Microautophagy is a mode of autophagy in which the lysosomal or vacuolar membrane invaginates and engulfs target components. Oku et al. show that, upon a diauxic shift, yeast microautophagy involves recruitment of ESCRT proteins to the vacuolar membrane, including clathrin-interacting Vps27, and uptake of lipid droplets by the vacuole.
Abstract
Microautophagy refers to a mode of autophagy in which the lysosomal or vacuolar membrane invaginates and directly engulfs target components. The molecular machinery of membrane dynamics driving microautophagy is still elusive. Using immunochemical monitoring of yeast vacuolar transmembrane proteins, Vph1 and Pho8, fused to fluorescent proteins, we obtained evidence showing an induction of microautophagy after a diauxic shift in the yeast Saccharomyces cerevisiae. Components of the endosomal sorting complex required for transport machinery were found to be required for this process, and the gateway protein of the machinery, Vps27, was observed to change its localization onto the vacuolar membrane after a diauxic shift. We revealed the functional importance of Vps27’s interaction with clathrin in this microautophagy that also contributed to uptake of lipid droplets into the vacuole. This study sheds light on the molecular mechanism of microautophagy, which does not require the core Atg proteins.
Introduction
Turnover of proteins underlies robust cell homeostasis by regulating the quantity and quality of proteins. The turnover of membrane proteins embedded in organelles is known to proceed through several mechanisms. In one case, the target proteins are dislocated from membrane structures, either by clipping at their transmembrane portions or by extraction with the aid of AAA-ATPases (Avci and Lemberg, 2015), before they are degraded by proteasome or intraorganelle proteases. In the other case, parts of membrane structures containing the target proteins are pinched off toward the lumen of the organelle through membrane vesiculations, leading to degradation of both lipids and proteins. The most well-known process for the latter case is formation of multivesicular bodies (MVBs) in the endosome driven by actions of the endosomal sorting complex required for transport (ESCRT) machinery, followed by degradation of the vesicles along with the cargo membrane proteins after transfer of the MVBs into the vacuole/lysosome (Henne et al., 2011).
The yeast V-ATPase complex functions as a proton pump on the vacuolar membrane, which is vital for maintaining low pH within the organelle and supporting the robustness of the organism against various stresses (Kane, 2007). Vph1, one of the transmembrane subunits of V-ATPase, constitutively resides in the vacuolar membrane, whereas the other peripheral subunits exhibit a conditional association with Vph1, depending on culture conditions or phosphoinositide dynamics (Kane, 1995; Li et al., 2014). To date, the mechanism by which Vph1 protein is subject to the turnover process is unknown. In this study, we found that Vph1 is degraded in the vacuolar lumen through microautophagy when yeast cells are subjected to a diauxic shift (metabolic shift upon a change in the carbon source from glucose to ethanol; Galdieri et al., 2010).
Microautophagy, constituting a mode of autophagy, refers to a transport system of cytosolic components, including organelles, into the lysosome or vacuole, via vacuolar engulfment and incorporation of the targets (Mijaljica et al., 2011). Recent studies from higher eukaryotes have revealed the physiological importance of microautophagy in the accumulation of anthocyanin-containing structures in the plant vacuole and in the progression of mouse embryonic development (Kawamura et al., 2012: Chanoca et al., 2015) and have also unveiled a novel mode of microautophagy, endosomal microautophagy (uptake of cytoplasmic components by the endosome; Sahu et al., 2011; Liu et al., 2015; Uytterhoeven et al., 2015). In yeasts, various organelles were identified as the targets of microautophagy: peroxisomes (Sakai et al., 1998), the nucleus (Roberts et al., 2003), the ER (Schuck et al., 2014), mitochondria (Kissová et al., 2007), lipid droplets (LDs; van Zutphen et al., 2014; Wang et al., 2014), and the cytoplasm (Müller et al., 2000). Some of these microautophagic pathways require autophagy-related (Atg) proteins that were responsible for another mode of autophagy, macroautophagy (Mukaiyama et al., 2002; Krick et al., 2008; van Zutphen et al., 2014). Although most of the Atg proteins act on de novo synthesis of double-membrane structures during macroautophagy (Xie and Klionsky, 2007), none of these structures have been identified in the Atg-dependent microautophagic pathways, except for the microautophagic membrane apparatus, which is formed during microautophagy of peroxisomes in the methylotrophic yeast Pichia pastoris (Mukaiyama et al., 2004). Thus, mechanistic details of the membrane dynamics for microautophagy are largely unknown.
In this study, we used Vph1, together with another integral vacuolar membrane protein, Pho8 (an alkaline phosphatase), in fusion with fluorescent proteins, to assess their transport into the vacuolar lumen through microautophagy. We found that the transport was induced after the diauxic shift, which depended on the ESCRT machinery, but not on the core Atg proteins, with the exception of Atg15 (an intravacuolar lipase). We further discovered an indispensable role of a clathrin-binding motif on the ESCRT protein Vps27 in microautophagy. We observed the localization change of fluorescent protein–tagged Vps27 onto the vacuolar membrane after the diauxic shift. Further studies with the vps27 mutants also suggested a role of microautophagy in degradation of LDs.
Results
Degradation of vacuolar membrane proteins Vph1 and Pho8 induced after the diauxic shift
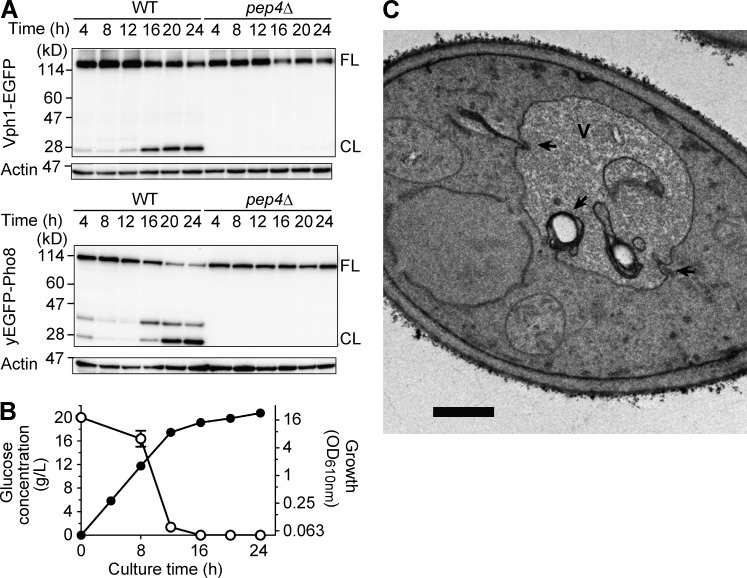
To detect the degradation of Vph1 biochemically, lysate from a WT strain expressing EGFP-tagged Vph1 (Vph1-EGFP) cultured in a nutrient-rich glucose medium (YEPD) was periodically acquired for an immunoblot analysis with an anti-GFP antibody. In addition to the full-length form, a cleaved form of this fusion protein mainly composed of EGFP portion was detected in the lysates from the cells cultured for 16 h or more (Fig. 1 A). Meanwhile, cells underwent a decrease in growth rate that was concomitant with glucose exhaustion in the medium at a time point between 12 and 16 h of culture, but they still exhibited a steady growth up to 24 h of culture (Fig. 1 B), indicating that the cleaved form of Vph1-EGFP was detected after the diauxic shift.
Figure 1.
Vacuolar dynamics after diauxic shift. (A) Immunoblot analyses of Vph1-EGFP and yEGFP-Pho8 expressed in either the WT or pep4Δ strain. Cells were taken from cultures in YEPD medium at 28°C for the indicated time period and subjected to immunoblot analysis with an anti-GFP antibody. FL represents a band position for the full-length form of the fusion proteins, and CL indicates that for the cleaved form containing EGFP or yEGFP moiety. The immunoblot data gained with anti–β-actin antibody are shown as loading controls. (B) Time course of glucose consumption (open circle) and growth (closed circle) of the WT strain in YEPD medium. The mean values from three independent cultures are plotted, and error bars indicate standard deviation. (C) A representative EM image of a WT strain cultured in YEPD medium at 28°C for 16 h. The vacuole invaginations are highlighted with black arrows. V, vacuole. Bar, 0.5 µm.
The cleavage of Vph1-EGFP was not detected in a PEP4-deleted mutant in which vacuolar proteases are inactivated (Fig. 1 A), showing that the cleavage of Vph1-EGFP is dependent on Pep4, reflecting a relatively stable feature of the GFP portion against vacuolar-protease activities (Cheong and Klionsky, 2008). Next, we assessed degradation of another vacuolar membrane protein, Pho8, in fusion with a codon-optimized variant of EGFP, yEGFP (Cormack et al., 1997). Unlike the transport of the V-ATPase complex including Vph1 to the vacuole through endosome membrane traffic (Rothman et al., 1989), the biosynthetic pathway of Pho8 depends on transport from the Golgi (Cowles et al., 1997; Vowels and Payne, 1998). Using the strain expressing yEGFP-Pho8, we found a pattern similar to that seen with Vph1-EGFP in which the cleaved form from yEGFP-Pho8, which was not in the pep4Δ background, was detected at 16 h and thereafter (Fig. 1 A). This indicates that Pho8 is also transported to the vacuole for degradation after the diauxic shift.
Vacuolar morphology after the diauxic shift implies induction of microautophagy
To elucidate the process leading to degradation of Vph1, we performed EM analysis of cells after the diauxic shift. A WT strain cultured in YEPD medium for 16 h contained vacuoles with invaginations whose diameters ranged from ∼100 nm to 350 nm (Fig. 1 C, arrows). The invaginations often surrounded electron-lucent structures, representing LDs (Fig. 1 C). Inside the vacuole, amorphous membrane structures were observed whose appearance resembles that of the vacuolar limiting membrane (Fig. 1 C). This observation strongly suggested the incorporation of portion of the vacuolar membrane into the lumen via invagination (i.e., microautophagy).
The apparent intravacuolar membrane structure observed during EM analysis was thought to represent a cross section of vacuolar invagination and/or that of an intravacuolar vesicle resulting from the pinch-off of the invagination. The intra-vacuolar population of Vph1-EGFP signal was observed to exhibit Brownian movement (Video 1), strongly suggesting detachment of the vesicle structures including Vph1-EGFP from the vacuolar membrane. These morphological data, in combination with biochemical observation of cleavage of Vph1-EGFP, altogether strongly suggested induction of microautophagy after the diauxic shift.
Gene requirements for transport of Vph1 after the diauxic shift
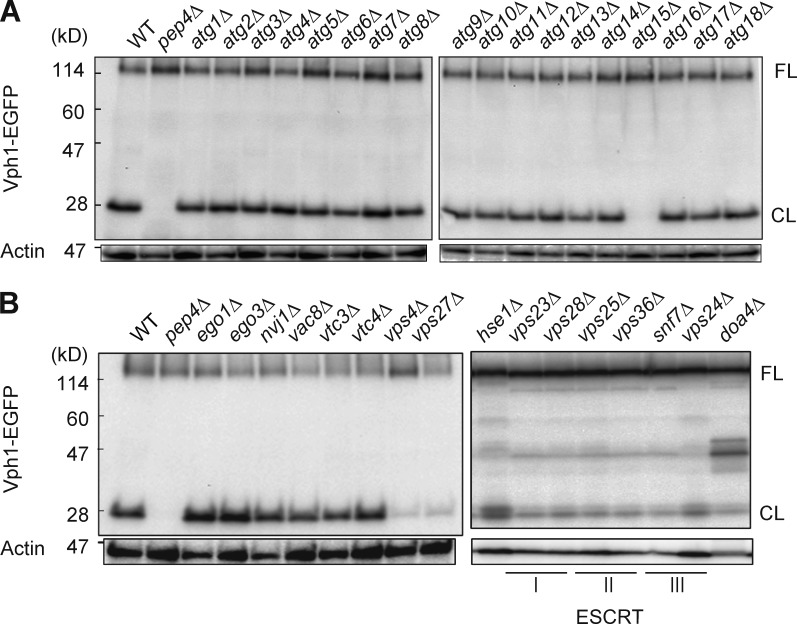
We investigated molecular requirements for the transport of Vph1 into the vacuole by analyzing whether the truncated form of Vph1-EGFP was detected in the lysates of various mutant cells. Notably, the cleaved form of Vph1-EGFP was detected in all of core atg mutants (Xie and Klionsky, 2007), except for atg15Δ mutant that lost an vacuolar lipase for disintegrations of autophagic bodies and MVBs (Epple et al., 2001; Teter et al., 2001; Fig. 2 A), showing that the transport of Vph1 into the vacuole is independent of the core Atg proteins necessary for macroautophagic transport system to the vacuole.
Figure 2.
Immunoblot analyses detecting cleavage of Vph1-EGFP expressed in mutant strains defective in Atg pathways. The denoted ATG gene–deleted mutant strains (A) or other gene-deletion mutants (B) were cultured at 28°C for 24 h in YEPD medium and subjected to immunoblot analysis as shown in Fig. 1 A.
We also examined factors that have been shown to be required for several types of microautophagy in Saccharomyces cerevisiae: Ego1 and Ego3 for microautophagy induced after removal of rapamycin (Dubouloz et al., 2005), Nvj1 and Vac8 for piecemeal microautophagy of the nucleus (Kvam and Goldfarb, 2007), and Vtc3 and Vtc4 for microautophagy through vacuolar tube formation (Uttenweiler et al., 2007). However, none of these mutants exhibited a defect in the cleavage of Vph1-EGFP (Fig. 2 B) after the diauxic shift, excluding involvements of these molecules.
A previous study on autophagy in mammalian cells showed that late endosome microautophagic activity is driven by the ESCRT-I and ESCRT-III complexes (Sahu et al., 2011). We tested whether ESCRT complex proteins are required for the transport of Vph1 into the vacuole induced after the diauxic shift. Cleavage of Vph1-EGFP was severely inhibited by loss of Vps27, the central factor of the ESCRT-0 complex (Piper et al., 1995), or by that of Vps4, the disassembly factor of the ESCRT-III complex (Babst et al., 1998; Fig. 2 B). Immunoblot analysis indicated that the factors of the other ESCRT complexes (I, II, and III) and Doa4, the downstream factor of the ESCRT machinery (Amerik et al., 2000), are also required for the cleavage, whereas loss of Hse1 caused only a minor defect (Fig. 2 B). These data demonstrate that the ESCRT machinery is required for transport of Vph1 into the vacuole after the diauxic shift.
Intramolecular domains of Vps27 required for microautophagy
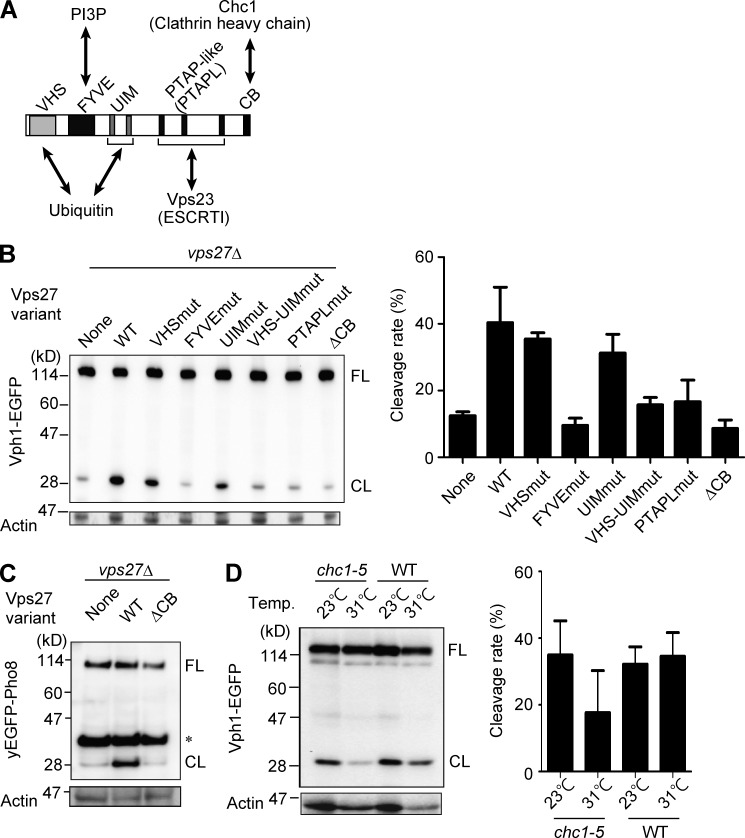
To clarify the functions of the ESCRT complexes, we focused on the analysis of Vps27, the gateway protein for the ESCRT machinery. Vps27 and its mammalian orthologue, Hrs, have multiple intramolecular domains whose actions have been extensively elucidated in previous studies (Fig. 3 A). Among them, VHS (named after Vps27p, Hrs, and STAM) and UIM (ubiquitin-interacting motif) function as ubiquitin-binding domains (Bilodeau et al., 2002), FYVE (Fab-1, YOTB, Vps27, and EEA1) as a phosphatidylinositol 3′ monophosphate (PI3P)–binding domain (Gaullier et al., 1998), and proline-threonine-alanine-proline–like (PTAPL) sequences as the key to recruitment of the downstream ESCRT-I complex (Bilodeau et al., 2003; Katzmann et al., 2003). All of these regions are required for the MVB pathway.
Figure 3.
Functional analysis of domain-mutated Vps27 derivatives. (A) Schematic drawing of Vps27 intramolecular domains. The binding partner of each domain is also shown. CB, clathrin-binding motif; PTAPL, PTAP-like amino-acid sequence. (B) Immunoblot analysis of Vph1-EGFP in vps27 mutants. The data are presented as in Fig. 1 A. In the VPS27-deleted (vps27Δ) strain, normal (WT) or mutant forms of Vps27 were expressed. The designation of mut indicates introduction of point mutations in the denoted domains. The cleavage rate for each variant [(band intensity of CL)/(band intensities of FL + CL) × 100 (%)] was calculated from three independent experiments (right). The error bars indicate standard deviation. (C) Immunoblot analysis of yEGFP-Pho8 expressed in vps27 mutant strains. The asterisk indicates a nonspecific band. (D) Immunoblot analysis of Vph1-EGFP expressed in chc1 (clathrin heavy chain) mutant or WT strain cultured at 23°C for 24 h (23°C) or cultured at 23°C for 16 h and subsequently cultured at 31°C for 8 h (31°C). The cleavage rates calculated as in B from three independent experiments are also shown. Error bars indicate standard deviation.
Vps27 derivatives mutated in these functional domains were introduced into the vps27Δ strain expressing Vph1-EGFP to investigate their ability to restore the cleavage of Vph1-EGFP (Fig. 3 B). Although a mutation in either the VHS or UIM domain caused a partial defect, mutations in both ubiquitin-binding domains or in other domains resulted in a severe defect in the cleavage of Vph1-EGFP (Fig. 3 B). This indicates that all of Vps27’s functions (i.e., binding to ubiquitin, PI3P, and ESCRT-I factors) are needed for transport of Vph1-EGFP into the vacuole.
At the C-terminal ends of yeast Vps27 and mammalian Hrs is conserved a clathrin-binding motif (Raiborg et al., 2001a; Bilodeau et al., 2003), but the physiological role of this motif in yeast has been unclear. Notably, Vps27 lacking this motif (Vps27ΔCB) failed to complement the cleavage of Vph1-EGFP (Fig. 3 B) and yEGFP-Pho8 (Fig. 3 C), indicating that the transport of Vph1-EGFP depends on the clathrin-binding motif of Vps27. We also examined HA-tagged Vps27 variants for their expression levels and compared cleavage of Vph1-EGFP to test their functionalities (Fig. S1). We observed unexpected enhancement of Vph1-EGFP cleavage in the strains expressing HA-tagged Vps27 variants; the cleavage rate for the strain expressing HA-tagged Vps27WT was ∼60%, whereas the rate in the strain with nontagged Vps27 was ∼40% (Figs. 3 B and S1). Nevertheless, we were also able to detect a reproducible defect in Vph1-EGFP cleavage in the strain expressing HA-tagged Vps27ΔCB mutant (Fig. S1).
The functionality of Vps27ΔCB in terms of endocytosis was assessed using several methods (Fig. S2). The fluorescence signal of yEGFP-tagged Cps1 (carboxypeptidase S1), a representative of membrane proteins transported via MVB pathway, was observed in the luminal part of the vacuole for the strains expressing Vps27 WT or ΔCB variant (Fig. S2 A), suggesting comparable Cps1 transport of Vps27ΔCB, which is consistent with data of the previous study (Bilodeau et al., 2003). Next, we compared the localization of pulse-labeled FM 4–64, as another indicator of endocytosis (Vida and Emr, 1995), between the strain expressing WT Vps27 and that expressing Vps27ΔCB (Fig. S2 B). The FM 4–64 fluorescence reached the vacuolar membrane in 1 h in the cells expressing Vps27ΔCB, whereas it did in 30 min in the cells expressing the WT Vps27, showing slowed rate of endocytosis in the mutant cells. The rate of endocytosis was also compared by chasing fluorescence of Mup1-pHluorin, a pH-sensitive probe of endocytosis that loses its fluorescence upon entering into the vacuole (Prosser et al., 2010; Fig. S2 C). The half-life of Mup1-pHluorin was ∼33 min in cells expressing HA-tagged WT Vps27 and ∼58 min in those expressing HA-Vps27ΔCB. These results altogether demonstrate that loss of the clathrin-binding motif of Vps27 affects the kinetics of endocytosis, although it does not completely inhibit it.
In line with the involvement of clathrin-binding motif on Vps27, a temperature-sensitive mutant of clathrin heavy chain (chc1-5; Chen and Graham, 1998) was also found to be defective in the cleavage of Vph1-EGFP at the nonpermissive temperature, but not at the permissive temperature (Fig. 3 D). Collectively, the present results support the collaborative action of clathrin heavy chain and Vps27 in microautophagy after the diauxic shift.
Fluorescent protein-tagged Vps27 changes its localization onto the vacuolar membrane after the diauxic shift
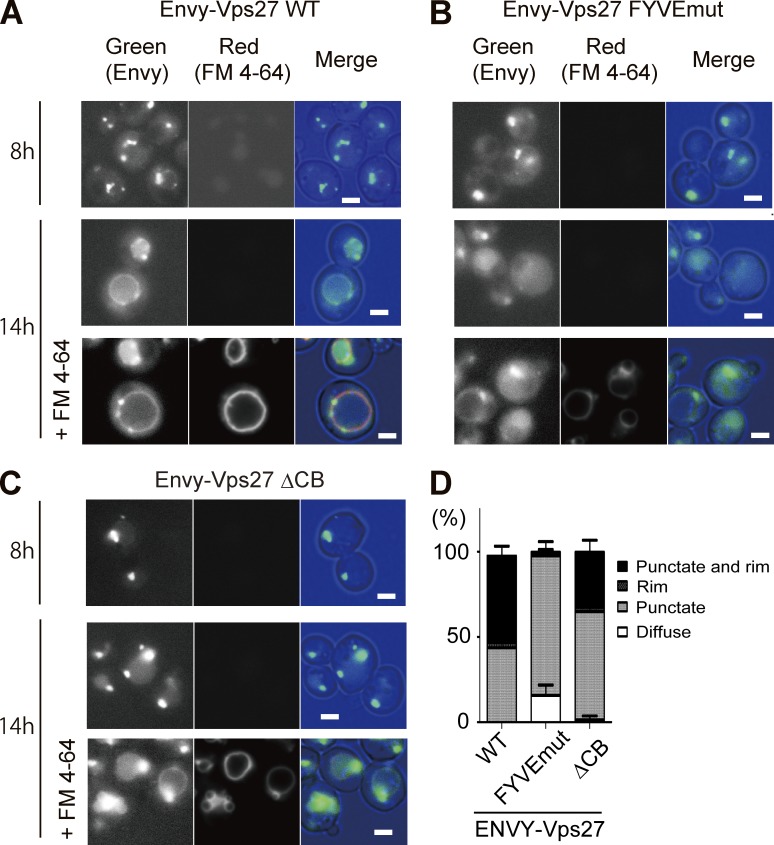
The observed vacuolar invaginations (Fig. 1 C), in combination with the functional requirements of ESCRT proteins (Figs. 2 and 3) for the transport of Vph1-EGFP and yEGFP-Pho8 into the vacuole after the diauxic shift, strongly suggest an induction of microautophagy that accompanies ESCRT-driven vacuole invagination, leading to the transport of these vacuolar membrane proteins into the lumen. To support this notion, it is pivotal to examine whether the ESCRT components are indeed translocated to the vacuolar membrane. To address this issue, we integrated into the genome of the vps27Δ strain a construct to express Vps27 N-terminally tagged with Envy, an EGFP variant with improved brightness (Slubowski et al., 2015), under the VPS27 promoter. The resultant strain was examined using fluorescence microscopy after culture in YEPD medium for 8 h (exponential growth phase) or 14 h (after the diauxic shift; Fig. 4).
Figure 4.
Localizations of Vps27 variants during different growth phases. VPS27-deleted (vps27Δ) strains expressing Envy-tagged WT (A), FYVE-domain–mutated Vps27 (B), or clathrin-binding motif–deleted Vps27 (C) were cultured in YEPD medium at 28°C for 8 h (at an early log phase) or 14 h (after diauxic shift). Both cells labeled with FM 4–64 and nonlabeled cells were analyzed by fluorescence microscopy with the same image-acquisition parameters. The merge image consists of green Envy image and red FM 4–64 image. Bars, 2 µm. (D) The numbers of cells exhibiting diffuse, punctate, and/or rim patterns of Envy-Vps27 were counted (n > 50) for each observation, and the percentages of the patterns are represented in the stacked bar chart. Error bars indicate standard deviation.
The fluorescence signal of Envy-Vps27 in cells cultured for 8 h was observed as several puncta (Fig. 4 A), most likely representing endosomes according to previous studies (Piper et al., 1995; Katzmann et al., 2003). In contrast, such puncta were less evident in the same strain cultured for 14 h, and notably, part of the Envy-Vps27 fluorescence signal exhibited rim patterns that colocalized with the FM 4–64 signal (Fig. 4 A). This observation demonstrates that Vps27 changes its localization onto the vacuolar membrane after the diauxic shift and reinforces the notion that ESCRT-driven microautophagy is induced at this time point.
When the FYVE domain within Envy-Vps27 was mutated, the rim pattern of the fluorescence signal was rarely detected, and instead, weak punctate patterns were dominantly observed (Fig. 4 B). In contrast, the frequency of Envy-Vps27 localization to the vacuolar membrane was partially diminished by the loss of the clathrin-binding motif (Fig. 4 C); the frequency of the rim pattern observation was 51% for Envy-Vps27 WT and 32% for Envy-Vps27ΔCB (Fig. 4 D), whereas their expression levels were comparable (Fig. S3 A). These results indicate that the translocation of Vps27 onto the vacuolar membrane depends on the PI3P-binding activity of the protein and is facilitated by the presence of the clathrin-binding motif.
Incorporation of LDs into the vacuole after the diauxic shift is mainly dependent on Vps27, which can interact with clathrin
Ultrastructure analysis of microautophagy (Fig. 1 C) indicated that the invaginating vacuole engulfed LDs, indicating the uptake of LDs via microautophagy (i.e., microlipophagy). To verify this, we initially used fluorescent proteins fused with Erg6 or Faa4 (authentic markers of LDs) to follow lipophagy. However, after 16 h of culture in YEPD, these fusion proteins exhibited ring-shaped ER patterns in addition to the dot pattern representing LDs (Fig. S3 B), which hampered reliable evaluation of lipophagy. After screening putative LD-resident proteins (Kohlwein et al., 2013), Osw5-EGFP (Suda et al., 2009) expressed under its original promoter was found to localize exclusively to LDs after the diauxic shift (Fig. S3 B).
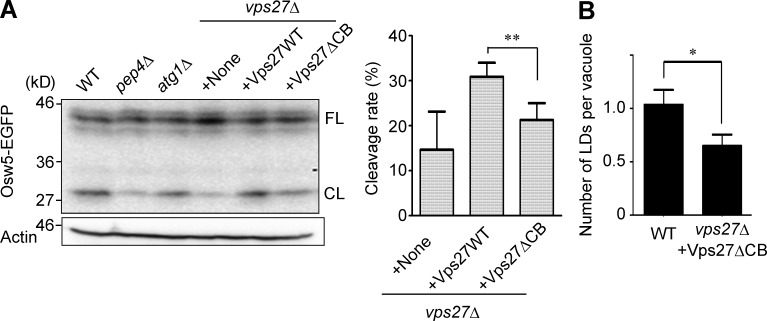
Immunoblot analysis of Osw5-EGFP showed that a cleaved form from this fusion protein was detected in the lysates from WT cells cultured for 16 h in YEPD medium. The intensity of the cleaved form was significantly lower in the lysate from the pep4Δ strain (Fig. 5 A), indicating that the cleavage of Osw5-EGFP depends, at least in part, on the activity of intravacuolar protease. Loss of Atg1, the pivotal kinase for macroautophagy, did not diminish the band intensity of the cleaved form of Osw5-EGFP (Fig. 5 A). In contrast, loss of Vps27 significantly reduced the rate of cleavage, and notably, Vps27ΔCB failed to fully restore the band intensity of the cleaved form, unlike the full-length form of Vps27 (Fig. 5 A). Morphometric EM analysis also indicated that the number of LDs found inside the vacuole was lower in cells expressing Vps27ΔCB than in WT cells (Fig. 5 B). These results strongly suggest that lipophagy after the diauxic shift is mainly dependent on microautophagy requiring Vps27 that can interact with clathrin.
Figure 5.
Efficient transport of LDs into the vacuole in cells after the diauxic shift depends on Vps27 harboring the clathrin-binding motif. (A) Immunoblot analysis of Osw5-EGFP in the designated mutant strains cultured in YEPD medium at 28°C for 16 h. The data for anti–actin antibody are also shown as a loading control. CL; cleaved form; FL, full-length form;. The graph indicates the cleavage rates calculated from three independent experiments. The error bars indicate standard deviation, and the double asterisk indicates statistical significance (**, P < 0.01, t test). (B) The numbers of LDs found inside the vacuoles in EM images of the designated strains (n > 50) were compared. Error bars indicate the standard error. The asterisk indicates the statistical significance of the comparison (*, P < 0.05, t test).
Discussion
In this study, we have provided several lines of evidence showing that microautophagy contributes to the degradation of the vacuolar transmembrane proteins Vph1 and Pho8 and LDs in a manner dependent on most of the ESCRT complex components, including Vps27, the gateway protein for the ESCRT machinery (Figs. 2 and 3). In the tested ESCRT mutants, hse1Δ strain alone showed a comparable cleavage of Vph1-EGFP to that in the WT strain (Fig. 2 B). Hse1 is localized to endosomes and associates with Vps27 as well as with a ubiquitin ligase, Rsp5 (Bilodeau et al., 2002; Ren et al., 2007), for efficient sorting of ubiquitinated proteins into MVBs. Together with our finding that the ubiquitin-binding ability of Vps27 is vital for transport of Vph1 into the vacuole (Fig. 3 B), the dispensability of Hse1 suggests that the recognition of ubiquitinated proteins by Vps27 alone mediates the transport of Vph1-EGFP or that other factors act in place of Hse1 to sort ubiquitinated proteins on the vacuolar membrane.
The association between Vps27 and the vacuolar membrane after the diauxic shift (Fig. 4) strongly supports the idea that invagination of the Vph1-containing vacuolar membrane is driven by the direct action of ESCRT proteins. Contributions of ESCRT proteins to membrane deformation were found not only on endosomes but also on the plasma membrane (Henne et al., 2011) and were successfully reproduced, even in vitro (Saksena et al., 2009), supporting the hypothesis that ESCRT proteins act directly on the vacuolar membrane.
Results of the cleavage assay with Vph1-EGFP strongly suggested that the clathrin heavy chain (Chc1) and its association with Vps27 are required for microautophagy (Fig. 3), but what is the role of clathrin in microautophagy? Because Envy-tagged Vps27ΔCB was translocated to the vacuolar membrane, albeit with an efficiency lower than that of the WT protein (Fig. 4 C), the defect in microautophagy found in cells with Vps27ΔCB (Fig. 3) may arise from other aspects of the molecular mechanism for membrane dynamics. Studies of mammalian Hrs (the orthologue of Vps27) demonstrated that this protein recruits clathrin onto endosomes (Raiborg et al., 2001a), and the lattice of clathrin formed on the organelle in turn supports the binding of Hrs to ubiquitinated membrane proteins (Raiborg et al., 2002). Therefore, we assume that the interaction between Vps27 and the clathrin heavy chain contributes to similar recruitment processes followed by vacuole invagination during microautophagy. If this is the case, then the clathrin subunit should be localized on the vacuolar membrane, which will be examined in future studies.
The membrane dynamics of microautophagy eventually generate intravacuolar vesicles derived from the limiting membrane of the vacuole. Thus, it is reasonable that this process required Atg15 (Fig. 2 A), an intravacuolar lipase that has been shown to be necessary for the disintegration of luminal vesicles (i.e., macroautophagic bodies and MVB vesicles; Teter et al., 2001; Epple et al., 2003). Although the precise mechanism by which this lipase specifically lyses these intravacuolar vesicles has not been elucidated, a recent study demonstrated an enzymatic activity of Atg15 toward phosphatidylserine, a phospholipid species accumulated on the cytoplasmic leaflet (Ramya and Rajasekharan, 2016), which might be crucial for its in vivo functions.
To examine whether membrane-fusion machinery also contributes to microautophagy induced after the diauxic shift, we used a temperature-sensitive mutant of PEP3/VPS18 encoding one of class C Vps proteins (Rieder and Emr, 1997) acting on a membrane-tethering process upstream of membrane fusion, as a component of CORVET (class C core vacuole/endosome tethering) and HOPS (homotypic fusion and protein sorting) complexes (Hickey and Wickner, 2010). This mutant strain expressing Vph1-EGFP was cultured for 16 h at a semipermissive temperature (28°C), transferred to a restrictive temperature (34°C) for 1 h, and subjected to EM. Vacuole invaginations were detected at higher frequencies after the temperature shift in this mutant than in the WT or the mutant strain kept at the semipermissive temperature (Fig. S4, A and B). The rate of Vph1-EGFP cleavage was significantly reduced, even at the semipermissive temperature (Fig. S4 C). This result indicates an important role of Vps18 in the transport of Vph1-EGFP into the vacuole. The topology of the invagination during microautophagy generates high-curvature (tip) parts of the vacuolar membrane that are in close proximity to each other and accessible by cytosolic factors. Thus, it is plausible that the class C/HOPS complex including Vps18 functions in the membrane fusion process to release the invaginated structures to the vacuolar lumen.
Recent studies demonstrated that homotypic fusion of the vacuolar membrane at the edge of the contact site of the apposing vacuoles (i.e., vertex fusion) functions in the incorporation of vacuolar membrane proteins into the organelle lumen in a process called the intraluminal fragment (ILF) pathway (Mattie et al., 2017; McNally et al., 2017). Because Vps27 was reported to be dispensable for the ILF pathway, the transport of Vph1 into the vacuole after the diauxic shift depending on Vps27 was suggested to be distinct from the ILF pathway. Indeed, the EM image of the vps18 mutant transferred to the restrictive temperature did not exhibit fragmented vacuoles (Fig. S4 A), the intermediate state of the ILF pathway. Furthermore, fluorescence microscopy of the vacuoles stained with Vph1-EGFP indicated no significant induction of vacuolar contacts or remarkable changes in the number of the vacuoles per cell after the diauxic shift (Fig. S4 D).
Other recent studies of the degradation of several vacuolar transporter proteins reported that these proteins were ubiquitinated, translocated out of the vacuolar membrane, incorporated into MVBs, and then transported into the vacuolar lumen in a process called vReD (vacuolar recycling and degradation) pathway, but Vph1 was reported to be excluded from the vReD pathway (Li et al., 2015a,b). In the present study, fluorescence microscopy analysis strongly suggested that the localization of Vph1-EGFP was restricted to the vacuolar membrane throughout the time periods after the diauxic shift (Fig. S4 D), unlike the targets of the vReD pathway, which have been reported to accumulate at foci near the vacuole (Li et al., 2015b). Thus, it is unlikely that Vph1-EGFP follows the vReD pathway after the diauxic shift.
By establishing dual markers for microautophagy and lipophagy (i.e., Vph1-EGFP and Osw5-EGFP), microautophagy has been shown to function in the degradation of LDs (Figs. 1 D and 5) in an ATG (macroautophagy)–independent manner. Previous studies of yeast microautophagic pathways identified Atg proteins responsible for microautophagy of LDs (microlipophagy; van Zutphen et al., 2014), that of the nuclear portion (piecemeal microautophagy of the nucleus; Krick et al., 2008), and that of peroxisomes in P. pastoris (micropexophagy; Mukaiyama et al., 2004). In contrast, another microautophagic pathway targeting the cytoplasm is partially dependent on the core Atg proteins (Sattler and Mayer, 2000), whereas that targeting ER is independent of the core Atg proteins (Schuck et al., 2014). A recent study of ER-phagy and lipophagy during ER stress (caused by phospholipid biosynthesis deficiency) showed that Atg proteins were also irrelevant for this pathway, whereas the pathway was dependent on ESCRT proteins (Vevea et al., 2015). Similarly, results of our immunoblot analysis (Fig. 2) revealed that microautophagy induced after the diauxic shift required the ESCRT machinery but was independent of the core Atg proteins (with the exception of Atg15). It is worth noting that all of the microautophagic pathways requiring Atg proteins in S. cerevisiae are induced by nitrogen starvation and thus undergo a concomitant induction of macroautophagy that leads to a massive influx of cytoplasmic components and macroautophagic membranes to the vacuole. Because the marker proteins of LDs are peripherally associated with the organelle surface, the possibility cannot be excluded that these marker proteins are dissociated and diffused into the cytoplasm and transported to vacuoles via macroautophagy under nitrogen-starved conditions. In this study, the transport of Osw5-EGFP into the vacuole after the diauxic shift was shown to be independent of Atg1 (Fig. 5 A), clearly indicating the existence of Atg-independent trafficking of LDs to the vacuole.
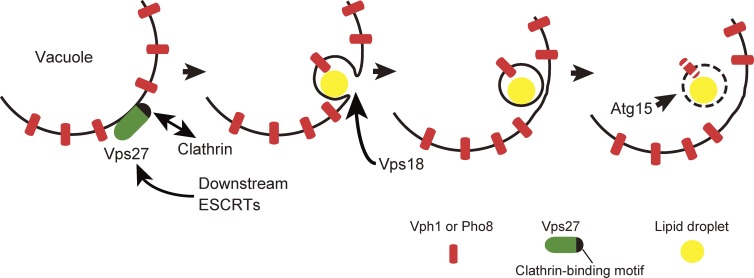
Fig. 6 summarizes the working mechanistic model of microautophagy induced after the diauxic shift in S. cerevisiae. In this model, Vps27 is recruited to the surface of the vacuole upon the diauxic shift (Fig. 4) and recruits other ESCRT proteins (similarly to the MVB pathway) and the clathrin heavy chain, which is necessary for the uptake of the vacuolar membrane portion containing Vph1-EGFP and yEGFP-Pho8 (Fig. 3), along with LDs (Figs. 1 C and 5), into the vacuolar lumen. It is possible that the class C fusion machinery including Vps18 contributes to vacuolar membrane fusion, leading to pinch-off of the invaginated structures into the vacuolar lumen (Fig. S4). The resultant microautophagic body is lysed within the vacuolar lumen in a manner dependent on the intravacuolar lipase Atg15 (Fig. 2 A). Although recent studies have revealed important functions of microautophagy in a wide range of eukaryotic kingdoms (Kawamura et al., 2012; Liu et al., 2015; Uytterhoeven et al., 2015), much remains to be elucidated in terms of molecular mechanism for microautophagy. The findings from this study offer a valuable molecular scheme for studies of higher eukaryotes.
Figure 6.
Schematic model of microautophagy proposed in this study.
Materials and methods
Construction of plasmids and yeast strains
S. cerevisiae W303-1A was used as the host strain, and other strains used in this study are listed in Table 1. All strains were generated for this study. PCR-base gene disruptions with SpHIS5 were done as described previously (Oku et al., 2013). Insertion of the EGFP-coding cassette at the end of VPH1 ORF in the yeast genome was done according to a PCR-based method described previously (Longtine et al., 1998), except that the sequence for GFPS65T was converted to that for EGFP. The Vph1-EGFP coding region in the genome, along with the 5′ UTR of VPH1 and TADH1, was amplified and cloned into the pRS305 vector, yielding pMO150. This plasmid was cut with NheI and introduced into LEU2 locus of the genome to create YMO411 through YMO454. For expression of Cps1 and Pho8 N-terminally tagged with yEGFP, the CgLEU2 gene cloned in BYP1419 (National BioResource Project, Japan) was amplified and cloned into pRS305, whose original ScLEU2 region was removed by treatment with AatII–SacI. Toward the resultant plasmid, 5′-UTR region of CUP1 and yEGFP-coding gene fragments were introduced, yielding pMO151. With this plasmid as the template, the CgLEU2-PCUP1-yEGFP gene fragment was amplified with flanking nucleotide sequences corresponding to the end of 5′ UTR and the start of CPS1 or PHO8 ORF, which were introduced into the genome at the locus of CPS1 or PHO8, respectively, in a manner analogous to that reported in a previous study (Janke et al., 2004). For the expression of Osw5-EGFP, the KanMX gene cassette within the EGFP-cloned vector was replaced by the CgLEU2 fragment, yielding pMO152, which was used as the template for the PCR-based gene tagging method in a matter similar to that used in a previous study (Longtine et al., 1998).
Table 1. Strains used in this study.
| Strain | Genotype |
|---|---|
| YMO400 | W303-1A, ADE2, HIS3, VPH1:: VPH1-EGFP (CgLEU2) |
| YMO411 | W303-1A, ADE2, HIS3, leu2::VPH1-EGFP (LEU2) |
| YMO412 | W303-1A, ADE2, pep4Δ::SpHIS5 (equivalent to ScHIS3), leu2::VPH1-EGFP (LEU2) |
| YMO413 | W303-1A, ADE2, atg1Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO414 | W303-1A, ADE2, atg2Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO415 | W303-1A, ADE2, atg3Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO416 | W303-1A, ADE2, atg4Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO417 | W303-1A, ADE2, atg5Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO418 | W303-1A, ADE2, atg6Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO419 | W303-1A, ADE2, atg7Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO420 | W303-1A, ADE2, atg8Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO421 | W303-1A, ADE2, atg9Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO422 | W303-1A, ADE2, atg10Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO423 | W303-1A, ADE2, atg11Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO424 | W303-1A, ADE2, atg12Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO425 | W303-1A, ADE2,atg13Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO426 | W303-1A, ADE2, atg14Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO427 | W303-1A, ADE2, atg15Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO428 | W303-1A, ADE2, atg16Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO429 | W303-1A, ADE2, atg17Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO430 | W303-1A, ADE2, atg18Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO431 | W303-1A, ADE2, ego1Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO432 | W303-1A, ADE2, ego3Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO433 | W303-1A, ADE2, nvj1Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO434 | W303-1A, ADE2, vac8Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO435 | W303-1A, ADE2, vtc3Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO436 | W303-1A, ADE2, vtc4Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO437 | W303-1A, ADE2, vps4Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO438 | W303-1A, ADE2, vps27Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO439 | W303-1A, ADE2, hse1Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO440 | W303-1A, ADE2, vps23Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO441 | W303-1A, ADE2, vps28Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO442 | W303-1A, ADE2, vps25Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO443 | W303-1A, ADE2,vps36Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO444 | W303-1A, ADE2, snf7Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO445 | W303-1A, ADE2, doa4Δ::SpHIS5, leu2::VPH1-EGFP (LEU2) |
| YMO446 | YMO438, ura3::pRS306, URA3 |
| YMO447 | YMO438, ura3::VPS27WT-pRS306(URA3) |
| YMO448 | YMO438, ura3::vps27VHSmut-pRS306(URA3) |
| YMO459 | YMO438, ura3::vps27FYVEmut-pRS306(URA3) |
| YMO450 | YMO438, ura3::vps27UIMmut-pRS306(URA3) |
| YMO451 | YMO438, ura3::vps27PTAPLmut-pRS306(URA3) |
| YMO452 | YMO438, ura3::vps27ΔCB-pRS306(URA3) |
| YMO453 | W303-1A, ADE2, chc1-5::HIS3, leu2::VPH1-EGFP (LEU2) |
| YMO454 | W303-1A, ADE2, vps18Δ::SpHIS5, leu2::VPH1-EGFP (LEU2), ura3::vps18ts–pRS306(URA3) |
| YMO455 | YMO438, ura3::HA-VPS27WT-pRS306(URA3) |
| YMO456 | YMO438, ura3::HA-vps27VHSmut-pRS306(URA3) |
| YMO457 | YMO438, ura3::HA-vps27FYVEmut-pRS306(URA3) |
| YMO458 | YMO438, ura3::HA-vps27UIMmut-pRS306(URA3) |
| YMO459 | YMO438, ura3::HA-vps27PTAPLmut-pRS306(URA3) |
| YMO460 | YMO438, ura3::HA-vps27ΔCB-pRS306(URA3) |
| YMO461 | W303-1A, ADE2, HIS3, PHO8:: PCUP1 yEGFP-PHO8 (CgLEU2) |
| YMO462 | W303-1A, ADE2, pep4Δ::SpHIS5, PHO8:: PCUP1-yEGFP-PHO8 (CgLEU2) |
| YMO463 | W303-1A, ADE2, vps27Δ::SpHIS5, PHO8:: PCUP1-yEGFP-PHO8 (CgLEU2), ura3::pRS306(URA3) |
| YMO464 | W303-1A, ADE2, vps27Δ::SpHIS5, PHO8:: PCUP1-yEGFP-PHO8 (CgLEU2), ura3::VPS27WT-pRS306(URA3) |
| YMO465 | W303-1A, ADE2, vps27Δ::SpHIS5, PHO8:: PCUP1 -yEGFP-PHO8 (CgLEU2), ura3::vps27ΔCB-pRS306(URA3) |
| YMO471 | W303-1A, ADE2, vps27Δ::SpHIS5, PHO8:: PCUP1 yEGFP-CPS1 (CgLEU2) |
| YMO472 | W303-1A, ADE2, vps27Δ::SpHIS5, PHO8:: PCUP1 yEGFP-CPS1 (CgLEU2), ura3::VPS27WT-pRS306(URA3) |
| YMO473 | W303-1A, ADE2, vps27Δ::SpHIS5, PHO8:: PCUP1 yEGFP-CPS1 (CgLEU2), ura3::vps27ΔCB-pRS306(URA3) |
| YMO481 | W303-1A, ADE2, vps27Δ::SpHIS5, ura3::PVPS27 Envy-VPS27WT-pRS306(URA3) |
| YMO482 | W303-1A, ADE2, vps27Δ::SpHIS5, ura3::PVPS27 Envy-vps27FYVEmut-pRS306(URA3) |
| YMO483 | W303-1A, ADE2, vps27Δ::SpHIS5, ura3::PVPS27 Envy-vps27FYVEΔCB–pRS306(URA3) |
| YMO501 | W303-1A, ADE2+, HIS3+, OSW5::OSW5-EGFP (CgLEU2) |
| YMO502 | W303-1A, ADE2+, pep4Δ::SpHIS5, OSW5::OSW5-EGFP (CgLEU2) |
| YMO503 | W303-1A, ADE2+, atg1Δ::SpHIS5, OSW5::OSW5-EGFP (CgLEU2) |
| YMO504 | W303-1A, ADE2+, vps27Δ::SpHIS5, OSW5::OSW5-EGFP (CgLEU2) ura3::VPS27WT-pRS306(URA3) |
| YMO505 | W303-1A, ADE2+, vps27Δ::SpHIS5, OSW5::OSW5-EGFP (CgLEU2), ura3::vps27DCB-pRS306(URA3) |
| YYK101 | W303-1A, ADE2+, OSW5::OSW5-EGFP (CgLEU2), ERG6-mCherry-KanMX |
| YYK102 | W303-1A, ADE2+, OSW5::FAA4-EGFP (CgLEU2), ERG6-mCherry-KanMX |
The pep3/vps18ts mutant was derived by introduction of a pRS306-based plasmid harboring the C826S point-mutated VPS18 gene (Rieder and Emr, 1997) to a vps18Δ strain expressing Vph1-EGFP.
The VPS27 gene fragment in the parental strain was amplified and cloned into pRS306. Mutations introduced into this original plasmid were VHSmut, L28A/L32D (Ren and Hurley, 2010); FYVEmut, R193A (Raiborg et al., 2001b); UIMmut, A266Q/S270D/A309Q/S313D (Ren and Hurley, 2010); PTAPLmut, P447A/S448A/D449A/P525A/S526A/D527A (Bilodeau et al., 2003); and ΔCB, L618Stop (Bilodeau et al., 2003). For expression of HA or yEGFP-tagged Vps27 variants, the plasmid harboring VPS27 or its mutant derivative was used as the template for inverse PCR, and the HA- or Envy-coding gene fragment (a gift from L.S. Weisman, University of Michigan, Ann Arbor, MI) was inserted between the end of the 5′ UTR and the start of the VPS27 ORF. Each of the resulting plasmids were introduced to the URA3 locus in the vps27Δ strain.
To generate the temperature-sensitive mutant of CHC1, a partial ORF fragment encoding the C-terminal region of Chc1 was cloned into pRS303, and mutagenesis was subsequently performed to eliminate a G nucleotide corresponding to position 4,831 of the ORF (Chen and Graham, 1998). The resultant plasmid, named pMO153, was treated with BamHI for single recombination at the CHC1 locus.
Expression of Mup1-pHluorin was analyzed as described previously (Prosser et al., 2010) using a pBW1679 template plasmid (a gift from B. Wendland, Johns Hopkins University, Baltimore, MD).
Culture conditions
Yeast cells were cultured in YEPD medium (2% [wt/vol] Bacto Yeast Peptone [BD Biosciences], 1% [wt/vol] Bacto Yeast Extract [BD Biosciences], and 2% [wt/vol] glucose) at 28°C for more than 16 h, diluted in fresh YEPD medium to 0.05 OD610, and cultured at 28°C with vigorous agitation.
EM
EM was done using a Pipes-KMnO4 fixation protocol described previously (Wright, 2000).
Fluorescence microscopy
An inverted microscope (IX81; Olympus) equipped with an UPLANSAPO 100× objective lens (numerical aperture 1.40; Olympus) and a charge-coupled device camera (DP30BW; Olympus) was used for fluorescence microscopy. The filter sets used to acquire fluorescence signals were GFP-4050B (Semrock) for GFP and a U-MRFPHQ unit (Olympus) for FM 4–64 signals. Acquired images were processed using MetaMorph version 7.0 (Molecular Devices).
Immunoblot analysis
Cells equivalent to 2 OD610 units were collected and resuspended in 1 ml buffer A (0.2 M NaOH and 0.5% [vol/vol] 2-mercaptoethanol) prechilled on ice. To the suspension, 1/10 vol trichloroacetic acid solution (100% wt/vol) was added. The samples were mixed well, kept on ice for 30 min, and centrifuged at 20,000 g for 5 min. The pellet was resuspended in 1 ml pure acetone prechilled to −20°C and again subjected to centrifugation. After the pellet was air-dried, it was resuspended in 40 µl sample buffer 1 (0.1 M NaCl, 25 mM Tris-HCl, pH 6.8, 15% [vol/vol] glycerol, 1.5% [wt/vol] SDS, 0.005% [wt/vol] bromophenol blue, and 0.5% [vol/vol] 2-mercaptoethanol) and heated at 65°C for 15 min to detect Vph1-EGFP, yEGFP-Cps1, and yEGFP-Pho8. For immunoblot analysis of Osw5-EGFP, the dried pellet was solubilized in 40 µl sample buffer 2 (100 mM Tris-HCl, pH 7.5, 1% [vol/vol] glycerol, 2% [wt/vol] SDS, 0.005% [wt/vol] bromophenol blue, and 0.5% [vol/vol] 2-mercaptoethanol) and heated at 80°C for 10 min. The solubilized and heated samples were subjected to centrifugation at 20,000 g for 5 min at ambient temperature, and 10 µl of the supernatant fractions was applied to SDS-PAGE. The proteins in the gel were transferred to an Immobilon-P PVDF membrane (Merck-Millipore) with Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad laboratories). Living Colors A.v. monoclonal antibody (JL-8) was used to detect EGFP or yEGFP after 2,000-fold dilution, and mouse monoclonal antibody (mAbcam 8224) to β-actin was also used at 2,000-fold dilution. As the secondary antibody, anti-IgG (H+L chain; mouse) pAB-HRP (MBL International) was used after 7,500-fold dilution.
Online supplemental material
Fig. S1 shows data from immunoblot analysis of Vph1-EGFP expressed in strains harboring HA-tagged Vps27 variants. Fig. S2 shows the results of fluorescence microscopy for the assessment of endocytosis functionality of a Vps27 variant devoid of its clathrin-binding motif. Fig. S3 show immunoblot data of expression levels of Envy-Vps27 variants and microscopic images comparing the localization of Erg6, Faa4, and Osw5 fluorescent protein fusions. Fig. S4 shows the results of EM and immunoblot analysis of Vph1-EGFP for vps18 mutants. Video 1 shows movement of the intravacuolar population of the Vph1-EGFP signal.
Supplementary Material
Acknowledgments
The authors thank Dr. Beverly Wendland for the gift of the pHluorin-coding plasmid, Dr. Lois Weisman for offering the Envy-containing plasmid, and Dr. Yoshinori Ohsumi (Tokyo Institute of Technology) for his encouragement.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (grant-in-aid for scientific research on priority areas 16H01200 to Y. Sakai) and the Japan Society for the Promotion of Science (grant-in-aid for scientific research C 16K07689 to M. Oku).
The authors declare no competing financial interests.
Author contributions: M. Oku performed experiments, analyzed data, and wrote the initial draft of manuscript. Y. Maeda, T. Kondo, Y. Kagohashi, and M. Yamada performed the experiments. T. Fujimoto designed and performed EM experiments. Y. Sakai supervised the research project, designed the experiments, and wrote and edited the manuscript.
Footnotes
Abbreviations used:
- Atg
- autophagy related
- ESCRT
- endosomal sorting complex required for transport
- ILF
- intraluminal fragment
- LD
- lipid droplet
- MVB
- multivesicular body
- PI3P
- phosphatidylinositol 3′ monophosphate
- PTAPL
- proline-threonine-alanine-proline–like
References
- Amerik A.Y., Nowak J., Swaminathan S., and Hochstrasser M.. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 11:3365–3380. 10.1091/mbc.11.10.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci D., and Lemberg M.K.. 2015. Clipping or extracting: Two ways to membrane protein degradation. Trends Cell Biol. 25:611–622. 10.1016/j.tcb.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., and Emr S.D.. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau P.S., Urbanowski J.L., Winistorfer S.C., and Piper R.C.. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539. [DOI] [PubMed] [Google Scholar]
- Bilodeau P.S., Winistorfer S.C., Kearney W.R., Robertson A.D., and Piper R.C.. 2003. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163:237–243. 10.1083/jcb.200305007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoca A., Kovinich N., Burkel B., Stecha S., Bohorquez-Restrepo A., Ueda T., Eliceiri K.W., Grotewold E., and Otegui M.S.. 2015. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell. 27:2545–2559. 10.1105/tpc.15.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., and Graham T.R.. 1998. An arf1Delta synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics. 150:577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., and Klionsky D.J.. 2008. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 451:1–26. 10.1016/S0076-6879(08)03201-1 [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Bertram G., Egerton M., Gow N.A., Falkow S., and Brown A.J.. 1997. Yeast-enhanced green fluorescent protein (yEGFP): A reporter of gene expression in Candida albicans. Microbiology. 143:303–311. 10.1099/00221287-143-2-303 [DOI] [PubMed] [Google Scholar]
- Cowles C.R., Snyder W.B., Burd C.G., and Emr S.D.. 1997. Novel Golgi to vacuole delivery pathway in yeast: Identification of a sorting determinant and required transport component. EMBO J. 16:2769–2782. 10.1093/emboj/16.10.2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., and De Virgilio C.. 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell. 19:15–26. 10.1016/j.molcel.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Epple U.D., Suriapranata I., Eskelinen E.L., and Thumm M.. 2001. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 183:5942–5955. 10.1128/JB.183.20.5942-5955.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple U.D., Eskelinen E.L., and Thumm M.. 2003. Intravacuolar membrane lysis in Saccharomyces cerevisiae: Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 278:7810–7821. 10.1074/jbc.M209309200 [DOI] [PubMed] [Google Scholar]
- Galdieri L., Mehrotra S., Yu S., and Vancura A.. 2010. Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS. 14:629–638. 10.1089/omi.2010.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier J.M., Simonsen A., D’Arrigo A., Bremnes B., Stenmark H., and Aasland R.. 1998. FYVE fingers bind PtdIns(3)P. Nature. 394:432–433. 10.1038/28767 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Hickey C.M., and Wickner W.. 2010. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol. Biol. Cell. 21:2297–2305. 10.1091/mbc.E10-01-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kane P.M. 1995. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270:17025–17032. [PubMed] [Google Scholar]
- Kane P.M. 2007. The long physiological reach of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 39:415–421. 10.1007/s10863-007-9112-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Stefan C.J., Babst M., and Emr S.D.. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423. 10.1083/jcb.200302136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N., Sun-Wada G.H., Aoyama M., Harada A., Takasuga S., Sasaki T., and Wada Y.. 2012. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 3:1071 10.1038/ncomms2069 [DOI] [PubMed] [Google Scholar]
- Kissová I., Salin B., Schaeffer J., Bhatia S., Manon S., and Camougrand N.. 2007. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 3:329–336. 10.4161/auto.4034 [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Veenhuis M., and van der Klei I.J.. 2013. Lipid droplets and peroxisomes: Key players in cellular lipid homeostasis or a matter of fat–Store ‘em up or burn ’em down. Genetics. 193:1–50. 10.1534/genetics.112.143362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.L., Millen J., Goldfarb D.S., and Thumm M.. 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 19:4492–4505. 10.1091/mbc.E08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam E., and Goldfarb D.S.. 2007. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 3:85–92. 10.4161/auto.3586 [DOI] [PubMed] [Google Scholar]
- Li M., Koshi T., and Emr S.D.. 2015a Membrane-anchored ubiquitin ligase complex is required for the turnover of lysosomal membrane proteins. J. Cell Biol. 211:639–652. 10.1083/jcb.201505062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Rong Y., Chuang Y.S., Peng D., and Emr S.D.. 2015b Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol. Cell. 57:467–478. 10.1016/j.molcel.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Li S.C., Diakov T.T., Xu T., Tarsio M., Zhu W., Couoh-Cardel S., Weisman L.S., and Kane P.M.. 2014. The signaling lipid PI(3,5)P2 stabilizes V1-V(o) sector interactions and activates the V-ATPase. Mol. Biol. Cell. 25:1251–1262. 10.1091/mbc.E13-10-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.M., Sun L.L., Hu W., Ding Y.H., Dong M.Q., and Du L.L.. 2015. ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Mol. Cell. 59:1035–1042. 10.1016/j.molcel.2015.07.034 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mattie S., McNally E.K., Karim M.A., Vali H., and Brett C.L.. 2017. How and why intralumenal membrane fragments form during vacuolar lysosome fusion. Mol. Biol. Cell. 28:309–321. 10.1091/mbc.E15-11-0759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally E.K., Karim M.A., and Brett C.L.. 2017. Selective lysosomal transporter degradation by organelle membrane fusion. Dev. Cell. 40:151–167. 10.1016/j.devcel.2016.11.024 [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., and Devenish R.J.. 2011. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy. 7:673–682. 10.4161/auto.7.7.14733 [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Oku M., Baba M., Samizo T., Hammond A.T., Glick B.S., Kato N., and Sakai Y.. 2002. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 7:75–90. 10.1046/j.1356-9597.2001.00499.x [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Baba M., Osumi M., Aoyagi S., Kato N., Ohsumi Y., and Sakai Y.. 2004. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell. 15:58–70. 10.1091/mbc.E03-05-0340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O., Sattler T., Flötenmeyer M., Schwarz H., Plattner H., and Mayer A.. 2000. Autophagic tubes: Vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 151:519–528. 10.1083/jcb.151.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M., Hoseki J., Ichiki Y., and Sakai Y.. 2013. A fluorescence resonance energy transfer (FRET)-based redox sensor reveals physiological role of thioredoxin in the yeast Saccharomyces cerevisiae. FEBS Lett. 587:793–798. 10.1016/j.febslet.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Piper R.C., Cooper A.A., Yang H., and Stevens T.H.. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603–617. 10.1083/jcb.131.3.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser D.C., Whitworth K., and Wendland B.. 2010. Quantitative analysis of endocytosis with cytoplasmic pHluorin chimeras. Traffic. 11:1141–1150. 10.1111/j.1600-0854.2010.01088.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Bache K.G., Mehlum A., Stang E., and Stenmark H.. 2001a Hrs recruits clathrin to early endosomes. EMBO J. 20:5008–5021. 10.1093/emboj/20.17.5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Bremnes B., Mehlum A., Gillooly D.J., D’Arrigo A., Stang E., and Stenmark H.. 2001b FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114:2255–2263. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache K.G., Gillooly D.J., Madshus I.H., Stang E., and Stenmark H.. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394–398. 10.1038/ncb791 [DOI] [PubMed] [Google Scholar]
- Ramya V., and Rajasekharan R.. 2016. ATG15 encodes a phospholipase and is transcriptionally regulated by YAP1 in Saccharomyces cerevisiae. FEBS Lett. 590:3155–3167. 10.1002/1873-3468.12369 [DOI] [PubMed] [Google Scholar]
- Ren X., and Hurley J.H.. 2010. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29:1045–1054. 10.1038/emboj.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Kee Y., Huibregtse J.M., and Piper R.C.. 2007. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol. Biol. Cell. 18:324–335. 10.1091/mbc.E06-06-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S.E., and Emr S.D.. 1997. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell. 8:2307–2327. 10.1091/mbc.8.11.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., and Goldfarb D.S.. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141. 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.H., Yamashiro C.T., Raymond C.K., Kane P.M., and Stevens T.H.. 1989. Acidification of the lysosome-like vacuole and the vacuolar H+-ATPase are deficient in two yeast mutants that fail to sort vacuolar proteins. J. Cell Biol. 109:93–100. 10.1083/jcb.109.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., and Santambrogio L.. 2011. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 20:131–139. 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Koller A., Rangell L.K., Keller G.A., and Subramani S.. 1998. Peroxisome degradation by microautophagy in Pichia pastoris: Identification of specific steps and morphological intermediates. J. Cell Biol. 141:625–636. 10.1083/jcb.141.3.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A.E., and Emr S.D.. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 136:97–109. 10.1016/j.cell.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler T., and Mayer A.. 2000. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J. Cell Biol. 151:529–538. 10.1083/jcb.151.3.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Gallagher C.M., and Walter P.. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127:4078–4088. 10.1242/jcs.154716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slubowski C.J., Funk A.D., Roesner J.M., Paulissen S.M., and Huang L.S.. 2015. Plasmids for C-terminal tagging in Saccharomyces cerevisiae that contain improved GFP proteins, Envy and Ivy. Yeast. 32:379–387. 10.1002/yea.3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y., Rodriguez R.K., Coluccio A.E., and Neiman A.M.. 2009. A screen for spore wall permeability mutants identifies a secreted protease required for proper spore wall assembly. PLoS One. 4:e7184 10.1371/journal.pone.0007184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter S.A., Eggerton K.P., Scott S.V., Kim J., Fischer A.M., and Klionsky D.J.. 2001. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 276:2083–2087. 10.1074/jbc.C000739200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttenweiler A., Schwarz H., Neumann H., and Mayer A.. 2007. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell. 18:166–175. 10.1091/mbc.E06-08-0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uytterhoeven V., Lauwers E., Maes I., Miskiewicz K., Melo M.N., Swerts J., Kuenen S., Wittocx R., Corthout N., Marrink S.J., et al. . 2015. Hsc70-4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron. 88:735–748. 10.1016/j.neuron.2015.10.012 [DOI] [PubMed] [Google Scholar]
- van Zutphen T., Todde V., de Boer R., Kreim M., Hofbauer H.F., Wolinski H., Veenhuis M., van der Klei I.J., and Kohlwein S.D.. 2014. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25:290–301. 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea J.D., Garcia E.J., Chan R.B., Zhou B., Schultz M., Di Paolo G., McCaffery J.M., and Pon L.A.. 2015. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell. 35:584–599. 10.1016/j.devcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., and Emr S.D.. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792. 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J.J., and Payne G.S.. 1998. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 17:2482–2493. 10.1093/emboj/17.9.2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206:357–366. 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510. [DOI] [PubMed] [Google Scholar]
- Xie Z., and Klionsky D.J.. 2007. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 9:1102–1109. 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.