Kourtidis et al. demonstrate that cadherin complexes at the zonula adherens recruit a diverse set of mRNAs to a junction-associated RISC, via PLEKHA7. In this way, PLEKHA7 regulates expression of critical regulators of epithelial homeostasis, such as JUN, MYC, and SOX2, through Ago2 and miRNA-mediated silencing.
Abstract
Cumulative evidence demonstrates that most RNAs exhibit specific subcellular distribution. However, the mechanisms regulating this phenomenon and its functional consequences are still under investigation. Here, we reveal that cadherin complexes at the apical zonula adherens (ZA) of epithelial adherens junctions recruit the core components of the RNA-induced silencing complex (RISC) Ago2, GW182, and PABPC1, as well as a set of 522 messenger RNAs (mRNAs) and 28 mature microRNAs (miRNAs or miRs), via PLEKHA7. Top canonical pathways represented by these mRNAs include Wnt/β-catenin, TGF-β, and stem cell signaling. We specifically demonstrate the presence and silencing of MYC, JUN, and SOX2 mRNAs by miR-24 and miR-200c at the ZA. PLEKHA7 knockdown dissociates RISC from the ZA, decreases loading of the ZA-associated mRNAs and miRNAs to Ago2, and results in a corresponding increase of MYC, JUN, and SOX2 protein expression. The present work reveals a mechanism that directly links junction integrity to the silencing of a set of mRNAs that critically affect epithelial homeostasis.
Introduction
The spatial distribution of RNAs is considered a widespread phenomenon with possibly critical functional consequences (Buxbaum et al., 2015). In the most comprehensive study to date, almost two-thirds of all mRNAs in Drosophila melanogaster exhibited subcellular localization patterns, including localization at areas of cell–cell contact (Lécuyer et al., 2007). In mammalian cells, specific mRNAs localize to cell protrusions (Mili et al., 2008) and promote their formation (Mardakheh et al., 2015). Dendritic mRNA transport and localized translation of select mRNAs at neuronal synapses has also been reported in vertebrates (Fernandez-Moya et al., 2014). Overall, it has been proposed that RNA localization may be facilitating localized protein expression, efficient protein complex formation, signaling regulation, or mRNA retention under conditions of stress (Anderson and Kedersha, 2009; Weatheritt et al., 2014). Nevertheless, the functional and mechanistic details of this phenomenon are still largely unexplored. Additionally, the presence and function of RNA transcripts at cell–cell junctions of mammalian epithelial cells has not been explored yet, with the exception of the junctional recruitment and local translation of the β-actin mRNA, which was shown to be necessary for adherens junction (AJ) assembly (Gutierrez et al., 2014).
AJs are adhesive structures essential for the development and maintenance of the epithelial phenotype (Harris and Tepass, 2010; Takeichi, 2014). Loss of AJ integrity results in developmental abnormalities and pathological conditions (Kourtidis et al., 2013; Yu and Elble, 2016). Mature AJs associate with a circumferential actin ring in polarized epithelial cells, forming the apical zonula adherens (ZA; Harris and Tepass, 2010; Takeichi, 2014). E-cadherin, the main AJ adhesion molecule in epithelial cells, associates with the protein family of catenins, which stabilize the structure, tether the cytoskeleton to the AJs, and mediate its downstream signaling (Harris and Tepass, 2010; Kourtidis et al., 2013; Takeichi, 2014; McCrea et al., 2015). p120 catenin (p120) was originally identified as a substrate of the Src oncogene (Reynolds et al., 1989), but soon afterward was recognized as a critical partner and stabilizer of E-cadherin and therefore of the AJs (Reynolds et al., 1994; Thoreson et al., 2000). Although E-cadherin and p120 localize at both apical and basolateral areas of cell–cell contact, the p120 binding partner PLEKHA7 localizes exclusively at the apical ZA (Meng et al., 2008; Pulimeno et al., 2010; Kourtidis et al., 2015), where it tethers the minus ends of microtubules (Meng et al., 2008) and promotes AJ integrity (Meng et al., 2008; Kourtidis et al., 2015). PLEKHA7 has a striking functional role in regulating the first step of miRNA biogenesis by recruiting the microprocessor complex to the ZA (Kourtidis et al., 2015). This way, PLEKHA7 promotes the processing of a small subset of miRNAs that suppress anchorage-independent growth (Kourtidis et al., 2015). In agreement, PLEKHA7 is consistently mislocalized or lost in breast and kidney patient tumor samples (Kourtidis et al., 2015; Tille et al., 2015).
The microprocessor complex regulates the processing of primary miRNAs to precursor miRNAs in what is the first step in the miRNA maturation pathway (Gregory et al., 2004; Ha and Kim, 2014). However, the RNAi machinery eventually fulfills its mRNA-silencing function via mature miRNAs that are incorporated in the RNA-induced silencing complex (RISC; Hammond et al., 2000, 2001; Krol et al., 2010). RISC targets mRNAs to either translational repression or degradation, resulting in suppression of protein expression (Hammond et al., 2000; Pillai et al., 2005, 2007; Kiriakidou et al., 2007). Therefore, beyond its action in regulating miRNA processing, we have now explored the interplay between PLEKHA7, RISC function, and the repertoire of mRNAs that are potentially regulated by PLEKHA7 and the ZA. Here, we show that PLEKHA7 associates with and recruits the RISC to the ZA. Furthermore, we demonstrate that specific sets of mRNAs and mature miRNAs associate with PLEKHA7 and the RISC at apical junctions, to regulate expression of critical regulators of epithelial cell behavior.
Results and discussion
PLEKHA7 associates with RISC and miRNA loading complex at the ZA
Immunoprecipitation (IP) of PLEKHA7 in Caco2 intestinal epithelial cells and subsequent mass-spectrometry analysis identified RNA posttranscriptional modification as the top interacting network (Table S1). Literature interrogation revealed that this PLEKHA7 interacting network includes most known members of RISC (Höck et al., 2007; Landthaler et al., 2008). In particular, the analysis suggested interaction of PLEKHA7 with Argonaute 2 (Ago2), the main enzymatic component of RISC that catalyzes miRNA-mediated silencing and degradation of mRNAs (Hammond et al., 2001; Liu et al., 2004; Meister et al., 2004; Table 1). It also revealed interactions of PLEKHA7 with the other essential RISC factors, TNRC6A (GW182), TNRC6B, and PABPC1 (Eulalio et al., 2008; Table 1). In addition, the proteomics revealed interactions with several accessory factors that have been previously associated with RISC and are also critical for its function, such as several HNRNP proteins, RNA helicases, heat shock proteins, and other RNA-binding proteins (Höck et al., 2007; Landthaler et al., 2008; Table 1). A functional RISC requires the prior and transient formation of the miRNA loading complex (miRLC) that delivers miRNAs to Ago2, triggering subsequent RISC assembly (Chendrimada et al., 2005; Maniataki and Mourelatos, 2005). Our proteomic analysis suggested that Dicer and TRBP, the main components of miRLC (Maniataki and Mourelatos, 2005), also interact with PLEKHA7 (Table 1). Combined, these data support the interaction of PLEKHA7 with all main components of the RNAi machinery required for miRNA-mediated mRNA silencing.
Table 1. PLEKHA7 IP and proteomics reveal association with the RISC.
| Gene ID | Gene name | Exclusive unique spectrum count | Protein identification probability |
|---|---|---|---|
| Adherens junctions controls | |||
| CDH1 | Cadherin-1; E-cadherin | 8 | 100.00% |
| CTNND1 | Catenin δ-1; p120 catenin | 8 | 100.00% |
| RISC-related hits | |||
| Core RISC proteins | |||
| EIF2C2 | Protein Argonaute-2 Ago2 | 18 | 100.00% |
| PABPC1 | Polyadenylate-binding protein 1 | 20 | 100.00% |
| TNRC6A | Trinucleotide repeat-containing gene 6A protein (GW182) | 5 | 100.00% |
| TNRC6B | Trinucleotide repeat-containing gene 6B | 16 | 100.00% |
| Accessory proteins | |||
| DDX5 | Probable ATP-dependent RNA helicase DDX5 | 9 | 100.00% |
| DDX6 | Probable ATP-dependent RNA helicase DDX6 | 6 | 100.00% |
| DDX17 | Probable ATP-dependent RNA helicase DDX17 | 20 | 100.00% |
| DDX20 | Probable ATP-dependent RNA helicase DDX20 | 14 | 100.00% |
| ELAVL1 | ELAV-like protein 1 | 4 | 100.00% |
| GEMIN4 | Gem-associated protein 4 | 6 | 100.00% |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 31 | 100.00% |
| HNRNPC | Heterogeneous nuclear ribonucleoprotein C1/C2 | 5 | 100.00% |
| HNRNPF | Heterogeneous nuclear ribonucleoprotein F | 17 | 100.00% |
| HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | 8 | 100.00% |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | 21 | 100.00% |
| HSP90AB1 | HSP 90-β | 21 | 100.00% |
| HSP90AA1 | HSP 90-α | 10 | 100.00% |
| HSPA1A | Heat shock 70-kD protein 1A/1B | 14 | 100.00% |
| HSPA8 | Heat shock cognate, 71 kD | 40 | 100.00% |
| IGF2BP1 | Insulin-like growth factor 2 mRNA-binding protein 1 | 28 | 100.00% |
| IGF2BP2 | Insulin-like growth factor 2 mRNA-binding protein 2 | 5 | 100.00% |
| IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 | 10 | 100.00% |
| MOV10 | Putative helicase MOV-10 | 12 | 100.00% |
| RALY | RNA-binding protein Raly | 3 | 100.00% |
| RBM4 | RNA-binding protein 4 | 12 | 100.00% |
| YBX1 | Nuclease-sensitive element-binding protein 1 | 2 | 100.00% |
| miRLC-related hits | |||
| DICER | Isoform 2 of endoribonuclease Dicer | 2 | 100.00% |
| TRBP | Isoform 2 of TAR DNA-binding protein | 12 | 100.00% |
Hits are in alphabetical order for each category. HSP, heat shock protein.
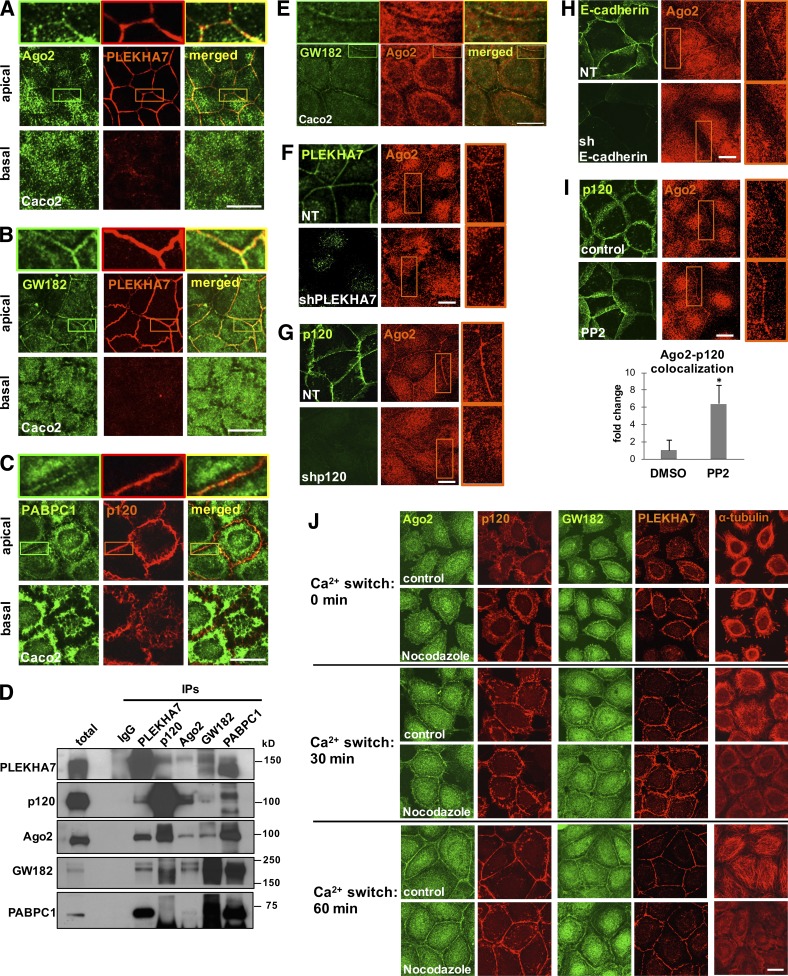
PLEKHA7 is a junctional protein that strictly localizes to the apical ZA in polarized epithelial cells (Meng et al., 2008; Pulimeno et al., 2010; Kourtidis et al., 2015). Therefore, PLEKHA7 proteomics suggest that the RISC components localize at the ZA, in addition to their widely reported cytoplasmic localization (Krol et al., 2010; Ha and Kim, 2014). To examine this and also confirm the proteomics results, we looked at the subcellular localization of RISC core proteins Ago2, GW182, and PABPC1 by costaining with PLEKHA7 or p120 in polarized monolayers of Caco2 and canine kidney MDCK cells, both classic models of epithelial cell morphology. Confocal microscopy revealed that Ago2, GW182, and PABPC1 indeed colocalize with PLEKHA7 or p120 at the apical ZA in both Caco2 and MDCK cells, in addition to their expected abundant cytoplasmic distribution, whereas they were absent from the basolateral junctions (Fig. 1, A–C; and Fig. S1, A–C). Furthermore, the junctional staining obtained with a different Ago2-specific antibody (Fig. S1 D), as well as the loss of junctional localization upon siRNA-mediated knockdown of Ago2 (Fig. S1 E), excluded the possibility of nonspecific staining and confirmed the presence of Ago2 at the ZA. In agreement with the proteomics and immunofluorescence data, co-IP of PLEKHA7 with Ago2, GW182, and PABPC1 was detected by Western blot analysis, further confirming the interactions between these proteins (Fig. 1 D and Fig. S1 F). Ago2 and GW182 costaining further confirmed their colocalization at areas of cell–cell contact (Fig. 1 E).
Figure 1.
Cadherin complexes recruit RISC to the apical junctions of polarized epithelial cells. Caco2 cells were grown to polarize and subjected to immunofluorescence for PLEKHA7 or p120 and the core proteins of the RISC complex Ago2 (A), GW182 (B), and PABPC1 (C). Images were obtained by confocal microscopy, and image stacks were acquired across the entire polarized monolayer; representative apical-basal images are shown. Enlarged parts of images on top of each stack indicate areas of cell–cell contact. (D) Western blots of PLEKHA7, p120, Ago2, GW182, and PABPC1 IPs of Caco2 cells for the same markers. IgG is the negative control. Molecular masses (kD) are indicated on the right. (E) Confocal microscopy images after immunofluorescence of Caco2 cells for GW182 and Ago2. Enlarged parts of images on top indicate areas of cell–cell contact. (F) Immunofluorescence of control (nontarget; NT) or PLEKHA7 knockdown (shPLEKHA7) Caco2 cells for Ago2, costained for PLEKHA7. PLEKHA7 background intracellular staining is an artifact of paraformaldehyde fixation. (G) Immunofluorescence of control (NT) or p120 knockdown (shp120) Caco2 cells for Ago2, costained for p120. (H) Immunofluorescence of control (NT) or E-cadherin knockdown (shE-cadherin) Caco2 cells for Ago2, costained for E-cadherin. (I) Caco2 cells treated with either vehicle control (DMSO) or the Src inhibitor PP2 were costained by immunofluorescence for p120 and Ago2. Colocalization of the Ago2 and p120 signals (bottom) was calculated using the Manders coefficient and expressed as fold change of the DMSO control (mean ± SD from n = 3 independent experiments; *, P < 0.02, Student’s two-tailed t test). (J) Calcium switch assay of Caco2 cells for the indicated time points after Ca2+ readdition (Ca2+: 0 min indicates Ca2+ depleted cells immediately before Ca2+ readdition), treated with vehicle (control) or 10 µM nocodazole and stained by immunofluorescence for α-tubulin, PLEKHA7, p120, Ago2, and GW182. Bars, 20 µM. Insets are magnified 3×.
Similar to the core RISC components, both Dicer and TRBP also exhibited apical junctional localization in polarized Caco2 cells (Fig. S1, G and H). Western blot analysis confirmed the presence of Dicer and TRBP in PLEKHA7 immunoprecipitates from Caco2 cells (Fig. S1 I). The ability of PLEKHA7 to coimmunoprecipitate with Ago2 and Dicer was unaffected by RNase treatment of the cell lysates, indicating that these interactions are RNA independent (Fig. S1 J). The use of a different Dicer-specific antibody and elimination of Dicer junctional staining after siRNA-mediated knockdown further validated the specificity of these findings (Fig. S1 K). Together, the data demonstrate that PLEKHA7 associates with the core components of RISC and miRLC at the apical ZA in polarized epithelial monolayers.
Localization of Ago2 at the ZA depends on E-cadherin, p120, and PLEKHA7
Next, we asked whether the junctional localization of the core RISC component Ago2 depends on PLEKHA7. Indeed, knockdown of PLEKHA7 in Caco2 cells resulted in loss of Ago2 junctional localization (Fig. 1 F). PLEKHA7 is tethered to the junctions via p120 and E-cadherin (Meng et al., 2008). Therefore, we examined whether the junctional localization of Ago2 is also dependent on p120 and E-cadherin. Knockdown of either p120 or E-cadherin resulted in loss of Ago2 junctional localization (Fig. 1, G and H), arguing that Ago2 is recruited to an E-cadherin–based junctional complex.
It is well established that activated Src negatively affects the integrity of E-cadherin–based AJs (Ozawa and Ohkubo, 2001; Irby and Yeatman, 2002). We have previously shown that inhibition of Src activity strengthens PLEKHA7 localization to the junctions (Kourtidis et al., 2015). Consequently, pharmacological inhibition of Src by PP2 also strengthened the junctional localization of Ago2 (Fig. 1 I), suggesting that Src activity disrupts localization of RISC to the ZA.
A calcium-switch assay showed that the membranous localization of Ago2 and GW182 was lost upon calcium depletion but was quickly recovered upon calcium readdition, behavior identical to that of the localizations of PLEKHA7 and p120 (Fig. 1 J). This observation further demonstrates that cadherin-based junctions are required for the membranous localization of RISC. Notably, RISC’s localization at the junctions was not affected by nocodazole treatment during the calcium switch (Fig. 1 J), indicating that it is microtubule independent and therefore not associated with the microtubule-binding function of PLEKHA7 (Meng et al., 2008). In agreement, the junctional localization of Ago2 was not affected after knockdown of Nezha (Fig. S1 L), a junctional protein that tethers PLEKHA7 to the microtubules (Meng et al., 2008). Collectively, these results demonstrate that localization of RISC at the ZA depends on E-cadherin and the junctional presence of both p120 and PLEKHA7.
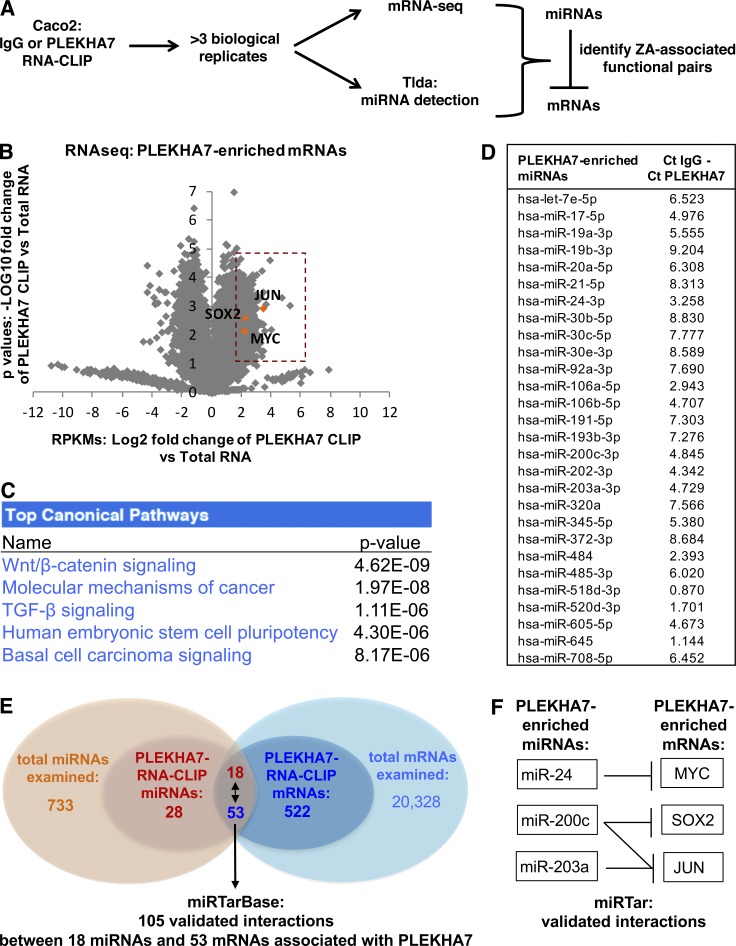
A diverse set of mRNAs and specific miRNAs associate with PLEKHA7 at the ZA
The recruitment of the RISC to the apical junctions suggested that mRNAs are present and silenced at the apical ZA. To investigate this in a comprehensive fashion, we sought to isolate all mRNAs and miRNAs that are potentially bound to the ZA-associated RISC. To accomplish this, we performed RNA IP of PLEKHA7 after intracellular cross-linking (RNA-CLIP) followed by stringent washes to minimize the opportunistic interactions of PLEKHA7 complexes with RNAs during IP. Because PLEKHA7 is not known to be an RNA-binding protein on its own but would coprecipitate with RNAs indirectly via its associated RISC, we performed formaldehyde cross-linking to maintain both the protein–protein and the protein–RNA interactions of the PLEKHA7-associated complexes (Niranjanakumari et al., 2002; Darnell, 2010; see also Materials and methods). We then used the eluted RNA from these RNA-CLIPs to identify mRNAs by performing RNA deep sequencing and to detect miRNAs with the TaqMan Low Density Array Human MicroRNA Panel (TLDA; Fig. 2 A). Because of the IP, we reasoned that mRNAs selectively associated to PLEKHA7 should be enriched in the PLEKHA7 CLIP compared with their level in total cell lysates. Therefore, to increase stringency, we compared the RPKMs of the PLEKHA7 CLIP with the whole-cell RNA-seq RPKM counts and selected mRNAs that were more than fourfold enriched in the PLEKHA7 CLIP. This analysis yielded a total of 522 mRNAs (Fig. 2 B and Table S2). Bioinformatic analysis showed that the top canonical pathways represented in this set of PLEKHA7-enriched mRNAs were the Wnt/β-catenin signaling pathway, TGF-β pathway, cancer signaling, and stem cell regulation (Fig. 2 C). Notably, one of the top PLEKHA7-enriched mRNAs was JUN (Fig. 2 B), a master regulator of oncogenic transformation (Vogt, 2001). Two other mRNAs that were highly enriched in both the PLEKHA7 CLIPs and the top pathways were the MYC oncogene, also a stem cell regulator, and SOX2 (Fig. 2 B), another major factor of pluripotency (Bernhardt et al., 2012).
Figure 2.
Analysis of PLEKHA7-associated RNAs reveals canonical pathways and candidate mRNAs regulated by PLEKHA7. (A) Overview of the experimental approach to identify junction-associated RNAs. (B) Volcano plot of the mRNA sequencing data, resulting from the PLEKHA7 RNA-CLIP. Only PLEKHA7-CLIP mRNAs with RPKMs that were above the IgG-CLIP RPKM background were included and compared with the total mRNA RPKMs to calculate fold enrichment (log2 values on the x axis) and p-values (n = 4 independent experiments; log10 values on the y axis). mRNAs showing fourfold enrichment (P < 0.05) in the PLEKHA7-CLIP are indicated in the orange box, highlighting JUN, SOX2, and MYC. See also Table S2. (C) Pathway analysis of the 522 PLEKHA7-enriched mRNAs, by using Ingenuity Pathway Analysis (Qiagen). The top pathways are shown. (D) RNA isolated as shown in A was subjected to TLDA. A total of 28 miRNAs exhibited Ct values in the PLEKHA7 RNA-CLIP that were above the negative control (IgG) RNA-CLIP; the mean is shown (Ct IgG/Ct PLEKHA7 column, n = 3 independent experiments). See also Table S3. (E and F) Schematics indicating the number and validated interactions (as reported in miRTar) of the PLEKHA7-enriched mRNAs and miRNAs. See also Table S4.
A functional RISC at the ZA entails silencing of mRNAs by associated miRNAs. Indeed, the TLDA analysis revealed 28 mature miRNAs to be enriched in the PLEKHA7 CLIPs of 758 examined (Fig. 2 D and Table S3). This set of miRNAs included members of the miR-30 family, in particular miR-30b, miR-30c, and miR-30e (Fig. 2 D); members of the miR-17-92 polycistronic family, namely miR-17, miR-19a, miR-19b, and miR-92a (He et al., 2005); the tumor suppressor miR-24; and the epithelial–mesenchymal transition (EMT) suppressor miR-200c (Mutlu et al., 2016; Fig. 2 D).
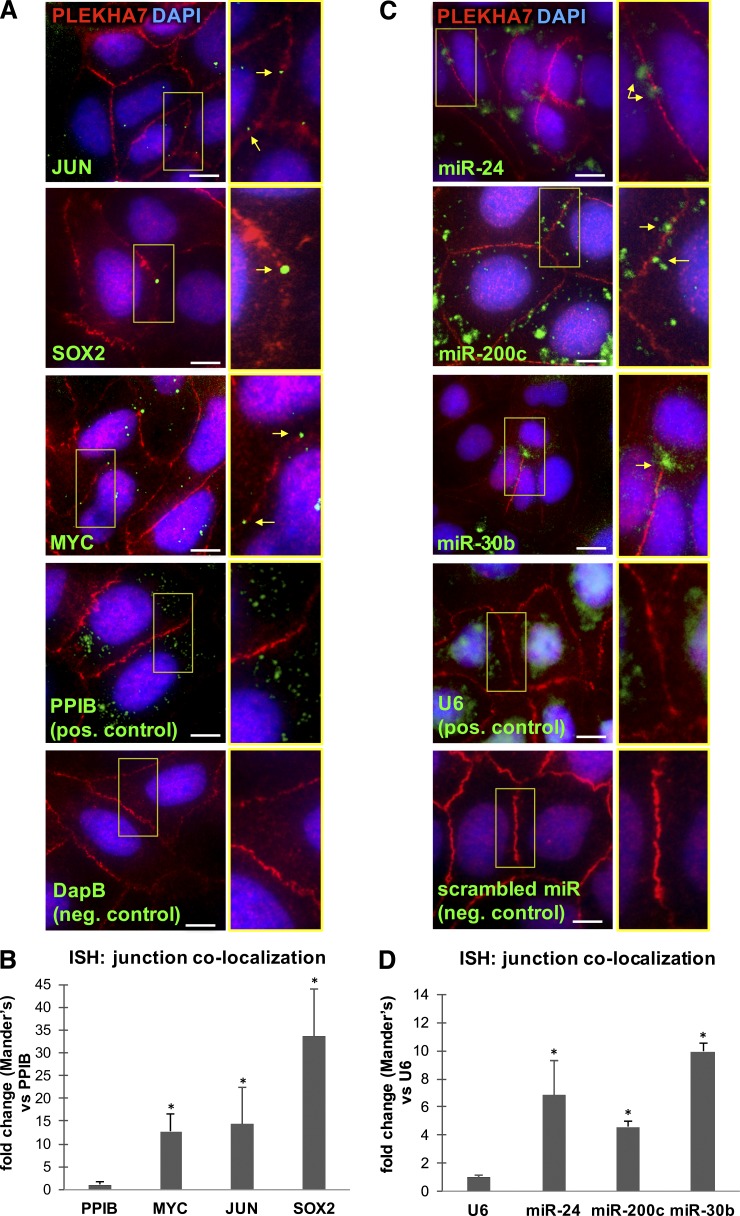
We then investigated the potential regulation of PLEKHA7-enriched mRNAs by miRNAs that also coprecipitated with PLEKHA7. Because the putative interactions between the 522 mRNAs and 28 miRNAs are numerous and beyond the scope of this study, we narrowed our search to the potential regulation of already validated mRNA-miRNA pairs by PLEKHA7, as a proof of concept, using miRTarBase (Chou et al., 2016). Although this approach is weighted toward heavily studied mRNAs and miRNAs, it allowed for faster identification of functionally relevant mRNA-miRNA interactions potentially mediated by PLEKHA7. Indeed, the search yielded more than a hundred validated interactions between 18 miRNAs and 53 mRNAs that were found to be enriched in the PLEKHA7 RNA-CLIPs (Fig. 2 E and Table S4). Notably, this search showed that JUN, MYC, and SOX2 are validated targets of miR-24, miR-200c, and miR-203a (Fig. 2 F and Table S4). In situ hybridization (ISH) analysis followed by PLEKHA7 co-immunofluorescence confirmed the presence of JUN, MYC, and SOX2 mRNAs, as well as miR-24, miR-200c, and miR-30b at the junctions (Fig. 3, A–D). In summary, PLEKHA7 coprecipitates and colocalizes with a select subset of cellular mRNAs and a small set of miRNAs that potentially regulate their expression at the ZA.
Figure 3.
PLEKHA7-associated RNAs localize at the ZA. (A) ISH of Caco2 cells for MYC, JUN, and SOX2 mRNAs; an ER-specific PPIB probe was used as the positive control and a bacterial DapB RNA probe as the negative control. (B) Quantification of mRNA colocalization to the junctions by using the Manders coefficient, expressed as fold difference of the junctional colocalization of the PPIB control (mean ± SEM from n = 6 fields; *, P < 0.01, Student’s two-tailed t test). (C) ISH of Caco2 cells for miR-24, miR-200c, and miR-30b miRNAs; a nuclear-specific U6 small RNA probe was used as the positive control and a scrambled miRNA probe as the negative control. (D) Quantification of miRNA colocalization to the junctions by using the Manders coefficient, expressed as fold difference of the junctional colocalization of the U6 control (mean ± SEM from n = 6 fields; *, P < 0.04, Student’s two-tailed t test). In all cases, ISH was followed by PLEKHA7 co-immunofluorescence and DAPI staining. Merged images are shown. Enlarged areas indicated by yellow boxes are shown to the right of each image. Arrows indicate hybridization signals. Bars, 20 µM. Insets are magnified 2×.
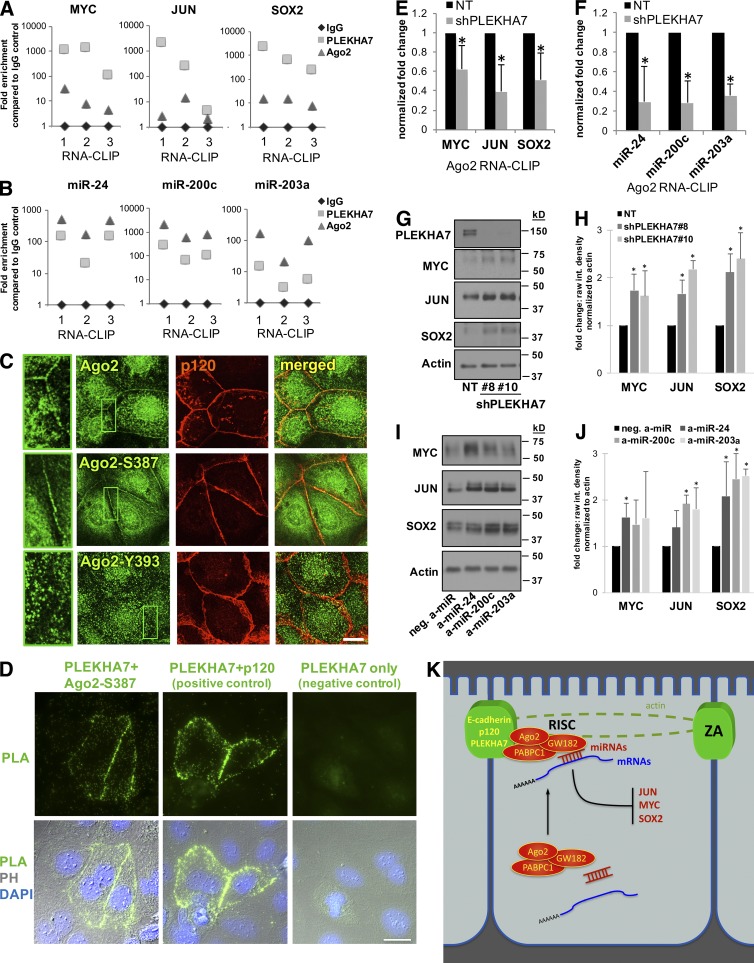
PLEKHA7 suppresses expression of JUN, MYC, and SOX2 mRNAs via Ago2 loading
PLEKHA7 coprecipitates with RISC and several mRNAs and miRNAs with previously validated interactions (Fig. 2 F and Table S4). However, for these miRNAs to suppress their target mRNAs, they should both be loaded onto Ago2 and the PLEKHA7-associated RISC. RNA-CLIP assays and subsequent quantitative PCR (qPCR) analysis verified the coprecipitation of miR-24, miR-200c, and miR-203a and JUN, MYC, and SOX2 mRNAs with PLEKHA7, but also showed that these RNAs coprecipitate with Ago2, indicating that they are indeed loaded into RISC (Fig. 4, A and B).
Figure 4.
PLEKHA7 promotes loading of junctional mRNAs and miRNAs to Ago2 and suppresses JUN, MYC, and SOX2 expression. (A and B) qRT-PCR of mRNAs (A) and of miRNAs (B) after RNA-CLIP of PLEKHA7 and Ago2. IgG is the negative control. Results are shown as individual points for each independent experiment. (C) Immunofluorescence of Caco2 cells costained for p120 and total Ago2, pS387-Ago2, or pY393-Ago2. Enlarged parts of images indicate areas of cell–cell contact. Bar, 20 µM. Insets are magnified 3×. (D) Caco2 cells transfected with a myc-tagged PLEKHA7–expressing construct were subjected to PLA using myc and Ago2-pS387 antibodies (PLEKHA7+ Ago2-S387), myc and p120 antibodies as the positive control (PLEKHA7+ p120), or myc (PLEKHA7) antibody alone, as the negative control. The PLA and combined PLA-phase contrast-DAPI (PLA-PH-DAPI) images are shown. (E) qRT-PCR for the indicated mRNAs after Ago2 RNA-CLIP of PLEKHA7 knockdown (shPLEKHA7) or control (nontarget; NT) Caco2 cells (mean ± SD from n = 3 independent experiments; *, P < 0.05, Student’s two-tailed t test). (F) qRT-PCR for the indicated miRNAs Ago2 RNA-CLIP of PLEKHA7 knockdown (shPLEKHA7) or control (NT) Caco2 cells (mean ± SD from n = 3 independent experiments; *, P < 0.005, Student’s two-tailed t test). (G) Western blot for the markers shown of control (NT) or PLEKHA7 knockdown Caco2 cells (shPLEKHA7; #8 and #10 indicate two different shRNAs; see also Materials and methods). Actin is the loading control. Molecular masses (kD) are indicated on the right. (H) Quantification of the raw band intensities of the blots, expressed as fold change of Actin loading control (mean ± SD from n = 3 independent experiments; *, P < 0.05, Student’s two-tailed t test). (I) Western blot of Caco2 cells transfected with the indicated anti-miRs (a-miR) for the displayed markers; Actin is the loading control. Molecular masses (kD) are indicated on the right. (J) Quantification of the raw band intensities of the blots, expressed as fold change of Actin loading control (mean ± SD from n = 3 independent experiments; *, P < 0.05, Student’s two-tailed t test). (K) Schematic summarizing the present results: the ZA recruit RISC and a specific set of miRNAs and mRNAs to suppress expression of growth markers, such as JUN, MYC, and SOX2.
It has been previously demonstrated that phosphorylation of Ago2 at S387 promotes miRNA-mediated translational repression of mRNAs (Zeng et al., 2008; Horman et al., 2013), whereas phosphorylation of Ago2 at Y393 inhibits its function (Shen et al., 2013). Detection of Ago2 phosphorylation at these sites by immunofluorescence showed that the junctional Ago2 is phosphorylated at S387 but not at Y393 (Fig. 4 C), suggesting that the junctional Ago2 is indeed active and primed to silence downstream mRNA targets. The specificity of the junctional localization of Ago2 phosphorylation at S387 was confirmed by siRNA-mediated knockdown (Fig. S2 A). As with total Ago2, the junctional localization of Ago2-S387 was lost after PLEKHA7 or p120 knockdown (Fig. S2 B) but was retained after knockdown of Nezha (Fig. S2 B). PLEKHA7 also coprecipitates with Ago2-S387 (Fig. S2 C). Interestingly, proximity ligation assay (PLA) showed that PLEKHA7 and Ago2-S387 are in close proximity specifically at areas of cell–cell contact (Fig. 4 D). These results demonstrate that PLEKHA7 associates and interacts with an active RISC specifically at the junctions.
PLEKHA7 knockdown results in dissociation of RISC from the ZA (Fig. 1 F). Therefore, we reasoned that loss of a functional RISC at the ZA would result in reduced loading of PLEKHA7-associated mRNAs and miRNAs to Ago2. Indeed, PLEKHA7 knockdown resulted in decreased loading of JUN, MYC, and SOX2 mRNAs, as well as miR-24, miR-200c, and miR-203a miRNAs, to Ago2 (Fig. 4, E and F). Knockdown of p120, which also affects junctional localization of Ago2 (Fig. 1 G), resulted in similar decreases of miRNA and mRNA loading to RISC (Fig. S2, D and E), showing that this regulation is indeed junction specific. The loss of functional RISC at the ZA and the decreased Ago2 loading after PLEKHA7 knockdown also imply that the silencing of these mRNAs would be alleviated and their protein levels should be increased upon PLEKHA7 loss. In agreement, PLEKHA7 knockdown resulted in up-regulation of JUN, MYC, and SOX2 at the protein level (Fig. 4, G and H). Finally, transfection of cells with anti-miRs targeting miR-24, miR-200c, and miR-203 also led to the up-regulation of the protein levels of (a) MYC by anti–miR-24, (b) SOX2 by anti–miR-200c, and (c) JUN by anti–miR-200c and anti–miR-203a, indicating that these mRNAs are indeed regulated by the above miRNAs in these cells (Fig. 4, I and J). SOX2 expression was also regulated by the stemness-inhibiting miR-203a (Wellner et al., 2009) and miR-24, suggesting additional direct or indirect regulation of SOX2 by these miRNAs (Fig. 4, I and J). Overall, these results demonstrate that the junctional RISC is active, mediating suppression of JUN, MYC, and SOX2 protein expression in a PLEKHA7-dependent manner (Fig. 4 K).
In summary, our data reveal a mechanism whereby cadherin complexes at the ZA of non-transformed epithelial cells recruit a set of mRNAs and an active RISC to regulate the levels of key proteins affecting cell behavior (Fig. 4 K). Combined with our recent work on the microprocessor (Kourtidis et al., 2015), the data provide comprehensive evidence for the presence of all essential components of the RNAi machinery in a single cellular compartment, the ZA. This expands our view of ZA-mediated cellular signaling but also uncovers a new function of the RNAi machinery in non-transformed epithelial cells. Until now, the subcellular localization of RISC components was primarily discussed in the context of cytoplasmic GW-bodies and processing bodies (p-bodies; Jakymiw et al., 2005, 2007) or stress granules, which mainly form under certain conditions of stress (Leung et al., 2006; Anderson and Kedersha, 2009). Both structures have been shown to be sites of mRNA translational silencing or degradation, as well as mRNA storage, exhibiting common and distinct features between them (Anderson and Kedersha, 2009; Stoecklin and Kedersha, 2013). However, here we present evidence that the specific localization of RISC at the ZA and the silencing of mRNAs occur under normal physiological conditions, in polarized monolayers of non-transformed epithelial cells. It has been proposed that p-bodies and stress granules act as hubs that facilitate the interaction of different RNA-related components to influence cell signaling (Anderson and Kedersha, 2009; Kedersha et al., 2013). We postulate that the ZA plays a similar role by facilitating the sequestration and silencing of particular mRNAs to regulate specific signaling pathways. Indeed, we identify only subsets of RISC proteins (Fig. 1, A–C; and Fig. S1, A–C), mRNAs (522 of >30,000 sequenced transcripts expressed in Caco2s, based on RNA-seq; Fig. 2 B and Table S2), and miRNAs (28 of 758 examined; Fig. 2 C and Table S3). Our data suggest that through this mechanism, PLEKHA7 and the apical ZA act as epithelial adhesion “sensors” to accordingly regulate cellular behavior through specific targets.
To identify PLEKHA7-associated miRNA-mRNA functional interactions, we mined miRTarBase, a database of previously validated miRNA-mRNA pairs. Of these interactions, we successfully confirmed that PLEKHA7 regulates at least three important mRNA targets, JUN, MYC, and SOX2. JUN and MYC are oncogenes and strong drivers of epithelial cell growth, migration, and tumor progression, which is consistent with the ability of PLEKHA7 to suppress cell growth and its common loss or mislocalization in human cancer (Kourtidis et al., 2015; Tille et al., 2015; Kourtidis and Anastasiadis, 2016a,b). Interestingly, previous studies have shown that depletion of E-cadherin or expression of a dominant negative E-cadherin mutant results in increased JUN expression and subsequent AP1 activation via an unidentified posttranscriptional mechanism (Andersen et al., 2005; Knirsh et al., 2009; Spangler et al., 2011). Our work now sheds light on this mechanism by showing that cadherin complexes at the ZA recruit JUN mRNA and silence it via a junction-associated RISC. MYC and SOX2 are also essential drivers of pluripotency (Bernhardt et al., 2012). Dedifferentiation of epithelial cells during EMT is common in cancer and promotes a stem cell–like phenotype (Bernhardt et al., 2012; Fabregat et al., 2016). Interestingly, our pathway analysis identified molecular mechanisms of cancer and human embryonic stem cell pluripotency as two of the major canonical pathways highly represented in the PLEKHA7-enriched mRNAs (Fig. 2 C). The two other pathways that were highly represented in this analysis were the Wnt/β-catenin and TGF-β signaling pathways (Fig. 2 C), which are major players in epithelial cell growth (Polakis, 2012; Katsuno et al., 2013) and are intimately linked to cell adhesion (Katsuno et al., 2013; McCrea and Gottardi, 2016). Although these data provide a first glimpse into all the potential signaling pathways affected by ZA-associated RISC, they argue that the latter may affect central pathways regulating epithelial monolayer homeostasis, cell growth, and stemness. The diverse set of PLEKHA7-associated mRNAs implies a large regulatory network affected by the integrity of apical cell–cell junctions and provides an unexpected mechanism by which cell–cell adhesion broadly affects cellular signaling and epithelial homeostasis. Although we validated regulation for a small set of the PLEKHA7-associated mRNAs (Fig. 2 B and Table S2), such as JUN, MYC, and SOX2, there are numerous putative interactions between the PLEKHA7-enriched mRNAs and miRNAs (Fig. 2 E and Table S4). The present work lays the foundation for the systematic interrogation of this regulatory network and its potential role in epithelial cell behavior and human disease.
Materials and methods
Cell culture, reagents, and chemicals
In all comparisons, cells were used strictly at the same confluences. All cell lines were obtained from ATCC, used at low passage (<20), and tested negative for mycoplasma contamination. Caco2 colon epithelial cells were cultured in MEM (Cellgro) supplemented with 10% FBS (Invitrogen), 1 mM sodium pyruvate (Invitrogen), and 1× nonessential amino-acid supplement (Mediatech). MDCK canine kidney epithelial cells and HEK 293FT human embryonic kidney cells were cultured in DMEM supplemented with heat-inactivated 10% FBS. PP2 was obtained from Calbiochem and used to treat cells at 10 µM for 24 h.
shRNAs, siRNAs, and anti-miRs
Cells were transfected using Lipofectamine 2000 (Invitrogen) or Lipofectamine RNAiMAX (Invitrogen) for anti-miR and siRNA transfection, according to the manufacturer’s protocols. Lentiviral shRNAs were derived from the pLKO.1-based TRC1 (Sigma-Aldrich/RNAi Consortium) shRNA library (pLKO.1-puro Non-Target shRNA Control, SHC016; PLEKHA7 #8, TRCN0000146289; PLEKHA7 #10, TRCN0000127584; E-cadherin #21, TRCN0000039664; and E-cadherin #23, TRCN0000039667) and the pLKO.5-based TRC2 library (pLKO.5-puro Non-Target shRNA Control, SHC216; NEZHA #72, TRCN0000268676; and NEZHA #73, TRCN0000283758). Lentiviruses were produced in HEK293FT cells and used to infect cells according to standard protocols. Retroviral pRetro-Super nontarget shRNA control and p120 shRNA were prepared and used as described previously (Kourtidis et al., 2015). SMARTpool (Dharmacon) siRNAs used were Ago2, M-004639-00-0005; Dicer, M-003483-00-0005; and nontarget, D-001206-14-05. mirVana (4464084; Life Technologies) anti-miRs used were anti–hsa-miR-24, ID MH10737; anti–hsa-miR-200c, ID MH11714; anti–hsa-miR-203a, ID MH10152; and negative control #1 (4464076).
Antibodies
The primary antibodies used in the present study were PLEKHA7 (HPA038610; Sigma-Aldrich), p120 (33-9600; Life Technologies), E-cadherin (610182; BD Transduction Labs), Ago2 (ab57113; Abcam), Ago2 (28550002; Novus Biologicals), Ago2 (AP5281; ECM Biosciences), pAgo2 S387 (AP5291; ECM Biosciences), pAgo2 Y393 (AP5311; ECM Biosciences), GW182 (sc-56314 and sc-377006; Santa Cruz Biotechnology), PABPC1 (04-1467), Dicer (SAB4200087; Sigma-Aldrich), Dicer (sc-136979; Santa Cruz Biotechnology), TRBP (ABE623; Millipore), SOX2 (2748; Cell Signaling Technology), JUN (9165; Cell Signaling Technology), MYC (13-2500; Life Technologies), Nezha (SAB4200415; Sigma-Aldrich), and Actin (A2066; Sigma-Aldrich). Working dilutions were 1:50 to 1:500 for immunofluorescence and 1:500 to 1:2,000 for Western blot.
The secondary antibodies used in the present study were HRP anti–mouse (715-035-150; Jackson ImmunoResearch Laboratories), HRP anti–rabbit (711-035-152; Jackson ImmunoResearch Laboratories), Alexa Fluor 488 anti–mouse (A-11029), Alexa Fluor 488 anti–rabbit (A11034; Life Technologies), Alexa Fluor 594 anti–mouse (A-11005; Life Technologies), and Alexa Fluor 594 anti–rabbit (A-11037; Life Technologies). Working dilutions were 1:500 for immunofluorescence and 1:2,000 for Western blot.
Immunofluorescence
MDCK and Caco2 cells were grown on Transwell inserts (Costar 3413) for 7 or 21 d, respectively, until they polarized or on sterile glass coverslips until they reach full confluence. Cells were washed once with PBS and fixed with either 100% methanol (Thermo Fisher Scientific) for 7 min or 4% formaldehyde (Electron Microscopy Sciences) for 20 min, followed by 0.02% Triton X-100 permeabilization for 10 min. Cells were blocked with either 3% nonfat milk (Carnation) in PBS or Protein-Block reagent (X090930-2; Dako) for 30 min and stained with primary antibodies diluted in either milk or Antibody Diluent (S302281-2; Dako) for 1 h. Cells were then washed three times with PBS, stained with the fluorescent-labeled secondary antibodies for 1 h, washed three times with PBS, costained with DAPI (Sigma-Aldrich) to visualize the nuclei, mounted (Aqua Poly/Mount; Polysciences), and imaged using a Zeiss LSM 510 META laser confocal microscope, under a 63× objective, with an additional 1.6× zoom. z-Stacks were acquired in 0.5-µM intervals. Images, stacks, and x–z representations of the image stacks were processed using Zen software (Zeiss). The Coloc 2 module of Fiji software was used to calculate the Manders coefficient (with Costes threshold regression) for colocalization measurements.
Immunoblotting
Whole-cell extracts were obtained using RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, and 0.1% SDS) supplemented with protease (cocktail III, RPI) and phosphatase inhibitors (Pierce). Lysates were homogenized through a 29-G needle and cleared by full-speed centrifugation for 5 min. Protein quantification was performed using the BCA assay (Pierce). Protein extracts were mixed with Laemmli sample buffer and separated by SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad), blotted according to standard protocols, detected by luminescence using ECL (GE Healthcare), and imaged using x-ray films (Pierce). Band quantifications were performed using the Raw Integrated Intensity module of Fiji and expressed as fold changes of the respected Actin loading controls.
IP and proteomics
Cells were grown on 10-cm plates until fully confluent, placed on ice, washed twice with ice-cold PBS, and lysed with ice-cold Triton X-100 lysis buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris, pH 7.4, and 1% Triton X-100) containing 2× protease (cocktail III, RPI) and phosphatase inhibitors (Pierce). Four 10-cm plates (∼3 to 4 × 106 cells) were used per IP. In parallel, 4 µg antibody or IgG control (011-000-003; Jackson ImmunoResearch Laboratories) was incubated with 40 μl Protein G Dynabeads (Invitrogen) overnight at 4°C with constant end-to-end rotation and then washed three times with IP lysis buffer. Cell lysates were incubated with the bead-conjugated antibodies overnight at 4°C with constant end-to-end rotation. Beads were then washed three times with IP lysis buffer and eluted using 50 mM DTT (Sigma-Aldrich) in lysis buffer at 37°C for 45 min, with constant agitation. Eluted proteins were separated by SDS-PAGE, as described above. RNase treatments of IPs were performed as previously described (Höck et al., 2007); in brief, beads were distributed equally after the final wash into two tubes with either 1.5 ml PBS or PBS containing 100 mg/ml RNase A (Sigma-Aldrich). After incubation at 4°C for 1.5 h, beads were washed three times with PBS, extracted using 50 mM DTT (Sigma-Aldrich), mixed with Laemmli sample buffer, and analyzed by Western blotting.
For proteomics, eluted proteins were subjected to SDS-PAGE and gels were silver-stained (Pierce SilverSnap kit), according to the manufacturer’s protocol. Gel slices were selected and excised using sterile scalpels and destained, reduced, and alkylated before digestion with trypsin (Promega). Samples were analyzed using nano-HPLC-electrospray tandem mass spectrometry in a ThermoFinnigan LTQ Orbitrap Hybrid Mass Spectrometer (ThermoElectron Bremen). Mascot (Matrix Sciences) was used to search the Swissprot database to identify isolated peptides. Common contaminants such as trypsin, casein, and keratin and microbial-specific proteins were removed from analysis.
Calcium switch assay
Caco2 cells were grown on coverslips until confluence, pretreated with either DMSO (control) or 10 µM nocodazole for 1 h to dissolve microtubules, and washed three times with calcium-free PBS and incubated in calcium-free Caco2 medium (11380-037; Life Technologies, supplemented with glutamine, 10% FBS, sodium pyruvate, and MEM) containing 4 mM EGTA for 30 min, until cells were rounded, while being kept in either DMSO or 10 µM nocodazole, respectively. Cells were then washed three times with PBS, returned to regular Caco2 medium, again with DMSO or nocodazole, and fixed for immunofluorescence for the indicated time points.
Total RNA isolation and qRT-PCR
Cells were lysed using Trizol (Invitrogen) and subjected to the Trizol Plus Total Transcriptome Isolation protocol of the PureLink RNA mini kit (Ambion; Life Technologies) specified to isolate both mRNAs and miRNAs. Final RNA concentrations were determined using a NanoDrop spectrophotometer. RNA was converted to cDNA using the High Capacity cDNA Reverse transcription kit (Applied Biosystems). qPCR reactions were performed using the Taqman FAST Universal PCR master mix (Applied Biosystems), in a 7900 HT or ViiA 7 Thermocycler (Applied Biosystems). Data were analyzed using RQ Manager (Applied Biosystems). U6 was used as a control for miRNA expression normalization and 18S for mRNA normalization. TaqMan assays used for miRNAs (4427975; Applied Biosystems) were hsa-miR-24, 000402; hsa-miR-200c, 002300; hsa-miR-203a, 000507; and U6, 001973. TaqMan assays for mRNAs (4331182; Applied Biosystems) were MYC, Hs00153408_m1; SOX2, Hs01053049_s1; JUN, Hs00277190_s1; and 18S, Hs99999901_s1.
RNA-CLIP
Cells were grown on 10-cm plates until fully confluent and washed once with PBS at RT. Protein–RNA complexes were then cross-linked using 0.75% PFA (EMS-Fisher) in PBS for exactly 5 min at RT with gentle agitation and neutralized with 250 mM glycine in PBS for another 5 min at RT with gentle agitation. Cells were then immediately placed on ice, washed twice with ice-cold PBS, and lysed for 20 min with ice-cold RIPA, containing 2× protease (cocktail III, RPI), phosphatase inhibitors (Pierce), and 100 U/ml RNase Inhibitor (Promega). Cells were scraped, passed through a 29-G needle, and sonicated using a waterbath sonicator (Fisher FS20) for 9 × 2 min, with half-minute intervals in which lysates were cooled on ice. The lysates were then cleared by full-speed centrifugation for 5 min and precleared with Protein G Dynabeads (Life) for 1 h at 4°C with constant end-to-end rotation. Lysate from a total of 4 × 10-cm plates was used per IP. Precleared lysates were subjected to protein quantification using BCA, and an equal protein amount was included in each IP. In parallel, 8 µg antibody or IgG control (011-000-003; Jackson ImmunoResearch Laboratories) were incubated with 40 μl Protein G Dynabeads (Invitrogen) overnight and then washed three times with RIPA buffer. Precleared lysates were incubated with the bead-conjugated antibodies overnight at 4°C with constant end-to-end rotation, with the exception of 100 μl kept at −20°C as the total lysate control. Beads were then washed at RT (a) once with RIPA and 5 mM EDTA; (b) twice with a stringent buffer containing RIPA, 5 mM EDTA, and 500 mM NaCl buffer; and (c) twice with RIPA and 5 mM EDTA buffer. Elution of immunoprecipitated complexes and reversal of the cross-linking was then performed by adding to the beads RIPA buffer containing 5 mM EDTA, 2× protease inhibitors, 2× phosphatase inhibitors, 100 U/ml RNasein RNase inhibitor, 50 mM DTT (Sigma-Aldrich), and 1% SDS, at 70°C for 1 h with occasional agitation. Similarly, 50 mM DTT and 1% SDS was added to the total lysate followed by incubation at 70°C for 1 h, to reverse the cross-linking. RNA was then extracted from the eluates using Trizol (Invitrogen), as described above. Immunoprecipitated RNAs were subjected to either RNA-seq analysis to identify mRNAs, as described below, or TLDA, also as described below. qRT-PCRs of precipitated RNAs were performed as above and the results were analyzed using RQ manager 1.2 (ABI/Life) and the Sigma RIP-qRT-PCR Data Analysis Calculation Shell, associated with the Sigma Imprint RIP kit (http://www.sigmaaldrich.com/life-science/epigenetics/imprint-rna.html) and the references therein. Results were normalized for U6 regarding miRNAs and for 18S regarding mRNAs, and all normalized numbers were expressed as fold difference of the negative control IP (IgG).
RNA sequencing
Immunoprecipitated RNAs from the RNA-CLIP were subjected to RNA sequencing according to the Illumina TruSeq Stranded Total RNA Sample Prep Protocol. In brief, RNA libraries were prepared using 200–500 ng RNA according to the manufacturer’s instructions for the TruSeq Stranded Total RNA Sample Prep kit (Illumina). The concentration and size distribution of the completed libraries was determined using an Agilent Bioanalyzer DNA 1000 chip and Qubit fluorometry (Invitrogen). Libraries were sequenced at eight samples per lane to generate 30–40 million reads per sample according to Illumina’s standard protocol using the Illumina cBot and cBot Paired end cluster kit version 3. The flow cells were sequenced as 51 × 2 paired-end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HCS v2.0.12 data collection software. Base-calling was performed using Illumina’s RTA version 1.17.21.3. Data were processed and analyzed using the Mayo Clinic mRNA-Seq analysis pipeline MAPRSeq (v1.2.1.4). Raw data in the form of FASTQ files was first processed with the quality control tool FASTQC (v0.10.1) to generate sequencing QC reports of the samples. Read alignment was performed with Tophat (v2). The aligned BAM file from Tophat was processed using HTSeq (v0.5.3p9) to summarize expression at the gene level. Exon quantification was obtained using customized Perl scripts that leverage BEDTools (v2.16.2). In addition to raw gene and exon expression counts, normalized RPKM values were obtained for the expression data. For accurate variant detection, GATK UnifiedGenotyper (v1.7) was used to call single nucleotide variants, which were further annotated with quality score, coverage, and additional criteria using GATK Variant Quality Score Recalibration (VQSR). In addition, detailed quality control data to estimate the distance between paired-end reads, evaluate the sequencing depth for alternate splicing analysis, determine the rate of duplicate reads, and evaluate coverage of reads across genes was obtained using RSeQC (v2.3.2) software.
RPKMs from the PLEKHA7 CLIP were compared with those of the negative control (IgG) CLIP, and nonspecific hits were excluded. The PLEKHA7 CLIP RPKMs were then compared with total RNA RPKMs and filtered based on bimodal distribution to include hits represented in both sets. Then, the fold change of PLEKHA7 CLIP RPKMs versus total RPKMs were calculated for all hits, as well as the averages and P values (using paired Student’s t test) from four independent experiments. Finally, hits that were at least fourfold enriched in the PLEKHA7 CLIP and statistically significant (P < 0.05) were further considered as positive hits (Fig. 2 B and Table S2).
TLDA miRNA analysis
Total RNA was isolated from the RNA-CLIPs as described above, and miRNA analysis was performed using TLDA. The Taqman Human MicroRNA A+B Cards Set v3.0 was used to assess miRNA expression. In brief, 500 ng total RNA was reverse transcribed with Megaplex RT primers and Taqman MicroRNA RT kit. Samples were then amplified with Megaplex Preamp primers and Taqman Preamp master mix. cDNAs were diluted 1:4.33 and loaded with Taqman universal PCR master mix on each low-density array according to the manufacturer’s instructions. Amplification kinetics were measured on an Applied Biosystems Prism 7900 FAST sequence detector and analyzed with Sequence Detection System software (Applied Biosystems). The Ct values of the PLEKHA7 RNA-CLIP for each miRNA were compared with the Ct values of the negative control RNA-CLIP (IgG). A Ct value of 40 was given to all runs that gave no amplification after 40 cycles (and value was marked “undetermined”). The miRNAs whose PLEKHA7 CLIP Ct value was consistently higher than the IgG CLIP background value, from three independent experiments, were further included in the study (Fig. 2 D and Table S3).
ISH
For ISH, Caco2 cells were grown on slides until confluent, fixed with 10% formalin (Protocol; Thermo Fisher Scientific), dehydrated and rehydrated in a series of 50/70/100/70/50% EtOH solutions, pretreated with peroxide and protease and hybridized. ISH for mRNAs used a RNAscope 2.5 HD Brown assay (322370; Advanced Cell Diagnostics), according to the manufacturer’s instructions; probes used were Hs-MYC (311761); Hs-JUN (470541), Hs-SOX2 (400871); positive control probe Hs-PPIB (313906); and negative control probe DapB (310048; all Advanced Cell Diagnostics). ISH for miRNAs used a Ventana Discovery Ultra system as follows: (a) denature probes at 80°C for 4 min; (b) probe hybridize at 60°C for 3 h; miRCURY LNA detection probes, 250 pmol, 5′-DIG and 3′-DIG labeled: hsa-miR-24-3p (610827-360); hsa-miR-200c (615855-360); hsa-miR-30b-5p (18143-15); U6 (99002-01); scrambled miR control (99004-01; all Exiqon); (c) three stringency washes using 0.5× SSC, at 65°C, 4 min each; (d) application of rabbit anti-DIG antibody (700772, clone 9H27L19; Invitrogen), 1:250 dilution, at 35°C for 30 min; (e) application of UltraMap anti-Rb Alk-Phos (760-4314; joche) for 12 min; and (f) application of ChromoMap Blue (760-161; Roche) for 24 min.
After ISH, in all cases, cells were rinsed twice with PBS and blocked with Protein-Block reagent (X090930-2; Dako) for 10 min. Immunofluorescence analysis was performed with PLEKHA7 (Sigma-Aldrich) or E-cadherin (BD) antibodies diluted in Antibody Diluent (S302281-2; Dako) for 30 min. Cells were then washed three times with PBS and stained with a fluorescence-labeled secondary antibody and DAPI for 30 min. Cells were finally washed three times with PBS, mounted (Aqua Poly/Mount) and imaged using a Leica DM5000B microscope under bright (ISH) and fluorescent (protein immunofluorescence) light. ISH images were pseudocolored, and merged overlays of each field were created using the microscope’s Leica suite software. The Coloc 2 module of Fiji software was used to calculate the Manders coefficient (with Costes threshold regression) of the ISH signal versus the junctional costaining (for this, the E-cadherin co-immunofluorescence was used because of background nuclear staining of PLEKHA7 caused by formalin fixation; see also Fig. 1 F).
PLA
Caco2 cells were transfected with a PLEKHA7-myc construct (Kourtidis et al., 2015), and a PLA was performed according to the manufacturer’s protocol. The assay was performed using Duolink PLA products (Sigma-Aldrich), including In situ Detection Reagents Green and anti-Mouse MINUS and anti-Rabbit PLUS probes. The antibodies against myc (13-2500, 9E10, 1:100; Invitrogen), pAgo2-S387 (AP5291, 1:50; ECM Biosciences), and p120 (1:2,000; F1aSH) were used. Cells were imaged using an ORCA-Flash4.0 LT camera (Hamamatsu Photonics) on an Olympus IX83 inverted microscope with an UPLSAPO Super Apochromatic 60XS2 silicone oil objective (NA 1.30). Image acquisition was controlled with cellSens software. DAPI and Alexa Fluor 488 were imaged with an AT-DAPI BX3 bandpass filter cube (excitation 375/28, emission 460/50, beamsplitter 415LP) and an ET-GFP bandpass filter cube (excitation 470/40, emission 525/50, beamsplitter T495LP), respectively.
Statistical analysis
For quantitative experiments, averages and standard deviations were calculated and presented as error bars, whereas the number of independent experiments performed and the related statistics are indicated in each figure legend. SEM was used for sample size more than 3. Student’s two-tailed t test was used for P value calculations because comparisons were between two groups: control and experimental conditions. For all other experiments, at least three independent experiments were performed, and either representative images or all individual points are shown.
Data deposition
The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct (Orchard et al., 2014) and assigned the identifier IM-25739. The complete set of the RNA sequencing data have been deposited to GEO under accession no. GSE86986.
Online supplemental material
Fig. S1 shows localization of RISC proteins at the ZA of MDCK cells and validation of the junctional specificity of Ago2 stainings. Fig. S1 also demonstrates localization of Dicer and TRBP at the ZA, validation of these stainings, and coprecipitation of Dicer and TRBP with PLEKHA7. Fig. S2 demonstrates validation of the specificity of the Ago2-S387 antibody and shows effects of p120 knockdown on miRNA and mRNA loading to Ago2. Table S1 includes the pathway analysis of the PLEKHA7 proteomics. Table S2 includes the full list of the mRNAs that were identified by the PLEKHA7 RNA-CLIP. Table S3 includes the full TLDA miRNA qPCR array data after PLEKHA7 RNA-CLIP. Table S4 shows all the validated interactions between the PLEKHA7-associated miRNAs and mRNAs, mined from MirTarBase. Tables S2–S4 are included as Excel files.
Supplementary Material
Acknowledgments
We thank Mayo Clinic’s Proteomics Core and Ben Madden for assistance with mass spectrometry; the Mayo Medical Genome Facility and Bruce Eckloff, Fariborz (Fred) Rakhshan Rohakhtar, Eileen Holicky, Aditya Vijay Bhagwate, Christine Wheatley, and Matt Bockol for RNA sequencing and data processing; Asha Nair and Pritha Chanana for bioinformatic analysis; Brandy Edenfield for assistance with ISH; Dr. Laura Lewis-Tuffin for assistance with confocal microscopy; Jeremy Barth and the MUSC Proteogenomics Facility for qPCR instrument use and assistance; and Dr. Isidore Rigoutsos for suggestions and comments.
This work was supported by the National Institutes of Health (R01 CA100467, R01 NS069753, P50 CA116201) and a Mayo Clinic Center for Biomedical Discovery award (P.Z. Anastasiadis); the Jay and Deanie Stein Career Development Award for Cancer Research at the Mayo Clinic, an Abney Foundation Scholarship, Hollings Cancer Center, and an American Cancer Society Institutional Research Grant awarded to the Hollings Cancer Center, Medical University of South Carolina (A. Kourtidis); and the 26.2 with Donna Foundation (E.A. Thompson).
The authors declare no competing financial interests.
Author contributions: A. Kourtidis and P.Z. Anastasiadis conceived and designed the study, conceived and designed experiments, analyzed data, and wrote the manuscript. A. Kourtidis performed all experiments except: W.-H. Lin performed the PLA assay, R. Lu performed antibody validations, and B. Necela and E.A. Thompson ran the Taqman miRNA array (TLDA) and assisted with RNA sequencing. R.W. Feathers provided technical support. Y.W. Asmann performed bioinformatic analysis.
Footnotes
Abbreviations used:
- AJ
- adherens junction
- CLIP
- intracellular cross-linking and IP
- IP
- immunoprecipitation
- ISH
- in situ hybridization
- miRLC
- miRNA loading complex
- PLA
- proximity ligation assay
- RISC
- RNA-induced silencing complex
- ZA
- zonula adherens
References
- Andersen H., Mejlvang J., Mahmood S., Gromova I., Gromov P., Lukanidin E., Kriajevska M., Mellon J.K., and Tulchinsky E.. 2005. Immediate and delayed effects of E-cadherin inhibition on gene regulation and cell motility in human epidermoid carcinoma cells. Mol. Cell. Biol. 25:9138–9150. 10.1128/MCB.25.20.9138-9150.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2009. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Bernhardt M., Galach M., Novak D., and Utikal J.. 2012. Mediators of induced pluripotency and their role in cancer cells—Current scientific knowledge and future perspectives. Biotechnol. J. 7:810–821. 10.1002/biot.201100347 [DOI] [PubMed] [Google Scholar]
- Buxbaum A.R., Haimovich G., and Singer R.H.. 2015. In the right place at the right time: Visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 16:95–109. 10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., and Shiekhattar R.. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 436:740–744. 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.H., Chang N.W., Shrestha S., Hsu S.D., Lin Y.L., Lee W.H., Yang C.D., Hong H.C., Wei T.Y., Tu S.J., et al. . 2016. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 44:D239–D247. 10.1093/nar/gkv1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R.B. 2010. HITS-CLIP: Panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip. Rev. RNA. 1:266–286. 10.1002/wrna.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., and Izaurralde E.. 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15:346–353. 10.1038/nsmb.1405 [DOI] [PubMed] [Google Scholar]
- Fabregat I., Malfettone A., and Soukupova J.. 2016. New insights into the crossroads between EMT and stemness in the context of cancer. J. Clin. Med. 5:e37 10.3390/jcm5030037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moya S.M., Bauer K.E., and Kiebler M.A.. 2014. Meet the players: Local translation at the synapse. Front. Mol. Neurosci. 7:84 10.3389/fnmol.2014.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., and Shiekhattar R.. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature. 432:235–240. 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- Gutierrez N., Eromobor I., Petrie R.J., Vedula P., Cruz L., and Rodriguez A.J.. 2014. The beta-actin mRNA zipcode regulates epithelial adherens junction assembly but not maintenance. RNA. 20:689–701. 10.1261/rna.043208.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., and Kim V.N.. 2014. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15:509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein E., Beach D., and Hannon G.J.. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 404:293–296. 10.1038/35005107 [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., and Hannon G.J.. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 293:1146–1150. 10.1126/science.1064023 [DOI] [PubMed] [Google Scholar]
- Harris T.J., and Tepass U.. 2010. Adherens junctions: From molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11:502–514. 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., and Hammond S.M.. 2005. A microRNA polycistron as a potential human oncogene. Nature. 435:828–833. 10.1038/nature03552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J., Weinmann L., Ender C., Rüdel S., Kremmer E., Raabe M., Urlaub H., and Meister G.. 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8:1052–1060. 10.1038/sj.embor.7401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman S.R., Janas M.M., Litterst C., Wang B., MacRae I.J., Sever M.J., Morrissey D.V., Graves P., Luo B., Umesalma S., et al. . 2013. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol. Cell. 50:356–367. 10.1016/j.molcel.2013.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irby R.B., and Yeatman T.J.. 2002. Increased Src activity disrupts cadherin/catenin-mediated homotypic adhesion in human colon cancer and transformed rodent cells. Cancer Res. 62:2669–2674. [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., and Chan E.K.. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7:1267–1274. 10.1038/ncb1334 [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Pauley K.M., Li S., Ikeda K., Lian S., Eystathioy T., Satoh M., Fritzler M.J., and Chan E.K.. 2007. The role of GW/P-bodies in RNA processing and silencing. J. Cell Sci. 120:1317–1323. 10.1242/jcs.03429 [DOI] [PubMed] [Google Scholar]
- Katsuno Y., Lamouille S., and Derynck R.. 2013. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 25:76–84. 10.1097/CCO.0b013e32835b6371 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., and Anderson P.. 2013. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 38:494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., and Mourelatos Z.. 2007. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 129:1141–1151. 10.1016/j.cell.2007.05.016 [DOI] [PubMed] [Google Scholar]
- Knirsh R., Ben-Dror I., Spangler B., Matthews G.D., Kuphal S., Bosserhoff A.K., and Vardimon L.. 2009. Loss of E-cadherin-mediated cell-cell contacts activates a novel mechanism for up-regulation of the proto-oncogene c-Jun. Mol. Biol. Cell. 20:2121–2129. 10.1091/mbc.E08-12-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtidis A., and Anastasiadis P.Z.. 2016a Bringing together cell-to-cell adhesion and miRNA biology in cancer research. Future Oncol. 12:1211–1214. 10.2217/fon-2016-0012 [DOI] [PubMed] [Google Scholar]
- Kourtidis A., and Anastasiadis P.Z.. 2016b PLEKHA7 defines an apical junctional complex with cytoskeletal associations and miRNA-mediated growth implications. Cell Cycle. 15:498–505. 10.1080/15384101.2016.1141840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtidis A., Ngok S.P., and Anastasiadis P.Z.. 2013. p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl. Sci. 116:409–432. 10.1016/B978-0-12-394311-8.00018-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtidis A., Ngok S.P., Pulimeno P., Feathers R.W., Carpio L.R., Baker T.R., Carr J.M., Yan I.K., Borges S., Perez E.A., et al. . 2015. Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat. Cell Biol. 17:1145–1157. 10.1038/ncb3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., and Filipowicz W.. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597–610. [DOI] [PubMed] [Google Scholar]
- Landthaler M., Gaidatzis D., Rothballer A., Chen P.Y., Soll S.J., Dinic L., Ojo T., Hafner M., Zavolan M., and Tuschl T.. 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 14:2580–2596. 10.1261/rna.1351608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., and Krause H.M.. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 131:174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., and Sharp P.A.. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 103:18125–18130. 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., and Hannon G.J.. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 305:1437–1441. 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- Maniataki E., and Mourelatos Z.. 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19:2979–2990. 10.1101/gad.1384005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardakheh F.K., Paul A., Kümper S., Sadok A., Paterson H., Mccarthy A., Yuan Y., and Marshall C.J.. 2015. Global analysis of mRNA, translation, and protein localization: Local translation is a key regulator of cell protrusions. Dev. Cell. 35:344–357. 10.1016/j.devcel.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea P.D., and Gottardi C.J.. 2016. Beyond β-catenin: Prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 17:55–64. 10.1038/nrm.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea P.D., Maher M.T., and Gottardi C.J.. 2015. Nuclear signaling from cadherin adhesion complexes. Curr. Top. Dev. Biol. 112:129–196. 10.1016/bs.ctdb.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., and Tuschl T.. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 15:185–197. 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Meng W., Mushika Y., Ichii T., and Takeichi M.. 2008. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 135:948–959. 10.1016/j.cell.2008.09.040 [DOI] [PubMed] [Google Scholar]
- Mili S., Moissoglu K., and Macara I.G.. 2008. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 453:115–119. 10.1038/nature06888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu M., Raza U., Saatci O., Eyupoglu E., Yurdusev E., and Sahin O.. 2016. miR-200c: A versatile watchdog in cancer progression, EMT, and drug resistance. J. Mol. Med. (Berl.). 94:629–644. 10.1007/s00109-016-1420-5 [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., and Garcia-Blanco M.A.. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 26:182–190. 10.1016/S1046-2023(02)00021-X [DOI] [PubMed] [Google Scholar]
- Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. . 2014. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42(D1):D358–D363. 10.1093/nar/gkt1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., and Ohkubo T.. 2001. Tyrosine phosphorylation of p120(ctn) in v-Src transfected L cells depends on its association with E-cadherin and reduces adhesion activity. J. Cell Sci. 114:503–512. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., and Filipowicz W.. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 309:1573–1576. 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., and Filipowicz W.. 2007. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 17:118–126. 10.1016/j.tcb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Polakis P. 2012. Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 4:a008052 10.1101/cshperspect.a008052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulimeno P., Bauer C., Stutz J., and Citi S.. 2010. PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PLoS One. 5:e12207 10.1371/journal.pone.0012207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A.B., Roesel D.J., Kanner S.B., and Parsons J.T.. 1989. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol. Cell. Biol. 9:629–638. 10.1128/MCB.9.2.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A.B., Daniel J., McCrea P.D., Wheelock M.J., Wu J., and Zhang Z.. 1994. Identification of a new catenin: The tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol. Cell. Biol. 14:8333–8342. 10.1128/MCB.14.12.8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Xia W., Khotskaya Y.B., Huo L., Nakanishi K., Lim S.O., Du Y., Wang Y., Chang W.C., Chen C.H., et al. . 2013. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 497:383–387. 10.1038/nature12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler B., Vardimon L., Bosserhoff A.K., and Kuphal S.. 2011. Post-transcriptional regulation controlled by E-cadherin is important for c-Jun activity in melanoma. Pigment Cell Melanoma Res. 24:148–164. 10.1111/j.1755-148X.2010.00787.x [DOI] [PubMed] [Google Scholar]
- Stoecklin G., and Kedersha N.. 2013. Relationship of GW/P-bodies with stress granules. Adv. Exp. Med. Biol. 768:197–211. 10.1007/978-1-4614-5107-5_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 2014. Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15:397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Thoreson M.A., Anastasiadis P.Z., Daniel J.M., Ireton R.C., Wheelock M.J., Johnson K.R., Hummingbird D.K., and Reynolds A.B.. 2000. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 148:189–202. 10.1083/jcb.148.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tille J.C., Ho L., Shah J., Seyde O., McKee T.A., and Citi S.. 2015. The expression of the zonula adhaerens protein PLEKHA7 is strongly decreased in high grade ductal and lobular breast carcinomas. PLoS One. 10:e0135442 10.1371/journal.pone.0135442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt P.K. 2001. Jun, the oncoprotein. Oncogene. 20:2365–2377. 10.1038/sj.onc.1204443 [DOI] [PubMed] [Google Scholar]
- Weatheritt R.J., Gibson T.J., and Babu M.M.. 2014. Asymmetric mRNA localization contributes to fidelity and sensitivity of spatially localized systems. Nat. Struct. Mol. Biol. 21:833–839. 10.1038/nsmb.2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., et al. . 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11:1487–1495. 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- Yu Y., and Elble R.C.. 2016. Homeostatic signaling by cell-cell junctions and its dysregulation during cancer progression. J. Clin. Med. 5:E26 10.3390/jcm5020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Sankala H., Zhang X., and Graves P.R.. 2008. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 413:429–436. 10.1042/BJ20080599 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.