.10, which seems to dissociate from chromatin with a slower kinetics than the other H1 subtypes (Fig. 2 b). Note that in the neighboring somatic cells the H1 and H3K9me3 signals remain relatively constant across the developmental stages assessed. After reprogramming, in late E11.5 PGCs, we first detected staining for all somatic H1 subtypes in the cytoplasm and subsequently at the nuclear periphery, where also the H3K9me3 signal can be first detected (Figs. 2 and S3).
Figure 2.
H1 subtypes are transiently lost from chromatin during epigenetic reprogramming in mouse germ cells. (a and b) Kinetics of H1.4 (a; green) and H1.10 (b; green) disappearance from PGCs isolated from genital ridges of embryos between E11.25 and E11.75 (according to the TS system; see Materials and methods) and stained with anti-Flag antibody or anti-H1.10 specific antibody. An anti-OCT4 antibody (red) was used as a germ cell–specific marker (arrowheads). Dotted circles indicate PGCs with undetectable H1 signal. Bars, 10 µm. Changes in H1 abundance and localization are shown in comparison to the previously described changes in H3K9me3 pattern (Hajkova et al., 2008). Representative images from at least four different genital ridges from two to three independent litters are shown. The box plots (right) in a and b show quantification of the immunostaining signal for the indicated H1 subtypes and of H3K9me3 in OCT4-positive cells relative to OCT4-negative cells (PGC/somatic cells) during the different phases (1–8) of epigenetic reprogramming. The kinetics of H3K9me3 disappearance is similar for all the H1 subtypes; hence only data for H1.4 is shown. The number (n) of cells analyzed is indicated. To highlight differences in the kinetics of disappearance of H1.10 compared with H1.4 (yellow shaded rectangle), only the p-values for stages 4 and 5 are shown; ns, not significant. Error bars correspond to the minimum and maximum values.
Together, our data show that the expression and localization of all studied H1 subtypes is dynamic during the epigenetic reprogramming in PGCs. Interestingly, our study documents that the germline reprogramming process is connected with the transient loss of all somatic H1 subtypes from germ cell chromatin.
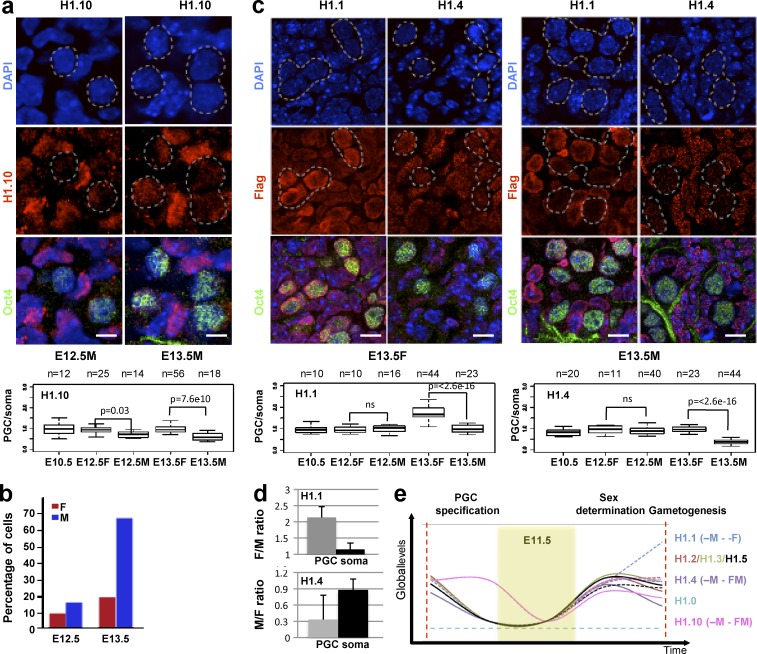
The relative abundance of H1 subtypes distinguishes male and female PGC chromatin after sex differentiation
In mice, the sex differentiation program starts in PGCs at E12.5, after gonadal reprogramming. At this stage, the expression of H1.1–H1.5 in germ cells is very similar to their surrounding somatic cells (Figs. S1 and S2, a–c). Interestingly, H1.10 signal decreased specifically in germ cells starting at E12.5 (Fig. 3 a). Quantification of H1.10 signal in isolated PGCs revealed that H1.10 levels are lower in male cells than in female cells (Fig. 3 b). Later on, at E13.5, the relative composition of H1.1, H1.2, and H1.4 subtypes differs between male and female PGCs: whereas female PGCs have higher levels of H1.1 signal, male PGCs have lower levels of H1.2 and H1.4 (Fig. 3, c and d). No major changes were observed for the other H1 subtypes relative to the surrounding somatic cells (Figs. S1 and S2). This is consistent with available RNA-seq data in PGCs, which shows increased mRNA levels of H1.1 in PGCs in females and lower levels of H1.2 and H1.4 mRNA in PGCs in males at later stages (Seisenberger et al., 2012).
Figure 3.
Male and female mouse germ cells differ in the H1 subtypes they express. (a) Immunofluorescence of cryosections of male genital ridges from E12.5 and E13.5 embryos using anti-H1.10 specific antibody (red). The box plot (bottom) shows the expression of H1.10 in OCT4-positive relative to OCT4-negative cells (PGC/somatic cells [soma]) from E10.5 to E13.5 in both males and females. The p-values for intragroup comparison between males and females were calculated using a pairwise t test with Bonferroni correction; p-values <0.05 were considered statistically significant; ns, not significant. The number (n) of cells analyzed is indicated. Samples from several embryos from at least three independent litters were analyzed. (b) Quantification of the H1.10 signal in isolated germ cells. For each developmental stage, the percentage value indicates the number of PGCs with lower H1.10 expression than that of somatic cells. Numbers of PGCs analyzed from at least four genital ridges from two independent litters were E12.5F, n = 50; E12.5M, n = 52; E13.5F, n = 23; and E13.5M, n = 54. (c) Immunofluorescence of cryosections from male and female genital ridges from Flag-HA-H1.1 and Flag-HA-H1.4 knockin embryos at E13.5 using anti-Flag antibodies (Flag; red). PGCs were identified with anti-OCT4 antibody staining (green). DNA was counterstained with DAPI. Bars, 10 µm. The box plots (bottom) show the quantification of the levels of the indicated H1 subtypes in OCT4-positive cells relative to OCT4-negative cells at the indicated developmental stages. Error bars correspond to the minimum and maximum values. (d) Bar graph showing the relative levels of H1.1 and H1.4 proteins normalized to H3 in isolated PGCs and soma between males and females. The expression of H1.1 and H1.4 was detected by immunoblot analysis using anti-Flag antibody and quantified using Image Lab software (Bio-Rad). Error bars are SD of the mean ratios of two independent experiments for PGCs and three independent experiments for somatic cells. At least 50 genital ridges from four to five independent litters were sorted by FACS in multiple rounds. Sorted cells were kept frozen at −80°C until reaching a minimum of n = 50,000 PGCs, sufficient to generate a signal in Western blot analysis. (e) Summary of the temporal dynamics of the expression of somatic H1 subtypes between E10.5 and E13.5. The mean values (based on the quantification in Figs. 2 and 3) of the ratio intensities in OCT4-positive and -negative cells at each developmental time point are plotted. Continuous and dotted lines of the same color refer to the H1 expression kinetics in female (F) and male (M) PGCs, respectively. The yellow panel highlights the epigenetic reprogramming window, for which only the time of H1 disappearance is shown. The number of PGCs analyzed is the same as in Figs. 2 and 3.
Together, our results reveal pronounced changes in global levels and subtype composition of histone H1 during epigenetic reprogramming in PGCs. Although all H1 subtypes are present upon exit from the reprogramming window in PGCs, our immunostaining and immunoblot data reveal that H1.1 and H1.4 display a different distribution pattern and that their levels of expression are gender specific (Fig. 3 e). Mechanistically, the observed progressive disappearance of H1 subtypes could facilitate a chromatin structure permissive for reprogramming.
Changes in H1 subtypes in preimplantation embryos demarcate embryonic genome activation and transitions in cell plasticity
Having established the different expression kinetics of H1 subtypes during germline development, we next examined the potential changes in histone H1 subtype composition after fertilization. We systematically analyzed H1 expression using preovulatory, fully grown oocytes and mouse embryos at distinct developmental stages, which were stained with an anti-HA antibody in toto. For all immunostainings performed, we included follicular cells of isolated preovulatory follicles as an internal positive control (Figs. 4 and 5). In addition, immunostaining of wild-type oocytes and embryos using anti-HA was used as a negative control (Fig. 5 e).
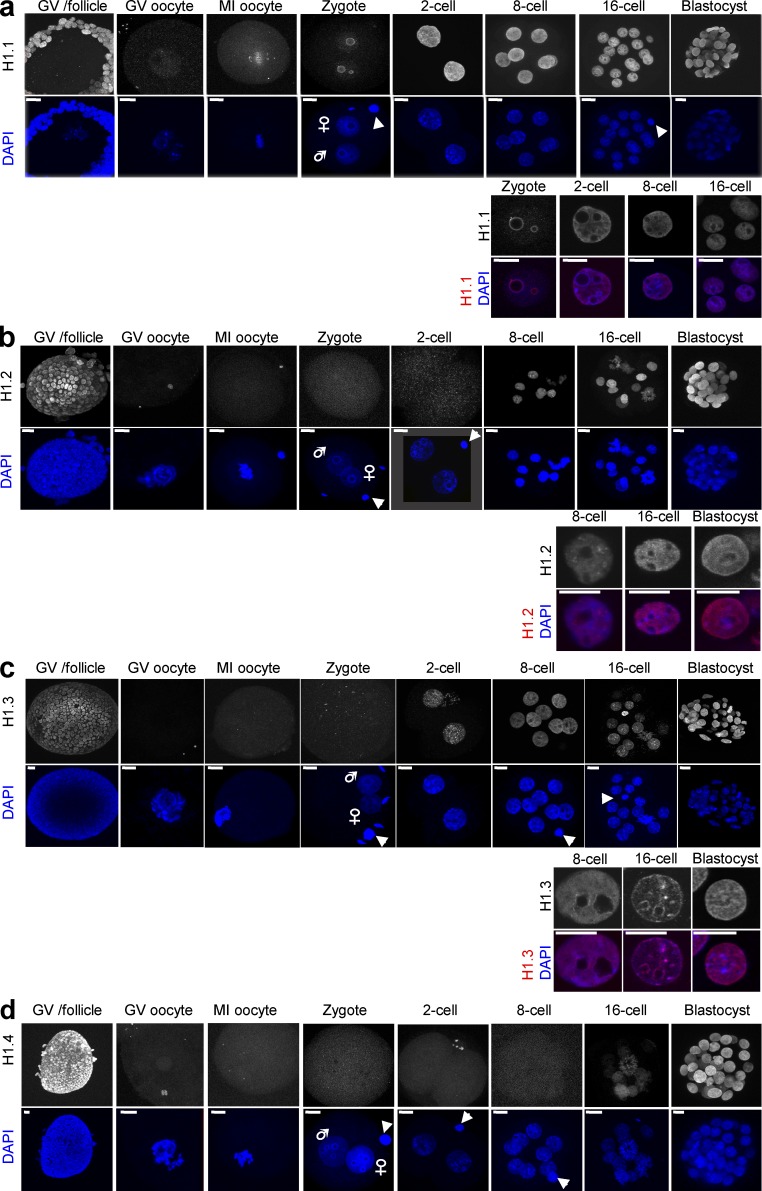
Figure 4.
Dynamics of H1.1, H1.2, and H1.3 subtypes during mouse preimplantation development. Immunofluorescence of embryos at the indicated developmental stages from Flag-HA-H1.1 (a), Flag-HA-H1.2 (b), Flag-HA-H1.3 (c), and Flag-HA-H1.4 (d) knockin mice using an anti-HA antibody (red). DNA was counterstained with DAPI (blue). Top, full projections of confocal z-sections taken every 1 µm. Bottom, higher magnification of a detail of the merge of a corresponding middle section with representative nuclei. Only the stages where H1 expression is detected are shown in the merged images. The arrowheads point to the polar body; male and female pronuclei are indicated. Bars, 10 µm. At least 10 oocytes or embryos per stage were analyzed, across three or more independent experiments. Note that for H1.3, there is some slight variation in the levels between nuclei in 16-cell-stage embryos and blastocysts, but all embryos analyzed displayed accumulation of H1.3.
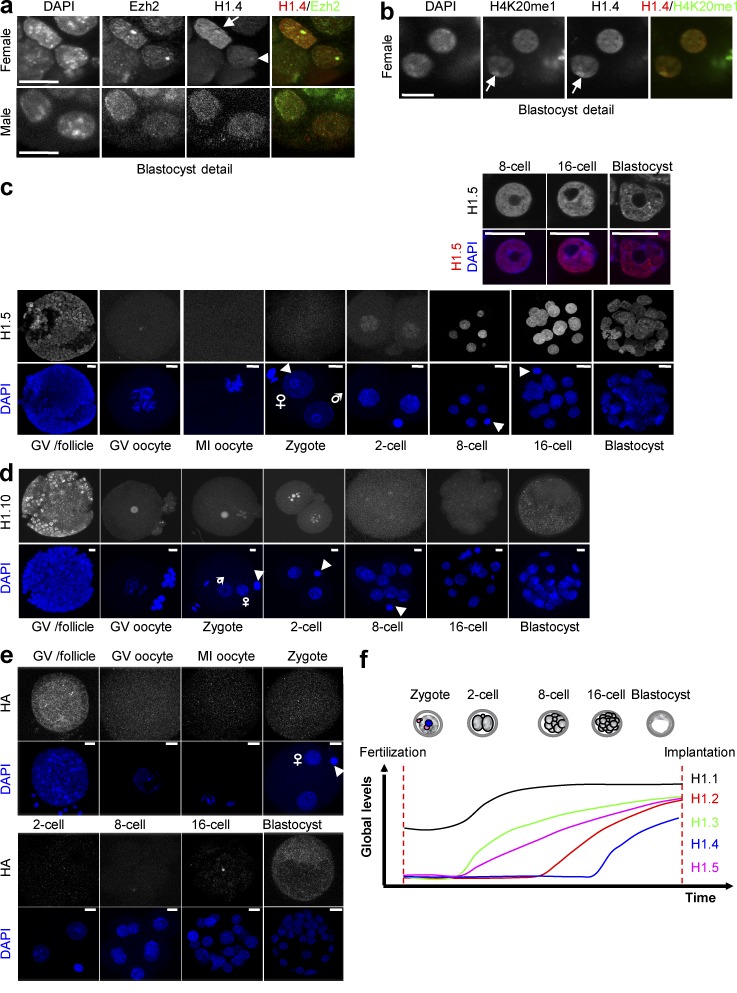
Figure 5.
Dynamics of H1.4 and H1.5 subtypes during mouse preimplantation development. (a) Detail of a merged confocal section of blastocysts stained with anti-HA (red), which recognizes H1.4 and EzH2 antibody (green). Ezh2 marks the inactive X chromosome (Xi) in female embryos (top) but not in males (bottom). (b) Detail of a merged confocal section of a representative female blastocyst stained with anti-HA antibody (red) and H4K20me1 antibody (green). The arrowhead points to Xi. (c) Full projections of confocal z-sections taken every 1 µm of embryos from Flag-HA-H1.5 knockin mice. (Top) Higher magnification of a merged image of representative nuclei taken from middle sections of the corresponding stages. Only the stages in which H1.5 is expressed are shown in the merged panels. The arrowheads point to the polar body; male and female pronuclei are indicated. Bar, 10 µm. At least 10 oocytes or embryos per stage were analyzed across three or more independent experiments. (d and e) Immunofluorescence of WT embryos at the indicated developmental stages with anti-H1.10 antibody (d) and anti-HA specific antibody as negative control (e). DNA was counterstained with DAPI (blue). The arrowheads point to the polar body; male and female pronuclei are indicated. Bar, 10 µm. At least 10 oocytes or embryos per stage were analyzed across three or more independent experiments. (f) Summary of the temporal dynamics of the expression of somatic H1 subtypes from fertilization to the blastocyst stage before implantation. At least 10 oocytes or embryos per stage were analyzed across three or more independent experiments. The panel is based on qualitative observations.
H1.1 staining was detected in the germinal vesicle of preovulatory-stage oocytes, albeit weakly, and remained present on the maternal chromatin during meiotic resumption in MI oocytes (Fig. 4 a, top). After fertilization, H1.1 localized to both pronuclei, where it was enriched in a ring-like pattern surrounding the nucleolar-like bodies (NLBs) that are known to harbor pericentric chromatin (Probst et al., 2007; Fig. 4 a, bottom). The levels of H1.1 were strongly increased from the two-cell stage until the blastocyst (Fig. 4 a). Overall, H1.1 was distributed throughout the nucleoplasm and was neither enriched nor depleted from DAPI-dense regions.
In contrast to H1.1, we did not detect H1.2 signal in fully grown preovulatory germinal vesicle oocytes or at fertilization (Fig. 4 b, top), and H1.2 was detected on embryonic chromatin only from the eight-cell stage onward. H1.2 was widely distributed throughout the nucleoplasm but enriched in some speckles in 8- and 16-cell-stage embryos (Fig. 4 b, bottom). In the blastocyst, where H1.2 was equally distributed in the inner cell mass (ICM) and the trophectoderm (TE), we noted a slight but visible enrichment of H1.2 in the nuclear periphery (Fig. 4 b, bottom).
Like that of H1.2, H1.3 staining was undetectable in mature preovulatory oocytes and at fertilization but appeared earlier than H1.2, from the two-cell stage onward (Fig. 4 c, top). The distribution of H1.3 was relatively homogeneous throughout the nucleus, with the exception of the 8- and 16-cell stages, during which H1.3 was enriched around the nucleoli (Fig. 4 c, bottom).
Regarding H1.4, we found it to be largely absent from embryonic chromatin during preimplantation development. We first detected weak levels of H1.4 in late 16-cell-stage embryos, but not in all blastomeres, suggesting that H1.4 becomes first expressed in the transition from the 16-cell to the blastocyst stage (Fig. 4 d). In the blastocyst, H1.4 was strongly expressed in both the ICM and the TE (Fig. 4 d). At that point, H1.4 signal accumulated in a dot in TE nuclei in approximately half of the embryos (n = 14), but never in nuclei from the ICM (unpublished data). This dot is reminiscent of the X chromosome that retains imprinted inactivation of the paternal X in the TE (Mak et al., 2004; Okamoto et al., 2004), which prompted us to investigate whether H1.4 is enriched at the inactive X chromosome (Xi) at this stage. To do so, we performed coimmunostaining with Ezh2, known to be enriched at the Xi in female embryos only (Erhardt et al., 2003). This analysis revealed that H1.4 is indeed enriched at the paternal Xi in the TE of female blastocysts (Fig. 5 a). Analysis of H4K20me1, which is also enriched on the Xi, further confirmed that H1.4 accumulates at the Xi (Fig. 5 b). Thus, H1.4 staining is absent in preovulatory oocytes and during cleavage stages and is present only in the blastocyst, where it decorates the paternal Xi.
H1.5 showed a temporal expression pattern similar to that of H1.3, being absent at fertilization and first detected at the two-cell stage, albeit weakly (Fig. 5 c). However, in contrast to the rather uniform distribution of H1.3 in the nucleoplasm, H1.5 showed some sites of accumulation throughout the nucleus from the eight-cell stage (Fig. 5 c, top). Finally, we also analyzed the expression and localization of H1.10, which was initially detected at the nucleoli precursors, the NLBs, and later on the nucleoli (Fig. 5 d), as has been shown for somatic cells (Stoldt et al., 2007).
Globally, our data indicate that the H1 subtypes H1.1–H1.5 and H1.10 are present in the blastocyst, with no obvious distinction between ICM and TE. These H1 subtypes show different temporal expression profiles (Fig. 5 f). As a result, each stage is characterized by a different composition of H1 subtypes, and changes in H1 subtype expression and localization could demarcate transitions in cell plasticity. H1.1 seems to be unique, as it persists in the mature preovulatory oocyte at fertilization, where it appears strongly enriched in the periphery of the NLBs with equal distribution in male and female chromatin.
Our study provides novel insights into the contribution of linker histone H1 subtypes to the chromatin landscape during epigenetic reprogramming, in both PGCs and the preimplantation embryo. It suggests that H1 and its subtypes might be critical for transitions in chromatin architecture during reprogramming. We found that in gonadal PGCs, all somatic H1 subtypes were detectable with the exception of the replacement subtype H1.0. The levels of histone H1 in PGCs and in the surrounding somatic cells are generally comparable. However, at E13.5, male and female germ cells display a distinct content of H1 subtypes. At E13.5, female germ cells enter meiotic prophase, whereas male germ cells undergo mitotic arrest until after birth, allowing male and female germ cells to be distinguished (Peters, 1970). Because H1.1 has the least chromatin-condensing capacity (Clausell et al., 2009), it is possible that higher levels of H1.1 detected in female PGCs (compared with surrounding somatic cells) allow chromatin to acquire the elasticity and flexibility necessary to sustain and coordinate the meiotic recombination process in female PGCs. In contrast, lower levels of H1.4 and H1.10 might promote cell cycle arrest in male PGCs and/or allow the selective activation of genes important for spermatogenesis. Indeed, it is known that somatic H1 expression decreases in tissue culture cells upon growth arrest (Clausell et al., 2009) and that loss of H1.4 results in the arrest of cell proliferation in breast cancer cells (Sancho et al., 2008).
A hallmark of PGCs undergoing specification and reprogramming is the erasure of multiple histone marks at ∼E11.5 (Hajkova et al., 2008) followed by the establishment of a new chromatin state. Our data show for the first time that signal for all somatic H1 subtypes (H1.1–H1.5 and H1.10) is transiently lost from chromatin during the period of epigenetic reprogramming and that H1.10 is the last among the histone H1 subtypes to dissociate from chromatin.
During the reprogramming phase, PGCs have been shown to be predominantly in G2 phase of the cell cycle (Hajkova et al., 2008). In this context, the mRNA of somatic H1 subtypes peaks in the S phase; however, their protein levels are mostly stable throughout the cell cycle (Happel et al., 2009). In line with this, we observed no significant changes in H1 subtype levels in G2 or any cell cycle phase analyzed in the knockin mouse embryonic fibroblasts (MEFs; unpublished data). Thus, rather than the dominant effect of the cell cycle, additional signaling or regulatory pathways may be involved in the observed disappearance of the H1 subtypes in PGCs.
Changes in H1 subtype composition in preimplantation mouse embryos demarcate embryonic genome activation and transitions in cell plasticity. Electron microscopy in the preimplantation embryo revealed a chromatin ultrastructure largely devoid of condensed, heterochromatic regions (Ahmed et al., 2010; Bošković et al., 2014). This global chromatin openness may in part be explained by the absence of most somatic H1 subtypes or their associated chromatin remodelers before the eight-cell stage (Fig. 5 f). Remarkably, there is a global marked increased in electron-dense regions in the transition from the two- to the eight-cell stage, which is concomitant with the appearance of H1.2 and H1.5 on embryonic chromatin, as well as an increased level of expression of H1.1. It will be interesting to determine whether the appearance of H1.2 and H1.5 and their combinations can explain the decrease in core histone mobility observed at the eight-cell stage. The combined knockout of H1.1, H1.2, and H1.3 will be necessary to address whether and how linker histones regulate developmental progression before implantation.
Among all H1 subtypes analyzed, only H1.1 remains detectable at the chromatin during reprogramming after fertilization, suggesting that the presence of the linker histone H1 itself is compatible with reprogramming. Therefore, H1 must not be seen merely as a repressive chromatin component, but instead as a potential fine-tuner of chromatin structure. Indeed, the existence of multiple H1 subtypes with diverse biochemical properties and specific histone modifications (Izzo et al., 2008; Izzo and Schneider, 2015) allows such a regulatory function. Our observation that each stage of mouse preimplantation development is characterized by a unique combination of H1 subtypes is also in support of a tightly regulated role for H1 in establishing embryonic chromatin at the beginning of development.
By focusing on the linker histone H1 subtypes, our analysis adds a new level of information to the current view of epigenetic reprogramming in both the preimplantation embryo and PGCs during germline formation. This process of extensive chromatin remodeling and epigenetic resetting exhibits intriguing similarities to other dedifferentiation or regeneration systems (Yamanaka and Blau, 2010; Gurdon, 2013), pointing toward the existence of common molecular pathways governing these processes. Therefore, a better understanding of the role of linker histone H1 in the epigenetic regulation of the genome during mouse development can improve our ability to manipulate cell fate and restore pluripotency in in vitro settings.
Materials and methods
Ethics statement
Animal work was approved by the Ethics Committee of the Université de Strasbourg and performed under the authorization and rules of the French legislation.
Generation of mouse lines
Chimeric mice were generated by injection of targeted ESCs into mouse blastocysts, and then crossed with C57BL/6 mice to obtain heterozygotes, which were further intercrossed to obtain homozygotes.
ESC culture and immunostaining
Mouse ESCs were cultured on irradiated MEFs in knockout DMEM (Gibco) supplemented with 15% KnockOut Serum Replacement (Gibco), 0.1 mM nonessential amino acids (Gibco), 2 mM GlutaMAX (Invitrogen), penicillin/streptomycin (Gibco), 0.1 mM β-mercaptoethanol, and 1000 U/ml leukemia inhibitory factor (EMD Millipore), at 37°C in 5% CO2. Immunostaining of ESCs was performed as described in Preparation and immunostaining of isolated PGCs.
Embryo and genital ridges collection
Genital ridges were dissected from F1 embryos (Flag-HA-H1.1 × C57BL/6, Flag-HA-H1.2 × C57BL/6, Flag-HA-H1.3 × C57BL/6, Flag-HA-H1.4 × C57BL/6, and Flag-HA-H1.5 × C57BL/6) at E11.5, E12.5, or E13.5. For embryos at E10.5, the entire trunk was used. Noon of the day of the vaginal plug was designated as E0.5. Embryos were staged according to the Theiler staging system (TS; http://www.emouseatlas.org). In particular, E11.5 embryos corresponded to TS 19, E11.25 were between TS 18 and 19, and E11.75 were between TS 19 and 20. Preimplantation mouse embryos were obtained from the same Flag-HA-H1 transgenic lines × C57BL/6 crosses after hormonal stimulation by intraperitoneal injection of 10 IU pregnant mare serum gonadotropin, followed by an injection of 10 IU human chorionic gonadotropin 48 h later. Embryos were collected at the indicated times and used immediately for immunostaining unless otherwise stated.
Preparation of samples for cryosections
Genital ridges were fixed in 4% PFA (Electron Microscopy Sciences)/2% sucrose in PBS for 4 h (E10.5 trunks) or 2 h (E12.5 and E13.5 genital ridges) at 4°C with gentle rocking, washed three times in PBS, cryoprotected in 20% sucrose in PBS overnight and embedded in OCT compound (Thermo Fisher Scientific) using histology molds. Frozen blocks were kept at −80°C up to several months. Sections were cut using a cryostat (Leica Microsystems; object temperature, −17°C; chamber temperature, −21°C) at 10-µm thickness, collected on Superfrost+ slides (Thermo Fisher Scientific), and air-dried for up to 3 h at room temperature.
Preparation of samples for paraffin embedding
Samples were fixed in 4% PFA/2% sucrose in PBS for 4 h (E10.5 trunks) or 2 h (E12.5 and E13.5 genital ridges) at 4°C with gentle rocking. Samples were then washed in PBS, dehydrated in ethanol starting at 25%, 50%, and 70% for 30 min each, and embedded in paraffin. Sections of 5-µm thickness were immersed twice for 5 min in Histosol (Shandon); rehydrated by incubating them two times for 3 min in 100% ethanol, once for 1 min in 90% ethanol, and once for 1 min in 70% ethanol; and washed for 5 min in water. Heat-induced antigen retrieval was performed in 0.01 M sodium citrate buffer, pH 6.0, for 15 min. Samples were then allowed to cool down to room temperature and further washed in PBS. The sections were then subjected to immunofluorescence staining.
Immunostaining of cryosections and paraffin-embedded sections
All incubations were performed using a humidified chamber. From incubation with secondary antibodies onward, all subsequent steps were performed in the dark. Cryosections of the dissected genital ridges (E10.5, E12.5, and E13.5) were postfixed with 2% PFA in PBS for 5 min, and slides were washed three times for 5 min in PBS. Paraffin-embedded sections were not subjected to postfixation. Cryosections and paraffin-embedded sections were permeabilized in 0.7% Triton X-100 in PBS for 1 h at room temperature and washed three times for 5 min in PBS. Cryosections were blocked in 1% BSA/0.1% Triton X-100 for 1 h. Paraffin-embedded sections were blocked in 5% BSA in PBS. Primary antibodies were incubated overnight in 1% BSA/0.1% Triton X-100 in PBS and washed three times for 5 min in PBS. Rabbit anti-Flag antibodies were used to stain genital ridges. The slides were subsequently incubated with Alexa Fluor–conjugated secondary antibodies (Jackson ImmunoResearch) in 1% BSA/0.1% Triton X-100 in PBS for 1–3 h at room temperature, washed three times for 5 min in PBS, mounted in Vectashield with DAPI (Vector Laboratories), and imaged using an SP2-UV confocal microscope (Leica Microsystems).
Preparation and immunostaining of isolated PGCs
For the preparation of single PGC cell suspensions, dissected genital ridges of the indicated developmental stages (E12.5 and E13.5) were collected in Eppendorf tubes on ice in PBS and centrifuged once at 1,200 rpm for 3 min. To obtain a sufficient number of cells, genital ridges from different embryos were pooled. Prewarmed 37°C trypsin-EDTA was added (e.g., ∼100 µl trypsin-EDTA per 10 E12.5 genital ridges), and samples were shaken gently at 300 rpm for 3–5 min at 37°C to dissociate the tissues. Dissociation was facilitated by pipetting the mixture up and down several times during the incubation period. When no cell clumps were visible, DMEM (volume equal to the volume of trypsin-EDTA added) was added to the suspension. Samples were then centrifuged at 3000 rpm for 3 min, resuspended in the appropriate volume of DMEM, and allowed to settle on slides treated with poly-l-lysine (Sigma-Aldrich) for 20 min. The cells were briefly washed with PBS and fixed in 4% PFA/2% sucrose in PBS for 10 min at room temperature (22°C), then permeabilized for 30 min using 0.7% Triton X-100 in PBS. The primary antibody staining was performed in 4% BSA and 0.1% Triton X-100 in PBS for 1 h at room temperature. The slides were washed three times in PBS for 5 min each, incubated with Alexa Fluor–conjugated secondary antibodies (Molecular Probes) for 1 h at room temperature in the dark, washed three times for 5 min in PBS, mounted in Vectashield with DAPI, and imaged using an SP2-UV confocal microscope.
Isolation of primordial germ cells by FACS and Western blot
Single-cell suspensions of genital ridges from E14.5 embryos were prepared. Viable PGCs were labeled with PE-conjugated anti–stage-specific embryonic antigen 1 (anti–SSEA-1) at 1:100 dilution in 1% BSA in PBS for 1 h at room temperature and briefly washed in PBS. Fluorescently labeled PGCs were sorted from the unlabeled somatic cells using FACS, and cell pellets were immediately snap-frozen in liquid nitrogen. Cells from different preparations were pooled together to reach a minimum of 50,000 cells, and a comparable amount of male and female PGCs and somatic cells were subjected to immunoblot staining using an anti-Flag antibody according to standard procedures.
Immunostaining of preimplantation embryos
Embryos were collected at the corresponding stage, fixed, and analyzed by confocal microscopy as described previously (Torres-Padilla et al., 2006). Embryo collection was performed at the same timing, per stage, for all H1 subtypes, so the analysis of their expression is comparable in terms of cell cycle. For all immunostainings performed, we included follicular cells of isolated preovulatory follicles as an internal positive control for the HA antibody.
Chromatin fractionation over sucrose gradient
Knockin cells (1–2 × 107 cells) were harvested and washed twice in hypotonic buffer (20 mM Hepes, pH 7, 20 mM NaCl, and 5 mM MgCl2). Nuclei were isolated by Dounce homogenization and recovered by centrifugation at 1,500 rpm and 4°C for 5 min. Isolated nuclei were resuspended in hypotonic buffer containing 0.5% NP-40 for 15 min on ice and centrifuged at 2,000 rpm and 4°C for 5 min. The chromatin pellet was washed twice in Ex100 buffer (20 mM Hepes, pH 7, 100 mM NaCl, and 0.5 mM MgCl2) to remove the MgCl2, resuspended in Ex100 at 500 µg/ml in the presence of 2 mM CaCl2, and digested with Mnase (10 units for 50 µg of chromatin) for 10 min at 25°C. Digestion was stopped by the addition of 8 mM EDTA, and digested chromatin was recovered by centrifugation at 14,000 rpm and 4°C for 20 min. The supernatant containing the digested chromatin was loaded on a 5–40% sucrose gradient in Ex100 buffer and centrifuged at 36,000 rpm and 4°C for 16 h in a SW40 rotor. The gradient was then separated in 600-µl fractions. Aliquots of each fraction were subjected to Western blotting and agarose gel electrophoresis under standard conditions.
Antibodies
The following antibodies were used: rabbit Flag (Sigma-Aldrich), 1:300 for immunostaining, 1:1,000 for Western blot; mouse HA (12CA5; Roche), 1:200; rabbit HA (Abcam), 1:200; rabbit H1.10 (Abcam), 1:100; mouse H1.0 (Santa Cruz), 1:200; mouse H3K9me3 (Active Motif), 1:100; rabbit H3K9ac (Cell Signaling Technologies), 1:100; mouse OCT4 (BD Biosciences), 1:200; goat OCT4 (N19; Santa Cruz), 1:200; mouse SSEA-1 (BD Biosciences); rabbit Lamin-B1 (Abcam), 1:100; and mouse H1 (AE4; Abcam), 1:100. For details about the antibody applicability and conditions, see Table 1.
Table 1. Antibodies used to detect the somatic H1 subtypes and their application.
| Antibody | ESCs, immunofluorescence | PGCs | ESCs and PGCs, Western blotting | Embryos, immunofluorescence | |
|---|---|---|---|---|---|
| Cryosections | Paraffin sections | ||||
| Rb Flag F7425 | 1:300 | 1:100 | Not tested | 1:1000 | Not tested |
| Ms HA 12CA5 | 1:100 | NA | Not tested | 1:1000 | 1:200 |
| Ms H1.0 sc-56695 | Not tested | NA | 1:100 | 1:1000 | Not tested |
| Rb H1x ab31972 | 1:100 | 1:100 | 1:100 | 1:1000 | 1:100 |
| H1foo (K14) sc-99918 | Not tested | NA | NA | Not tested | Not tested |
NA, no specific signal under the conditions tested.
Microscope image acquisition
Images were acquired at room temperature using a TCS SP5/AOBS inverted confocal microscope (Leica Microsystems) for the embryos and a TCS SP2/UV inverted confocal microscope (Leica Microsystems) for PGCs and ESCs with a Plan Apo CS 63× (NA 1.4) oil-immersion objective (Leica Microsystems). The following fluorescence settings were used: DAPI (excitation 405; emission 410–470), GFP (489; 492–550), Cy3 (558; 560–600), and Cy5 (650; 652–700) when needed. The acquisition software was LAS AF Lite 2.6.3, build 8173 (Leica Microsystems). No gamma adjustments, deconvolution, or reconstitutions were performed. Recording was sequential to avoid bleach-through. Images for this manuscript were formatted using Adobe Photoshop CS4 or ImageJ.
Statistical analysis
The fluorescence intensity from the Flag immunostaining signal in OCT4-positive and -negative cells was measured using Fiji software and calculated as the mean pixel intensity divided by the nuclear area. The ratios between OCT4-positive and -negative cells were calculated and displayed as box plots using R. p-values were calculated using a pairwise t test with Bonferroni correction. Only p-values <0.05 were considered statistically significant. The sample sizes for each developmental stage and H1 subtype analyzed are shown throughout the figures and figure legends.
Online supplemental material
Fig. S1 shows the immunofluorescence staining of cryosections from genital ridges at the indicated developmental stages for H1.1, H1.2, H1.3, and H1.4 subtypes and the corresponding statistical analysis for the H1.2 and H1.3 subtypes. Fig. S2 shows the immunofluorescence staining of cryosections from genital ridges at the indicated developmental stages for H1.5 and the corresponding statistical analysis and for H1.10 and H1.0. Also included is the characterization of H1.0 and H1.10 antibodies used in this study and the staining of isolated PGCs with H1 and laminB1 antibodies. Fig. S3 shows the kinetics of H1 disappearance during epigenetic reprogramming of H1.1, H1.2, H1.3, and H1.5 variants and the corresponding statistical analyses.
Supplementary Material
Acknowledgments
We thank Marie-Christine Birling (Institut Charles Sadron, Strasbourg) for helpful discussions during the design of the knockin mice.
Work in the Schneider laboratory is supported by the Helmholtz Gesellschaft and the EpiTrio project. Work in the Hajkova laboratory is supported by the Medical Research Council, European Research Council Consolidator Grant “dynamic modifications,” and the EpiGeneSys Network. P. Hajkova is a member of the European Molecular Biology Organization Young Investigator Program; P.W.S. Hill was a recipient of the Medical Research Council Doctoral award. M.-E. Torres-Padilla acknowledges funding from EpiGeneSys, European Research Council Starting Grant “nuclear potency,” European Molecular Biology Organization Young Investigator Program, the Foundation Schlumberger for Education and Research, and the Helmholtz Gesellschaft.
The authors declare no competing financial interests.
Author contributions: A. Izzo performed the experiments in Figs. 1–3 and S1–S3. C. Ziegler-Birling performed the experiments in Figs. 4 and 5. P.W.S. Hill provided key expertise and supervision on PGC work. L. Brondani provided technical assistance with the mice and contributed to the data in Fig. 1 D. A. Izzo, P. Hajkova, M.-E. Torres-Padilla, and R. Schneider designed the study and wrote the manuscript. All authors read and approved the manuscript.
Footnotes
Abbreviations used:
- E
- embryonic day
- ESC
- embryonic stem cell
- ICM
- inner cell mass
- MEF
- mouse embryonic fibroblast
- NLB
- nucleolar-like body
- PGC
- primordial germ cell
- TE
- trophectoderm
- TS
- Theiler staging
References
- Ahmed K., Dehghani H., Rugg-Gunn P., Fussner E., Rossant J., and Bazett-Jones D.P.. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 5:e10531 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami R., Fan Y., Pack S., Sonbuchner T.M., Besse A., Lin Q., Greally J.M., Skoultchi A.I., and Bouhassira E.E.. 2003. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc. Natl. Acad. Sci. USA. 100:5920–5925. 10.1073/pnas.0736105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Hartman P.G., Crane-Robinson C., and Aviles F.X.. 1980. The structure of histone H1 and its location in chromatin. Nature. 288:675–679. 10.1038/288675a0 [DOI] [PubMed] [Google Scholar]
- Bošković A., Eid A., Pontabry J., Ishiuchi T., Spiegelhalter C., Raghu Ram E.V., Meshorer E., and Torres-Padilla M.E.. 2014. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 28:1042–1047. 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A., and Torres-Padilla M.E.. 2014. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15:723–734. 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- Cao K., Lailler N., Zhang Y., Kumar A., Uppal K., Liu Z., Lee E.K., Wu H., Medrzycki M., Pan C., et al. 2013. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 9:e1003417 10.1371/journal.pgen.1003417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausell J., Happel N., Hale T.K., Doenecke D., and Beato M.. 2009. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One. 4:e0007243 10.1371/journal.pone.0007243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S., Su I.H., Schneider R., Barton S., Bannister A.J., Perez-Burgos L., Jenuwein T., Kouzarides T., Tarakhovsky A., and Surani M.A.. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 130:4235–4248. 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- Fan Y., Sirotkin A., Russell R.G., Ayala J., and Skoultchi A.I.. 2001. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol. Cell. Biol. 21:7933–7943. 10.1128/MCB.21.23.7933-7943.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Nikitina T., Morin-Kensicki E.M., Zhao J., Magnuson T.R., Woodcock C.L., and Skoultchi A.I.. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23:4559–4572. 10.1128/MCB.23.13.4559-4572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia B.A., Busby S.A., Barber C.M., Shabanowitz J., Allis C.D., and Hunt D.F.. 2004. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J. Proteome Res. 3:1219–1227. 10.1021/pr0498887 [DOI] [PubMed] [Google Scholar]
- Groth A., Corpet A., Cook A.J., Roche D., Bartek J., Lukas J., and Almouzni G.. 2007. Regulation of replication fork progression through histone supply and demand. Science. 318:1928–1931. 10.1126/science.1148992 [DOI] [PubMed] [Google Scholar]
- Gunjan A., Alexander B.T., Sittman D.B., and Brown D.T.. 1999. Effects of H1 histone variant overexpression on chromatin structure. J. Biol. Chem. 274:37950–37956. 10.1074/jbc.274.53.37950 [DOI] [PubMed] [Google Scholar]
- Gurdon J.B. 2013. The egg and the nucleus: A battle for supremacy. Development. 140:2449–2456. 10.1242/dev.097170 [DOI] [PubMed] [Google Scholar]
- Hajkova P. 2010. Epigenetic reprogramming—Taking a lesson from the embryo. Curr. Opin. Cell Biol. 22:342–350. 10.1016/j.ceb.2010.04.011 [DOI] [PubMed] [Google Scholar]
- Hajkova P., Ancelin K., Waldmann T., Lacoste N., Lange U.C., Cesari F., Lee C., Almouzni G., Schneider R., and Surani M.A.. 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 452:877–881. 10.1038/nature06714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N., Warneboldt J., Hänecke K., Haller F., and Doenecke D.. 2009. H1 subtype expression during cell proliferation and growth arrest. Cell Cycle. 8:2226–2232. 10.4161/cc.8.14.8982 [DOI] [PubMed] [Google Scholar]
- Hemberger M., Dean W., and Reik W.. 2009. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 10:526–537. 10.1038/nrm2727 [DOI] [PubMed] [Google Scholar]
- Izzo A., and Schneider R.. 2015. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim. Biophys. Acta 1859:486–495. [DOI] [PubMed] [Google Scholar]
- Izzo A., Kamieniarz K., and Schneider R.. 2008. The histone H1 family: Specific members, specific functions? Biol. Chem. 389:333–343. 10.1515/BC.2008.037 [DOI] [PubMed] [Google Scholar]
- Izzo A., Kamieniarz-Gdula K., Ramírez F., Noureen N., Kind J., Manke T., van Steensel B., and Schneider R.. 2013. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Reports. 3:2142–2154. 10.1016/j.celrep.2013.05.003 [DOI] [PubMed] [Google Scholar]
- Kamakaka R.T., and Biggins S.. 2005. Histone variants: Deviants? Genes Dev. 19:295–310. 10.1101/gad.1272805 [DOI] [PubMed] [Google Scholar]
- Liao L.W., and Cole R.D.. 1981. Condensation of dinucleosomes by individual subfractions of H1 histone. J. Biol. Chem. 256:10124–10128. [PubMed] [Google Scholar]
- Mak W., Nesterova T.B., de Napoles M., Appanah R., Yamanaka S., Otte A.P., and Brockdorff N.. 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science. 303:666–669. 10.1126/science.1092674 [DOI] [PubMed] [Google Scholar]
- Medrzycki M., Zhang Y., Cao K., and Fan Y.. 2012. Expression analysis of mammalian linker-histone subtypes. J. Vis. Exp. (61):3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I., Otte A.P., Allis C.D., Reinberg D., and Heard E.. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 303:644–649. 10.1126/science.1092727 [DOI] [PubMed] [Google Scholar]
- Peters H. 1970. Migration of gonocytes into the mammalian gonad and their differentiation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 259:91–101. 10.1098/rstb.1970.0048 [DOI] [PubMed] [Google Scholar]
- Ponte I., Vidal-Taboada J.M., and Suau P.. 1998. Evolution of the vertebrate H1 histone class: Evidence for the functional differentiation of the subtypes. Mol. Biol. Evol. 15:702–708. 10.1093/oxfordjournals.molbev.a025973 [DOI] [PubMed] [Google Scholar]
- Probst A.V., Santos F., Reik W., Almouzni G., and Dean W.. 2007. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma. 116:403–415. 10.1007/s00412-007-0106-8 [DOI] [PubMed] [Google Scholar]
- Sancho M., Diani E., Beato M., and Jordan A.. 2008. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 4:e1000227 10.1371/journal.pgen.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., and Reik W.. 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 48:849–862. 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y., Yamaji M., Yabuta Y., Sano M., Shigeta M., Matsui Y., Saga Y., Tachibana M., Shinkai Y., and Saitou M.. 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 134:2627–2638. 10.1242/dev.005611 [DOI] [PubMed] [Google Scholar]
- Shen X., and Gorovsky M.A.. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 86:475–483. 10.1016/S0092-8674(00)80120-8 [DOI] [PubMed] [Google Scholar]
- Stoldt S., Wenzel D., Schulze E., Doenecke D., and Happel N.. 2007. G1 phase-dependent nucleolar accumulation of human histone H1x. Biol. Cell. 99:541–552. 10.1042/BC20060117 [DOI] [PubMed] [Google Scholar]
- Surani M.A., Hayashi K., and Hajkova P.. 2007. Genetic and epigenetic regulators of pluripotency. Cell. 128:747–762. 10.1016/j.cell.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Syed S.H., Goutte-Gattat D., Becker N., Meyer S., Shukla M.S., Hayes J.J., Everaers R., Angelov D., Bednar J., and Dimitrov S.. 2010. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. USA. 107:9620–9625. 10.1073/pnas.1000309107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Th’ng J.P., Sung R., Ye M., and Hendzel M.J.. 2005. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 280:27809–27814. 10.1074/jbc.M501627200 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Bannister A.J., Hurd P.J., Kouzarides T., and Zernicka-Goetz M.. 2006. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50(Next):455–461. 10.1387/ijdb.052073mt [DOI] [PubMed] [Google Scholar]
- Wisniewski J.R., Zougman A., Krüger S., and Mann M.. 2007. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol. Cell. Proteomics. 6:72–87. 10.1074/mcp.M600255-MCP200 [DOI] [PubMed] [Google Scholar]
- Yamanaka S., and Blau H.M.. 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 465:704–712. 10.1038/nature09229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.