Axonal lysosomes accumulate abnormally in Alzheimer’s disease brains. However, whether and how such lysosomes contribute to disease pathology has been unclear. Gowrishankar et al. show that the JIP3-dependent transport of axonal lysosomes negatively regulates amyloid precursor protein processing into amyloidogenic peptides.
Abstract
Lysosomes robustly accumulate within axonal swellings at Alzheimer’s disease (AD) amyloid plaques. However, the underlying mechanisms and disease relevance of such lysosome accumulations are not well understood. Motivated by these problems, we identified JNK-interacting protein 3 (JIP3) as an important regulator of axonal lysosome transport and maturation. JIP3 knockout mouse neuron primary cultures accumulate lysosomes within focal axonal swellings that resemble the dystrophic axons at amyloid plaques. These swellings contain high levels of amyloid precursor protein processing enzymes (BACE1 and presenilin 2) and are accompanied by elevated Aβ peptide levels. The in vivo importance of the JIP3-dependent regulation of axonal lysosomes was revealed by the worsening of the amyloid plaque pathology arising from JIP3 haploinsufficiency in a mouse model of AD. These results establish the critical role of JIP3-dependent axonal lysosome transport in regulating amyloidogenic amyloid precursor protein processing and support a model wherein Aβ production is amplified by plaque-induced axonal lysosome transport defects.
Introduction
Amyloid plaques, a defining feature of Alzheimer’s disease (AD) brain pathology, have long been recognized to contain an extracellular aggregate of the β-amyloid peptide that is surrounded by microglia and an abundance of swollen axons (Terry et al., 1964; Itagaki et al., 1989; Tsai et al., 2004; Fiala, 2007; Condello et al., 2011). These axons contain a massive accumulation of organelles that resemble lysosomes and/or hybrid organelles arising from the fusion of lysosomes with late endosomes and autophagosomes (subsequently referred to as lysosomes for simplicity; Terry et al., 1964; Su et al., 1993; Cataldo et al., 1994; Nixon et al., 2005; Condello et al., 2011; Gowrishankar et al., 2015). Despite their long-known and robust occurrence, the disease relevance of these lysosome-filled axonal swellings has not been established.
Our current understanding of AD has been greatly influenced by the identification of mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) genes as causes of early onset, familial forms of the disease (Selkoe and Hardy, 2016). These advances in the understanding of human AD genetics have supported the development of transgenic mice that develop amyloid plaque pathology due to the expression of mutant forms of APP and presenilin. Even in these mouse models, the amyloid plaques are surrounded by swollen, lysosome-filled axons (Yu et al., 2005; Dikranian et al., 2012; Kandalepas et al., 2013; Gowrishankar et al., 2015). The possible disease relevance of these lysosome-filled axonal swellings is supported by human and mouse studies that revealed striking enrichment for both APP and BACE1 within the swollen axons at amyloid plaques (Cras et al., 1991; Cummings et al., 1992; Kandalepas et al., 2013; Yoon et al., 2013; Gowrishankar et al., 2015; Yuan et al., 2016). Furthermore, a recent investigation of the subcellular localization of PSENs revealed lysosomal localization of AD-related PSEN1 mutants and the constitutive localization of WT PSEN2 to lysosomes (Sannerud et al., 2016). The presence of PSEN1 within lysosome-like organelles at plaques is further supported by observations in the human AD brain (Yu et al., 2005). Although such observations strongly suggest that the lysosome-filled axonal swellings at amyloid plaques could act as important sites of Aβ peptide production, this model is largely based on descriptive studies and lacks direct evidence of causality.
The high abundance of lysosomes within axonal swellings at amyloid plaques and their potential role as sites of APP processing also raise questions concerning the fundamental mechanisms that govern axonal lysosome abundance. Multiple studies have identified late endosomes and autophagosomes within distal regions of axons that likely play key housekeeping functions by sequestering old or damaged proteins and organelles (Hollenbeck, 1993; Maday et al., 2012; Maday and Holzbaur, 2014; Cheng et al., 2015). However, to degrade and recycle their contents, these organelles must acquire lysosomal properties such as hydrolytic enzymes and a highly acidic lumen. To this end, these organelles are thought to undergo a maturation process within axons that is coupled with their retrograde axonal transport toward the neuronal cell body (Hollenbeck, 1993; Overly et al., 1995; Maday et al., 2012). We previously identified such lysosome intermediates in healthy axons based on their low content of lysosomal proteases (Gowrishankar et al., 2015). Because of the similarly low protease content of axonal lysosomes at amyloid plaques, we furthermore proposed that a plaque-triggered defect in retrograde transport and maturation causes local accumulation of immature lysosomes at sites of contact between axons and amyloid fibrils (Gowrishankar et al., 2015).
To test the contribution of axonal lysosomes to amyloid plaque pathology, we first sought to develop a genetic strategy to perturb axonal lysosome abundance. To this end, we identified an important role for mouse JNK-interacting protein 3 (JIP3) in regulating the abundance and maturation state of axonal lysosomes. Of particular interest, immature lysosomes accumulated in the axons of JIP3 knockout (KO) neurons in a manner that recapitulated the key molecular and morphological properties of plaque-proximal axonal lysosomes in AD, including the buildup of APP-processing machinery. Such changes in the abundance and/or localization of APP-processing proteins were accompanied by increased Aβ peptide production. We then tested the in vivo effect of depleting JIP3 in a mouse model of AD and found a dramatic worsening in the severity of amyloid plaque pathology. These observations support a model wherein the accumulation of lysosomes within local axonal swellings at plaques actively contributes to APP processing and plaque development and suggest that restoration of normal axonal lysosome transport and maturation could help to suppress the development and progression of AD brain pathology.
Results
Lysosomes accumulate in axons of JIP3 KO neurons
Lysosome-filled axonal swellings are a major but poorly understood feature of AD amyloid plaques. To test the relevance of such axonal lysosomes to amyloid plaque pathology, we sought to establish a strategy to perturb axonal lysosome abundance. Relatively little is known about the factors that selectively regulate such organelles. However, a literature search for mutations in model organisms that increased axonal lysosome abundance yielded studies on the Caenorhabditis elegans and zebrafish orthologues of mammalian JIP3 (Drerup and Nechiporuk, 2013; Edwards et al., 2013). Although conclusions about the underlying mechanisms varied between these studies, the similar axonal lysosome phenotypes in JIP3 mutants in such distantly related organisms suggested an evolutionarily conserved role for JIP3 as a negative regulator of axonal lysosome abundance. JIP3 KO mice were previously reported to lack the telencephalic commissure and die shortly after birth because of impaired breathing (Kelkar et al., 2003) but had not been characterized from the perspective of neuronal lysosome cell biology.
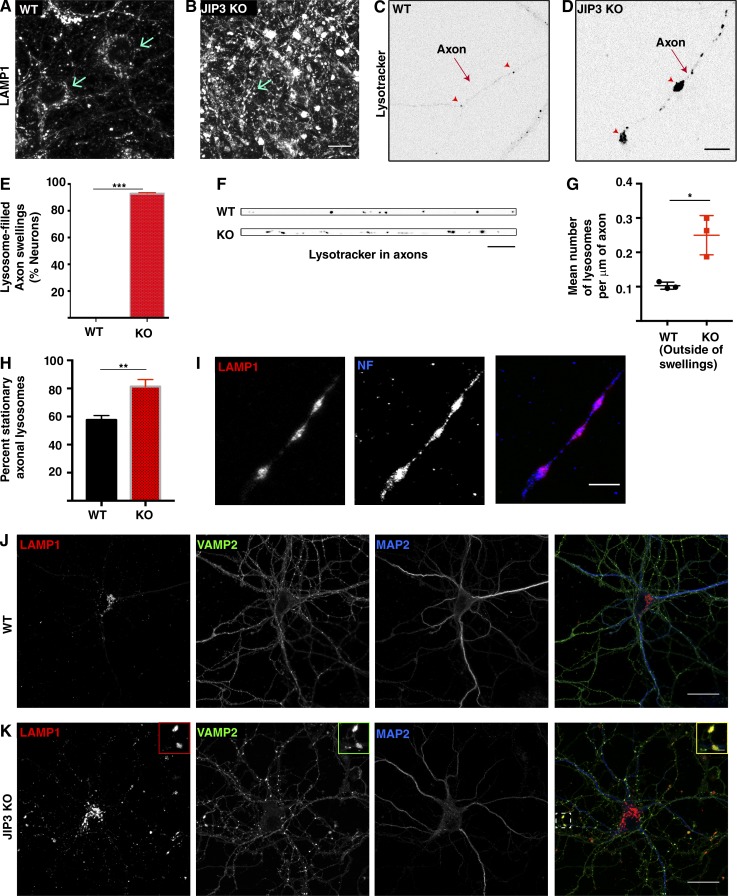
To specifically address the relationship between mammalian JIP3 and axonal lysosomes, we obtained this JIP3 KO mouse strain and generated primary cultures of cortical neurons from JIP3 KO embryos (E16–E18) and their littermate controls. After confirming the absence of JIP3 protein in the KO cultures (Fig. S1 A), lysosomes were first examined by analysis of lysosome-associated membrane protein 1 (LAMP1) immunofluorescence. Although lysosomes in control neurons were most concentrated within neuronal cell bodies (Fig. 1, A and J), with a much smaller number distributed throughout neurites, LAMP1 localization in the JIP3 KO neurons was strikingly different because of the presence of numerous large LAMP1-positive foci outside of cell bodies (Fig. 1, B and K). Colocalization with neurofilament (an axonal marker; Fig. 1 I) and VAMP2 (a synaptic vesicle protein; Fig. 1 K) and lack of overlap with a dendritic marker (MAP2B, Fig. 1 K) established that these lysosome accumulations are axonal.
Figure 1.
Loss of JIP3 results in the axonal accumulation of lysosomes. (A and B) LAMP1 immunofluorescence in WT and JIP3 KO cortical neuron primary cultures (12 DIV). Arrows indicate position of cell bodies. Bar, 10 µm. (C and D) Live-cell images of LysoTracker fluorescence in WT and JIP3 KO axons of primary neurons grown at low density. Arrowheads highlight individual lysosomes in the WT axon and focal axonal swellings filled with lysosomes in the JIP3 KO. Bar, 10 µm. (E) Quantification of the percentage of neurons with lysosome-filled axonal swellings (10 DIV, low-density cultures, mean ± SD from three independent KO and control cultures, >30 neurons per genotype; ***, P < 0.001, unpaired t test). (F) Axonal LysoTracker signals in WT and JIP3 KO (outside of swellings) neurons. Bar, 5 µm. (G) Quantification of lysosome density in WT and JIP3 KO axons (outside of swellings; 15–17 neurons per genotype from 3 independent cultures; mean ± SD; *, P = 0.0118, unpaired t test). (H) Quantification of fraction of stationary axonal lysosomes in WT and JIP3 KO (outside of swellings) neurons (mean ± SD); n = 3 independent WT and JIP3 KO littermate cultures; ∼90 WT lysosomes and 150 JIP3 KO lysosomes analyzed; **, P < 0.01, unpaired t test). (I) LAMP1 and neurofilament immunofluorescence confirm the axonal localization of lysosome accumulations in the JIP3 KO neurons. Bar, 10 µm. (J and K) Stitched images of VAMP2 (green; presynaptic protein), LAMP1 (red; lysosomes), and MAP2 (blue; dendrites) immunofluorescence in WT and JIP3 KO neurons reveals that sites of lysosome accumulation are positive for the presynaptic marker. Bars: 25 µm; (inset) 1 µm.
As previously reported (Sato et al., 2015), JIP3 KO neurons showed signs of axonal degeneration by 21 days in vitro (DIV; unpublished data). Importantly, we observed axonal lysosomal accumulations in these neurons by 7 DIV (Fig. 1 D). This lag between the appearance of axonal lysosome accumulations and axonal degeneration suggests that lysosome transport defects are not a secondary consequence of degeneration. Although this lysosome localization phenotype was robust in neurons, JIP3 KO astrocytes did not exhibit altered LAMP1 staining (Fig. S1, B and C). The differential effect of JIP3 depletion on neurons versus astrocytes was paralleled by higher JIP3 levels in neurons (Fig. S1 B).
We used LysoTracker (a fluorescent dye that preferentially identifies acidic organelles) labeling to examine the dynamics and localization of lysosomes in primary neurons. For these experiments, the neurons were cultured at low density to enable morphological discrimination between the axons and dendrites of individual neurons. In both WT and JIP3 KO neurons, LysoTracker-positive puncta were most concentrated in the cell body. Lysosomes in dendrites exhibited bidirectional movement (Video 1). Meanwhile, axons had relatively few LysoTracker puncta in WT neurons (Fig. 1 C, arrowheads), and these moved in a predominantly retrograde direction (Video 1). In contrast to the low abundance of axonal lysosomes in WT neurons, by 10 DIV, 92.8% of the JIP3 KO neurons exhibited one or more lysosome-filled axonal swellings (Fig. 1, D [arrowheads] and E). Consistent with measurements from a recent study of axonal lysosomes (Allison et al., 2017), we observed approximately one lysosome per 10 μm of axon in WT neurons (Fig. 1 F). Even outside of the swellings, the density of lysosomes was higher in JIP3 KO axons (Fig. 1, F and G).
We also found that there is a higher fraction of stationary lysosomes in JIP3 KO axons (measured outside of lysosome-enriched swellings) compared with WT axons (Fig. 1 H). Lysosomes move locally within each swelling in the JIP3 KO but only occasionally escape out into the axonal process (Video 2). The lysosome accumulation in JIP3 axons is selective, as mitochondria (visualized by both MitoTracker labeling and cytochrome c staining) do not similarly accumulate in these swellings (Fig. S1, E–K). There is an ∼30-fold increase in lysosome abundance within JIP3 axonal swellings as compared with WT axons (Fig. S1 H), whereas mitochondrial abundance changed less than twofold (Fig. S1 I), and this modest change likely reflects the increase in volume associated with the swelling. In addition to LAMP1 and LysoTracker, additional lysosome proteins such as GFP-Rab7 and GFP-Arl8 also showed robust enrichment within these axonal swellings (Fig. S1, D and E).
JIP3 KO axonal swellings resemble amyloid plaque-proximal dystrophic axons
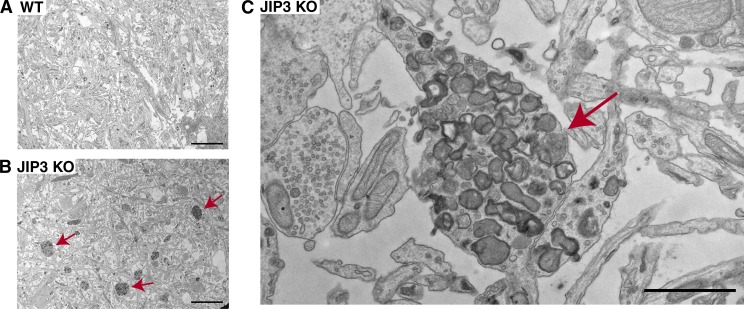
Axonal lysosome accumulations in the JIP3 KO neurons were also robustly detected by EM (Fig. 2, A–C). Low-magnification images revealed numerous clusters of electron-dense organelles that were selectively abundant in the axons of JIP3 KO cultures (Fig. 2 B). Higher-magnification observations showed numerous organelles with lysosome morphology (lamellar, multivesicular, and electron-dense properties) within such clusters without any similarly major accumulation of other prominent axonal organelles such as mitochondria or synaptic vesicles. The selective accumulations of lysosomes in these axonal swellings strongly resemble those found around amyloid plaques in AD mice (Yu et al., 2005; Dikranian et al., 2012; Kandalepas et al., 2013; Gowrishankar et al., 2015) and in human AD brain tissue (Terry et al., 1964; Cataldo et al., 1994; Nixon et al., 2005).
Figure 2.
Ultrastructure of axonal lysosome accumulations in JIP3 KO neurons. (A and B) Low-magnification EM images of WT and JIP3 KO neurons showing increased abundance of axonal lysosomes in JIP3 KO neurons (arrows). Bars, 5 µm. (C) Higher-magnification image of one such JIP3 KO axonal swelling (arrow) reveals the selective accumulation of organelles with lysosome-like morphology. Bar, 1 µm.
JIP3 KO axonal lysosome accumulations contain low levels of lysosomal proteases
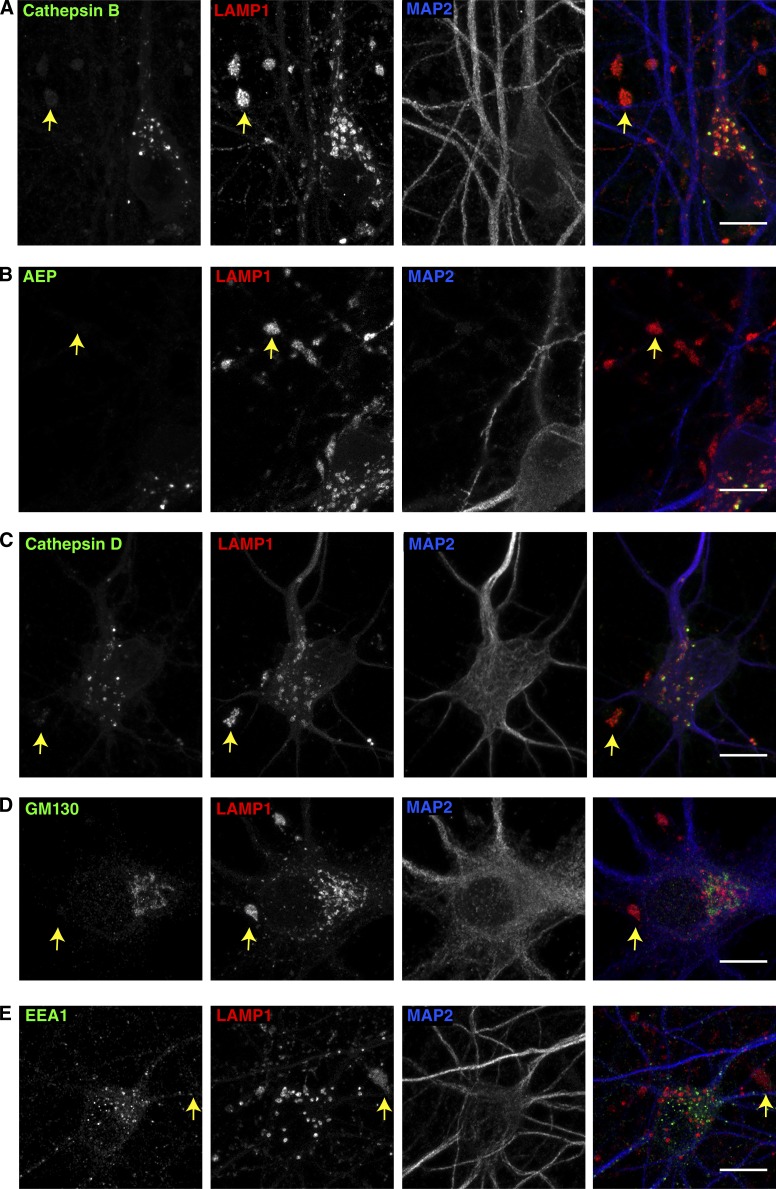
Having previously defined the axonal lysosomes that accumulate at amyloid plaques as being distinct from the lysosomes of the somatodendritic compartment based on their low content of luminal proteases (Gowrishankar et al., 2015), we next investigated the enrichment of such proteins within the JIP3 KO axons. In both WT and JIP3 KO axons, lysosomal proteases such as cathepsin B, cathepsin D, and asparagine endopeptidase (AEP) were strongly enriched within lysosomes of neuronal cell bodies (Fig. 3, A–C; Fig. S2, A–C; and Fig. S3 C). However, the highly abundant lysosomes within JIP3 KO axonal swellings contained only low or undetectable levels of these lysosomal proteases (Fig. 3, A–C, arrows). Thus, the lysosomes accumulating in JIP3 KO axons are lacking in multiple lysosomal proteases in a pattern that is similar to that of the dystrophic axons found at AD neuritic plaques (Gowrishankar et al., 2015). The low protease levels in these axonal lysosomes distinguish them from lysosomes of neuronal cell bodies and argue against anterograde transport of cell-body lysosomes into the axon as the major source of these organelles. These observations instead favor a model wherein the retrograde transport and maturation of lysosome precursors that form distally are impaired in the absence of JIP3.
Figure 3.
Analysis of organelle distribution and lysosomal protein composition of JIP3 KO axonal lysosomes. (A–C) JIP3 KO neurons triple stained for cathepsin B (A), AEP (B), and cathepsin D (C), along with both LAMP1 and MAP2. Arrows highlight representative axonal LAMP1 accumulations that have low levels of the respective lysosomal proteases compared with lysosomes of nearby neuronal cell bodies. (D and E) JIP3 KO neurons triple labeled for LAMP1 and MAP2 along with GM130 (D) or EEA1 (E), showing that axonal swellings filled with lysosomes (LAMP1 staining; arrows) are devoid of Golgi (D) and early endosomes (E). Images are representative of results from at least three independent JIP3 KO neuron cultures grown for 10–12 DIV. Bars, 10 µm.
In addition to the ultrastructural evidence supporting a selective accumulation of lysosomes versus other organelles within JIP3 KO axonal swellings, the specificity of this phenotype was further established by a light microscopy examination of the impact of JIP3 KO on the abundance and subcellular localization of other organelles. In contrast to studies that reported axonal early endosome and Golgi accumulation in C. elegans UNC-16/JIP3 mutants (Brown et al., 2009; Edwards et al., 2013), mouse JIP3 KO axonal swellings did not show enrichment for early endosome antigen 1 (EEA1; Fig. 3 E and Fig. S2 E). There was also no enrichment of sortilin 1–positive endosomes in the JIP3 KO axonal swellings (Fig. S3, A and B). We also observed that the Golgi (GM130) was restricted to the somatodendritic region in both WT and KO neurons and undetectable in the lysosome-filled axonal swellings (Fig. 3 D and Fig. S2 D). Coupled with our mitochondrial localization results (Fig. S1, J and K), our data thus defines a selective role for mammalian JIP3 in the regulation of axonal lysosome abundance.
JIP3 localizes to axonal lysosomes and is necessary for normal axonal lysosome distribution
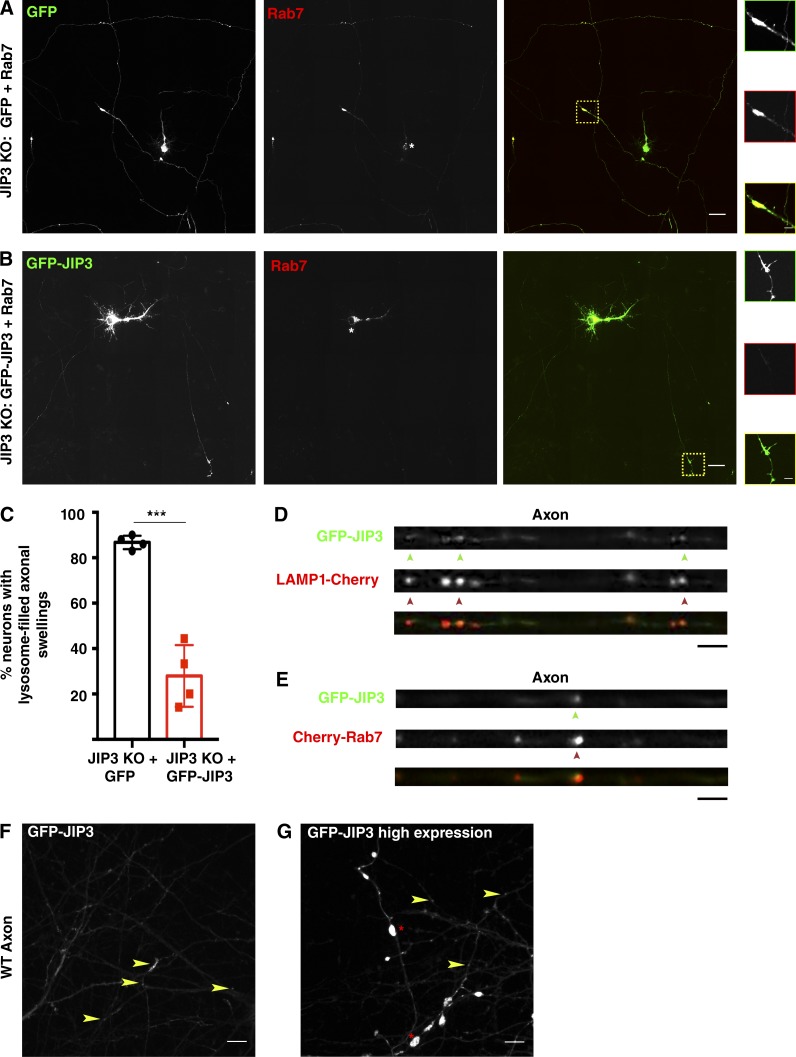
The axonal swellings filled with lysosomes combined with defects in their axonal transport raised questions about the underlying mechanisms. The possibility of a direct role for JIP3 in controlling axonal lysosome transport is consistent with past studies of interactions between JIP3 and kinesin as well as dynein motor subunits (Cavalli et al., 2005; Arimoto et al., 2011; Huang et al., 2011; Sun et al., 2011; Sato et al., 2015). As a first step toward addressing the mechanisms that support JIP3-dependent regulation of axonal lysosome abundance, we sought to validate the use of a GFP-JIP3 fusion protein by investigating its subcellular localization and its ability to rescue lysosome accumulation in JIP3 KO neurons. Consistent with our previous observations of untransfected neurons (Fig. 1), transfection of JIP3 KO neurons with GFP (negative control) and mCherry-Rab7 (lysosome marker) revealed lysosome-filled axonal swellings in 87.16% of neurons (Fig. 4, A and C). This phenotype was significantly rescued following cotransfection with GFP-JIP3 and mCherry-Rab7 (Fig. 4, B and C). The strong reduction in the JIP3 KO phenotype following expression of GFP-JIP3 supports the functionality of this fusion protein. In the few JIP3 KO neurons that still exhibited swellings following GFP-JIP3 expression, the extent of the lysosome accumulation was relatively mild compared with that of the cells transfected with the GFP (negative control) plasmid, suggesting a partial rescue even in these cells (Fig. S3, F and G). Having established the functionality of the GFP-JIP3 fusion protein, we examined its subcellular localization. In both WT and KO neurons, GFP-JIP3 was diffusely cytosolic in the cell bodies and dendrites but was distinctly enriched within axon endings and at branch points (Fig. 4 F, arrowheads). Additionally, GFP-JIP3 was also present on axonal lysosomes (defined by colocalization with either Cherry-tagged LAMP1 or Rab7) in WT (Fig. 4, D and E) and KO neurons (Fig. S3 H). We observed that the lysosome distribution in axons was sensitive to the levels of JIP3 expression. Similar to the JIP3 KO, excessively high levels of JIP3 overexpression also resulted in axonal swellings even in WT neurons (Fig. 4 G, asterisks), suggesting that the pathway in which JIP3 functions is very sensitive to JIP3 expression levels. This sensitivity to JIP3 levels may also help to explain the lack of full rescue of the JIP3 KO phenotype with exogenous GFP-JIP3 (Fig. 4 C).
Figure 4.
GFP-JIP3 localizes to axonal lysosomes and rescues axonal lysosomes over abundance in JIP3 KO neurons. (A and B) Representative stitched image of 9 DIV JIP3 KO neurons transfected at 7 DIV with GFP and Cherry-Rab7 (A) or GFP-JIP3 and Cherry-Rab7 (B) showing that GFP-transfected KO neurons have large axonal swellings filled with lysosomes (A; yellow box with dashed lines), whereas the majority of GFP-JIP3–transfected KO neurons do not exhibit this phenotype. Bars: 25 µm; (inset) 5 µm. (C) Quantification of fraction of neurons showing one or more lysosome-filled axonal swellings in GFP or GFP-JIP3–transfected JIP3 KO neurons (n = 5 independent experiments, mean ± SD; 35–45 neurons per treatment/transfection type; ***, P < 0.001). (D and E) Portions of WT axons transfected with either GFP-JIP3 and Cherry-LAMP1 (D) or GFP-JIP3 and Cherry-Rab7 (E) showing enrichment of GFP-JIP3 (green arrowheads) on lysosomes (red arrowheads). Bars, 5 µm. (F) Image of GFP-JIP3–transfected WT axon showing GFP-JIP3 distributed in vesicles and local accumulation at axon tips and branches (arrowheads highlight JIP3-positive vesicles). Bar, 10 µm. (G) High levels of GFP-JIP3 overexpression induce the formation of abnormal axonal swellings (asterisk) even in WT neurons. Bar, 10 µm.
APP-processing machinery is enriched within JIP3 KO axonal swellings
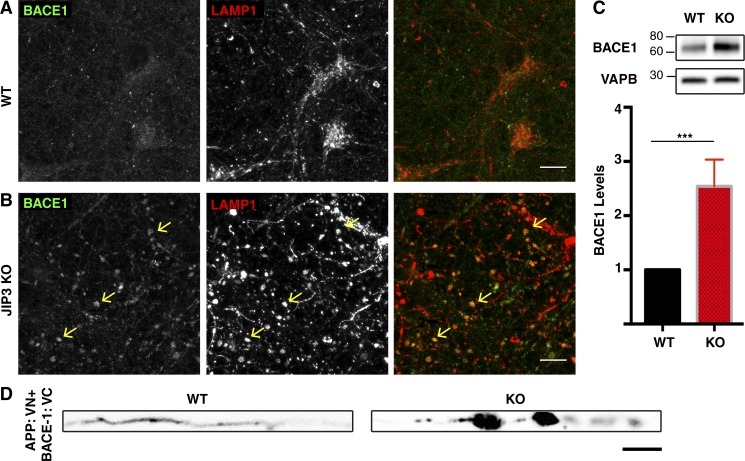
High levels of BACE1 are a striking feature of the plaque-associated dystrophic axons (Zhao et al., 2007; Kandalepas et al., 2013; Gowrishankar et al., 2015). However, the pathophysiological relevance of this abnormal BACE1 abundance and localization has not been well established. We thus sought to determine whether JIP3 KO could serve as a tool for dissecting this problem. Consistent with what has been previously reported (Vassar et al., 1999), we observed a punctate pattern for BACE1 that is distinct from that of lysosomes in WT neurons (Fig. 5 A and Fig. S4 A). In contrast, BACE1 strongly coaccumulated with LAMP1 in axonal swellings in the JIP3 KO neurons (Fig. 5 B and Fig. S4 B, arrows). Furthermore, there was an increase in total BACE1 protein abundance in the JIP3 KO cultures (Fig. 5 C), whereas levels of multiple canonical lysosome proteins remained unchanged (Fig. S4 C). Interestingly, this observation from JIP3 KO neurons parallels the increased overall BACE1 protein abundance in mouse AD models (Kandalepas et al., 2013) and human AD brains (Ewers et al., 2008).
Figure 5.
Impact of JIP3 KO on the abundance and localization of BACE1. (A and B) Representative images of WT and JIP3 KO primary cortical neurons stained for BACE1 and LAMP1. BACE1 is seen in LAMP1-negative puncta in cell bodies of both genotypes, whereas JIP3 KO neurons also contain large amounts of BACE1 coaccumulating with LAMP1 in the axonal swellings (arrows, n = 5 independent JIP3 KO cultures). Bars, 10 µm. (C) Immunoblotting reveals increased levels of BACE1 protein in JIP3 KO neurons compared with WT neurons (VAPB used as loading control; n = 4; mean ± SD; ***, P < 0.001; unpaired t test). (D) WT and JIP3 KO primary axons (straightened) expressing APP and BACE proteins tagged with split Venus fragments (APP: VN+BACE-1: VC). Note the accumulation of the complemented (interacting proteins) at axonal swellings in JIP3 KO. Bar, 5 µm.
It was recently reported that BACE1 and APP are cotransported in axons based on observations of reporter proteins containing split Venus fragments (APP: VN+BACE-1: VC) that depend on BACE1–APP interactions for reconstitution of Venus protein fluorescence (Das et al., 2016). This suggested the existence of interactions within axons between these proteins that are required for Aβ production. By extension, defects in axon transport, such as those that occur following JIP3 depletion, could prolong this convergence of APP-processing machinery and thus result in increased Aβ production. We tested this model by examining the axonal distribution of the split Venus-tagged APP and BACE1 reporter proteins. Compared with WT axons, where only occasional discrete puncta were observed (Fig. 5 D), there was robust accumulation of the reconstituted Venus signal in JIP3 KO neuron axonal swellings (Fig. 5 D). The high endogenous BACE1 levels (Fig. 5, B and C) and the BACE1–APP split construct signal at axonal lysosome accumulations (Fig. 5 D) suggested that they might act as sites of APP processing. This raised questions about PSEN localization.
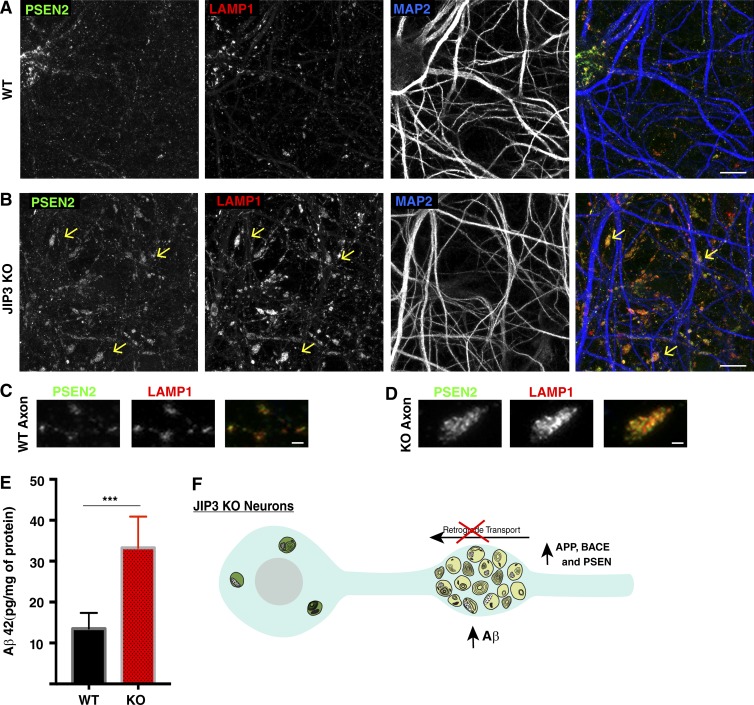
The subcellular localization and site of action of the PSENs have long been controversial (Cupers et al., 2001). Some studies suggested that PSENs and other γ secretase components (nicastrin, APH-1, and PEN-2) are enriched in lysosomes (Pasternak et al., 2003) or autophagosomes (Yu et al., 2005). Meanwhile, others found an ER, Golgi, or plasma membrane localization for PSEN1 (Kovacs et al., 1996; Annaert et al., 1999; Kim et al., 2000; Area-Gomez et al., 2009). A major limiting factor in conclusively establishing PSEN localization has been the lack of specific antibodies and knowledge concerning trafficking mechanisms that control PSEN subcellular distribution. However, PSEN2 was recently shown to contain a sorting motif that targets it to late endosomes and lysosomes within cell bodies and dendrites of WT neurons (Sannerud et al., 2016). We took advantage of the PSEN2 antibody that was described by Sannerud et al. (2016) to robustly detect the endogenous mouse PSEN2 protein and observed that it colocalized with LAMP1 in both WT and JIP3 KO neuronal cell bodies (Fig. 6, A and B). Interestingly, we also noted that PSEN2 puncta additionally colocalized with LAMP1 in both WT axons and dendrites (Fig. 6, A and B). Most importantly, we detected a striking buildup of PSEN2 in the LAMP1-positive axonal swellings of JIP3 KO neurons (Fig. 6, B and D). Thus, a consequence of the axonal lysosome transport defects in JIP3 KO is the convergence of high local levels of BACE1 and PSEN2 within the lysosome-filled swellings.
Figure 6.
PSEN2 accumulates within axonal swellings in JIP3 KO neurons. (A and B) PSEN2, LAMP1, and MAP2 immunofluorescence shows that PSEN2 colocalizes with lysosomes in neuronal cell bodies, axons, and dendrites of both WT and KO neurons. Large accumulations of PSEN2 on lysosomes in axonal swellings are selectively observed in JIP3 KO neurons (arrows; images are representative of results from five independent cultures). Bars, 10 µm. (C and D) Magnified images showing PSEN2 (green) and LAMP1 (red) colocalization in both WT (C) and JIP3 KO (D) axons. Bars, 1 µm. All results obtained from DIV 10–12 neuronal cultures. (E) Quantification of endogenous mouse Aβ42 based on ELISA measurements (mean ± SD) from n = 5 independent WT and KO cultures (***, P < 0.001, unpaired t test). (F) Schematic diagram summarizing that axonal lysosome accumulations in the JIP3 KO neurons are accompanied by the local buildup of APP, BACE1, and PSEN2 and increased production of Aβ42.
As the localization of PSEN2 at amyloid plaques has not been previously established, we next examined PSEN2 localization in the mouse 5xFAD model of AD (Oakley et al., 2006) and observed that PSEN2 is present within a subset of the swollen axons present at amyloid plaques (Fig. S4 D). We had previously hypothesized that a defect in the retrograde movement of immature axonal lysosomes leads to a local buildup of APP-processing enzymes that could result in enhanced Aβ peptide production (Gowrishankar et al., 2015). In this study, we have recapitulated key aspects of this phenotype using JIP3 KO neurons (Fig. 6 F). The convergence of APP, BACE1, and PSEN2 within JIP3 KO neurons suggested the possibility of increased APP processing and Aβ42 generation. Consistent with this prediction, ELISA assays revealed significantly elevated Aβ42 levels in JIP3 KO neurons (Fig. 6 E).
JIP3 haploinsufficiency exacerbates amyloid plaque pathology
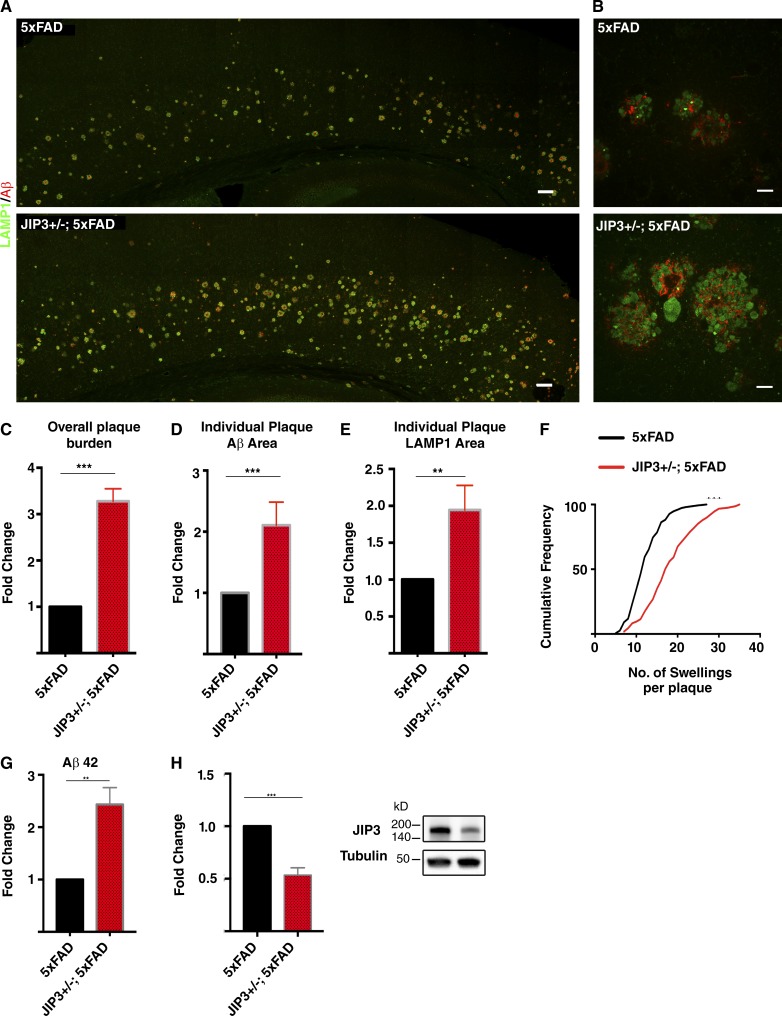
Having established that JIP3 plays a critical role in controlling the abundance and maturation state of axonal lysosomes, we next used the JIP3 genetic perturbation to assess the contribution of such lysosomes to the in vivo development of amyloid plaque pathology. As the JIP3 KO mice die perinatally (Kelkar et al., 2003; unpublished data), we examined the impact of JIP3 haploinsufficiency on the development of amyloid plaque pathology in the 5xFAD mouse model of AD. Analysis of amyloid plaque pathology via LAMP1 and Aβ immunofluorescence revealed a striking increase in overall plaque burden (area occupied by plaques) in the cerebral cortex of JIP3+/−; 5xFAD mice compared with their respective gender-matched 5xFAD control littermates (Fig. 7, A and C). Additionally, examination of individual plaques revealed a greater mean area per plaque that was occupied by axonal dystrophies (LAMP1 staining) and amyloid deposits (Aβ staining; Fig. 7 B). These observations are supported by quantification of the mean area occupied by each of these markers per plaque (Fig. 7, D and E; and Fig. S5 A). There was also an increased number of lysosome-filled axonal swellings per plaque (Fig. 7 F and Fig. S5 B). Lastly, in addition to the worsened amyloid plaque pathology, there were also increased amounts of soluble Aβ42 in JIP3+/−; 5xFAD brains compared with their 5xFAD littermates (Fig. 7 G). Consistent with their genotype, JIP3 heterozygous 5xFAD mice had half the levels of JIP3 protein of their 5xFAD littermates (Fig. 7 H). Collectively, these results identify JIP3-dependent regulation of axonal lysosome transport as protective against the development of amyloid plaque pathology.
Figure 7.
JIP3 haploinsufficiency exacerbates in vivo Aβ42 levels and the development of amyloid plaque pathology. (A) Lysosomes (LAMP1, green) and amyloid plaque (Aβ, red) labeling in stitched confocal images of the cerebral cortex of 5xFAD and JIP3+/−; 5xFAD littermates. Bars, 100 µm. (B) High-magnification images of Aβ deposits and their surrounding lysosome-filled dystrophic axons from 5xFAD and JIP3+/−; 5xFAD littermates. Bars, 10 µm. (C) Quantification of the plaque burden (area occupied by plaques, normalized to 5xFAD sample) in three pairs of 5xFAD controls and JIP3+/−; 5xFAD gender-matched littermates. ***, P < 0.001, unpaired t test. (D) Area of individual plaques as defined by Aβ signal in JIP3+/−; 5xFAD animals normalized to their corresponding gender-matched 5xFAD littermates (n = 5 pairs of gender-matched littermates, 45–50 plaques measured per animal per genotype; ***, P < 0.001, unpaired t test). (E) Quantification of the area of individual dystrophic axon cross sections (LAMP1 signal) in JIP3+/−; 5xFAD animals normalized to their corresponding gender-matched littermates (45–50 accumulations per genotype per animal were analyzed; **,P < 0.01, unpaired t test). (F) Cumulative frequency distribution of the number of axonal swellings per plaque in both 5xFAD and JIP3+/−; 5xFAD littermates (***, P < 0.001, Kolmogorov–Smirnov test). (G) Quantification of soluble Aβ42 (outside of plaques) levels in brain homogenates from three pairs of 5xFAD and JIP3+/−; 5xFAD gender-matched littermates (**, P < 0.01, unpaired t test). (H) Quantification of JIP3 protein levels from three pairs of 5xFAD and JIP3+/−; 5xFAD littermates (***, P < 0.001, unpaired t test). Data presented as mean ± SD in C–E, G, and H.
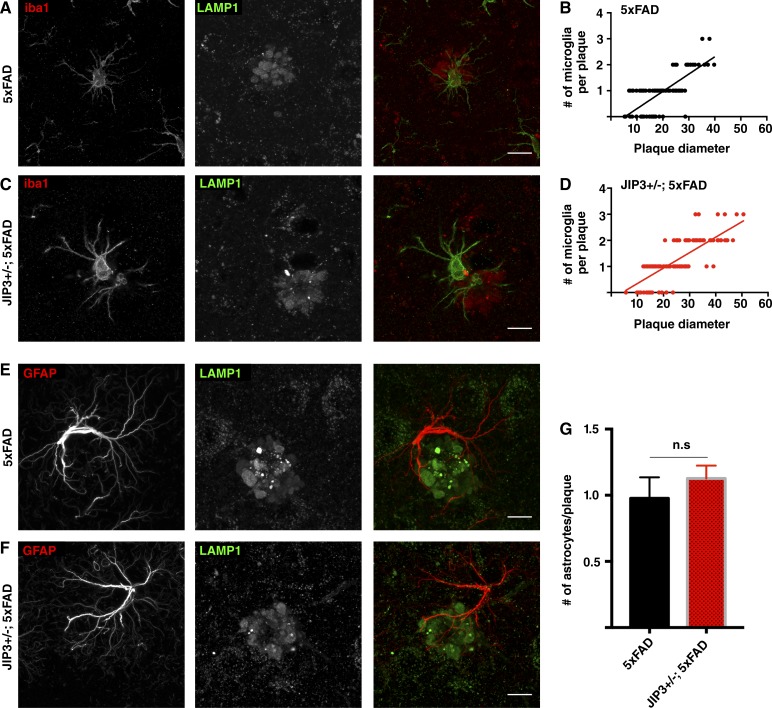
Our neuron primary culture and in vivo data are consistent with the conclusion that JIP3 depletion results in intraneuronal APP processing and Aβ42 generation. However, given that microglia can also play a role as negative regulators of amyloid plaque pathology (Wang et al., 2016; Yuan et al., 2016), we also examined the microglia in JIP3+/−; 5xFAD animals but did not detect any changes in activated (Iba1-positive) microglia abundance and/or recruitment to plaques that could explain the increased amyloid plaque abundance in the JIP3 KO mice (Fig. 8, A–D). As observed previously (Gowrishankar et al., 2015), we found that there is a positive correlation between plaque size (diameter) and the number of activated microglia associated with each plaque (Fig. 8, B and D). The higher frequency of larger plaques in JIP3+/−; 5xFAD mice was accompanied by proportionally more microglia at these plaques (Fig. 8, C and D). The number of astrocytes (GFAP staining) at these plaques also did not differ (Fig. 8, E–G). Thus, based on the Aβ42 increases in both cultured JIP3 KO neurons and JIP3+/−; 5xFAD brain homogenates, as well as the microglial and astrocyte staining in JIP3+/−; 5xFAD animals, we propose that the enhanced plaque pathology in JIP3+/−; 5xFAD mice is a result of increased intraneuronal APP processing rather than a secondary consequence of altered glial responses to amyloid plaque pathology.
Figure 8.
JIP3 haploinsufficient 5xFAD animals do not exhibit defects in glial recruitment to plaques. (A–D) Representative images of 5xFAD (A) controls and JIP3+/−; 5xFAD (C) gender-matched littermate brain sections stained for LAMP1 (plaque; red) and Iba-1 (activated microglia; green. Bars, 10 µm. (B and D) Correlation between number of microglia at plaque with plaque size from 5xFAD (B) and JIP3+/−; 5xFAD (D) mice showing that the number of activated microglia at a plaque is similarly correlated with the plaque size in both genotypes (n = 3 pairs of gender-matched littermates; 30–40 plaques per animal per genotype analyzed). (E and F) Representative images from 5xFAD (E) and JIP3+/−; 5xFAD (F) cerebral cortices stained for GFAP (astrocytes; red) and LAMP1 (lysosomes; green) showing similar astrocyte recruitment to plaques in both genotypes. Bars, 10 µm. (G) Quantification of mean number of astrocytes per plaque (mean ± SD) in three pairs of 5xFAD controls and JIP3+/−; 5xFAD gender-matched littermates. 30–45 plaques and their associated astrocytes were analyzed per animal per genotype.
Discussion
Lysosome-filled axonal swellings are a major but poorly understood feature of amyloid plaques in both human AD patients and mouse models of the disease (Terry et al., 1964; Cataldo et al., 1994; Nixon et al., 2005; Dikranian et al., 2012; Kandalepas et al., 2013; Gowrishankar et al., 2015). Based on our previous study, which identified an immature, protease-deficient population of lysosomes that builds up in axons at amyloid plaques (Gowrishankar et al., 2015), we proposed a model where these axonal lysosomes accumulate because of a local defect in their retrograde transport and maturation and represent pathologically meaningful sites of Aβ production. In the current study, we sought to test this model. To this end, we identified JIP3 as a candidate for regulating axonal lysosome abundance and validated such a function through extensive cell biological characterization of JIP3 KO mouse neurons. We found that JIP3 is an important regulator of the transport, abundance, and maturation state of axonal lysosomes. Furthermore, we documented multiple similarities between the axonal lysosomes that accumulate at amyloid plaques and in JIP3 KO neurons. This included a buildup of APP, BACE1, and PSEN2 that suggested that the loss of JIP3 could be accompanied by an increase in the processing of APP into Aβ peptides. Indeed, we detected significantly elevated Aβ levels in the JIP3 KO neurons. We then tested the in vivo effect of decreasing JIP3 gene dosage by assessing the development of amyloid plaque pathology in a mouse model of AD and found that reduced JIP3 expression resulted in an increase in soluble Aβ levels, plaque size, abundance, and axonal dystrophy. These results collectively support a model wherein efficient retrograde axonal lysosome transport acts as a negative regulator of APP processing and development of amyloid plaque pathology.
Regulation of axonal lysosome abundance by JIP3
From a neuronal cell biology perspective, the high abundance of lysosomes within axons at amyloid plaques raises questions about the normal processes that control the biogenesis, transport, and abundance of lysosomes within this specific subcellular compartment. Recent years have yielded advances in the understanding of mechanisms that control the subcellular distribution of lysosomes (Pu et al., 2016), as well as an appreciation that distinct subtypes of lysosomes can exist within different subcellular compartments (Johnson et al., 2016). However, there is still little known about how such processes are adapted to the unique demands of neurons. Although previous studies of C. elegans and zebrafish both concluded that the loss of JIP3 resulted in increased axonal lysosome abundance (Drerup and Nechiporuk, 2013; Edwards et al., 2013), they differed in their interpretation of the underlying mechanisms. The C. elegans JIP3 (UNC-16) loss-of-function mutant was proposed to have a defect in the ability of axon initial segments to inhibit export of lysosomes from the cell body (Edwards et al., 2013). Meanwhile, analysis of zebrafish JIP3 mutants instead concluded that JIP3 is critical for the removal of lysosomes from axons via dynein-mediated retrograde transport (Drerup and Nechiporuk, 2013).
Our observations of the mouse JIP3 KO neurons fit best with the retrograde transport model for JIP3 function, as the lysosomes that accumulated in the JIP3 KO axons were distinguishable from cell body lysosomes by their low levels of cathepsins. They are thus more likely to reflect intermediates in the axonal biogenesis of lysosomes rather than cell body lysosomes that have escaped into the axon. This interpretation builds on a growing appreciation that there are high rates of autophagy in the distal regions of axons but that such autophagosomes must fuse with endosomes and acquire lysosomal properties such as acidification before undergoing efficient retrograde back toward the cell body for the eventual degradation and recycling of their cargos (Hollenbeck, 1993; Maday et al., 2012; Maday and Holzbaur, 2014; Cheng et al., 2015). We also observed an increased fraction of stationary lysosomes in JIP3 KO axons and localized JIP3 to axonal lysosomes. Regulation of retrograde axonal transport of lysosomes by JIP3 is additionally supported by previous observations that it interacts with dynein subunits such as p150glued (Cavalli et al., 2005) and the dynein light intermediate chain (DLIC; Huang et al., 2011). Furthermore, JIP3 coenrichment with p150glued at sites distal to sciatic nerve injury (Cavalli et al., 2005) also fits with putative direct functions for JIP3 in retrograde axonal transport.
The robust, evolutionarily conserved axonal lysosome phenotype in JIP3 KO neurons raises important questions about the spatiotemporal regulation of JIP3 interactions with motors, small GTPases, and signaling proteins and their respective contributions to regulation of axonal lysosome transport. In other cellular contexts, the small GTPase Rab7 links dynein to late endosomes and lysosomes via the RILP adaptor protein (Jordens et al., 2001). Although JIP3 has not been found to bind to Rab7, interactions have been detected between JIP3 and Rab36, and Rab36 has been implicated in the retrograde movement of melanosomes in melanocytes (Matsui et al., 2012). However, the physiological relevance of the JIP3–Rab36 interaction is not known, and little is known about the Rab36 function in neurons. Another GTPase, Arf6, binds the leucine zipper region on JIP3 (Montagnac et al., 2009). This JIP3–Arf6 interaction was implicated in early parts of the endocytic pathway (Montagnac et al., 2009) in nonneuronal cells, but Arf6 is not known to have a direct role in the axonal transport of lysosomes. Thus, although known interactions with these small GTPases may warrant further investigation, there is also a need for the unbiased identification of physiologically relevant neuronal binding partners for JIP3.
Convergence of Aβ−processing machinery within axonal lysosomes
Initial observations of organelle accumulation in swollen axons around amyloid plaques in human AD brains were made over five decades ago (Terry et al., 1964). Subsequent studies noted that the dystrophic axons that surround amyloid plaques in human AD brains contain BACE1, PSEN1, and APP (Cummings et al., 1992; Yu et al., 2005; Kandalepas et al., 2013; Gowrishankar et al., 2015; Yuan et al., 2016). These observations previously led us to propose that such dystrophic axons at amyloid plaques could represent sites of APP processing to generate Aβ peptides and thus actively contribute to further disease progression (Gowrishankar et al., 2015). Our new results support an important role for efficient retrograde transport of axonal lysosomes in limiting APP processing and the development of amyloid plaque pathology. While this article was under revision, additional support for this model was provided by a study that targeted the snapin gene and also observed similar effects on BACE1 transport and Aβ production in response to impaired axonal transport of lysosomes (Ye et al., 2017). Snapin was previously proposed to link late endosomes/lysosomes to the dynein intermediate chain and thus promote their retrograde, dynein-dependent axonal transport (Cai et al., 2010). However, as snapin was subsequently also identified as a subunit of the BORC complex that promotes the kinesin-dependent transport of lysosomes (Pu et al., 2015), it remains to be determined how the proposed role for snapin in retrograde axonal transport of lysosomes is coordinated with its role within the BORC complex in promoting anterograde transport of axonal lysosomes (Farías et al., 2017). Given the emerging importance of axonal lysosome transport in regulating amyloidogenic processing of APP, further dissection and understanding of the complex interplay of adaptors and scaffolding proteins (such as JIP3) that control the anterograde and retrograde movement of axonal lysosomes is critically required.
Axonal transport and AD vulnerability
In this study, we found that increasing the abundance of axonal lysosomes had striking consequences for the localization and levels of APP-processing machinery and exacerbated the amyloid plaque pathology in a mouse model of AD. These results suggest that other genetic and nongenetic perturbations that impair axonal transport could influence APP processing and thus confer AD risk. Indeed, reducing kinesin levels was previously reported to increase the abundance of axonal swellings, Aβ deposition, and plaque burden in a mouse model of AD (Stokin et al., 2005). Traumatic brain injury (TBI) may be an AD risk factor (Johnson et al., 2010), and controlled cortical impact TBI enhances intra-axonal Aβ levels in 3x-Tg AD mice (Tran et al., 2011). TBI also recapitulates many of these factors, including axonal swellings, axonal organelle accumulations, and enhanced Aβ production (Sherriff et al., 1994; Johnson et al., 2010), as well as rapid and abundant accumulation of APP (Gentleman et al., 1993). Buildup of APP-processing machinery (BACE1 and PSENs) also occurs in axons in both animal and human models of TBI (Roberts et al., 1991). These studies offer broad support for a model wherein axonal organelles are a source of Aβ and perturbation of axonal transport through injury or genetic means can potentially promote the development of amyloid plaque pathology.
Conclusion
Collectively, our results indicate that the axonal accumulations of lysosomes at amyloid plaques are not innocent bystanders but instead are important contributors to APP processing and amyloid plaque growth. These findings emphasize the need for a more comprehensive elucidation of the mechanisms that regulate the abundance and transport of axonal lysosomes as well as the mechanisms whereby contact between axons and amyloid fibrils locally disrupts such processes at amyloid plaques. The robust axonal lysosome phenotype arising in JIP3 KO neurons raises questions about how this protein functions as part of a pathway that controls axonal lysosome abundance and protects against the amyloidogenic processing of APP. The identification of other proteins that function alongside JIP3 and the elucidation of rate-limiting steps in this process will be critical for developing strategies that enhance the axonal transport and maturation of lysosomes. Our characterization of JIP3 KO neurons resulted in the development of an in vitro model that will support future studies of mechanisms that regulate axonal lysosome transport and maturation as well as the coordination of both β- and γ-secretase activities at such sites. Increased understanding of these problems could ultimately lead to strategies to modulate the axonal abundance of lysosomes for therapeutic purposes.
Materials and methods
Mouse strains
Animal procedures were approved by and performed in accordance with guidelines established by the Institutional Animal Care and Use Committee at Yale University. 5xFAD mice (Oakley et al., 2006) were provided by R. Vasser (Northwestern University, Evanston, IL). The JIP3 KO (MAPK8ip3tm1Rjd/J strain) mice (Kelkar et al., 2003) were acquired from The Jackson Laboratory. 5xFAD heterozygous male transgenic mice were bred with JIP3 heterozygous females. The resulting 5xFAD;JIP3+/− offspring were compared with age- and sex-matched 5xFAD littermates.
Primary neuron culture
Embryos from JIP3+/− matings were obtained at embryonic day 16–18. The brains were stored in ice-cold Hibernate E medium (Thermo Fisher Scientific) while PCR-based genotyping was performed to identify WT and KO embryos.
Neuron culture
Growth of neurons at low density was performed in the following manner: Divots were made in 12-well plates using a soldering iron to create micropedestals. These dishes were then coated with poly-d-lysine (PDL; 20 µg/ml; Sigma Aldrich). In parallel, No. 1.5 18-mm coverslips (Assistant Brand; Carolina Biological) were added to other 12-well plates and also coated with PDL. Neurons were subsequently plated at high density (300,000/well) on the bottom of the dish containing the pedestals and at low density (5,000–8,000/coverslip) on the coverslips. Neurons on the coverslips were allowed to grow overnight before they were inverted over the high-density support. This was accompanied by replacement of the plating medium with Neurobasal + 2% B27, 1% glutamax, and penicillin/streptomycin (Neuronal Medium). Neurons were grown in this media until experiments were performed. For imaging of neurons grown at high density, 100,000–300,000 neurons were directly plated onto PDL-coated, 18-mm coverslips in 12-well dishes and grown in Neuronal Medium.
Microscopy
16-bit images (512 × 512 pixels) were acquired with a laser scanning confocal microscope (LSM 710; ZEISS) equipped with a 63× plan Apo (NA 1.4) oil immersion objective or a 20× (NA 0.8) objective and a QUASAR (quiet spectral array) photomultiplier detector. Z-stacks with a step size of 0.49 µm and 1.0 µm were routinely acquired with the 63× and 20× objectives, respectively. Spinning disk confocal images were obtained using the UltraVIEW VoX system (PerkinElmer), including an inverted microscope (Nikon Ti-E Eclipse equipped with a spinning disk confocal scan head; CSU-X1; Yokogawa) driven by Volocity (PerkinElmer) software. Images were acquired without binning with a 14-bit (1,000 × 1,000) electron-multiplying charge-coupled device camera (Hamamatsu Photonics). Spinning disk confocal images were obtained with a 60× (NA 1.3) objective.
Live imaging of lysosomes in primary neurons
Neurons were incubated with 10 nM LysoTracker Red alone or LysoTracker Deep Red (Invitrogen) and MitoTracker green (Invitrogen; 10 nM each) for 2 min and then rinsed gently twice with imaging medium (136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, and 10 mM Hepes, pH 7.4, supplemented with BSA and glucose) and imaged in the same buffer for 10 min.
Quantification of neurons with axonal swellings
For quantification of the percentage of neurons with lysosome-filled axonal swellings, DIV 10 primary cortical neurons grown at low density were incubated with LysoTracker Red (10 nm; 2 min) and imaged using the spinning disk confocal microscope. Stitched images of entire neurons were obtained to capture the full axonal length, and only those neurons whose axons were uniquely identifiable were used for the analysis.
Analysis of lysosome density and mobility in axons
Lysosome density and mobility in axons were analyzed in young (DIV 4) primary cortical neurons cultured from WT and JIP3 KO embryos, as this age preceded the major development of lysosome-filled axonal swellings. Neurons grown at low density were incubated with LysoTracker Red (10 nM; 2 min) and subsequently imaged by spinning disk confocal microscopy. Lysosome dynamics were imaged at a rate of 120 frames/min for 2 min. Individual axons in the first frame were traced to measure their lengths, and the number of lysosomes in that length was counted and expressed as the number of lysosomes per micrometer axon length. To measure the mobility of axonal lysosomes, kymographs were generated using the “reslice” function on “straightened” axons (ImageJ), and the percentage of lysosomes that were stationary (moved <2 µm in 2 min) in each axon was measured. At least four axons per culture per genotype were analyzed from three independent WT and JIP3 KO cultures (90 and 150 lysosomes, respectively).
Rescue experiments with GFP-JIP3
For expression of N-terminal GFP-tagged JIP3, a mouse JIP3 cDNA (GenBank accession no. BC068312) was purchased from Thermo Fisher Scientific and cloned into pEGFP-C1 (Clontech). Primary neurons from JIP3 KO embryos were cultured and transfected using Lipofectamine 2000 (Invitrogen) with Cherry-Rab7 (from G. Voeltz, University of Colorado Boulder, Boulder, CO), Addgene plasmid no. 61804; Rowland et al., 2014) and either control GFP or GFP-JIP3 on 7 DIV and imaged live on 9 DIV on a spinning disk microscope. All transfected neurons (with Cherry-Rab7 plus GFP or GFP-JIP3) whose axons could be traced were used for evaluation, and the fraction of total neurons that contained at least one lysosome-filled axonal swelling was calculated. Five independent experiments (cultures) were performed, with a total of 35–45 neurons per transfection condition.
Immunofluorescence analysis of neurons
Primary cultures of WT and JIP3 KO embryonic cortical neurons were prepared and used between DIV 9 and 14. Cells were fixed with 4% PFA in 0.1 M sodium phosphate buffer, pH 7.2, and 4% sucrose for 20 min, rinsed with 50 mM NH4Cl, and permeabilized with PBS containing 0.1% Triton X-100 (PBST) for 10 min. Cells were then blocked with PBST + 3% BSA. Primary (overnight) and secondary antibody (1 h) incubations were performed in the same buffer with three 10-min washes in PBST before and after secondary antibody treatment. Antibodies used in this study are summarized in Table S1.
EM of primary neurons
Neurons were grown on coverslips at high density and processed for EM at 12–14 DIV as described previously (Wu et al., 2014).
Immunoblotting
Primary neurons (WT and JIP3 KO) were grown on six-well plates (600,000 cells/well). DIV 14 neurons were washed with ice-cold PBS and then lysed in lysis buffer (150 mM NaCl, 20 mM Hepes, 0.5% SDS, and protease inhibitor cocktail) and spun at 13,000 g for 5 min. The supernatant was collected and incubated at 95°C for 5 min in SDS sample buffer before SDS-PAGE, transfer to nitrocellulose membranes, and immunoblotting. See Table S1 for antibody information.
Immunofluorescence on mouse brain sections
Immunofluorescence on free-floating coronal mouse brain sections was performed as described previously (Gowrishankar et al., 2015).
ELISA-based measurement of endogenous Aβ
Primary neurons from JIP3 KO and WT embryos (littermates) were cultured as described in the Neuron culture section for 2 wk. Aβ42 ELISA measurements were performed as per a previously validated assay for the measurement of mouse and human Aβ42 (Teich et al., 2013). In brief, neurons were lysed in Tris-EDTA with protease inhibitor and processed double-blind for ELISA measurements using a high-sensitivity ELISA kit (catalog no. 292–64501; Wako). Standard curves were generated using Aβ1-42 peptide standards for each set of experiments. Such ELISA measurements were made from five independent WT and KO cultures.
ELISA-based measurement of Aβ42 from mouse brains
Brains from 3.5- to 4-mo-old JIP3+/−; 5xFAD/+ animals and their gender-matched littermate controls (5xFAD/+) were homogenized in Tris buffer (10 mM Tris, 1 mM EDTA, 250 mM sucrose) with protease inhibitor and spun at 13,000 g for 5 min. Supernatant was used for ELISA measurements using a high-sensitivity ELISA kit (catalog no. 292–64501; Wako). Standard curves were generated using Aβ1-42 peptide standards for each experiment.
Quantification of axonal lysosome accumulation and amyloid plaque abundance
Quantification of the area of axonal lysosome accumulations and amyloid plaque size was done as described previously (Gowrishankar et al., 2015). In brief, coronal brain sections from 3.5- to 4-mo-old JIP3+/−; 5xFAD/+ animals and their gender-matched control littermates (5xFAD/+) were costained for LAMP1 and Aβ and imaged on a spinning disk microscope to acquire both lysosome and amyloid plaque signals. Regions of interest were identified based on the distinct patterns of both Aβ and LAMP1 signals and manually outlined for area measurements.
Quantification of plaque burden in AD mice
Coronal brain sections from 3.5- to 4-mo-old JIP3+/−; 5xFAD/+ animals and their gender-matched littermate controls (5xFAD/+) were costained for LAMP1 and Aβ and imaged with the 20× objective on the LSM710 confocal microscope using the Zen software “Tile” option to obtain a stitched image of amyloid plaques throughout the entire neocortex region. Plaques were identified (based on Aβ staining) in an automated fashion using the “threshold” and “analyze particle” functions of ImageJ software, and the area of the entire cortical tissue that was outlined was calculated. The percentage area of tissue occupied by amyloid plaques in JIP3+/−; 5xFAD mice was normalized to that of the gender-matched 5xFAD littermate controls.
Statistical analysis
Data are represented as mean ± SD unless otherwise specified. Statistical analysis was performed using Prism 7 software. Detailed statistical information (statistical test performed, number of independent experiments, and p-values) is described in the respective figure legends.
Online supplemental material
Fig. S1 shows lysosome distribution in astrocytes (WT and JIP3 KO) and mitochondrial distribution in neurons. Fig. S2 summarizes the distribution of multiple organelles and lysosomal enzymes in WT neurons. Fig. S3 demonstrates the organelle composition of axonal swellings in JIP3 KO neurons and rescue following GFP-JIP3 expression. Fig. S4 presents the distribution and levels of APP-processing machinery in JIP3 KO neurons. Fig. S5 shows characteristics of amyloid plaque pathology in JIP3 haploinsufficient mice. Table 1 lists antibodies (source and dilution) used in the study. Table 2 describes plasmids used in the study. Videos 1 and 2 show lysosome dynamics in a WT and JIP3 KO neuron, respectively.
Supplementary Material
Acknowledgments
We thank Pietro De Camilli for his generous advice, Agnes Roczniak-Ferguson and Frank Wilson for superb management and maintenance of laboratory infrastructure, Wendolyn Hill for illustration assistance, and Heather Wheeler for outstanding technical assistance.
This work was supported in part by grants from the National Institutes of Health (NIH; GM105718 and AG047270) to S.M. Ferguson; S.M. Ferguson was also supported by the Ellison Medical Foundation. S. Gowrishankar is a recipient of a BrightFocus postdoctoral fellowship. Support for S. Gowrishankar and Y. Wu was also provided by grants to Pietro De Camilli from the NIH (NS036251 and DK45735). Microscopy studies were supported by the Yale University Program in Cellular Neuroscience, Neurodegeneration, and Repair imaging facility.
The authors declare no competing financial interests.
Author contributions: S. Gowrishankar and S.M. Ferguson conceived of and designed all experiments. S. Gowrishankar and S.M. Ferguson analyzed and interpreted data and wrote the manuscript. S. Gowrishankar carried out all experiments with the exception of the EM studies that are presented in Fig. 2 (Y. Wu).
Footnotes
Abbreviations used:
- AD
- Alzheimer’s disease
- APP
- amyloid precursor protein
- DIV
- days in vitro
- KO
- knockout
- PSEN
- presenilin
- TBI
- traumatic brain injury
References
- Allison R., Edgar J.R., Pearson G., Rizo T., Newton T., Günther S., Berner F., Hague J., Connell J.W., Winkler J., et al. 2017. Defects in ER–endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 216:1337–1355. 10.1083/jcb.201609033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annaert W.G., Levesque L., Craessaerts K., Dierinck I., Snellings G., Westaway D., George-Hyslop P.S., Cordell B., Fraser P., and De Strooper B.. 1999. Presenilin 1 controls gamma-secretase processing of amyloid precursor protein in pre-golgi compartments of hippocampal neurons. J. Cell Biol. 147:277–294. 10.1083/jcb.147.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Area-Gomez E., de Groof A.J., Boldogh I., Bird T.D., Gibson G.E., Koehler C.M., Yu W.H., Duff K.E., Yaffe M.P., Pon L.A., and Schon E.A.. 2009. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 175:1810–1816. 10.2353/ajpath.2009.090219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto M., Koushika S.P., Choudhary B.C., Li C., Matsumoto K., and Hisamoto N.. 2011. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31:2216–2224. 10.1523/JNEUROSCI.2653-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H.M., Van Epps H.A., Goncharov A., Grant B.D., and Jin Y.. 2009. The JIP3 scaffold protein UNC-16 regulates RAB-5 dependent membrane trafficking at C. elegans synapses. Dev. Neurobiol. 69:174–190. 10.1002/dneu.20690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Lu L., Tian J.H., Zhu Y.B., Qiao H., and Sheng Z.H.. 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 68:73–86. 10.1016/j.neuron.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo A.M., Hamilton D.J., and Nixon R.A.. 1994. Lysosomal abnormalities in degenerating neurons link neuronal compromise to senile plaque development in Alzheimer disease. Brain Res. 640:68–80. 10.1016/0006-8993(94)91858-9 [DOI] [PubMed] [Google Scholar]
- Cavalli V., Kujala P., Klumperman J., and Goldstein L.S.. 2005. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 168:775–787. 10.1083/jcb.200410136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.T., Zhou B., Lin M.Y., Cai Q., and Sheng Z.H.. 2015. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209:377–386. 10.1083/jcb.201412046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello C., Schain A., and Grutzendler J.. 2011. Multicolor time-stamp reveals the dynamics and toxicity of amyloid deposition. Sci. Rep. 1:19 10.1038/srep00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cras P., Kawai M., Lowery D., Gonzalez-DeWhitt P., Greenberg B., and Perry G.. 1991. Senile plaque neurites in Alzheimer disease accumulate amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 88:7552–7556. 10.1073/pnas.88.17.7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings B.J., Su J.H., Geddes J.W., Van Nostrand W.E., Wagner S.L., Cunningham D.D., and Cotman C.W.. 1992. Aggregation of the amyloid precursor protein within degenerating neurons and dystrophic neurites in Alzheimer’s disease. Neuroscience. 48:763–777. 10.1016/0306-4522(92)90265-4 [DOI] [PubMed] [Google Scholar]
- Cupers P., Bentahir M., Craessaerts K., Orlans I., Vanderstichele H., Saftig P., De Strooper B., and Annaert W.. 2001. The discrepancy between presenilin subcellular localization and gamma-secretase processing of amyloid precursor protein. J. Cell Biol. 154:731–740. 10.1083/jcb.200104045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U., Wang L., Ganguly A., Saikia J.M., Wagner S.L., Koo E.H., and Roy S.. 2016. Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat. Neurosci. 19:55–64. 10.1038/nn.4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikranian K., Kim J., Stewart F.R., Levy M.A., and Holtzman D.M.. 2012. Ultrastructural studies in APP/PS1 mice expressing human ApoE isoforms: Implications for Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 5:482–495. [PMC free article] [PubMed] [Google Scholar]
- Drerup C.M., and Nechiporuk A.V.. 2013. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 9:e1003303 10.1371/journal.pgen.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S.L., Yu S.C., Hoover C.M., Phillips B.C., Richmond J.E., and Miller K.G.. 2013. An organelle gatekeeper function for Caenorhabditis elegans UNC-16 (JIP3) at the axon initial segment. Genetics. 194:143–161. 10.1534/genetics.112.147348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers M., Zhong Z., Bürger K., Wallin A., Blennow K., Teipel S.J., Shen Y., and Hampel H.. 2008. Increased CSF-BACE 1 activity is associated with ApoE-ε4 genotype in subjects with mild cognitive impairment and Alzheimer’s disease. Brain. 131:1252–1258. 10.1093/brain/awn034 [DOI] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C. 2007. Mechanisms of amyloid plaque pathogenesis. Acta Neuropathol. 114:551–571. 10.1007/s00401-007-0284-8 [DOI] [PubMed] [Google Scholar]
- Gentleman S.M., Nash M.J., Sweeting C.J., Graham D.I., and Roberts G.W.. 1993. β-amyloid precursor protein (β APP) as a marker for axonal injury after head injury. Neurosci. Lett. 160:139–144. 10.1016/0304-3940(93)90398-5 [DOI] [PubMed] [Google Scholar]
- Gowrishankar S., Yuan P., Wu Y., Schrag M., Paradise S., Grutzendler J., De Camilli P., and Ferguson S.M.. 2015. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl. Acad. Sci. USA. 112:E3699–E3708. 10.1073/pnas.1510329112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P.J. 1993. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 121:305–315. 10.1083/jcb.121.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.H., Duan S., Sun T., Wang J., Zhao L., Geng Z., Yan J., Sun H.J., and Chen Z.Y.. 2011. JIP3 mediates TrkB axonal anterograde transport and enhances BDNF signaling by directly bridging TrkB with kinesin-1. J. Neurosci. 31:10602–10614. 10.1523/JNEUROSCI.0436-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itagaki S., McGeer P.L., Akiyama H., Zhu S., and Selkoe D.. 1989. Relationship of microglia and astrocytes to amyloid deposits of Alzheimer disease. J. Neuroimmunol. 24:173–182. 10.1016/0165-5728(89)90115-X [DOI] [PubMed] [Google Scholar]
- Johnson D.E., Ostrowski P., Jaumouillé V., and Grinstein S.. 2016. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212:677–692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V.E., Stewart W., and Smith D.H.. 2010. Traumatic brain injury and amyloid-β pathology: A link to Alzheimer’s disease? Nat. Rev. Neurosci. 11:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kandalepas P.C., Sadleir K.R., Eimer W.A., Zhao J., Nicholson D.A., and Vassar R.. 2013. The Alzheimer’s β-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 126:329–352. 10.1007/s00401-013-1152-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar N., Delmotte M.H., Weston C.R., Barrett T., Sheppard B.J., Flavell R.A., and Davis R.J.. 2003. Morphogenesis of the telencephalic commissure requires scaffold protein JNK-interacting protein 3 (JIP3). Proc. Natl. Acad. Sci. USA. 100:9843–9848. 10.1073/pnas.1733944100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lah J.J., Thinakaran G., Levey A., and Sisodia S.S.. 2000. Subcellular localization of presenilins: association with a unique membrane pool in cultured cells. Neurobiol. Dis. 7:99–117. 10.1006/nbdi.1999.0280 [DOI] [PubMed] [Google Scholar]
- Kovacs D.M., Fausett H.J., Page K.J., Kim T.W., Moir R.D., Merriam D.E., Hollister R.D., Hallmark O.G., Mancini R., Felsenstein K.M., et al. 1996. Alzheimer-associated presenilins 1 and 2: neuronal expression in brain and localization to intracellular membranes in mammalian cells. Nat. Med. 2:224–229. 10.1038/nm0296-224 [DOI] [PubMed] [Google Scholar]
- Maday S., and Holzbaur E.L.. 2014. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell. 30:71–85. 10.1016/j.devcel.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Wallace K.E., and Holzbaur E.L.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417. 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Ohbayashi N., and Fukuda M.. 2012. The Rab interacting lysosomal protein (RILP) homology domain functions as a novel effector domain for small GTPase Rab36: Rab36 regulates retrograde melanosome transport in melanocytes. J. Biol. Chem. 287:28619–28631. 10.1074/jbc.M112.370544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G., Sibarita J.B., Loubéry S., Daviet L., Romao M., Raposo G., and Chavrier P.. 2009. ARF6 interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19:184–195. 10.1016/j.cub.2008.12.043 [DOI] [PubMed] [Google Scholar]
- Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., and Cuervo A.M.. 2005. Extensive involvement of autophagy in Alzheimer disease: An immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 64:113–122. 10.1093/jnen/64.2.113 [DOI] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overly C.C., Lee K.D., Berthiaume E., and Hollenbeck P.J.. 1995. Quantitative measurement of intraorganelle pH in the endosomal-lysosomal pathway in neurons by using ratiometric imaging with pyranine. Proc. Natl. Acad. Sci. USA. 92:3156–3160. 10.1073/pnas.92.8.3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak S.H., Bagshaw R.D., Guiral M., Zhang S., Ackerley C.A., Pak B.J., Callahan J.W., and Mahuran D.J.. 2003. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J. Biol. Chem. 278:26687–26694. 10.1074/jbc.M304009200 [DOI] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Guardia C.M., Keren-Kaplan T., and Bonifacino J.S.. 2016. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129:4329–4339. 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G.W., Gentleman S.M., Lynch A., and Graham D.I.. 1991. β A4 amyloid protein deposition in brain after head trauma. Lancet. 338:1422–1423. 10.1016/0140-6736(91)92724-G [DOI] [PubMed] [Google Scholar]
- Rowland A.A., Chitwood P.J., Phillips M.J., and Voeltz G.K.. 2014. ER contact sites define the position and timing of endosome fission. Cell. 159:1027–1041. 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R., Esselens C., Ejsmont P., Mattera R., Rochin L., Tharkeshwar A.K., De Baets G., De Wever V., Habets R., Baert V., et al. 2016. Restricted location of PSEN2/γ-secretase determines substrate specificity and generates an intracellular Aβ pool. Cell. 166:193–208. 10.1016/j.cell.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Ishikawa M., Mochizuki M., Ohta M., Ohkura M., Nakai J., Takamatsu N., and Yoshioka K.. 2015. JSAP1/JIP3 and JLP regulate kinesin-1-dependent axonal transport to prevent neuronal degeneration. Cell Death Differ. 22:1260–1274. 10.1038/cdd.2014.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D.J., and Hardy J.. 2016. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8:595–608. 10.15252/emmm.201606210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherriff F.E., Bridges L.R., and Sivaloganathan S.. 1994. Early detection of axonal injury after human head trauma using immunocytochemistry for β-amyloid precursor protein. Acta Neuropathol. 87:55–62. 10.1007/BF00386254 [DOI] [PubMed] [Google Scholar]
- Stokin G.B., Lillo C., Falzone T.L., Brusch R.G., Rockenstein E., Mount S.L., Raman R., Davies P., Masliah E., Williams D.S., and Goldstein L.S.. 2005. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 307:1282–1288. 10.1126/science.1105681 [DOI] [PubMed] [Google Scholar]
- Su J.H., Cummings B.J., and Cotman C.W.. 1993. Identification and distribution of axonal dystrophic neurites in Alzheimer’s disease. Brain Res. 625:228–237. 10.1016/0006-8993(93)91063-X [DOI] [PubMed] [Google Scholar]
- Sun F., Zhu C., Dixit R., and Cavalli V.. 2011. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J. 30:3416–3429. 10.1038/emboj.2011.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teich A.F., Patel M., and Arancio O.. 2013. A reliable way to detect endogenous murine β-amyloid. PLoS One. 8:e55647 10.1371/journal.pone.0055647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry R.D., Gonatas N.K., and Weiss M.. 1964. Ultrastructural studies in Alzheimer’s presenile dementia. Am. J. Pathol. 44:269–297. [PMC free article] [PubMed] [Google Scholar]
- Tran H.T., LaFerla F.M., Holtzman D.M., and Brody D.L.. 2011. Controlled cortical impact traumatic brain injury in 3xTg-AD mice causes acute intra-axonal amyloid-β accumulation and independently accelerates the development of τ abnormalities. J. Neurosci. 31:9513–9525. 10.1523/JNEUROSCI.0858-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J., Grutzendler J., Duff K., and Gan W.B.. 2004. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat. Neurosci. 7:1181–1183. 10.1038/nn1335 [DOI] [PubMed] [Google Scholar]
- Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., Teplow D.B., Ross S., Amarante P., Loeloff R., et al. 1999. β-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 286:735–741. 10.1126/science.286.5440.735 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., O’Toole E.T., Girard M., Ritter B., Messa M., Liu X., McPherson P.S., Ferguson S.M., and De Camilli P.. 2014. A dynamin 1-, dynamin 3- and clathrin-independent pathway of synaptic vesicle recycling mediated by bulk endocytosis. eLife. 3:e01621 10.7554/eLife.01621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Feng T., Tammineni P., Chang Q., Jeong Y.Y., Margolis D.J., Cai H., Kusnecov A., and Cai Q.. 2017. Regulation of synaptic amyloid-β generation through BACE1 retrograde transport in a mouse model of Alzheimer’s disease. J. Neurosci. 37:2639–2655. 10.1523/JNEUROSCI.2851-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.Y., Choi J.U., Cho M.H., Yang K.M., Ha H., Chung I.J., Cho G.S., and Kim D.H.. 2013. α-secretase cleaved amyloid precursor protein (APP) accumulates in cholinergic dystrophic neurites in normal, aged hippocampus. Neuropathol. Appl. Neurobiol. 39:800–816. 10.1111/nan.12032 [DOI] [PubMed] [Google Scholar]
- Yu W.H., Cuervo A.M., Kumar A., Peterhoff C.M., Schmidt S.D., Lee J.H., Mohan P.S., Mercken M., Farmery M.R., Tjernberg L.O., et al. 2005. Macroautophagy—A novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. J. Cell Biol. 171:87–98. 10.1083/jcb.200505082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Condello C., Keene C.D., Wang Y., Bird T.D., Paul S.M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron. 90:724–739. (published erratum appears in Neuron 2016. 92:252–264) 10.1016/j.neuron.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Fu Y., Yasvoina M., Shao P., Hitt B., O’Connor T., Logan S., Maus E., Citron M., Berry R., et al. 2007. β-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: Implications for Alzheimer’s disease pathogenesis. J. Neurosci. 27:3639–3649. 10.1523/JNEUROSCI.4396-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.