End-binding proteins regulate the dynamics and function of microtubule plus ends by recruiting a plethora of diverse factors. Yang et al. show that EB1 and EB3 also affect microtubule minus ends by participating in their attachment to Golgi membranes. This function is important for cell polarity and migration.
Abstract
End-binding proteins (EBs) are the core components of microtubule plus end tracking protein complexes, but it is currently unknown whether they are essential for mammalian microtubule organization. Here, by using CRISPR/Cas9-mediated knockout technology, we generated stable cell lines lacking EB2 and EB3 and the C-terminal partner-binding half of EB1. These cell lines show only mild defects in cell division and microtubule polymerization. However, the length of CAMSAP2-decorated stretches at noncentrosomal microtubule minus ends in these cells is reduced, microtubules are detached from Golgi membranes, and the Golgi complex is more compact. Coorganization of microtubules and Golgi membranes depends on the EB1/EB3–myomegalin complex, which acts as membrane–microtubule tether and counteracts tight clustering of individual Golgi stacks. Disruption of EB1 and EB3 also perturbs cell migration, polarity, and the distribution of focal adhesions. EB1 and EB3 thus affect multiple interphase processes and have a major impact on microtubule minus end organization.
Introduction
Microtubules (MTs) are dynamic polymers essential for cell division, organization of intracellular compartments, and cell polarity. Many MT-associated processes critically depend on the proteins that bind to the two MT ends, the plus and minus ends, which have different growth rates and cellular functions (Howard and Hyman, 2003; Akhmanova and Steinmetz, 2015). MT plus ends polymerize rapidly in vitro; in cells, they are the sites where MT elongation takes place. MT minus ends grow slowly in vitro and in cells are often anchored to MT organizing centers (MTOCs). Until recently, it was believed that in cells, MT minus ends do not grow. However, studies of calmodulin-regulated spectrin-associated (CAMSAP)/Patronin proteins showed that they can recognize and protect free MT minus ends by decorating stretches of MT lattice formed by minus end polymerization (Goodwin and Vale, 2010; Hendershott and Vale, 2014; Jiang et al., 2014), thus demonstrating that MT minus end growth is a physiologically important process.
Growing MT ends accumulate a rich collection of proteins that are termed MT plus end tracking proteins, or +TIPs (Schuyler and Pellman, 2001; Akhmanova and Steinmetz, 2008). The core components of +TIP complexes are the members of end-binding (EB) family, which recognize growing MT ends by sensing the nucleotide state of tubulin through their N-terminal calponin homology (CH) domains (Maurer et al., 2012). The C-terminal part of EBs consists of a dimeric parallel coiled coil, which ends with a four-helix bundle, and an acidic tail similar to the tail of α-tubulin (Akhmanova and Steinmetz, 2008). The C-terminal EB domain is responsible for binding to numerous partners, which fall into two major structural classes: cytoskeleton-associated protein glycine-rich (CAP-Gly) domain proteins and proteins containing SxIP (Ser-any amino acid-Ile-Pro) motifs (Kumar and Wittmann, 2012; Akhmanova and Steinmetz, 2015). EB partners can either promote or restrict MT growth and regulate MT interactions with different cell components (Kumar and Wittmann, 2012; Akhmanova and Steinmetz, 2015).
Mammalian cells typically coexpress three members of the EB family, EB1, EB2, and EB3, and although these proteins have been knocked down individually or in combinations (Straube and Merdes, 2007; Toyoshima and Nishida, 2007; Komarova et al., 2009; Nakamura et al., 2012; Ferreira et al., 2013; Yue et al., 2014), the impact of disruption of all three EBs on MT organization and dynamics has not been described. Here, we made use of the CRISPR/Cas9 technology to stably mutate all three mammalian EB-encoding genes. These mutations disrupted the CH domains of EB2 and EB3 and abrogated C-terminal partner-binding half of EB1. The EB mutant cell lines displayed only minor defects in cell division and MT plus end polymerization but had strongly perturbed organization of noncentrosomal MTs. In the studied cell lines, MT minus ends that are not attached to the centrosome are stabilized by CAMSAP2, and many of them are tethered to the Golgi apparatus. Disruption of EB1 and EB3 led to shortening of CAMSAP2-decorated MT minus end stretches, their detachment from the Golgi, and Golgi compaction. Furthermore, we found that the mutation of EB1 and EB3 affected cell migration on 2D substrates and invasion in 3D matrix. Our results thus show that EB proteins control different aspects of interphase mammalian cell architecture and have an unexpectedly large impact on the organization of MT minus ends.
Results
Human cell lines with disrupted EB1 and EB3 are viable
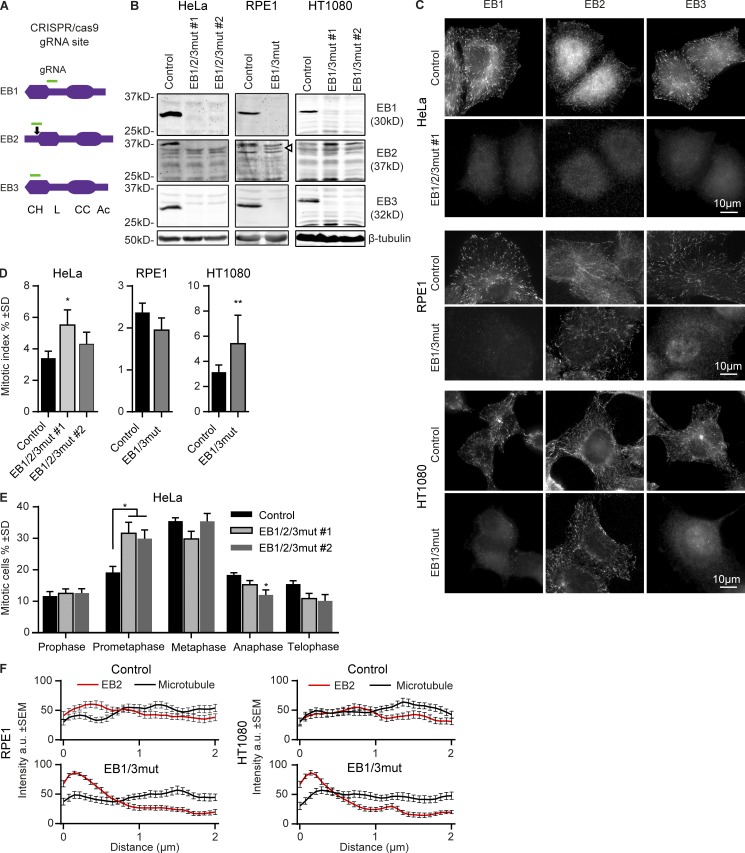
To generate triple EB mutant cells, we first tested the efficiency of individual gRNAs targeting EB-encoding genes in HeLa cells using staining with antibodies against the C-terminal halves of EBs (Stepanova et al., 2003; Komarova et al., 2005). Complete loss of EB2 and EB3 reactivity was found with gRNA constructs targeting the N-terminal extension preceding the CH domain of EB2 and the N-terminal part of the CH domain of EB3 (Fig. 1 A and Table S1). In contrast, the EB1-specific gRNA constructs targeting the CH domain were not effective, and we therefore used a construct with a target site after the CH domain (Fig. 1 A and Table S1). We then simultaneously transfected the three EB-targeting constructs into HeLa cells; among 51 clones, we obtained four stable lines that were negative for all three EBs (termed EB1/2/3mut) and analyzed further two of these lines (Fig. 1, B and C). Sequencing showed the presence of deletions or frameshift mutations close to the gRNA targeting sites in all EB-encoding genes (Table S1). The disruption of EB1-encoding gene occurred downstream of the CH domain, thus deleting the C-terminal half of the protein, which is responsible for dimerization, partner binding, and reactivity with the existing EB1-specific antibodies, while the residual CH-domain-containing EB1 fragment might still be expressed and exert some MT-dependent or independent functions. When examined as a GFP fusion, this fragment displayed weak MT binding (Fig. S1 A). Mutations that perturbed EB2 and EB3 sequences were located upstream or within the N-terminal part of the CH domain and can be expected to prevent MT localization (see Fig. S1 A for the illustration of a diffuse distribution of an N-terminal EB3 fragment with an incomplete CH domain). Because the antibodies we used react with the C-terminal EB regions, these clones likely completely lack EB2 and EB3 function.
Figure 1.
Characterization of cell lines with disrupted EB-encoding genes. (A) Schemes of EB1, EB2, and EB3 proteins and the position of gRNA sequences. Arrow indicates the in-frame Met at the beginning of the EB2 CH domain. (B and C) Western blot analysis and immunostaining of control and the EB mutant cells with the indicated antibodies. (D and E) Quantification of the mitotic index and mitotic stages in the indicated cell lines. (D) n ≥ 3 and (E) n = 4 experiments, with 3,000 cells each. (F) Mean intensity of EB2 and α-tubulin staining starting from the MT tip (0); 26–28 MT ends analyzed per condition from 6–10 cells. *, P < 0.05; **, P < 0.01 (Mann-Whitney U test).
We have also attempted to simultaneously perturb all three EB-encoding genes in retinal pigment epithelium (RPE1) and HT1080 fibrosarcoma cells but obtained no triple mutants. From 39 clones, we obtained a single RPE1 line lacking both EB1 and EB3 reactivity (termed EB1/3mut; Fig. 1 B). Although the expression of MT plus end–associated EB2 was maintained in this cell line (Fig. 1, B and C), both EB2 gene copies contained a single nucleotide insertion (Table S1), and we observed an EB2-specific band of a smaller size (Fig. 1 B). The presence of this band could be explained by translation initiation from the start codon at the N terminus of the CH domain (Fig. 1 A), as the N-terminal EB2-specific extension has no major impact on MT plus end tracking (Komarova et al., 2009). Furthermore, by simultaneously targeting only the EB1 and EB3-encoding genes, we obtained two HT1080 cell lines lacking both EB1 and EB3 reactivity (Fig. 1, B and C). In both RPE1 and HT1080 cells with mutated EB1 and EB3, EB2 was concentrated more strongly at the outmost MT tips (Fig. 1, C and F), in agreement with the data obtained using RNAi (Komarova et al., 2009). These data support the notion that EBs compete for MT plus ends, with EB2 having lower affinity than EB1 and EB3.
Mitotic index was mildly elevated in HeLa EB1/2/3mut and HT1080 EB1/3mut cells but not in EB1/3mut RPE1 cells (Fig. 1 D and Fig. S1 B). siRNA-mediated depletion of EB2 in EB1/3mut RPE1 cells revealed no additional effects on cell proliferation or division (Fig. S1, C–E). We thus have no explanation for our failure to obtain a triple EB mutant in this cell line.
Analysis of the HeLa EB1/2/3mut cells revealed no striking deviations in the morphology of mitotic spindles and showed that the elevation of the mitotic index was likely due to a prometaphase delay (Fig. 1 E and Fig. S2 A). These results are consistent with previous studies, which showed that knockdown or abnormal expression of individual EB proteins can cause spindle abnormalities but does not block mitotic progression (Green et al., 2005; Draviam et al., 2006; Toyoshima and Nishida, 2007; Ban et al., 2009; Xia et al., 2012; Ferreira et al., 2013; Iimori et al., 2016). In our cell lines, we observed some, but not all, of these abnormalities. For example, although we found no delay in cytokinesis, we did observe that the time interval between spreading of two daughter cells after mitotic exit was significantly increased (Fig. S2, B and C), as described previously for EB3 depletion (Ferreira et al., 2013). However, the EB1/2/3mut HeLa cells showed no reduction of Ska1 localization the mitotic spindle (Fig. S2, D and E), in contrast to the phenotype of acute EB1 depletion (Thomas et al., 2016). These differences are likely due to the fact that the isolation of stable cell lines can result in selection for compensatory mutations. Collectively, our results suggest that proliferating mammalian cells can adapt to the loss of function of all three full-length EBs.
EB1 and EB3 regulate +TIP accumulation at MT plus ends
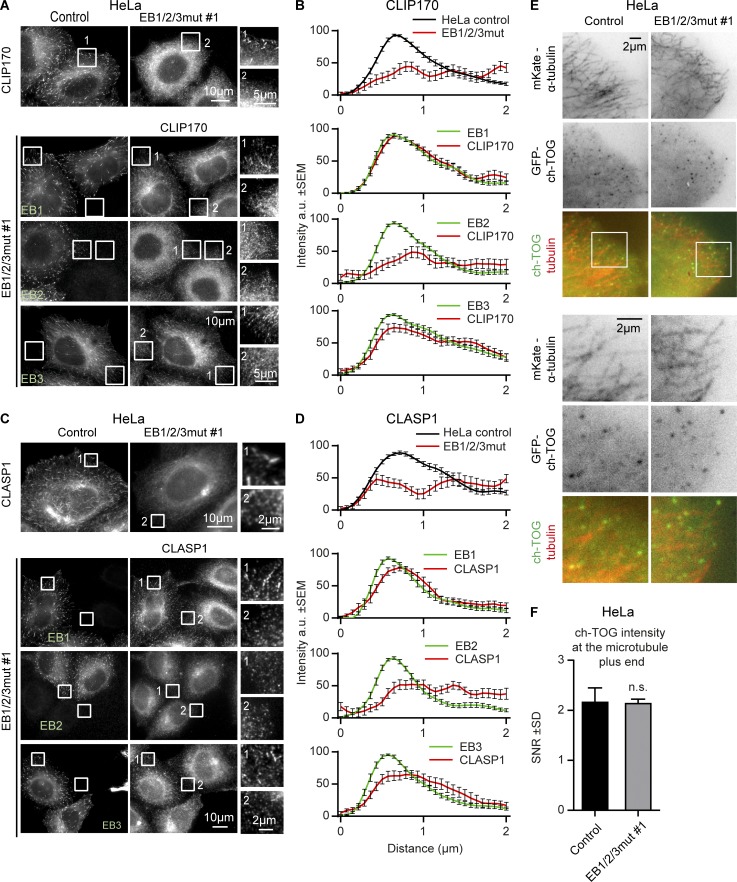
EB proteins are well known to recruit other +TIPs to MT plus ends, with EB1 and EB3 having higher affinity for some partner proteins compared with EB2 (Akhmanova and Steinmetz, 2008; Bjelić et al., 2012; Jiang et al., 2012; Liu et al., 2015). However, because SxIP and CAP-Gly binding sites are conserved in all three EBs, the contribution of EB2 to +TIP localization remained unclear. The generation of a HeLa EB1/2/3mut cells allowed us to address this question unambiguously. In contrast to control cells, EB1/2/3mut cells lacked enrichment of a CAP-Gly domain–containing protein CLIP-170 and an SxIP-containing protein CLASP1 at MT plus ends; this enrichment could be restored by expressing untagged EB1 or EB3, but not EB2, although all three EBs showed nice MT plus end localization (Fig. 2, A–D). EB2 is thus unable to promote localization of canonical EB-dependent +TIPs CLIP-170 and CLASP1 to MT plus ends.
Figure 2.
Effect of EB disruption on +TIP localization. (A and C) Immunostaining for CLIP170 and CLASP1 in control and EB1/2/3mut HeLa cells expressing the indicated constructs. Enlargements of the boxed areas indicated by numbers are shown on the right. In C, cells were incubated with the GSK3 inhibitor SB415286 (20 µM) for 30 min before fixation. (B and D) Averaged intensities of staining for the indicated +TIP obtained for 20–24 MT ends per condition. MT end position (0) was determined by staining for β-tubulin. (E and F) Live images of the indicated cell lines transfected with ch-TOG-GFP together with mKate-α-tubulin (larger and smaller fields of view shown; E) and quantification of ch-TOG signal-to-noise ratio at MT tips (F). n = 30 cells in three independent experiments. Mann-Whitney U test.
We also investigated the localization of the MT polymerase ch-TOG, which can recognize MT plus ends autonomously but can also be recruited to them by the SxIP protein SLAIN2 (Brouhard et al., 2008; van der Vaart et al., 2011). ch-TOG localizes as dots at the outmost MT plus ends (Nakamura et al., 2012; Maurer et al., 2014), and this localization is easier to observe when the ch-TOG–GFP fusion is expressed in cells depleted of endogenous ch-TOG (Gutiérrez-Caballero et al., 2015). Using this approach, we found that ch-TOG-GFP can be clearly detected at the plus ends of both control and EB1/2/3mut HeLa cells (Fig. 2, E and F), in agreement with the previous results on EB1 and EB3 depletion (Gutiérrez-Caballero et al., 2015). These data confirm that ch-TOG binding to MT plus ends does not require EB-dependent +TIPs, although the association with SLAIN2 might offer ch-TOG a competitive advantage when MT ends are crowded by EBs and their partners.
EB1 and EB3 support rapid processive MT growth
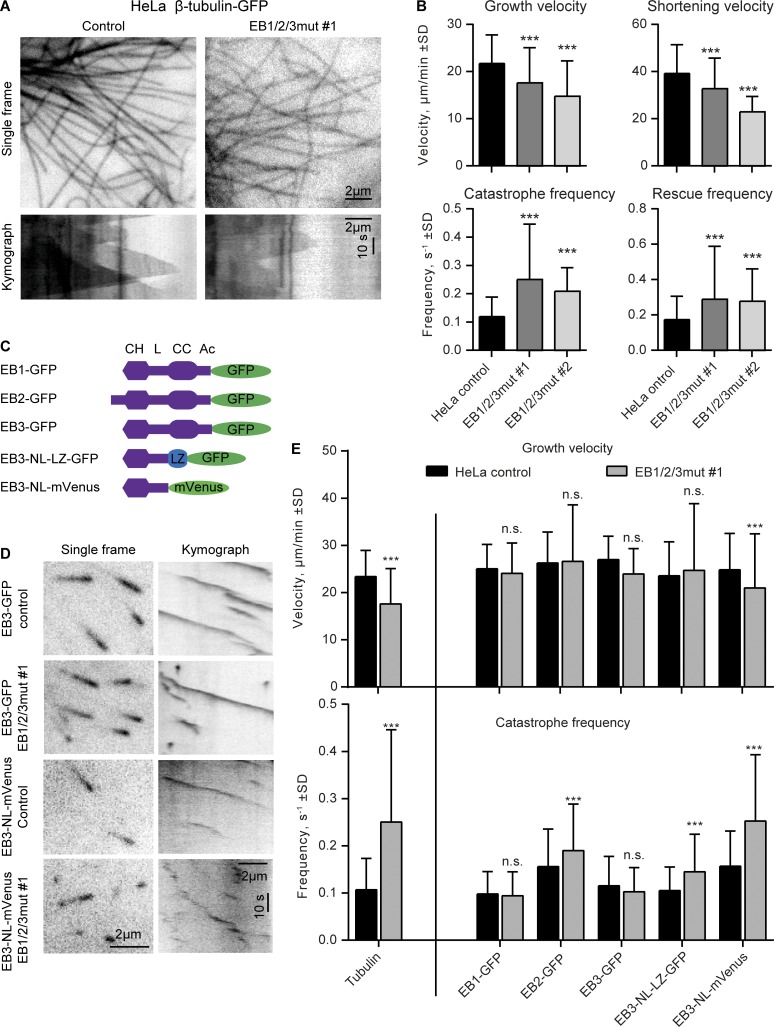
Previous work in CHO cells showed that simultaneous depletion of EB1 and EB3 had no effect on MT growth rate but increased catastrophe frequency, while in vitro, EBs increase MT growth rate, promote catastrophes, and can strongly synergize with the ch-TOG homologue XMAP215 to increase MT polymerization rate (Komarova et al., 2009; Zanic et al., 2013). Analysis of MT dynamics using β-tubulin–GFP as a marker in HeLa cells partially confirmed our previous observations by showing that the mutation of all three EBs led to increased catastrophe frequency (Fig. 3, A and B). Furthermore, we did observe an ∼25% reduction in MT growth rate (Fig. 3, A and B). Increased MT catastrophe frequency was accompanied by a higher rescue frequency (Fig. 3, A and B), similar to what we previously observed in cells depleted of SLAIN2 or ch-TOG, in which both growth and shortening episodes became more interrupted (van der Vaart et al., 2011). MT depolymerization rate was reduced (Fig. 3, A and B), possibly because the most rapid depolymerization rates are observed during long shrinkage events, and such events were less frequent at higher rescue frequency.
Figure 3.
Effect of EB disruption on MT dynamics. (A) Single frames and kymographs of MT dynamics in the indicated cell lines transiently transfected with β-tubulin–GFP. (B) Parameters of MT dynamics in the indicated cell lines. 74–240 MT plus ends were analyzed per condition. (C and D) Scheme of the used EB constructs (C) and single frames and kymographs illustrating MT plus end dynamics visualized with the indicated constructs (D). (E) Plots of MT growth velocity and catastrophe frequency. 158–264 MT plus ends were analyzed per condition. ***, P < 0.001 (Mann-Whitney U test).
To test which EBs and which domains of EBs are important for regulating MT growth, we performed rescue experiments with EB–GFP fusions and used comets formed by these proteins as readout (Fig. 3, C and D). We observed a good match between the data obtained with fluorescent tubulin and EB1-GFP or EB3-GFP in control cells (Fig. 3 E). MT polymerization rate could be restored by all three EBs, including EB2, as well as by a dimeric EB3 construct, EB3-NL-LZ, in which the whole C-terminal partner-binding domain was substituted with a dimeric coiled coil (leucine zipper) of GCN4 (Fig. 3, C–E). These data suggest that the N-terminal CH domain of EB3 with the adjacent linker region (termed NL), which is sufficient for efficient MT binding (Komarova et al., 2009; Zimniak et al., 2009), can regulate MT growth rate in cells. The protein does need to be in a dimeric configuration, as the monomeric version of the same protein could not fully rescue growth velocity (Fig. 3, C–E), possibly because CH domain dimerization increases protein affinity for MT tips or affects their conformation. It should be noted that because our EB1/2/3mut cells are possibly already expressing monomeric CH domain of EB1, the expression of EB3-NL monomer might have little additional impact.
Strikingly, although EB2 could restore MT growth rate, it could not rescue growth processivity (Fig. 3, C–E), in agreement with our previous data (Komarova et al., 2009). EB3-NL-LZ also failed to suppress catastrophes, and even more interrupted MT growth was observed upon expression of EB3-NL monomer (Fig. 3, C–E). Together, these data suggest that the acceleration of MT growth might mostly depend on the efficient EB binding to the MT tip, whereas catastrophe suppression requires recruitment of EB partners.
EB1 and EB3 are required for coorganization of MTs and the Golgi apparatus
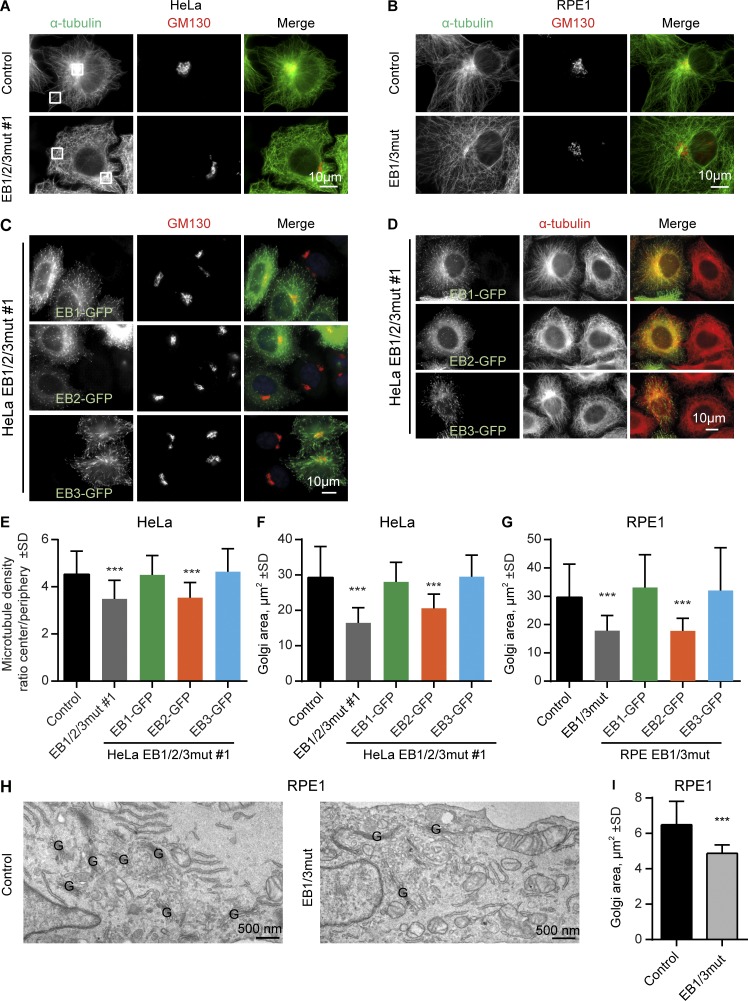
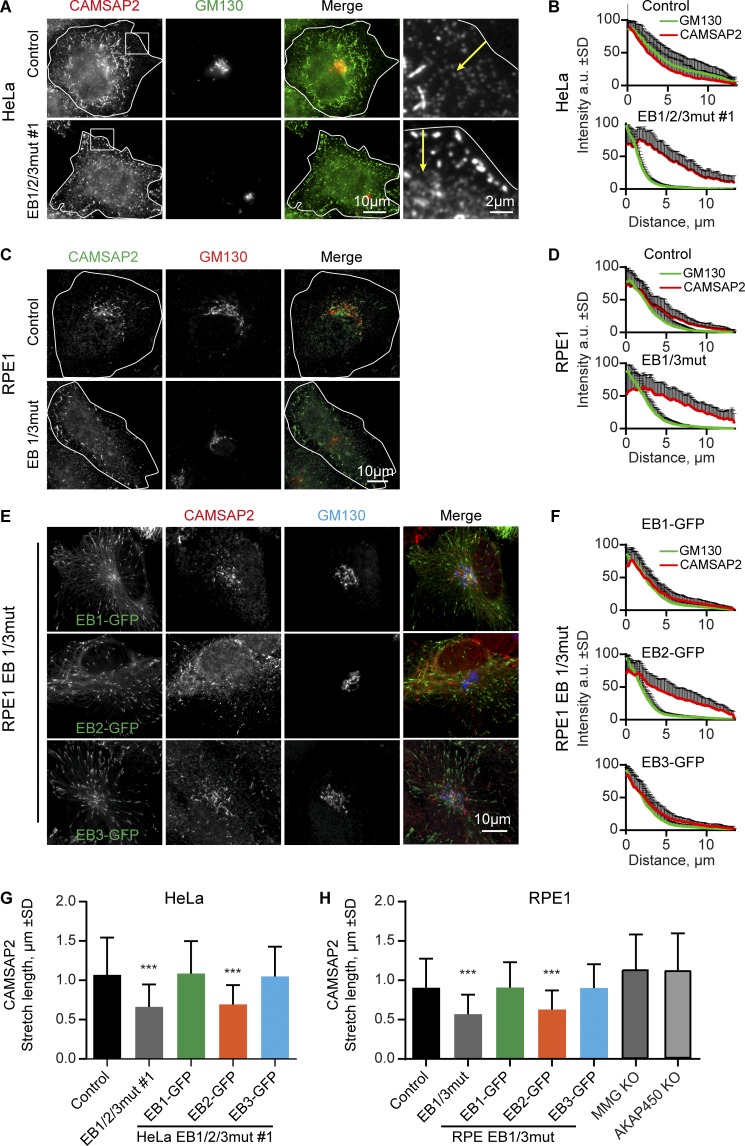
Next, we examined the organization of MT network in EB1/2/3mut cells. Importantly, in HeLa cells the centrosome does not play a dominant role in organizing interphase MTs, except for the G2 phase (Lansbergen et al., 2006; Jeffery et al., 2013), but a high MT density is observed in the Golgi area (Mimori-Kiyosue et al., 2005; Sato et al., 2014). We found that the major impact of the triple EB mutation was on the arrangement of MT minus ends, as the Golgi-associated accumulation of noncentrosomal MTs was lost and the MTs became randomly distributed in the cytoplasm (Fig. 4 A). Loss of MT density at the Golgi could also be clearly observed in RPE1 and HT1080 EB1/3mut cells (Fig. 4 B and Fig. S3, A and B). However, in these cell lines MT disorganization was not as profound, because unlike HeLa cells, they have many centrosomally anchored MTs that were retained in EB1/3mut cells. The loss of Golgi-associated MTs had a strong impact on the area occupied by the Golgi apparatus, which was significantly diminished in both EB1/2/3mut HeLa cells and EB1/3mut RPE1 cells. Both the disorganization of MTs and overcompaction of the Golgi could be fully reversed by expressing EB1-GFP and EB3-GFP but not by EB2-GFP (Fig. 4, C–G; rescue with EB2-GFP was included also in EB1/3mut cells because EB2 gene is mutated in this clone [Fig. 1 B]). To characterize Golgi overcompaction in more detail, we used EM and found that the morphology and size of the individual Golgi stacks were very similar in control and EB1/3mut RPE1 cells, but the area occupied by the Golgi stacks was reduced in the mutant (Fig. 4, H and I; and Fig. S3 C).
Figure 4.
Effect of EB disruption on MT and Golgi organization. (A–D) Immunostaining for α-tubulin and GM130 in the indicated cell lines; in C and D, cells were transfected with the indicated EB–GFP fusions. Boxes in A show examples of areas used for quantification in E. (E–G) Quantification of the ratio of MT intensity in the cell center and cell periphery (boxes in A; E) or the Golgi area in the indicated cell lines, transfected with the indicated constructs. n = 24–41 cells per condition (E), 31–66 cells (F), and 28–47 cells (G). (H and I) EM images of the Golgi area (Golgi stacks indicated with a G; H) and quantification of the total area occupied by in Golgi stacks (I). n = 3 experiments with 50 cells each. ***, P < 0.001 (Mann-Whitney U test).
Our recent study showed that the attachment of noncentrosomal MTs to the Golgi depends on the interaction of the minus end decorating protein CAMSAP2 with the complex of myomegalin (MMG) and AKAP450 (Wu et al., 2016), a large scaffolding protein recruited to the Golgi membranes by the Golgi matrix protein GM130 (Hurtado et al., 2011). We examined the distribution of CAMSAP2 in EB1/2/3mut cells and found that it was affected in two ways: the stretches of CAMSAP2-decorated MT lattice became significantly shorter and were redistributed from the central part of the cell to the cell periphery (Fig. 5, A, B, and G). A similar phenotype was also observed in RPE1 and HT1080 EB1/3mut cells (Fig. 5, C, D, and H; and Fig. S3 D). Shortening of CAMSAP2-decorated stretches in EB1/3mut cells was not due to the reduction of CAMSAP2 expression (Fig. S3 E) or their detachment from Golgi, because in cells lacking AKAP450 or MMG, two proteins required for MT minus end tethering to Golgi membranes (Wu et al., 2016), CAMSAP2-bound MT segments were even slightly longer than in control (Fig. 5 H). Importantly, both the length and the localization of CAMSAP2 stretches could be fully rescued by expressing EB1-GFP or EB3-GFP, but not by EB2-GFP in both HeLa 1/2/3mut cells and RPE1 EB1/3mut cells (Fig. 5, E–H; and Fig. S3, F and G). EB1 and EB3, but not EB2, thus control the length of CAMSAP2-decorated MT minus ends and their attachment to the Golgi.
Figure 5.
Distribution of CAMSAP2-decorated MT minus ends in cells with mutated EBs. (A and C) Immunostaining for CAMSAP2 and GM130 in the indicated cells. Enlargements on the right in A show cell periphery; yellow arrows indicate the 3-µm-broad region used for quantification in Fig. S3 G. (B, D, and F) Averaged intensity distributions for GM130 and CAMSAP2 in the indicated cell lines, either untransfected or transfected with the indicated constructs. Distance from the center of the Golgi is shown on the horizontal axis and normalized fluorescence intensity on the vertical axis. n = 23–43 cells per condition. (E) Immunostaining for CAMSAP2 and GM130 in EB1/3mut RPE1 cells expressing the indicated constructs. (G and H). Quantification of CAMSAP2 stretch length. 1000 stretches in 10 cells were measured per condition, except for MMG and AKAP450 knockout cells (H), for which 200 stretches in 8 and 10 cells were measured. ***, P < 0.001 (Mann-Whitney U test).
Coorganization of the Golgi and noncentrosomal MTs depends on the EB–MMG complex
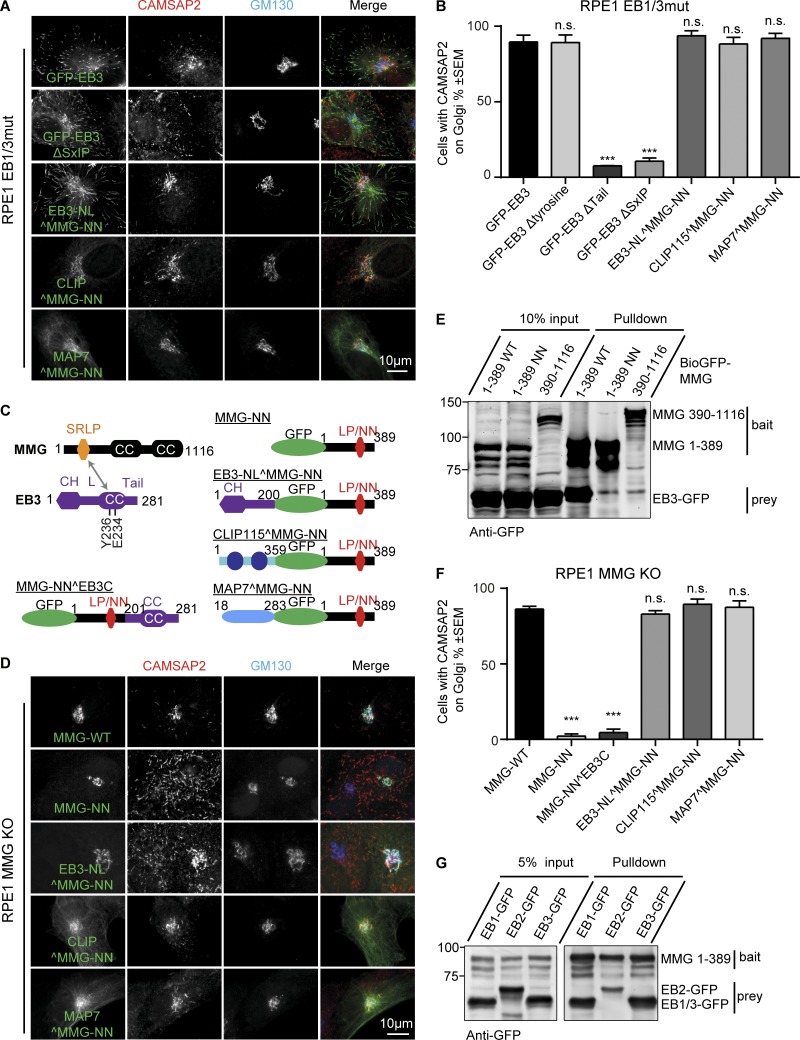
To test which part of the EBs is required for the attachment of noncentrosomal MTs to the Golgi, we used different EB3 mutants. We found that the localization of CAMSAP2 stretches to the Golgi could be rescued by an EB3 mutant lacking the C-terminal tyrosine, which is essential for EB binding to CAP-Gly domains (Honnappa et al., 2006), but not by EB3 mutants, which were unable to bind to SxIP partners either because of the substitutions in the SxIP-binding pocket (the double Y226A, E234A mutant, termed ΔSxIP) or the deletion of the whole acidic tail region (ΔTail; Montenegro Gouveia et al., 2010; Fig. 6, A and B; and Fig. S4 A). Consistently, GFP-tagged EB1 and EB3, but not EB2, were recruited to Golgi membranes in nocodazole-treated RPE1 EB1/3mut cells in a manner dependent on an intact SxIP- but not the CAP-Gly–binding site (Fig. S4 B).
Figure 6.
EB interaction with MMG is required to recruit CAMSAP2 stretches to the Golgi. (A and D) Immunostaining for CAMSAP2 and GM130 of EB1/3mut (A) or MMG knockout (D) RPE1 cells expressing the indicated constructs. (B and F) Percentage of cells with CAMSAP2 stretches in the Golgi area in EB1/3mut (B) or MMG knockout (F) RPE1 cells expressing the indicated constructs. n = 77–112 transfected cells in B and 41–81 transfected cells in F. **, P < 0.01; ***, P < 0.001 (Student’s t test). (C) A scheme of MMG–EB3 interaction and the fusion constructs used. (E and G) Streptavidin pull-down assays with the extracts of HEK293T cells coexpressing the indicated biotinylation tag (Bio)-GFP–MMG fusions, the indicated EB–GFP fusions, and biotin ligase BirA, analyzed by Western blotting with anti-GFP antibodies.
In vitro binding assays with purified proteins showed that CAMSAP2 does not interact with EBs (Fig. S4 C), excluding direct binding between CAMSAP2 and EBs as the mechanism of MT minus end tethering to the Golgi. We therefore focused our attention on MMG, which is required for MT minus end attachment to the Golgi (Wu et al., 2016), directly interacts with EB1, and recruits it to the Golgi through an SxIP motif SRLP (Roubin et al., 2013; Wang et al., 2014; Fig. 6 C). Knockout of MMG caused detachment of CAMSAP2 stretches from the Golgi, and this defect could be rescued by expressing the N-terminal SRLP-containing portion of MMG (Wu et al., 2016; Fig. 6 D). We disrupted the SRLP site in MMG N terminus by mutating leucine and proline residues to asparagines (LP/NN mutant) and found that these mutations blocked the binding of MMG to EB3 (Fig. 6 E), in agreement with the previously published data on EB1 (Wang et al., 2014). In contrast, the interactions between MMG and AKAP450, which recruits MMG to the Golgi, and the binding of the AKAP450–MMG complex to CAMSAP2 could still occur (Fig. S4, D and E). However, the LP/NN mutant of the MMG N terminus (MMG-1-389-NN) completely failed to rescue localization of CAMSAP2 stretches at the Golgi in MMG knockout cells (Fig. 6, D and F).
We hypothesized that by binding to MMG, EBs create an additional MT-binding site through their CH domain. To prove this idea, we fused either the N- or C-terminal part of EB3 to the MMG N terminus with the mutated SRLP motif (Fig. 6 C). Both fusions localized to the Golgi, and in addition, the N-terminal EB fusion also accumulated at MT plus ends (Fig. 6, A and D; and Fig. S4 A). The Golgi localization of this construct was more obvious in MMG knockout cells, while MT tips were better visible in EB1/3mut cells (Fig. 6, A and D), as could be expected because of the competition with endogenous MMG or EB proteins, respectively. Remarkably, the fusion bearing the N-terminal part of EB3, but not the EB3 C terminus, restored the localization of CAMSAP2 stretches to the Golgi both in MMG knockout and EB1/EB3mut cells (Fig. 6, A, B, D, and F; and Fig. S4 A). Furthermore, a complete rescue of CAMSAP2 stretch localization was also observed by fusing the LP/NN mutant of the MMG N terminus to MT-binding domains of CLIP-115 (a double CAP-Gly motif; Hoogenraad et al., 2000) or the MT-decorating protein MAP7/ensconsin (Faire et al., 1999). Because the latter protein is not a +TIP, these data indicate that the binding of CAMSAP2 stretches to the Golgi requires additional affinity of Golgi membranes for MT shafts but not to their growing ends. This notion was supported by the experiment in which we inhibited MT growth by applying the MT-stabilizing drug paclitaxel and found that the colocalization of CAMSAP2 stretches at the Golgi was preserved (Fig. S5 A). Finally, because EB2 could not be recruited to the Golgi in nocodazole treated cells (Fig. S4 B) and could not restore MT minus end tethering to the Golgi and Golgi morphology in EB knockout cells (Fig. 4, C–G; Fig. 5 E; and Fig. S3, F and G), we hypothesized that EB2 has a lower affinity for MMG compared with EB1 and EB3 and found that this was indeed the case (Fig. 6 G). Collectively, our results indicate that EB1 and EB3 bind to Golgi membranes because their C-terminal domains strongly interact with MMG. At the Golgi, EB1 and EB3 contribute to the tethering of CAMSAP2-decorated MTs, a function that does not strictly depend on the recognition of growing MT ends.
EB-dependent Golgi–MT attachment controls Golgi morphology
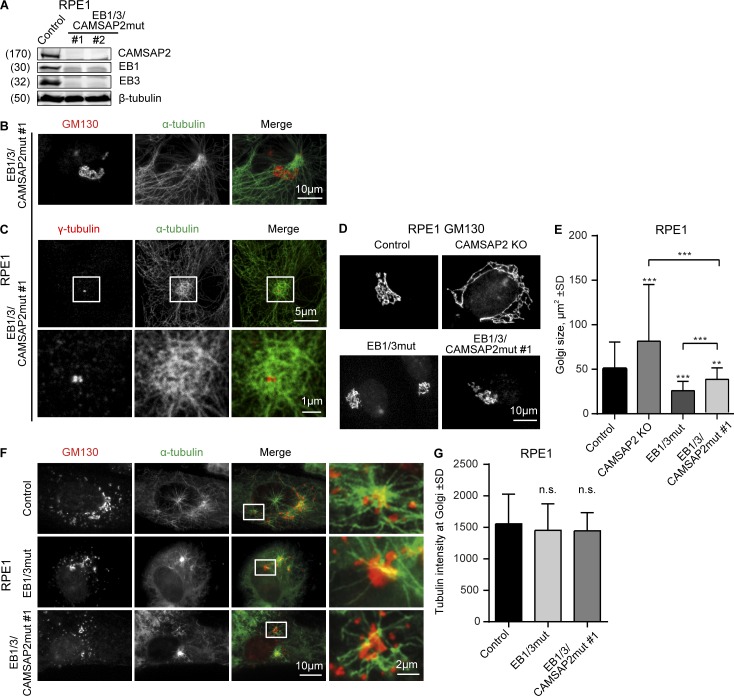
The data described above indicate that MT binding of EBs contributes to Golgi–MT tethering. This binding might counteract dynein-mediated Golgi compaction and thus explain why the disruption of EB1 and EB3 leads to reduction of the area occupied by Golgi stacks. In contrast, the Golgi was enlarged in CAMSAP2 knockout cells (Wu et al., 2016), possibly because Golgi-attached free MT minus ends facilitate Golgi self-assembly (Vinogradova et al., 2012). To further explore how EBs and CAMSAP2 regulate Golgi compaction, we generated a triple-mutant line by targeting CAMSAP2 in EB1/3mut RPE1 cells (termed EB1/3/CAMSAP2mut). Also these cells could be stably propagated (Fig. 7 A). They had a centrosomally organized MT system and no MT density in the Golgi area (Fig. 7 B). Interestingly, the centrosome in these cells was surrounded by short compacted MTs (Fig. 7 C), the origin of which is unclear. The Golgi area in EB1/3/CAMSAP2mut cells was smaller than in CAMSAP2 knockout but larger than in EB1/3mut cells (Fig. 7, D and E). The changes in the Golgi area were due to the loss of MT tethering but not because of the inability of Golgi membranes to nucleate MTs, as this function was fully preserved in EB1/3mut and EB1/3/CAMSAP2mut cells (Fig. 7, F and G).
Figure 7.
Effect of EB1, EB3, and CAMSAP2 disruption on Golgi organization and MT nucleation from Golgi. (A) Western blots of the extracts of EB1/3/CAMSAP2mut RPE1 cells. (B and C) Immunostaining for α-tubulin and GM130 (B) or γ-tubulin (C) in EB1/3/CAMSAP2mut RPE1 cells. In C, enlarged images of the boxed areas are shown below. (D and E) Immunostaining for GM130 and quantification of the Golgi area in the indicated cell lines. n = 50 cells per condition. (F) Immunostaining of GM130 and α-tubulin in the indicated cell lines after a 3-min recovery from nocodazole treatment. (G) Tubulin intensity in the vicinity of Golgi membranes in the indicated cell lines treated as in F. n = 30 cells per condition. n.s., no significant differences; **, P < 0.01; ***, P < 0.001 (Mann-Whitney U test).
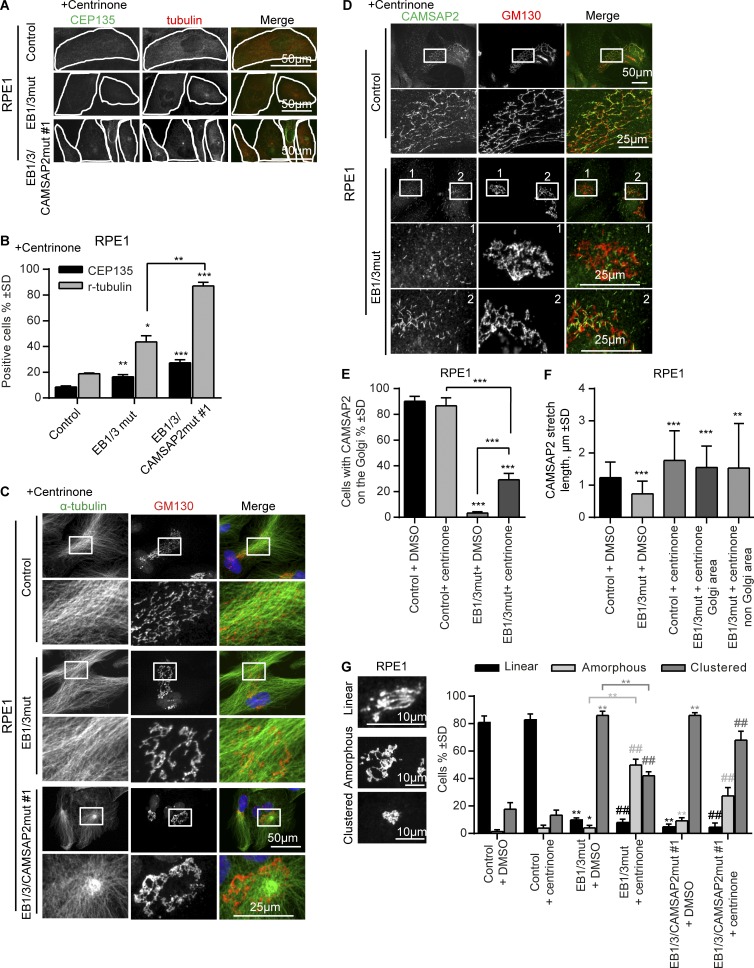
Previously, we observed that MT-dependent extension of Golgi ribbons became more obvious in cells treated with centrinone (Wu et al., 2016), a Plk4 inhibitor that prevents duplication of centrioles and causes centrosome loss (Wong et al., 2015). In RPE1 cells, this drug inhibits cell cycle progression, leading to cell enlargement and Golgi expansion. Centrinone treatment caused centriole loss in >90% of control cells and in 80% of EB1/3mut cells (Fig. 8, A and B). Our previous work showed that centriole depletion led to formation of acentrosomal MT arrays, the organization of which depended on the presence of MT minus end regulators (Wu et al., 2016). In control and CAMSAP2 knockout cells, the Golgi apparatus became the dominant MTOC, likely because of AKAP450-dependent recruitment of γ-tubulin released from the centrosome (Wu et al., 2016). In cells lacking AKAP450 or MMG, loss of centrosomes led to the formation of noncentrosomal MT arrays with the minus ends stabilized by CAMSAP2, while the Golgi was strongly compacted and completely detached from CAMSAP2 stretches and MT density (Wu et al., 2016).
Figure 8.
Effect of EB1, EB3, and CAMSAP2 disruption on MT and Golgi organization after centriole depletion. (A and B) Immunostaining for CEP135 and γ-tubulin (A) and percentage of γ-tubulin– and CEP135-positive cells in the indicated cell lines after 11 d of treatment with 125 nM centrinone (B). n = 366–391 cells per condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). (C and D) Immunostaining for α-tubulin (C) or CAMSAP2 (D) and GM130 in the indicated cell lines treated with centrinone. Enlarged portions of the boxed areas are shown at the bottom. (E and F) Percentage of cells with CAMSAP2 stretches at the Golgi (E) and quantification of the CAMSAP2 stretch length (F) for the indicated cell lines and treatments. n = 248–418 cells per condition in E and 199–260 CAMSAPP2 stretches in F. **, P < 0.01; ***, P < 0.001 (Mann-Whitney U test). (G) Quantification of Golgi morphology for the indicated PRE1 cell lines and treatments. Three types of Golgi organization are shown on the left. 335–642 control and 153–593 centrinone-treated cells were analyzed. Significant differences between values are indicated: *, control (DMSO) treatment; #, centrinone treatment. * or #, P < 0.05; ** or ##, P < 0.01 (Mann-Whitney U test).
In EB1/3mut cells, a γ-tubulin–positive MTOC was still present in 44% of cells, indicating that a significant part of the cell population had an acentriolar MTOC (Fig. 8, A and B). In the remaining 56% of the cells, a γ-tubulin–positive MTOC was lost, and MTs acquired a semiparallel organization (Fig. 8 C), very similar to what we observed AKAP450 and MMG knockouts (Wu et al., 2016). However, in contrast to AKAP450 and MMG knockouts, Golgi membranes partly overlapped with the area of high MT density (Fig. 8 C). Furthermore, although in centrinone-untreated EB1/3mut cells we observed strong detachment of CAMSAP2 stretches from the Golgi (Fig. 5, A–D), this defect was partially restored after centrinone treatment (Fig. 8, D and E). We noticed that CAMSAP2 stretches became significantly longer after centrinone treatment, and their length became much less sensitive to the loss of EB1 and EB3 (Fig. 8 F). Such longer CAMSAP2 stretches likely have a higher affinity for the AKAP450/MMG-bound Golgi membranes and may be less dependent on additional interactions provided by EBs.
In the triple EB1/3/CAMSAP2mut cells, the efficiency of centriole depletion was 73%, but most of them still retained a γ-tubulin–positive MTOC (Fig. 8, A and B). Previously, we observed a similar phenomenon in cells simultaneously lacking CAMSAP2 and AKAP450: the majority of surviving centrinone-treated AKAP450/CAMSAP2 knockout cells maintained an acentriolar γ-tubulin–containing MTOC (Wu et al., 2016). The fact that cells with acentriolar MTOCs were also predominant in the centrinone-treated EB1/3/CAMSAP2mut cell population is consistent with the view that, similar to AKAP450, EB1 and EB3 are part of an important link between MTs and the Golgi, and when this link is removed along with CAMSAP2, cell survival depends on the clustering of pericentriolar material (PCM) even though centrioles are absent.
Because centrinone-treated EB1/3/CAMSAP2mut cells retained a γ-tubulin–positive cluster, they had a radial MT array, and the Golgi was organized around the MTOC (Fig. 8 C). In control cells, Golgi stacks were often arranged into linear structures, which were particularly prominent after centrinone treatment (Fig. 8, D and G). In contrast, in all cells with disrupted EB1 and EB3, Golgi elements had a much more amorphous and clustered appearance (Fig. 8 G). Collectively, our data show EB1 and EB3 participate in MT minus end organization and in controlling the extension of Golgi ribbons along MTs, while in the absence of these two EBs, Golgi stacks acquire a more compact arrangement.
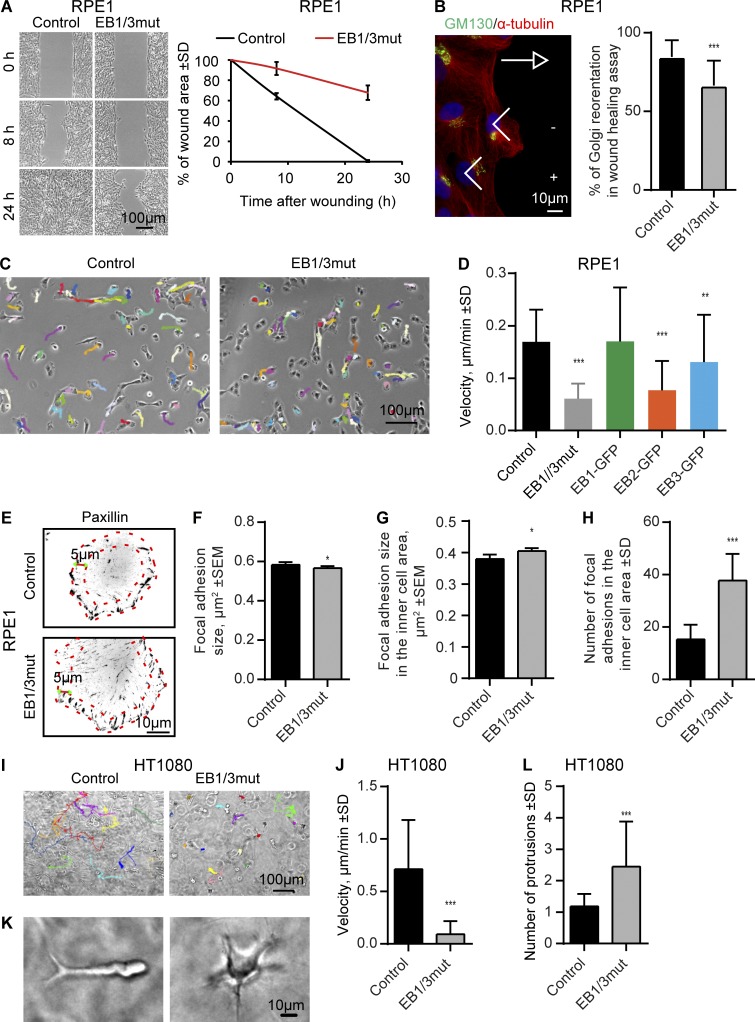
EB1 and EB3 are required for cell migration in 2D and 3D
To get further insight into the function of EBs in interphase cells, we next analyzed cell movement. Migration in EB1/2/3mut cells could not be tested, because of the poor motility of the parental HeLa strain. EB1/3mut RPE1 knockout cells moved significantly slower than control cells in monolayer wound healing assays (Fig. 9 A). The ability of cells to polarize by positioning the Golgi in the direction of migration was perturbed in the mutants (Fig. 9 B), similar to what was previously observed for the knockouts of AKAP450 and CAMSAP2, which are also required for Golgi–MT attachment (Rivero et al., 2009; Wu et al., 2016). Also, when RPE1 cell motility was observed in sparse cultures, the velocity of cell movement was significantly reduced, a defect that could be rescued by expressing EB1-GFP and to some extent by EB3-GFP, but not by EB2-GFP (Fig. 9, C and D). However, the ability of cells to move in one direction and the polarized organization of the actin cytoskeleton (i.e., formation of actin-rich lamellae at the leading edge and localization of myosin IIB at the retracting edge) were not significantly affected (Fig. S5, B and C). We reasoned that slower motility could be due to a defect in cell adhesion. Indeed, although the mean size of focal adhesions (FAs) was not affected, EB1/3mut RPE1 cells had more FAs in inner cell regions, and their size was slightly larger (Fig. 9, E–H). We next examined MT-dependent trafficking, focusing on the motility of Rab6-positive exocytotic carriers because they were previously implicated FA remodeling (Stehbens et al., 2014), and found that although the velocity of vesicle motility in EB1/3mut cells was normal, the radial organization of vesicle tracks was strongly perturbed (Fig. S5, D–F). The strong reduction of motility on 2D substrate observed in EB1/3mut cells is thus likely caused by aberrant FA turnover, possibly due to their abnormal regulation by EB-dependent +TIPs or trafficking defects (Stehbens and Wittmann, 2012).
Figure 9.
Effects of EB1 and EB3 disruption on cell migration. (A) Phase-contrast images of monolayer wound-healing assays in control and EB1/3mut RPE1 cells at the indicated time points and percentage of wound area closure. n = 3 independent experiments. (B) Immunostaining for α-tubulin and GM130 in a wound healing assay and the quantification of the Golgi reorientation. n = 163 control and 152 EB1/3mut RPE1 cells. (C). Phase contrast images and tracks of control and EB1/3mut cells during 7-h migration in sparse culture (C). (D) Quantification of cell migration velocity in control and EB1/3mut RPE1 cells expressing the indicated constructs. n = 30–60 cells. (E–H) Immunostaining for paxillin in the indicated cell lines (E), quantification of FA size in the whole cell (F), and FA size (G) and number (H) in the inner cell area (with 5-µm-broad cell rim excluded, red dotted lines in E). n = 44 control and 55 EB1/3mut RPE1 cells; per condition, 3,000 and 1,000 FAs were analyzed in the whole cell and the inner cell area, respectively. (I and J) Phase contrast images and tracks of control and EB1/3mut HT1080 cells in 3D matrix during 24-h migration (I) and quantification of their migration velocity (J). n = 24 control and 26 EB1/3mut HT1080 cells. (K and L) Morphology of control and EB1/3mut HT1080 cells in 3D matrix and the mean number of protrusions in these cells. n = 65 control and 63 EB1/3mut HT1080 cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Mann-Whitney U test).
We also examined the ability of EB1/3mut cells to invade 3D collagen matrix using HT1080 cells as a model and found that the velocity of their movement was very strongly reduced (Fig. 9, I and J). This defect was likely due at least partly to the fact that these cells were unable to acquire a polarized morphology with a single long invasive pseudopod at the front and instead extended multiple short protrusions (Fig. 9, K and L). This phenotype was reminiscent of that previously observed for AKAP450, MMG, and CAMSAP2 depletion (Wu et al., 2016), suggesting that the loss of Golgi–MT attachment might affect cell polarity and thus motility in 3D matrix.
Discussion
In this study we showed that some mammalian cell lines, such as HeLa, can survive when all the members of the EB protein family are stably disrupted. This result is not entirely surprising given that loss of EB homologues in other eukaryotes, such as budding yeast, is not lethal (Berlin et al., 1990), and Arabidopsis thaliana plants with strongly reduced expression of all three EB homologues have surprisingly mild phenotypes (Bisgrove et al., 2008). However, we were unable to disrupt all three EB-encoding genes in RPE1 and HT1080 background, even though the depletion of EB2 in RPE1 on EB1/3mut background revealed no striking additional defects. Recent work showed that acute simultaneous knockout of EB1 and EB3 in HeLa cells causes very significant spindle abnormalities (McKinley and Cheeseman, 2017). We favor the idea that mammalian cells normally require EBs for different aspects of cell division but can adapt to their absence through compensatory mutations, the selection for which can occur with different efficiency dependent on cell background.
The most striking and unexpected finding of our study is the important role of the EB1 and EB3 in controlling the organization of MT minus ends. This function comprises two effects (Fig. 10). First, EB1 and EB3 promote elongation of CAMSAP2-decorated MT stretches, and this may depend on the binding of EBs to growing MT plus or minus ends, as the elongation of CAMSAP2 stretches requires minus end polymerization, while their disassembly can occur from both ends (Jiang et al., 2014). Second, EBs can directly promote the interaction between MTs and Golgi membranes by binding to the Golgi-associated SxIP protein MMG. In MMG knockout cells, the length of CAMSAP2 stretches is normal, but they are not recruited to the Golgi. CAMSAP2 localization to the Golgi can be restored by directly targeting the MT-binding CH domain of EB3 or other MT-binding domains to Golgi membranes. MT minus end attachment to Golgi membranes does not require the high-affinity binding of EBs to growing, GTP cap-bearing MT tips. Instead, this interaction appears to rely on the weaker EB binding to the GDP-MT lattice, and can be also taken over by a MT-associated protein lacking MT tip preference. Such “zippering” between MT shafts and membranes associated with an EB partner might be similar to MT–cortex interactions in the axon initial segment, which require EBs and the SxIP motif-containing protein ankyrin-G (Fréal et al., 2016). The EB-dependent Golgi–MT connection becomes less important when CAMSAP2 stretches are elongated in centrinone-treated cells. These data support the view that Golgi–MT coorganization is based on the presence of multiple weak interactions and that an increase in CAMSAP2 decoration of MTs can compensate for the absence of EBs. Weak multisite binding also helps explain why other proteins, such as CLASPs and MTCL1, contribute to Golgi–MT association (Efimov et al., 2007; Sato et al., 2014).
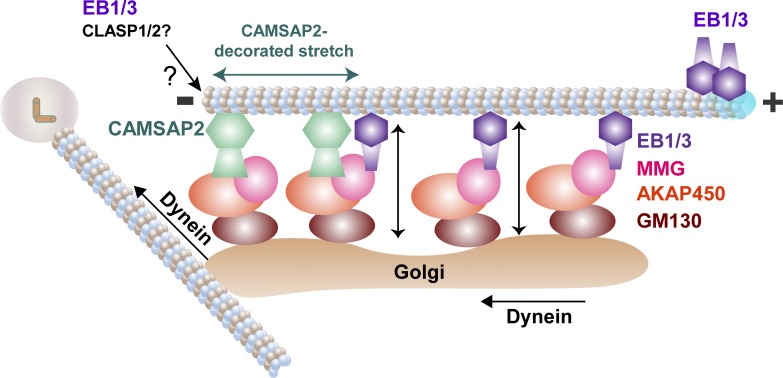
Figure 10.
Model for EB-dependent Golgi–MT coorganization. MMG associates with Golgi membranes through AKAP450 and GM130 and recruits EB1 and EB3, which bind to MT shafts. CAMSAP2 forms MT minus end-localized stretches, which associate with Golgi membranes by binding to the complex of AKAP450 and MMG. EB1 and EB3 also regulate the length of CAMSAP2-decorated MT stretches, likely by controlling MT end dynamics together with some of their partners such as CLASPs. Association of Golgi membranes with free, CAMSAP2-decorated MT minus ends can promote their clustering through dynein-driven minus end–directed transport. Golgi-associated EB1 and EB3 promote spreading of Golgi ribbons along MTs and counteract their dynein-mediated compaction (double-headed arrows).
The importance of the EBs for connecting Golgi membranes to MTs is supported by the observation that in the absence of EBs, Golgi ribbons are more compact. We think that this is due to the fact that MT attachments to the Golgi do not only allow this membrane structure to serve as an MTOC which promotes its own assembly (Efimov et al., 2007; Miller et al., 2009; Rivero et al., 2009; Zhu and Kaverina, 2013; Rios, 2014; Sanders and Kaverina, 2015) but also prevent membrane overcompaction caused by dynein-mediated minus end directed transport. When the static links between the Golgi membranes and MTs are removed, as is the case when AKAP450, MMG, or EB1/3 is disrupted, individual Golgi stacks, despite maintaining their normal morphology at the ultrastructural level (Wu et al., 2016), are brought together into a compact aggregate. The function of CAMSAP2 in this context is more complex, because although it can promote Golgi membrane spreading along the MT stretches that it decorates, it can also enhance the overall compaction of the Golgi, because CAMSAP2-bound MT minus are free to be clustered by dynein activity (Fig. 10).
Cells with disrupted AKAP450 and EB1/EB3 display another similarity: when CAMSAP2 is also knocked out, they require an MTOC for survival. Therefore, centriole depletion, which causes formation of nonradial MT networks in control cells, leads to selection of cells with acentriolar but γ-tubulin–positive MTOCs in AKAP450/CAMSAP2 or EB1/3/CAMSAP2 mutants. We propose that while in normal cells, centrioles serve as a high-affinity site for PCM assembly, in centriole-depleted cells, PCM components are recruited to the Golgi, which then serves as an alternative MTOC. When MTs cannot properly attach to the Golgi because of AKAP450 or EB1/3 disruption, semiparallel acentrosomal MT arrays, the minus ends of which are stabilized by CAMSAP2, are formed. However, when CAMSAP2 is also absent, PCM assembles into a single MTOC, likely by dynein-mediated processes (Balczon et al., 1999; Purohit et al., 1999; Young et al., 2000; Dammermann and Merdes, 2002; Kim et al., 2004; Casenghi et al., 2005), and when this fails to happen, MT minus ends become unstable and cell viability is impaired (Wu et al., 2016). This suggests that although CAMSAP2 can stabilize individual MT minus ends, effective minus end stabilization by PCM requires its clustering. Our data also indicate that the interactions between EB1 and EB3 and their C-terminal partners are not essential for interphase centrosome-based MT organization, although we do not exclude the participation of EB2 in this process.
What are the functional consequences of the interactions between MTs and Golgi membranes? First, it is likely that spreading of the Golgi ribbons along MTs facilitates in- and outgoing vesicle trafficking. Consistently, we previously observed that in AKAP450 knockout cells, the overcompacted Golgi located in cell regions with low MT density was surrounded by an abnormally large number of vesicles, suggesting a trafficking defect (Wu et al., 2016). This notion is supported by the observation that ER-to-Golgi trafficking is perturbed in MMG-depleted cells (Wang et al., 2014). Second, detachment of MTs from the Golgi and a complete loss of noncentrosomal MTs impeded cell migration (Hurtado et al., 2011; Vinogradova et al., 2012; Roubin et al., 2013), with a particularly prominent defect observed during cell invasion in 3D matrix (Wu et al., 2016; this study). It is likely that Golgi-anchored MTs are important for cell polarity, for example, by supporting directional vesicle transport to the leading cell edge (Vinogradova et al., 2012) or by promoting cell asymmetry due to concentration of MT density on one side of the nucleus. The defects in MT growth-dependent extension of protrusions that we recently described for SLAIN2-, ch-TOG–, and CLASP1-depleted 3D-cultured mesenchymal cells (Bouchet et al., 2016), as well abnormalities in the turnover of cell–matrix adhesions (Stehbens et al., 2014), likely also contribute to the observed phenotype. Still, it is striking that a major MT-related function of the most abundant and ubiquitous +TIPs in mammalian cells, EB1 and EB3, is to control the organization of MT minus ends, a function that at least partly relies on the association of EBs with the MT shafts and is typically attributed to conventional MT-associated proteins.
Materials and methods
Constructs
Human EB1-GFP, EB2-GFP, and EB3-GFP are described in Stepanova et al. (2003); EB3-NL-LZ-GFP and EB3-NL-mVenus are described in Komarova et al. (2009); and GFP-EB3-Δtyrosine, GFP-EB3-ΔSxIP, and GFP-EB3-ΔTail are described in Montenegro Gouveia et al. (2010). pBrain-chTOGKDP-GFP-shchTOG (Gutiérrez-Caballero et al., 2015) was a gift of S. Royle (University of Warwick, Coventry, England, UK). mKate-α-tubulin was obtained from Evrogen. We used the following previously described constructs: Bio-mCherry-CAMSAP2, GFP-AKAP450 and GFP-MMG (Wu et al., 2016), human βIVb-tubulin-GFP (Bouchet et al., 2016), and GFP-Rab6A (Grigoriev et al., 2007). Truncated GFP-tagged EB1 and EB3, MMG point mutants and MMG–EB3, CLIP-115, and MAP7 chimeras were generated by PCR-based strategies; to generate the two latter clones, cDNAs encoding CLIP-115 (Hoogenraad et al., 2000) and MAP7 (Faire et al., 1999; a gift of C. Bulinski, Columbia University Medical Center, New York, NY) were used. GFP-Ska1 construct was a gift of S. Lens (University Medical Center Utrecht, Utrecht, Netherlands).
A list of primers that were used to generate the DNA constructs is given in Table 1.
Table 1. Primers used to generate the DNA constructs.
| Construct | Primer Sequence (5′–3′) |
|---|---|
| GFP-MMG 1-389 WT | CCGGACTCAGATCTCGCGGCCGCATGAAGGAGATTTGCAGGATCTGTG |
| GATCAGTTATCTAGATCCGGTGGATCCTTATGAATTCAGCTTTTGTTGTTTCAGC | |
| GFP MMG 1-389 SxNN | AATAACATTCCAGTGAAATCCAGCCTACCTGG |
| GCTGGATTTCACTGGAATGTTATTCCTGCTCCCTCGGGGGCTC | |
| GFP-MMG 1-389 SxNN-EB3 201-281 | GTTGCTGAAACAACAAAAGCTGAATTCAGGCAGCGGACAAATTCTTGAACTCAACCAACAGCTG |
| CTAGATCCGGTGGATCCTTATGATTAGTACTCGTCCTGGTCTTCTTGTTG | |
| EB3-GFP-MMG 1-389 SxNN | CGTCAGATCCGCTAGCGCTACCGGTCATGGCCGTCAATGTGTACTCCAC |
| CTTGCTCACCATGGTGGCGACGGATCCGCCTCCAGAACCAGCTCCGGCATCAGTCTCATG | |
| CLIP115 1-359-GFP-MMG 1-389 SxNN | TCAGATCCGCTAGCGCTACCGGTCATGCAGAAGCCCAGTGGC |
| CTCACCATGGTGGCGACGGATCCGCCTCCAGAACCAGCTCCGCCTGAGATCTTCCGGGC | |
| MAP7 18-283-GFP-MMG 1-389 SxNN | TCAGATCCGCTAGCGCTACCGGTCATGGTGCGAAGCGAAACAGCAC |
| CTCACCATGGTGGCGACGGATCCGCCTCCAGAACCAGCTCCAGAAGAGCCCTCAGGTGGT |
The pSpCas9-2A-Puro (PX459) vector that was used for the CRISPR/Cas9 knockout was purchased from Addgene (Ran et al., 2013). Guide RNAs for human EB1, EB2, and EB3 (also known as MAPRE1, MAPRE2, and MAPRE3) were designed using the CRISPR design webpage tool (http://crispr.mit.edu). The targeting sequences for gRNAs were as follows (coding strand sequence indicated): EB1, 5′-TGGAAAAGACTATGACCCTG-3′; EB2, 5′-CCGGAAGCACACAGTGCGCG-3′; EB3, 5′-TGCACCTCAACTATACCAAG-3′; and CAMSAP2, 5′-CATGATCGATACCCTCATGA-3′ (Wu et al., 2016).
Cell culture, transfection, and rescue experiments
HeLa, human embryonic kidney 239T (HEK293T), HT1080, and RPE1 cell lines were cultured in medium that consisted of 45% DMEM, 45% Ham’s F10, and 10% FCS supplemented with penicillin and streptomycin (Akhmanova et al., 2001). The cell lines were routinely checked for mycoplasma contamination using LT07-518 Mycoalert assay (Lonza). For 3D migration experiments, HT1080 cell cultures were set up by cell suspension seeding in 2 mg/ml rat trail collagen I gels (VMR) neutralized according to the manufacturer’s protocol, in 8-well chambered cover glasses (Thermo Fisher Scientific).
FuGENE 6 (Promega) was used to transfect HeLa, HT1080, and RPE1 cells with plasmids for generating knockout lines, immunofluorescence, and live cell imaging; polyethylenimine (PEI; Polysciences) was used to transfect HEK293T cells for streptavidin pull-down assays. For rescue experiments, cells were fixed and stained 24–48 h after transfection. Lipofectamine RNAiMAX (Thermo Fisher Scientific) was used to transfect RPE1 cells with siRNA EB2 (target sequence 5′-AATGAACGTTGATAAGGTA-3′) and luciferase (target sequence 5′-CGTACGCGGAATACTTCGA-3′) at 20 nM.
Generation and characterization of EB knockout cell lines
The CRISPR/Cas9-mediated EB1, EB2, and EB3 knockout was performed according to the protocol of Ran et al. (2013). In brief, HeLa, RPE1, and HT1080 cell lines were transfected with the vectors bearing the appropriate targeting sequences using FuGENE 6. 1 d after transfection, RPE1 cells were subjected to selection with 25 µg/ml puromycin for 3 d; HeLa and HT1080 cells were subjected to selection with 2 µg/ml puromycin for 2 d. After selection, cells were allowed to recover in normal medium for ∼7 d, and knockout efficiency was checked by immunofluorescence staining. Depending on the efficiency, 39–51 individual clones were isolated and characterized by Western blotting and immunostaining. EB1/2/3mut HeLa cells and EB1/3mut RPE1 cells were generated by targeting EB1-, EB2-, and EB3-encoding genes simultaneously, and EB1/3mut HT1080 cells were generated by targeting EB1- and EB3-encoding genes simultaneously. EB1/3/CAMSAP2mut RPE1 cells were generated on the basis of the EB1/3mut cell line. MMG and AKAP450 knockout RPE1 cells were described previously (Wu et al., 2016). The mutated portions of the EB-encoding genes were sequenced by PCR using primers located in the vicinity of the corresponding gRNA target sites.
Antibodies, pull down assays and immunofluorescence cell staining
We used rabbit antibodies against CAMSAP2 (NBP1-21402, Novus; 17880-1-AP, Proteintech), EB3 (Stepanova et al., 2003), CLIP170 and CLASP1 (Akhmanova et al., 2001), GM130 (ab52649; Abcam), GFP (ab290; Abcam), CEP135 (SAB4503685-100UG; Sigma-Aldrich), and goat anti–myosin IIB antibody (sc-47205; Santa Cruz). Alexa-Fluor-594-conjugated phalloidin was purchased from Life Technologies (A12381). We used mouse monoclonal antibodies against GM130 and paxillin (610822 and 610823; BD Biosciences), β-tubulin, γ-tubulin (T5201 and T6557; Sigma-Aldrich), and rat monoclonal antibodies against EB2 (Komarova et al., 2005) and α-tubulin YL1/2 (MA1-80017; Pierce). The following secondary antibodies were used: IRDye 800CW/680LT goat anti–rabbit, anti–rat, and anti–mouse and streptavidin IRDye 680RD (Li-Cor Biosciences) and Alexa Fluor 350–, 405–, 488–, and 594–conjugated goat antibodies against rabbit, rat, and mouse IgG (Molecular Probes).
Streptavidin pull-down assays were performed from extracts of HEK293T cells coexpressing biotin ligase BirA, biotinylation- and GFP-tagged constructs used as bait, and GFP-tagged constructs used as prey, as described previously (Lansbergen et al., 2006). In brief, constructs were transfected separately into HEK293 cells by using PEI (Polysciences). After 48 h, protein extracts were prepared using the lysis buffer containing 20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 1% Triton X-100, and a protease inhibitor cocktail (Roche). The pull-down assays were performed using streptavidin beads (Invitrogen).
GST pull-down assays were performed as described previously using EB–GST fusions purified from Escherichia coli (Komarova et al., 2005). In brief, individual GST fusion proteins were bound to glutathione Sepharose beads (Amersham Biosciences) in a buffer containing 20 mM Tris HCl, pH 7.5, 300 mM NaCl, and 1% Triton X-100. After incubation for 1 h on a rotating wheel at 4°C, beads were separated from the supernatant by centrifugation and washed four times in the same buffer. The beads were then incubated with purified GFP-CAMSAP2 or GFP-MACF43 and analyzed on Coomassie-stained gels. GFP-CAMSAP2 was purified from HEK293 cells. Cells were transfected using PEI and lysed in the lysis buffer (50 mM Hepes, 300 mM NaCl, and 0.5% Triton X-100, pH 7.4) supplemented with protease inhibitors (Roche). After clearing debris by centrifugation, cell lysates were incubated with StrepTactin beads (28-9355-99; GE Healthcare), and the beads were washed with the lysis buffer without protease inhibitors and with the wash buffer (50 mM Hepes, 300 mM NaCl, and 0.5% Triton X-100, pH 7.4). The proteins were eluted with the elution buffer (50 mM Hepes, 150 mM NaCl, 0.05% Triton X-100, and 2.5 mM desthiobiotin). The N-terminally 6xHis-tagged GFP-MACF43 purified from E. coli by immobilized metal-affinity chromatography on Ni-NTA agarose (QIAGEN Benelux) was described previously (Preciado López et al., 2014).
For immunofluorescence cell staining, cultured cells were fixed in –20°C methanol for 10 min followed by 4% paraformaldehyde for 10 min in the case of EB1, EB2, EB3, and CLIP170. In the case of CAMSAP2, GM130, and α-tubulin, cells were only fixed with –20°C methanol for 10 min. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 10 min; subsequent washing and labeling steps were performed in PBS supplemented with 2% BSA and 0.05% Tween-20. At the end, slides were rinsed in 70% and 100% ethanol, air-dried, and mounted in Vectashield mounting medium (Vector Laboratories).
Drugs and drug treatments
We used the following drugs: SB 415286 and nocodazole (Sigma-Aldrich), paclitaxel (Enzo Life Sciences), and centrinone (a gift of A. Shiau and T. Gahman, Ludwig Institute for Cancer Research, San Diego, CA). To enhance CLASP1 localization at MT tips, cells were incubated with the GSK3 inhibitor SB415286 at a concentration of 20 µM for 30 min. MT stabilization was performed by treating cells with 5 µM paclitaxel for 30 min at 37°C. MT disassembly was performed by treating the cells with 10 µM nocodazole for 1 h at 37°C, followed by 1 h at 4°C, to achieve both Golgi dispersion and complete disassembly of stable MTs. To deplete centrioles, RPE1 cells were treated with 125 nM centrinone for 11 d, and the drug-containing medium was refreshed every 24 h.
Image acquisition
Images of fixed cells were collected with an Eclipse Ni upright fluorescence microscope and DS-Qi2 CMOS camera (Nikon), using a Plan Apo Lambda 40×, NA 1.30, oil objective (Nikon) or Plan Apo Lambda 100×, NA 1.45, oil objective (Nikon) and Nikon NIS (Br) software. Intensilight C-HGFI (Nikon) was used as a light source. Images of fixed cells were also collected with spinning disk microscopy, which was performed on an inverted research microscope Eclipse Ti-E with the Perfect Focus System (Nikon), equipped with Nikon Apo total internal reflection fluorescence (TIRF) 100×, NA 1.49, oil objective, Yokogawa CSU-X1-A1 confocal head with 405-491-561 triple-band mirror and GFP, mCherry, and GFP/mCherry emission filters (Chroma), ASI motorized stage MS-2000-XYZ with Piezo Top Plate (ASI), a Photometrics Evolve 512 electron-multiplying charge-coupled device (CCD) camera (Photometrics), and controlled by MetaMorph 7.7 software (Molecular Devices). The microscope was equipped with a custom-ordered illuminator (MEY10021; Nikon) modified by Roper Scientific France/PICT-IBiSA, Institut Curie. Cobolt Calypso 491 nm (100 mW) and Cobolt Jive 561 nm (100 mW) lasers (Cobolt) were used as light sources. For simultaneous imaging of green and red fluorescence, we used ET-mCherry/GFP filter set (59022; Chroma) together with the DualView (DV2; Roper) equipped with the dichroic filter 565dcxr (Chroma) and HQ530/30m emission filter (Chroma).
Live-cell fluorescent TIRF imaging was performed on an inverted research microscope Eclipse Ti-E(Nikon) with the Perfect Focus System (Nikon), equipped with Nikon Apo TIRF 100×, NA 1.49, oil objective (Nikon) and iLas2 system (Dual Laser illuminator for azimuthal spinning TIRF [or Hilo] illumination and Simultaneous Targeted Laser Action) from Roper Scientific. The system was also equipped with ASI motorized stage MS-2000-XY and a Photometrics Evolve Delta 512 electron-multiplying CCD camera and controlled by MetaMorph 7.8 software. Stradus 488 nm (150 mW; Vortran) and OBIS 561 nm (100 mW; Coherent) lasers were used as light sources. For simultaneous imaging of green and red fluorescence, we used Optosplit III (Cairn Research) equipped with the filters from ET-GFP (49002), ET-mCherry (49008), and ET-GFPmCherry (59022) filter sets (Chroma). To keep the cells at 37°C, a stage top incubator model INUBG2E-ZILCS (Tokai Hit) was used. Images were acquired at 15.4 pixels/µm with exposure time 500 ms at 2 frames/s for MT plus end imaging and with exposure 100 ms at 10 frames/s for GFP-Rab6 imaging (in both cases, the total duration of a movie was 50 s). Cells were plated on round 25-mm coverslips, which were mounted in Attofluor Cell Chamber (Thermo Fisher Scientific) and maintained at 37°C.
Live-cell phase contrast imaging was performed on inverted research microscope Eclipse Ti-E with the Perfect Focus System, equipped with Nikon Plan Fluor 10×, NA 0.30, Ph1 and Nikon Plan Apo 20×, NA 0.75, Ph2 Phase Contrast objectives, Lambda SC Smart Shutters (Sutter), motorized stage MS-2000-XY (ASI), and a CoolSNAP HQ2 CCD camera (Photometrics) and controlled with Micro-Manager Open Source Microscopy Software (https://micro-manager.org). To keep the cells at 37°C, a stage top incubator model INUBG2E-ZILCS was used. Images were acquired at 3.1 and 1.6 pixels/µm. Cells were plated on round 25-mm coverslips, which were mounted in Attofluor Cell Chamber and maintained at 37°C and 5% CO2. Cells were imaged every 5–10 min for 8–24 h.
Preparation and analysis of fluorescence microscopy images
Images were prepared for publication using MetaMorph, ImageJ, and Adobe Photoshop. All images were modified by adjustments of levels and contrast. ImageJ was used for quantification of the immunofluorescence signal intensity. For measurement of parameters of MT dynamics, HeLa EB1/2/3mut cell lines were used for the analysis 24 h after transfection with β-tubulin–GFP or EB–GFP fusions. Parameters of MT growth were analyzed by using kymographs in MetaMorph software (Kymograph function) to distinguish episodes of growth and shortening (in case of imaging of fluorescently labeled tubulin) or the episodes of growth (in case of imaging of fluorescently labeled EB constructs). For measurements of instantaneous rates or transition frequencies, MT end displacements longer than 0.5 µm were taken into account.
The slopes of MT growth and shortening were analyzed in MetaMorph software using Line Region tool and Region Measurements function (Distance and Angle options), resulting in a distance and an angle value per episode of growth or shortening. Velocity of growth and shortening was calculated in SigmaPlot software (version 7.101; SPSS) as the tangent of the episode angle. The duration of growth and shortening was calculated as the distance, multiplied by the cosine of the angle. Transition frequencies were calculated as inverted episode durations.
Analysis of fluorescence intensity distribution of EB2, CLIP170, and CLASP1 along MT ends was performed using ImageJ. In brief, MTs with clearly visible ends at the cell periphery were analyzed. Fixed Line Length tool was used to obtain the profile of MT or +TIP intensity, and the intensity profiles were then normalized and averaged.
For quantification of ch-TOG intensity at the MT plus ends, the mean signal intensity of ch-TOG signal at a MT tip in a free-shaped region of interest was measured in MetaMorph software (Region Measurements function, Average Intensity option), and the mean intensity and SD of ch-TOG signal in cytoplasm were also measured (SD was measured in MetaMorph software using Region Measurements function as Intensity Standard Deviation option). The signal-to-noise ratio was calculated as [(mean intensity at the tip) − (mean intensity in cytoplasm)]/(SD in the cytoplasm).
CAMSAP2 and GM130 distributions were analyzed using the ImageJ Radial Profile plugin. A circle of 13.3 µm was drawn with the center positioned in the center of the Golgi. Golgi size was determined by Quick Selection tool in Photoshop. The Analyze Particles plugin was used to quantify FA size and number. MTrackJ tool was used to extract cell migration tracks and measure their speed and direction.
GFP-Rab6 vesicle movements were analyzed in ImageJ using our previously developed tracking methods (“method 11” in Chenouard et al., 2014). The tracks were then processed using another custom-made ImageJ plugin, which removes the nonprocessive movements within each track and splits trajectories into multiple segments of processive motion. Finally, the resulting tracks were processed using another published ImageJ plugin (Yao et al., 2017) to quantify the velocities and track radiality with respect to a manually set reference point. For this analysis, we used as the reference point location the center of the Golgi apparatus. The track radiality is the angle between the line connecting the start and end points of this track and the line from the starting point of the track to the reference point. The angle ranges from 0° to 180°, where 0° corresponds to a particle moving along the radius away from the reference point to the cell periphery, and 180° corresponds to a particle moving radially toward the cell center.
Transmission EM sample preparation and analysis
Cells were fixed at room temperature by adding Karnovsky fixative (2.5% glutaraldehyde and 2% formaldehyde [Electron Microscopy Sciences] in 0.2 M cacodylate buffer, pH 7.4) 1:1 to culture medium for 10 min. This was replaced by fresh fixative for 2 h at room temperature. Cells were then postfixed with 1% OsO4/1.5% K3Fe(III)(CN)6 in 0.065 M phosphate buffer for 2 h at 4°C and finally 1 h with 0.5% uranyl acetate. After fixation, cells were dehydrated and embedded in epon epoxy resin (Polysciences). Ultrathin sections of 60 nm were contrasted with uranyl acetate and lead citrate using the AC20 (Leica) and examined with a 1010 electron microscope (Jeol Europe). Images were taken from three different sets of cells per condition. 50 random images per set were taken (150 per condition).
All measurements were done using ImageJ. For the quantification of the area within which Golgi stacks are confined in the cell, the smallest possible ellipse to enclose all the Golgi stacks was drawn in ImageJ, and then the area of that ellipse was measured. This was done for three different independent sets of cells per condition, and then 50 cells within each set (in total 150 cells were measured per experimental condition). Independently, the length of individual Golgi stacks was measured with the Segmented Line tool, the thickness with the Straight Line tool, and the area with the Freehand Selection tool.
Statistical analysis
Statistical significance was analyzed either using the Mann-Whitney U test or two-tailed t test, as indicated in figure legends. For the t test, data distribution was assumed to be normal, but this was not formally tested. Statistical significance was determined using GraphPad Prism software (version 6.07).
Online supplemental material
Figs. S1 and S2 illustrate analyses of cell division in EB1/3mut RPE1 and EB1/2/3mut HeLa cells. Fig. S3 provides information on MTs, Golgi, and CAMSAP2 stretch organization in different knockout lines. Fig. S4 shows the recruitment of EBs and CAMSAP2 to the Golgi and the biochemical characterization of the interactions between AKAP450, MMG, CAMSAP2, and EB1. Fig. S5 illustrates the distribution of EB1 in paclitaxel-treated cells and shows that disruption of EB1 and EB3 has no significant impact on certain aspects of cell polarity and directionality of cell movement but does affect the organization of tracks of GFP-Rab6–positive exocytotic vesicles. Table S1 provides information on mutations in sequences of EB-encoding genes in different knockout lines.
Supplementary Material
Acknowledgments
We thank A. Shiau, T. Gahman, S. Royle, C. Bulinski, and S. Lens for gifts of materials.
This study was supported by China Scholarship Council scholarships to C. Yang and J. Wu, a H2020 European Research Council Synergy grant 609822 to A. Akhmanova, and Netherlands Organisation for Scientific Research STW grant OTP13391 to E. Meijering and A. Akhmanova.
The authors declare no competing financial interests.
Author contributions: C. Yang and J. Wu designed and conducted the experiments and wrote the paper. I. Grigoriev performed live cell imaging and image analysis. C. de Heus, N. Liv, and J. Klumperman performed and analyzed EM experiments. Y. Yao, I. Smal, and E. Meijering established live cell imaging analysis procedures and helped with image analysis. R.Z. Qi contributed essential reagents. A. Akhmanova designed and supervised the study and wrote the paper.
Footnotes
Abbreviations used:
- CAMSAP
- calmodulin-regulated spectrin-associated protein
- CH
- calponin homology
- EB
- end-binding
- FA
- focal adhesion
- HEK
- human embryonic kidney
- MMG
- myomegalin
- MT
- microtubule
- MTOC
- MT organizing center
- PCM
- pericentriolar material
- RPE
- retinal pigment epithelium
References
- Akhmanova A., and Steinmetz M.O.. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., and Galjart N.. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935. 10.1016/S0092-8674(01)00288-4 [DOI] [PubMed] [Google Scholar]
- Balczon R., Varden C.E., and Schroer T.A.. 1999. Role for microtubules in centrosome doubling in Chinese hamster ovary cells. Cell Motil. Cytoskeleton. 42:60–72. [DOI] [PubMed] [Google Scholar]
- Ban R., Matsuzaki H., Akashi T., Sakashita G., Taniguchi H., Park S.Y., Tanaka H., Furukawa K., and Urano T.. 2009. Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J. Biol. Chem. 284:28367–28381. 10.1074/jbc.M109.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V., Styles C.A., and Fink G.R.. 1990. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J. Cell Biol. 111:2573–2586. 10.1083/jcb.111.6.2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove S.R., Lee Y.R., Liu B., Peters N.T., and Kropf D.L.. 2008. The microtubule plus-end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis. Plant Cell. 20:396–410. 10.1105/tpc.107.056846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelić S., De Groot C.O., Schärer M.A., Jaussi R., Bargsten K., Salzmann M., Frey D., Capitani G., Kammerer R.A., and Steinmetz M.O.. 2012. Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued). J. Struct. Biol. 177:160–167. 10.1016/j.jsb.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Bouchet B.P., Noordstra I., van Amersfoort M., Katrukha E.A., Ammon Y.C., Ter Hoeve N.D., Hodgson L., Dogterom M., Derksen P.W.B., and Akhmanova A.. 2016. Mesenchymal cell invasion requires cooperative regulation of persistent microtubule growth by SLAIN2 and CLASP1. Dev. Cell. 39:708–723. 10.1016/j.devcel.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., and Hyman A.A.. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casenghi M., Barr F.A., and Nigg E.A.. 2005. Phosphorylation of Nlp by Plk1 negatively regulates its dynein-dynactin-dependent targeting to the centrosome. J. Cell Sci. 118:5101–5108. 10.1242/jcs.02622 [DOI] [PubMed] [Google Scholar]
- Chenouard N., Smal I., de Chaumont F., Maška M., Sbalzarini I.F., Gong Y., Cardinale J., Carthel C., Coraluppi S., Winter M., et al. 2014. Objective comparison of particle tracking methods. Nat. Methods. 11:281–289. 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V.M., Shapiro I., Aldridge B., and Sorger P.K.. 2006. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 25:2814–2827. 10.1038/sj.emboj.7601168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 12:917–930. 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faire K., Waterman-Storer C.M., Gruber D., Masson D., Salmon E.D., and Bulinski J.C.. 1999. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J. Cell Sci. 112:4243–4255. [DOI] [PubMed] [Google Scholar]
- Ferreira J.G., Pereira A.J., Akhmanova A., and Maiato H.. 2013. Aurora B spatially regulates EB3 phosphorylation to coordinate daughter cell adhesion with cytokinesis. J. Cell Biol. 201:709–724. 10.1083/jcb.201301131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fréal A., Fassier C., Le Bras B., Bullier E., De Gois S., Hazan J., Hoogenraad C.C., and Couraud F.. 2016. Cooperative interactions between 480 kDa ankyrin-G and EB proteins assemble the axon initial segment. J. Neurosci. 36:4421–4433. 10.1523/JNEUROSCI.3219-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S.S., and Vale R.D.. 2010. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 143:263–274. 10.1016/j.cell.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Wollman R., and Kaplan K.B.. 2005. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell. 16:4609–4622. 10.1091/mbc.E05-03-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., Splinter D., Keijzer N., Wulf P.S., Demmers J., Ohtsuka T., Modesti M., Maly I.V., Grosveld F., Hoogenraad C.C., and Akhmanova A.. 2007. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell. 13:305–314. 10.1016/j.devcel.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Caballero C., Burgess S.G., Bayliss R., and Royle S.J.. 2015. TACC3-ch-TOG track the growing tips of microtubules independently of clathrin and Aurora-A phosphorylation. Biol. Open. 4:170–179. 10.1242/bio.201410843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershott M.C., and Vale R.D.. 2014. Regulation of microtubule minus-end dynamics by CAMSAPs and patronin. Proc. Natl. Acad. Sci. USA. 111:5860–5865. 10.1073/pnas.1404133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F.K., and Steinmetz M.O.. 2006. Key interaction modes of dynamic +TIP networks. Mol. Cell. 23:663–671. 10.1016/j.molcel.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Akhmanova A., Grosveld F., De Zeeuw C.I., and Galjart N.. 2000. Functional analysis of CLIP-115 and its binding to microtubules. J. Cell Sci. 113:2285–2297. [DOI] [PubMed] [Google Scholar]
- Howard J., and Hyman A.A.. 2003. Dynamics and mechanics of the microtubule plus end. Nature. 422:753–758. 10.1038/nature01600 [DOI] [PubMed] [Google Scholar]
- Hurtado L., Caballero C., Gavilan M.P., Cardenas J., Bornens M., and Rios R.M.. 2011. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 193:917–933. 10.1083/jcb.201011014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimori M., Watanabe S., Kiyonari S., Matsuoka K., Sakasai R., Saeki H., Oki E., Kitao H., and Maehara Y.. 2016. Phosphorylation of EB2 by Aurora B and CDK1 ensures mitotic progression and genome stability. Nat. Commun. 7:11117 10.1038/ncomms11117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery J.M., Grigoriev I., Poser I., van der Horst A., Hamilton N., Waterhouse N., Bleier J., Subramaniam V.N., Maly I.V., Akhmanova A., and Khanna K.K.. 2013. Centrobin regulates centrosome function in interphase cells by limiting pericentriolar matrix recruitment. Cell Cycle. 12:899–906. 10.4161/cc.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Toedt G., Montenegro Gouveia S., Davey N.E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L.B., Bezstarosti K., et al. 2012. A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22:1800–1807. 10.1016/j.cub.2012.07.047 [DOI] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F., Heck A.J., Hoogenraad C.C., and Akhmanova A.. 2014. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 28:295–309. 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Kim J.C., Badano J.L., Sibold S., Esmail M.A., Hill J., Hoskins B.E., Leitch C.C., Venner K., Ansley S.J., Ross A.J., et al. 2004. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 36:462–470. 10.1038/ng1352 [DOI] [PubMed] [Google Scholar]
- Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G.G., and Akhmanova A.. 2005. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell. 16:5334–5345. 10.1091/mbc.E05-07-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., De Groot C.O., Grigoriev I., Gouveia S.M., Munteanu E.L., Schober J.M., Honnappa S., Buey R.M., Hoogenraad C.C., Dogterom M., et al. 2009. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184:691–706. 10.1083/jcb.200807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., and Wittmann T.. 2012. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22:418–428. 10.1016/j.tcb.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A.B., Grosveld F., and Akhmanova A.. 2006. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell. 11:21–32. 10.1016/j.devcel.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Liu H., Yue J., Huang H., Gou X., Chen S.Y., Zhao Y., and Wu X.. 2015. Regulation of focal adhesion dynamics and cell motility by the EB2 and Hax1 protein complex. J. Biol. Chem. 290:30771–30782. 10.1074/jbc.M115.671743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer S.P., Fourniol F.J., Bohner G., Moores C.A., and Surrey T.. 2012. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 149:371–382. 10.1016/j.cell.2012.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer S.P., Cade N.I., Bohner G., Gustafsson N., Boutant E., and Surrey T.. 2014. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 24:372–384. 10.1016/j.cub.2013.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2017. Large-scale analysis of CRISPR/Cas9 cell-cycle knockouts reveals the diversity of p53-dependent responses to cell-cycle defects. Dev. Cell. 40:405–420. 10.1016/j.devcel.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.M., Folkmann A.W., Maia A.R., Efimova N., Efimov A., and Kaverina I.. 2009. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 11:1069–1080. 10.1038/ncb1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., and Akhmanova A.. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153. 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia S., Leslie K., Kapitein L.C., Buey R.M., Grigoriev I., Wagenbach M., Smal I., Meijering E., Hoogenraad C.C., Wordeman L., et al. 2010. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 20:1717–1722. 10.1016/j.cub.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Grigoriev I., Nogi T., Hamaji T., Cassimeris L., and Mimori-Kiyosue Y.. 2012. Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS One. 7:e51442 10.1371/journal.pone.0051442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preciado López M., Huber F., Grigoriev I., Steinmetz M.O., Akhmanova A., Koenderink G.H., and Dogterom M.. 2014. Actin-microtubule coordination at growing microtubule ends. Nat. Commun. 5:4778 10.1038/ncomms5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit A., Tynan S.H., Vallee R., and Doxsey S.J.. 1999. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147:481–492. 10.1083/jcb.147.3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios R.M. 2014. The centrosome-Golgi apparatus nexus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:369 10.1098/rstb.2013.0462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S., Cardenas J., Bornens M., and Rios R.M.. 2009. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28:1016–1028. 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubin R., Acquaviva C., Chevrier V., Sedjaï F., Zyss D., Birnbaum D., and Rosnet O.. 2013. Myomegalin is necessary for the formation of centrosomal and Golgi-derived microtubules. Biol. Open. 2:238–250. 10.1242/bio.20123392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A.A., and Kaverina I.. 2015. Nucleation and dynamics of Golgi-derived microtubules. Front. Neurosci. 9:431 10.3389/fnins.2015.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Hayashi K., Amano Y., Takahashi M., Yonemura S., Hayashi I., Hirose H., Ohno S., and Suzuki A.. 2014. MTCL1 crosslinks and stabilizes non-centrosomal microtubules on the Golgi membrane. Nat. Commun. 5:5266 10.1038/ncomms6266 [DOI] [PubMed] [Google Scholar]
- Schuyler S.C., and Pellman D.. 2001. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 105:421–424. 10.1016/S0092-8674(01)00364-6 [DOI] [PubMed] [Google Scholar]
- Stehbens S., and Wittmann T.. 2012. Targeting and transport: how microtubules control focal adhesion dynamics. J. Cell Biol. 198:481–489. 10.1083/jcb.201206050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S.J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T.. 2014. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16:561–573. 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C.C., Lansbergen G., Dortland B., De Zeeuw C.I., Grosveld F., van Cappellen G., Akhmanova A., and Galjart N.. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A., and Merdes A.. 2007. EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr. Biol. 17:1318–1325. 10.1016/j.cub.2007.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G.E., Bandopadhyay K., Sutradhar S., Renjith M.R., Singh P., Gireesh K.K., Simon S., Badarudeen B., Gupta H., Banerjee M., et al. 2016. EB1 regulates attachment of Ska1 with microtubules by forming extended structures on the microtubule lattice. Nat. Commun. 7:11665 10.1038/ncomms11665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., and Nishida E.. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26:1487–1498. 10.1038/sj.emboj.7601599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B., Manatschal C., Grigoriev I., Olieric V., Gouveia S.M., Bjelic S., Demmers J., Vorobjev I., Hoogenraad C.C., Steinmetz M.O., and Akhmanova A.. 2011. SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J. Cell Biol. 193:1083–1099. 10.1083/jcb.201012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova T., Paul R., Grimaldi A.D., Loncarek J., Miller P.M., Yampolsky D., Magidson V., Khodjakov A., Mogilner A., and Kaverina I.. 2012. Concerted effort of centrosomal and Golgi-derived microtubules is required for proper Golgi complex assembly but not for maintenance. Mol. Biol. Cell. 23:820–833. 10.1091/mbc.E11-06-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang C., and Qi R.Z.. 2014. A newly identified myomegalin isoform functions in Golgi microtubule organization and ER-Golgi transport. J. Cell Sci. 127:4904–4917. 10.1242/jcs.155408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., de Heus C., Liu Q., Bouchet B.P., Noordstra I., Jiang K., Hua S., Martin M., Yang C., Grigoriev I., et al. 2016. Molecular pathway of microtubule organization at the Golgi apparatus. Dev. Cell. 39:44–60. 10.1016/j.devcel.2016.08.009 [DOI] [PubMed] [Google Scholar]
- Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., and Yao X.. 2012. EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. USA. 109:16564–16569. 10.1073/pnas.1202639109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Smal I., Grigoriev I., Martin M., Akhmanova A., and Meijering E.. 2017. Automated analysis of intracellular dynamic processes. Methods Mol. Biol. 1563:209–228. 10.1007/978-1-4939-6810-7_14 [DOI] [PubMed] [Google Scholar]
- Young A., Dictenberg J.B., Purohit A., Tuft R., and Doxsey S.J.. 2000. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell. 11:2047–2056. 10.1091/mbc.11.6.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J., Xie M., Gou X., Lee P., Schneider M.D., and Wu X.. 2014. Microtubules regulate focal adhesion dynamics through MAP4K4. Dev. Cell. 31:572–585. 10.1016/j.devcel.2014.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M., Widlund P.O., Hyman A.A., and Howard J.. 2013. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15:688–693. 10.1038/ncb2744 [DOI] [PubMed] [Google Scholar]
- Zhu X., and Kaverina I.. 2013. Golgi as an MTOC: making microtubules for its own good. Histochem. Cell Biol. 140:361–367. 10.1007/s00418-013-1119-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., and Westermann S.. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391. 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.