The elevated CDK2 activity of oncogene-expressing cells induces DNA replication stress. Yang et al. show that the DNA repair protein RAD18 facilitates damage-tolerant DNA synthesis via the DNA polymerase κ in cells with aberrantly high CDK2 activity, suggesting an important new role for RAD18 in sustaining neoplastic cell survival.
Abstract
The mechanisms by which neoplastic cells tolerate oncogene-induced DNA replication stress are poorly understood. Cyclin-dependent kinase 2 (CDK2) is a major mediator of oncogenic DNA replication stress. In this study, we show that CDK2-inducing stimuli (including Cyclin E overexpression, oncogenic RAS, and WEE1 inhibition) activate the DNA repair protein RAD18. CDK2-induced RAD18 activation required initiation of DNA synthesis and was repressed by p53. RAD18 and its effector, DNA polymerase κ (Polκ), sustained ongoing DNA synthesis in cells harboring elevated CDK2 activity. RAD18-deficient cells aberrantly accumulated single-stranded DNA (ssDNA) after CDK2 activation. In RAD18-depleted cells, the G2/M checkpoint was necessary to prevent mitotic entry with persistent ssDNA. Rad18−/− and Polκ−/− cells were highly sensitive to the WEE1 inhibitor MK-1775 (which simultaneously activates CDK2 and abrogates the G2/M checkpoint). Collectively, our results show that the RAD18–Polκ signaling axis allows tolerance of CDK2-mediated oncogenic stress and may allow neoplastic cells to breach tumorigenic barriers.
Introduction
During tumorigenesis, neoplastic cells must endure DNA damage from environmental, metabolic, and other intrinsic sources (Bartkova et al., 2006; Halazonetis et al., 2008). Oncogene-induced DNA replication stress can be a major cause of intrinsic DNA damage and represents a potential source of genome instability in cancer cells. Many oncogenes, including v-RAS, cyclin E, and others, induce DNA replication defects that trigger DNA damage signaling (including ATM–CHK2, ATR–CHK1, and p53) and lead to irreversible cell cycle exit often termed oncogene-induced senescence (OIS; Bartkova et al., 2006; Di Micco et al., 2006).
The precise mechanisms by which oncogenes induce DNA damage are incompletely understood. Oncogene-induced DNA damage has been attributed to induction of genotoxic reactive oxygen species (ROS; DeNicola et al., 2011), depletion of nucleotide pools (Bester et al., 2011), collisions between the DNA replication and transcriptional machinery (Jones et al., 2013), or aberrant reinitiation of DNA synthesis multiple times each per cell cycle—a process usually termed “rereplication” or “hyperreplication” (Di Micco et al., 2006). Rereplication likely generates “onion skin” DNA structures in which head-to-tail collisions between replication forks produce double-strand breaks (DSBs; Davidson et al., 2006). It is unknown whether oncogene-induced rereplication is caused by inappropriate activation of DNA replication licensing factors, initiation factors, or deregulation of both licensing and initiation phases of DNA synthesis. It is also unclear whether common mechanisms mediate rereplication and DNA damage in response to all oncogenes. It is possible that the constitutive mitogenic signals induced by oncogenes culminate in aberrant cyclin-dependent kinase 2 (CDK2) activation, in turn leading to DNA rereplication and other replication defects. Indeed, oncogene-induced DNA replication stress is often modeled experimentally by overexpression of CDK2 activators (Cyclin E and CDC25A) or inhibition of the WEE1 kinase to remove negative constraints over CDK2 (Sogo et al., 2002; Bartkova et al., 2006; Beck et al., 2010, 2012; Jones et al., 2013).
Despite our limited mechanistic understanding of how oncogenes dysregulate DNA synthesis and cause DNA damage, there is general consensus that OIS poses a barrier to tumorigenesis. Clearly, however, the OIS barrier is imperfect and can be breached. The precise mechanisms by which oncogene-expressing cells withstand replication stress and DNA damage are poorly understood. DNA repair and/or DNA damage tolerance capacity could potentially impact whether DNA synthesis and viability are sustained when cells experience oncogenic stress. Interestingly, the DNA polymerase δ subunits POLD3 and POLD4 can facilitate DNA replication in cyclin E–overexpressing cells (Costantino et al., 2014). Moreover, the ATR–CHK1 pathway can promote oncogene-induced carcinogenesis (Schoppy et al., 2012). Therefore, DNA damage signaling and genome maintenance might critically influence whether oncogene-expressing cells breach the OIS barrier. However, there has been no systematic analysis of how DNA damage signaling and repair mechanisms impact DNA replication and cell cycle progression of oncogene-expressing cells. It remains to be investigated whether all genome maintenance mechanisms or only specific subpathways of the DNA damage response confer oncogenic stress tolerance. Importantly, many cancer chemotherapeutic agents act by causing DNA replication stress and DNA damage. The selective pressures for preneoplastic cells to acquire DNA damage tolerance during tumorigenesis could also provide a mechanism for chemoresistance. Therefore, the mechanisms by which cancer cells tolerate oncogenic DNA replication stress represent therapeutic targets whose inhibition could sensitize tumors to intrinsic and therapy-induced DNA damage.
We recently found that many cancer cells co-opt an aberrantly expressed meiotic protein, the cancer/testes antigen MAGE-A4, to pathologically activate trans-lesion synthesis (TLS; Gao et al., 2016a). Cancer cell–specific RAD18 pathway activation by MAGE-A4 first suggested to us a possible role for TLS in the tolerance of replicative stresses that are unique to neoplastic cells. TLS is a specialized mode of DNA replication involving the DNA damage–tolerant and error-prone Y family DNA polymerases η (Polη), κ (Polκ), and ι (Polι) as well as REV1 (Prakash et al., 2005). Individual TLS polymerases perform replicative bypass of preferred cognate DNA lesions. Collectively, the TLS polymerases support DNA replication and viability in cells harboring damaged genomes. Y family polymerase-deficient cells are often sensitive to agents that cause replication fork–stalling and DNA replication stress, demonstrating a key role for the TLS pathway in DNA damage tolerance. However, owing to the low fidelity of Y family polymerases, TLS can be mutagenic (Prakash et al., 2005). Thus, TLS must be regulated stringently and used sparingly to maintain genome stability.
The E3 ubiquitin ligase RAD18 is a proximal activator of all four Y family TLS DNA polymerases (Hedglin and Benkovic, 2015). In response to DNA replication fork stalling, RAD18 monoubiquitinates proliferating cell nuclear antigen (PCNA) at the conserved residue K164. Y family DNA polymerases possess ubiquitin-binding (ubiquitin-binding motif [UBM] and ubiquitin-binding zinc finger [UBZ]) domains and PCNA-interacting peptide boxes that facilitate their preferential association with PCNA in its monoubiquitinated form (Bienko et al., 2005). The DNA damage–inducible interaction between TLS polymerases and PCNA is the basis for the polymerase switch that replaces replicative DNA polymerases ε and δ with Y family polymerases at stalled DNA replication forks. Polη exists in complex with RAD18 and is probably the default Y family TLS polymerase recruited to stalled DNA replication forks (Watanabe et al., 2004; Day et al., 2010). Polη is versatile and can perform replicative bypass of many bulky lesions, possibly explaining why this TLS polymerase is preferentially recruited to all stalled replication forks.
In addition to its central role in TLS, RAD18 contributes to activation of other genome maintenance pathways, including the Fanconi Anemia pathway (Palle and Vaziri, 2011; Williams et al., 2011), interstrand cross-link repair (Räschle et al., 2015), and DSB repair (Huang et al., 2009). As the apical component of the TLS polymerase switch and other DNA repair pathways, RAD18 expression levels and activity critically impact DNA damage sensitivity. In neoplastic cells, pathological activation of RAD18 by its stabilizing binding partner MAGE-A4 has the potential to confer both DNA damage tolerance and mutability, two important hallmarks and enabling characteristics of tumor cells. Because oncogenic stress is a major source of intrinsic DNA damage experienced by cancer cells, we hypothesized a role for the RAD18 pathway in the tolerance of oncogene-induced DNA damage. We show in this study that RAD18 is activated in response to oncogenic stimuli that induce aberrant CDK2 activity. Moreover, RAD18 and its effector Y family DNA polymerase Polκ (but surprisingly not the default TLS polymerase Polη) are necessary for S phase progression and viability in the face of excess CDK2 activity. Collectively, this work suggests that TLS may be an important driver of carcinogenesis that promotes both tolerance of oncogenic stress and mutagenesis.
Results
Acute oncogene expression promotes RAD18-mediated PCNA monoubiquitination in untransformed cells
We determined whether the TLS branch of the DNA damage response is activated in response to acute oncogene expression in human cells. We selected representative stimuli that are known to induce DNA replication stress and OIS, including Ha-RASV12, Cyclin E, c-MYC, and CDT1 (Bartkova et al., 2006; Liontos et al., 2007; Srinivasan et al., 2013). Oncogenes were transiently expressed in untransformed normal human fibroblasts (NHFs) using adenovirus vectors. 48 h after infection, overexpressed Cyclin E, C-MYC, and CDT1 proteins were readily detectable by immunoblotting, and MAPK phosphorylation was used to validate RAS pathway activation (Fig. 1 A).
Figure 1.
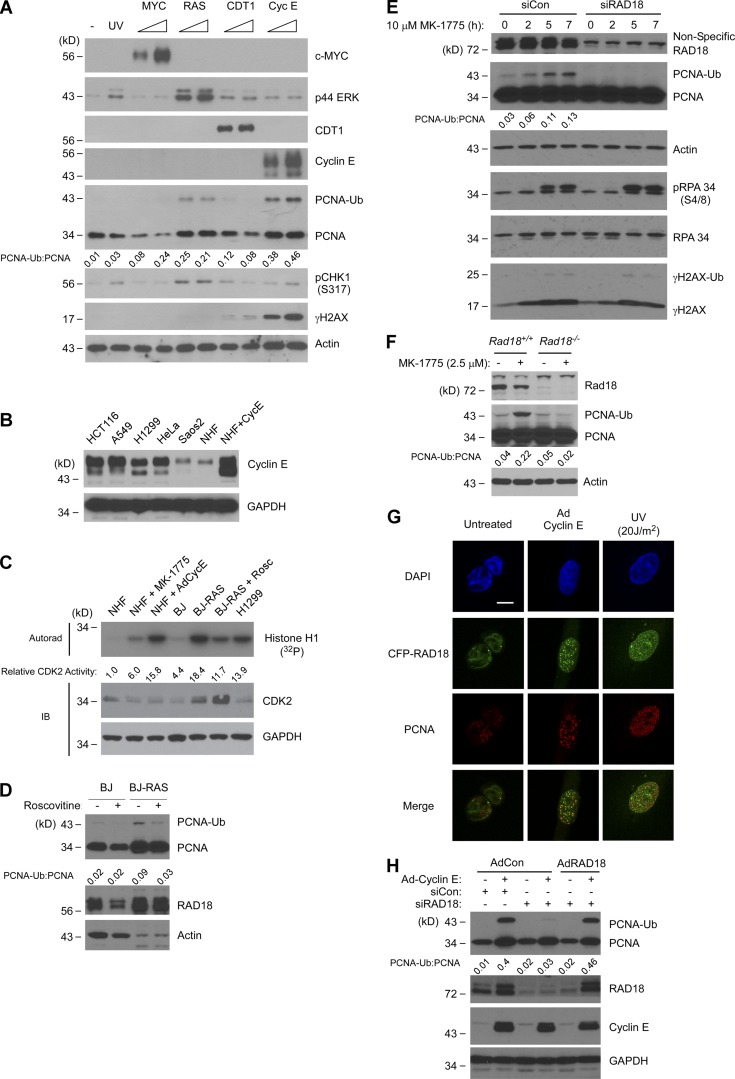
Oncogenic stimuli promote RAD18-mediated PCNA monoubiquitination. (A) Adenoviral vectors were used to express c-MYC, Ha-RASV12, CDT1, and Cyclin E in cultured NHF. After 48 h, cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (B) Immunoblot showing relative Cyclin E protein levels in AdCyclin E–infected NHFs and various cancer cell lines. Soluble extracts from the indicated cells were normalized for protein concentration and then analyzed by SDS-PAGE and immunoblotting using antibodies against Cyclin E and GAPDH (for loading controls). (C) IP–kinase assay showing relative CDK2 activities in RAS-transformed cells (BJ-RAS) and isogenic parental fibroblasts (BJ). Also shown are CDK2 activities in control, MK-1775–treated, and Cyclin E–overexpressing NHFs and in H1299 lung adenocarcinoma cells. (D) Immunoblot showing the effects of 24 h roscovitine treatment on PCNA monoubiquitination in BJ and BJ-RAS cells. (E) Control and RAD18-depleted NHF cells were treated for 0–7 h with MK-1775. At different times, cell lysates were analyzed for expression of the indicated DNA damage markers using SDS-PAGE and immunoblotting. (F) Rad18+/+ and Rad18−/− MEFs were treated with MK-1775 for 7 h. Lysates from the resulting cells were analyzed by SDS-PAGE and immunoblotted with indicated antibodies. (G) U2OS cells were coinfected with AdCFP-RAD18 and AdCyclin E (or with control adenovirus) for 24 h. Some cultures were UV irradiated (20 J/m2), and 2 h later, CFP-RAD18 subcellular distribution was analyzed by confocal microscopy. Bar, 10 µm. (H) NHFs were transfected with siRAD18 or with nontargeting control siRNA. Transfected cells were infected with adenoviruses encoding siRNA-resistant RAD18, Cyclin E, or with an “empty” virus for control. Lysates from the resulting cells were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
As expected from previous work, DNA damage markers including CHK1 pS317 and γH2AX were induced by ectopically expressed Cyclin E and RAS (Fig. 1 A; Bartkova et al., 2006; Di Micco et al., 2006; Neelsen et al., 2013a). PCNA monoubiquitination was not significantly affected by MYC or CDT1. However, levels of monoubiquitinated PCNA were increased 25-fold in response to overexpressed Ha-RAS and 46-fold after Cyclin E expression. The fold changes in PCNA monoubiquitination after Ha-RAS and Cyclin E expression substantially exceeded the threefold increase in PCNA modification induced by UV irradiation in NHFs (Fig. 1 A, second lane from left). The levels of ectopically expressed cyclin E that induced PCNA monoubiquitination in untransformed NHF cells were similar to steady-state Cyclin E levels in various cancer cell lines including HCT116 colon cancer cells, A549 and H1299 lung carcinoma cells, and HeLa cervical cancer cells (Fig. 1 B). In immunoprecipitation (IP) kinase assays, the CDK2 activity of Cyclin E–overexpressing cells was comparable to CDK2 activity in the representative lung cancer cell line H1299 (Fig. 1 C). Therefore, our standard Cyclin E overexpression conditions are appropriate for modeling the aberrant Cyclin E–CDK2 signaling of cancer cells.
In RAS-transformed BJ human fibroblasts (Hahn et al., 1999), CDK2 activity was approximately fourfold higher than in isogenic parental BJ fibroblasts (Fig. 1 C) and was comparable to CDK2 activity of AdCyclin E–transduced fibroblasts and H1299 lung cancer cells (Fig. 1 C). PCNA was constitutively monoubiquitinated in BJ-RAS cells when compared with parental BJ fibroblasts (Fig. 1 D). Moreover, acute treatment with the CDK2 inhibitor roscovitine attenuated PCNA monoubiquitination in BJ-RAS cells (Fig. 1 D). We conclude that Cyclin E– or RAS-induced CDK2 activity stimulates PCNA monoubiquitination.
As an additional approach to test the effect of CDK2 activation on PCNA monoubiquitination, we used a pharmacological inhibitor of WEE1 kinase (MK-1775) to de-repress CDK2. Using IP–kinase assays, we confirmed that CDK2 was activated in MK-1775–treated cells (Fig. 1 C) as expected from previous work (Beck et al., 2012; Sørensen and Syljuåsen, 2012; Sakurikar et al., 2016). Similar to ectopic Cyclin E or RAS expression, MK-1775 treatment induced PCNA monoubiquitination in NHFs (Fig. 1 E). The MK-1775–induced increases in CDK2 activity and PCNA monoubiquitination (sixfold and 4.3-fold increases, respectively) were modest compared with AdCyclin E–induced CDK activity and PCNA monoubiquitination, indicating that MK-1775–induced CDK2 activity is limited by endogenous Cyclin E levels.
Because MK-1775 de-represses both CDK1 and CDK2, we sought to determine which CDK contributes to PCNA monoubiquitination when WEE1 is inhibited. In CDK1-depleted cells, MK-1775–induced PCNA monoubiquitination was attenuated when compared with CDK1-replete cells (Fig. S1 A), possibly suggesting that CDK1 contributes to PCNA monoubiquitination. However, CDK1-depleted cells were arrested in G2/M (Fig. S1 B), potentially causing attenuation of PCNA monoubiquitination indirectly after cell cycle arrest. Therefore, as an alternative approach to determining the potential roles of CDK2 and CDK1 in stimulating PCNA monoubiquitination, we tested the effects of specific CDK2 activators (Cyclins E and A) or CDK1 activators (Cyclin B and CDC25C) on PCNA. As shown in Fig. S1 (C and D), PCNA monoubiquitination was induced by Cyclin E and more modestly by Cyclin A, but not by Cyclin B or CDC25C. We infer that MK-1775–induced PCNA monoubiquitination is primarily caused by de-repression of CDK2 and is not CDK1 mediated. Collectively, Fig. 1 (A–E) and Fig. S1 show that deregulated CDK2 signaling triggers PCNA monoubiquitination in untransformed human cells. PCNA was also monoubiquitinated in response to aberrant CDK2 activity in other cell types that have been used extensively to model cellular responses to oncogenic stress, including mouse embryonic fibroblasts (MEFs) and U2OS cells (as shown in Figs. 1 F and 4).
Figure 4.
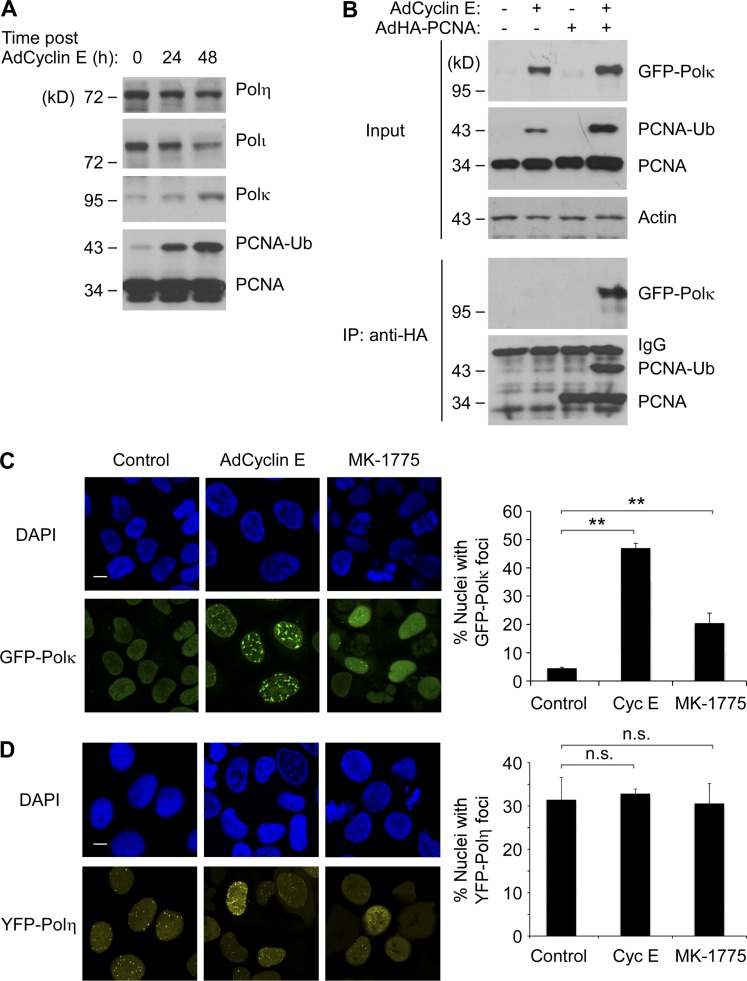
Cyclin E overexpression and WEE1 inhibition promote nuclear redistribution and PCNA binding of Polκ. (A) U2OS cells were infected with 0, 108, and 1010 pfu/ml of AdCyclin E. 48 h later, chromatin extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (B) U2OS cells were infected with AdCyclin E, AdHA-PCNA, and AdGFP-Polκ as indicated. 24 h later, solubilized chromatin fractions were prepared and analyzed directly by SDS-PAGE (“input”) or were first immunoprecipitated with anti-HA (IP) to recover PCNA complexes, which were analyzed for the presence of associate GFP-Polκ. (C and D) Replicate cultures of U2OS cells were infected with GFP-Polκ (C) or YFP-Polη (D) adenovirus for 24 h. The resulting cultures were infected with AdCyclin E for 24 h, were treated with MK-1775 for 5 h, or were left untreated for controls. The subcellular distributions of GFP-Polκ and YFP-Polη were analyzed by fluorescence microscopy, and representative images of TLS polymerase–expressing nuclei are shown. Bars, 10 µm. For quantitative analysis of DNA polymerase foci, at least 100 nuclei were scored for each experimental condition. The results are compiled from three independent experiments. Error bars represent SEM. Statistical analyses were performed using Student’s t test. The differences in numbers of foci between experimental conditions were significant for GFP-Polκ (**, P < 0.01) but not significant for YFP-Polη (P > 0.05).
PCNA is a substrate for monoubiquitination by several E3 ligases including RAD18 (Watanabe et al., 2004), CRL4Cdt2 (Terai et al., 2010), and HLTF (Lin et al., 2011). However, Cyclin E overexpression induced the formation of RAD18 nuclear foci (Fig. 1 G). Moreover, MK-1775–induced PCNA monoubiquitination was attenuated in RAD18-depleted NHFs and in Rad18−/− MEFs relative to isogenic RAD18-replete cells (Fig. 1, E and F). Ectopically expressed siRNA-resistant RAD18 fully rescued the PCNA monoubiquitination defect of RAD18-depleted cells (Fig. 1 H). Therefore, we conclude that PCNA monoubiquitination in response to elevated CDK2 is RAD18 mediated.
Cyclin E–CDK2–induced PCNA monoubiquitination requires replication licensing and origin firing
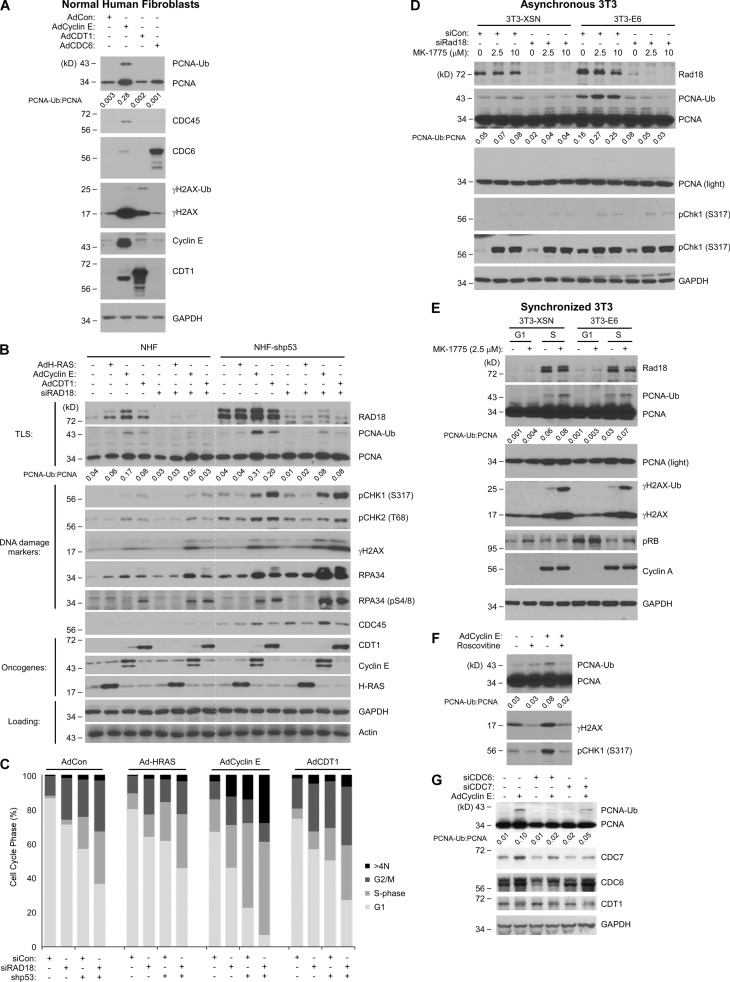
Cyclin E–CDK2 is active in both G1 and S phase, and RAD18-mediated PCNA monoubiquitination can occur throughout the cell cycle (Daigaku et al., 2010; Zlatanou et al., 2011; Diamant et al., 2012; Yang et al., 2013). Therefore, we investigated the cell cycle stage specificity and proximal trigger of PCNA monoubiquitination in response to aberrant Cyclin E–CDK2 activity. First, we asked whether PCNA monoubiquitination was associated with the imbalance of licensing or initiation phases of DNA replication. We ectopically overexpressed DNA replication licensing factors (CDT1 and CDC6) or Cyclin E to stimulate replication licensing or initiation of DNA synthesis, respectively. As expected, Cyclin E led to increased chromatin binding of CDC45 (a distal and rate-limiting step in the initiation of DNA synthesis) coincident with increased PCNA monoubiquitination (Fig. 2 A). Overexpressed licensing factors did not promote chromatin binding of CDC45 and induced negligible changes in PCNA monoubiquitination (Fig. 2 A). Therefore, RAD18 activation is associated with aberrant initiation of DNA synthesis, not altered replication licensing.
Figure 2.
Passage through the G1/S restriction point and initiation of DNA synthesis are necessary for CDK2-induced PCNA monoubiquitination. (A) NHFs were infected with the indicated adenoviral vectors for 24 h, and cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (B) Replicate cultures of NHFs and shp53-NHF cells were transfected with siRAD18 or control nontargeting RNA. The resulting cultures were infected with the indicated viruses. One plate of each replicate culture was analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The other plate of each replicate pair was pulse labeled with BrdU for 1 h. The resulting cultures were costained with propidium iodide and BrdU and then analyzed for cell cycle distribution as shown in C. The white dotted line serves to separate the NHF and shp53-NHF data for easier comparison of protein levels between the two cell lines. (C) Graphs summarizing the distribution of cells between different cell cycle phases for each experimental condition. (D) Swiss 3T3-XSN and 3T3-E6 cells were transfected with siRAD18 or with nontargeting siRNA and then treated with the indicated concentrations of MK-1775 for 5 h. Cell lysates were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (E) Quiescent (G0) cultures of Swiss 3T3-XSN and 3T3-E6 cells were stimulated with 10% serum. At 8 and 16 h after serum stimulation (time points corresponding to mid-G1 and S phase, respectively), cells were treated with MK-1775 for 5 h before harvest, SDS-PAGE, and immunoblot analysis of the indicated proteins. (F) NHFs were infected with AdCyclin E (or empty vector for control) for 24 h. The resulting cultures were treated with roscovitine for 2 h before harvest for SDS-PAGE and immunoblot analysis. (G) NHFs were transfected with siCDC6, siCDC7, or nontargeting RNA duplexes. The resulting cultures were infected with AdCyclin E (or empty adenoviral vector). 48 h later, cells were harvested for SDS-PAGE and immunoblotting with the indicated antibodies.
In a complementary approach to test the relationship between PCNA monoubiquitination and initiation of DNA synthesis, we used p53-directed shRNA or expressed the high-risk HPV-E6 oncoprotein to abrogate the G1 checkpoint and increase initiation of DNA synthesis. As expected, p53 depletion promoted chromatin binding of CDC45 (Fig. 2 B) and S phase entry (Fig. 2 C) while also coincidently inducing PCNA monoubiquitination (Fig. 2 B). Similarly, acute Wee1 inhibition in asynchronous HPV-E6–expressing 3T3 cells led to a 4.7-fold increase in PCNA monoubiquitination when compared with p53-replete 3T3 cells (Fig. 2 D). The results of Fig. 2 (B–D) suggest that loss of the G1 checkpoint and the ensuing increases in S phase entry and origin firing lead to increased PCNA monoubiquitination. We considered the possibility that p53 loss might directly promote PCNA monoubiquitination. However, when we inhibited Wee1 in S phase–synchronized cultures, HPV E6 expression did not affect PCNA monoubiquitination (Fig. 2 E). Moreover, Wee1 inhibition before S phase did not induce PCNA monoubiquitination (Fig. 2 E). Therefore, p53 limits CDK2-induced PCNA monoubiquitination by enforcing a G1/S checkpoint and preventing S phase entry.
To test whether Cyclin E–induced PCNA monoubiquitination was dependent on initiation of DNA replication, we used pharmacological inhibition of CDK2 and gene silencing of CDC7 to inhibit protein kinase activities that are essential for origin firing. Fig. 2 (F and G) show that CDK2 and CDC7 are necessary for Cyclin E–induced PCNA monoubiquitination. As expected, silencing of CDC6, which limits the number of licensed origins available for initiation of DNA synthesis, also abrogated Cyclin E–induced PCNA monoubiquitination (Fig. 2 G). We conclude that CDK2-induced PCNA monoubiquitination requires passage through the G1/S restriction point and initiation of DNA synthesis.
MUS81 and replication protein A (RPA) contribute to RAD18 pathway activation
The mechanism of RAD18 activation during S phase has been attributed to its direct association with p95/NBS1 (Yanagihara et al., 2011), with RPA-coated single-stranded DNA (ssDNA; Davies et al., 2008; Tsuji et al., 2008), or with ubiquitinated chromatin generated by the E3 ligase RNF8 (Huang et al., 2009) at damaged replication forks.
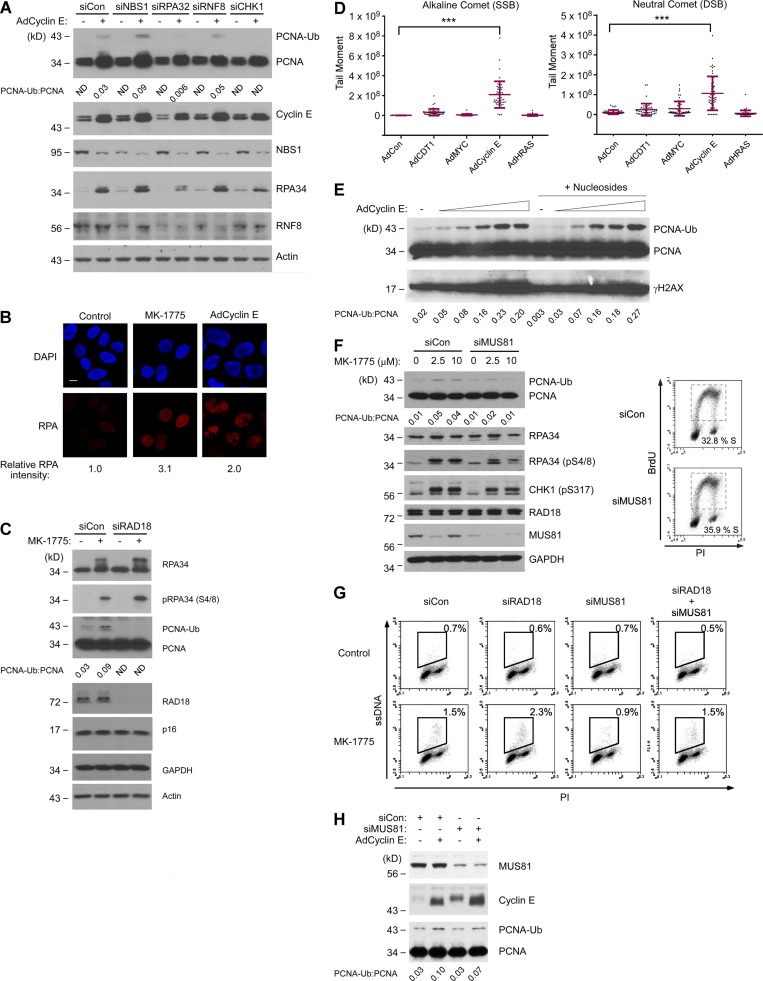
Levels of chromatin-bound p95/NBS1 and RNF8 were not increased upon Cyclin E overexpression (Fig. 3 A). Moreover, depletion of p95/NBS1 or RNF8 did not affect Cyclin E–induced PCNA monoubiquitination (Fig. 3 A), suggesting that p95/NBS1 and RNF8 do not mediate RAD18 pathway activation in response to excess CDK2. However, Cyclin E overexpression and MK-1775 treatment both led to increased numbers of nuclear RPA foci (Fig. 3 B) and increases in chromatin-bound RPA (Fig. 3, A and C), coincident with PCNA monoubiquitination. Moreover, Cyclin E–induced PCNA monoubiquitination was attenuated in RPA-depleted cells (Fig. 3 A). WEE1 inhibition was previously shown to cause an increase in levels of ssDNA (Beck et al., 2010) and in alkaline COMET assays, Cyclin E overexpression led to a 27-fold increase in ssDNA levels relative to control cultures (Fig. 3 D). Collectively, our results are most consistent with an RPA-ssDNA–mediated RAD18 activation mechanism in response to excess CDK2 activity.
Figure 3.
CDK2-induced PCNA monoubiquitination is p95/NBS1-independent but requires MUS81. (A) NHFs were transfected with siNBS1, siRPA32, siRNF8, siCHK1, or control nontargeting siRNA. The resulting cultures were infected with AdCyclin E or control adenovirus, and 48 h later, cells were harvested for SDS-PAGE and immunoblot analysis. (B) Replicate cultures of NHFs were infected with AdCyclin E for 24 h, treated with MK-1775 for 5 h, or left untreated for controls. The resulting cells were fixed, stained with anti-RPA 32 and DAPI, and analyzed by confocal microscopy. The images represent representative fields of RPA- and DAPI-stained nuclei. At least 100 cells were scored for each experimental condition. Bar, 10 µm. (C) NHFs were transfected with siRAD18 or nontargeting siRNA and then treated with MK-1775 for 5 h. Cells were harvested and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (D) NHFs were infected with the indicated adenoviruses for 48 h. Nuclei from the resulting cultures were analyzed using alkaline and neutral comet assays. 50 tail moments were measured for each experimental condition. To determine the statistical significance of the differences in single-strand break (SSB) and DSB levels, we performed ANOVA between groups followed by Tukey’s multiple comparison of means test. Results of the Tukey test indicated significant differences between AdCon and AdCyclin E–overexpressing cells (P < 0.001), indicating that Cyclin E expression induces single-strand breaks and DSBs. The result shown is representative of two independent experiments that yielded qualitatively similar results. ***, P < 0.001. (E) NHFs were grown for 48 h in complete medium or medium supplemented with 50 µM of all four nucleosides (A, U, C, and G) and increasing doses of AdCyclin E (ranging from 0.05−1.0 × 1010 pfu/ml). Chromatin fractions from the resulting cells were analyzed by immunoblotting with the indicated antibodies. (F) NHFs were transfected with siMUS81 and nontargeting control siRNAs for 48 h. The resulting cultures were treated with the indicated doses of MK-1775 for 5 h before harvest for SDS-PAGE and immunoblot analysis with the indicated antibodies. For siMUS81 and siCon-transfected cultures, one replicate plate was pulse labeled with 10 µM BrdU and analyzed by flow cytometry to quantify S phase–positive populations. (G) Replicate plates of NHFs were transfected with the indicated siRNAs for 48 h. One plate of each replicate pair was treated with MK-1775 for 5 h before flow cytometry analysis of ssDNA. (H) U2OS cells were transfected with siMUS81 or nontargeting control (siCon) RNAs. 24 h after transfection, cultures were infected with AdCyclin E or AdCon. After 24 h, cells were collected and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
Nucleoside depletion has been postulated as a mechanism for replication defects in response to certain oncogenic stimuli (Bester et al., 2011). However, exogenously added nucleosides did not suppress the Cyclin E–induced PCNA monoubiquitination (Fig. 3 E). Therefore, we consider it most likely that the PCNA monoubiquitination induced by elevated CDK2 activity does not result from limited availability of nucleotide pools.
CDK2-induced aberrant DNA replication structures are substrates for nucleolytic cleavage by MUS81 (Neelsen et al., 2013a). Beck et al. (2012) showed that MUS81 is recruited to stalled forks when WEE1 kinase is inhibited. Moreover, MUS81 is known to process stalled replication forks to generate the RPA-ssDNA that triggers CHK1 pathway activation (Regairaz et al., 2011). Therefore, we used gene silencing to test a possible role for MUS81 in RAD18 pathway activation. As expected from a previous study (Regairaz et al., 2011), levels of chromatin-bound pRPA (pS4/8) and pChk1 (S317) were reduced after MUS81 depletion (Fig. 3 F). Interestingly, MK-1775–induced PCNA monoubiquitination was attenuated in MUS81-depleted cells (Fig. 3 F). BrdU labeling rates and cell cycle profiles were unaffected by MUS81 depletion (Fig. 3 F). Therefore, the reduced PCNA monoubiquitination of MUS81-depleted cells was not secondary to an altered S phase.
We used a flow cytometry–based assay to determine relative levels of MK-1775–induced ssDNA in control and MUS81-depleted cells. The flow-based ssDNA assay is a modified version of a previously published protocol for visualizing ssDNA regions (Raderschall et al., 1999) and was validated as described in Fig. S2. As shown in Fig. 3 G, MK-1775 led to an approximately twofold increase in ssDNA levels in MUS81-replete cells. However, WEE1 inhibition did not induce ssDNA in MUS81-depleted cultures (Fig. 3 G). We conclude that certain CDK2-induced aberrant DNA replication intermediates can be processed by MUS81 to generate RPA-ssDNA, which in turn serves as a common trigger for both RAD18 and CHK1 pathways.
In Cyclin E–expressing cells (in which CDK2 activity was ∼2.6-fold higher than in MK-1775–treated cells under our standard experimental conditions; see Fig. 1 C), PCNA monoubiquitination was very modestly reduced after MUS81 depletion (Fig. 3 H). As noted previously (Neelsen et al., 2013a), the standard conditions used to express Cyclin E generate severe DNA replication defects, irreversible cell cycle arrest, and massive amounts of ssDNA in all cells. Therefore, MUS81-independent processing of DNA replication intermediates also contributes to PCNA monoubiquitination after the severe replication stress from overexpressed Cyclin E.
Cyclin E–induced PCNA monoubiquitination specifically regulates Polκ
To identify relevant effectors of genome maintenance in response to excess CDK2 signaling, we investigated the regulation of Y family DNA polymerases. Of the Y family DNA polymerases examined (Polη, Polκ, and Polι), only Polκ was redistributed to chromatin in response to overexpressed Cyclin E (Fig. 4 A). Cyclin E overexpression also induced association of Polκ with PCNA as determined by coimmunoprecipitation experiments (Fig. 4 B). In immunofluorescence microscopy studies, Cyclin E overexpression and WEE1 inhibition both led to an increase in GFP-Polκ foci (Fig. 4 C). As expected from previous work, YFP-Polη formed constitutive nuclear foci in replicating cells, but numbers of YFP-Polη nuclear foci were unaffected by Cyclin E expression or WEE1 inhibition (Fig. 4 D; Durando et al., 2013). Collectively, the results of Fig. 4 suggest that Polκ is recruited to CDK2-induced aberrant DNA replication intermediates.
Rad18 and Polκ sustain ongoing DNA replication and cell viability after Wee1 inhibition
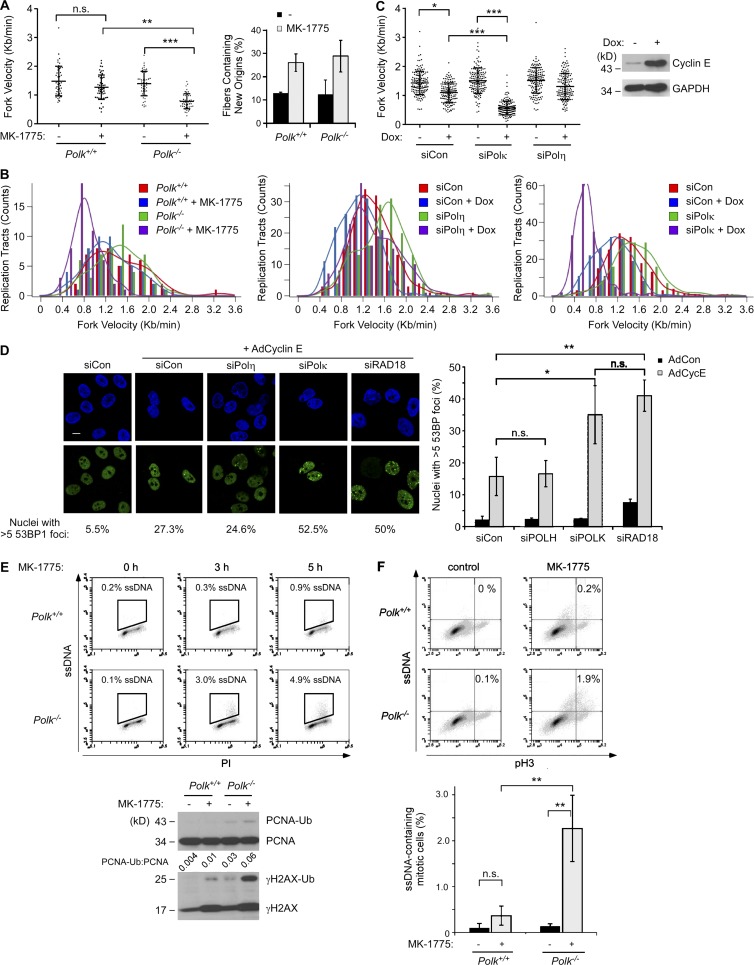
The results of Figs. 1, 2, 3, and 4 suggested a possible role for TLS in tolerating the aberrant S phase induced by excessive CDK2 activity. To elucidate the contribution of TLS to ongoing DNA synthesis in cells harboring excess CDK2, we compared DNA replication dynamics of MK-1775–treated Polk−/− MEFs with isogenic (Polk+/+) control cells.
As shown by the DNA fiber analyses in Fig. 5 (A and B), DNA replication fork velocities were reduced by 15% after 5 h of Wee1 inhibition in the Polk+/+ MEFs. However, in Polk−/− cells, Wee1 inhibition led to a 44% reduction of fork velocity. New origin firing was increased approximately twofold after WEE1 inhibition in both Polk+/+ and Polk−/− MEFs. In NHFs, increased Cyclin E expression (driven by a stably integrated doxycycline-inducible expression vector) led to a 23% decrease in replication fork velocities (Fig. 5 C). However, in Polκ-depleted fibroblasts, replication fork velocities were reduced by 63% after Cyclin E induction (Fig. 5 C). Replication fork velocities were unaffected by Polη depletion (Fig. 5 C). We conclude that Polκ-dependent TLS sustains DNA replication fork progression in cells harboring elevated CDK activity.
Figure 5.
Polκ facilitates DNA replication and prevents ssDNA accumulation in cells with elevated CDK2 activity. (A and B) DNA fiber analyses showing effects of MK-1775 treatment on DNA replication dynamics in cultures of isogenic WT and Polk−/− MEFs. Panel A shows results of a representative experiment in which fork velocities were determined for 50 individual replication tracts under each experimental condition. Based on the results of ANOVA and Tukey’s multiple comparison of means test, MK-1775 treatment led to statistically significant decreases in fork velocity in Polk−/− MEFs (P < 0.001) but not Polk+/+ cells (P > 0.05). To quantify new origin firing, 300 fibers were counted for each experimental condition. There were no significant differences in numbers of new origins when comparing between Polk−/− and Polk+/+ MEFs. In panel B, lengths of ongoing replication forks (containing both CldU and IdU) were measured to determine the distribution of replication fork speeds. 50 fibers were quantified for each experimental condition. The experiment described in A and B was repeated twice with similar results. (C) DNA fiber analyses showing effects of doxycycline (Dox)-inducible Cyclin E expression on DNA replication dynamics of Polκ- or Polη-depleted NHFs. DNA replication fork velocities were determined for 150 individual replication tracts from each experimental condition. Based on the results of ANOVA and Tukey’s multiple comparison of means test, doxycycline-inducible Cyclin E expression led to statistically significant reduction of fork velocity by in siPOLK-transfected NHFs (P < 0.001) but not siPOLH-transfected cells (P > 0.05). The data presented are representative of an experiment that was repeated twice with similar results. (D) Replicate cultures of U2OS cells were transfected with the indicated siRNAs. After 48 h, cells were treated with 2.5 µM MK-1775 (or vehicle for control) for 5 h. Cells were fixed, and 53BP distribution was analyzed by immunofluorescence confocal microscopy. Representative images of 53BP1-stained nuclei are shown. Bar, 10 µm. For quantitative analysis of DNA, at least 100 nuclei were scored for each experimental condition. The graph shows results of three independent experiments, and error bars represent SEM. Statistical analyses were performed using a Student’s t test. The resulting p-values indicate significant differences in numbers of 53BP1 foci between siCon and siPOLK (P < 0.05) and between siCon and siRAD18 (P < 0.01), but no significant differences between siCon and siPOLH-transfected cells. (E) Replicate cultures of isogenic Polk+/+ and Polk−/− MEFs were treated with 2.5 µM MK-1775 for the indicated times and then analyzed for ssDNA content using flow cytometry. For the 0- and 5-h time points, one plate of each replicate culture was collected for SDS-PAGE and immunoblotting with the indicated antibodies. PI, propidium iodide. (F) Polk+/+ and Polk−/− MEFs were prelabeled with BrdU for 24 h and then treated with 2.5 µM MK-1775 (or vehicle) for 5 h. Cultures were analyzed by flow cytometry to identify phospho–histone H3– and ssDNA-positive populations. The histogram shows the results from three independent experiments in which the columns represent means and error bars represent the range. From the results of ANOVA and Tukey’s multiple comparison of means test, MK-1775 treatment induced statistically significant increases in ssDNA levels in Polk−/− MEFs (P < 0.001) but not in Polk+/+ cells (P = 0.8). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In genotoxin-treated cells, TLS prevents DNA replication fork collapse and DNA DSB accumulation (Bi et al., 2005, 2006). To determine whether TLS also protects against DSB formation caused by excessive CDK2, we enumerated 53BP1 nuclear foci (as a surrogate for unrepaired DSBs) in Cyclin E–overexpressing cells that were depleted of RAD18, Polκ, or Polη. As shown in Fig. 5 D, silencing of RAD18 or Polκ (but not Polη) led to an approximately twofold increase in the number of 53BP1 foci in Cyclin E–overexpressing cells. The results of Fig. 5 (A–D) suggest that RAD18 and Polκ (but not Polη) sustain DNA replication and prevent DSB accumulation in Cyclin E–overexpressing cells.
TLS operates postreplicatively and patches 5′ ssDNA gaps generated behind nascent leading strands (Lopes et al., 2006; Daigaku et al., 2010). We predicted therefore that TLS deficiency would lead to persistent ssDNA when there is excess CDK2. To test this prediction, we used flow cytometry to compare ssDNA levels in Polk+/+ and Polk−/− cells basally and after Wee1 inhibition. As shown in Fig. 5 E, basal ssDNA levels were indistinguishable between exponentially growing Polk+/+ and Polk−/− cells. However, after Wee1 inhibition, Polk−/− MEFs accumulated up to 10-fold higher levels of ssDNA than isogenic Polk+/+ cells (Fig. 5 E). The ssDNA induced by Wee1 inhibition in Polk−/− cells coincided with 4N (G2 + M) populations identified by propidium iodide staining (Fig. 5 E). Using anti–phospho–histone H3 (to more precisely define mitotic populations), we demonstrated that mitotic Polk-deficient MEFs accumulated approximately fivefold higher levels of ssDNA after Wee1 inhibition when compared with Polk+/+ cells (Fig. 5 F). We conclude that Polκ is important to prevent mitotic ssDNA accumulation in response to Wee1 inhibition.
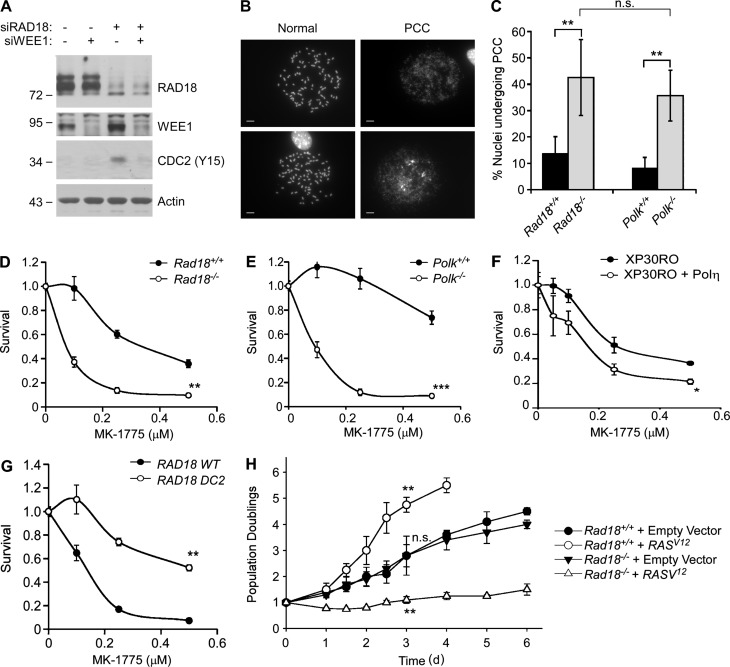
Persistent ssDNA triggers the “replication checkpoint,” a CHK1-mediated mechanism that inhibits CDC2/CDK1 via Y15 phosphorylation and prevents mitosis when the genome is incompletely replicated (Canman, 2001). In NHFs, RAD18 depletion led to an increase in Y15-phosphorylated CDC2 (Fig. 6 A), indicating that the replication checkpoint is activated when TLS is compromised. As expected, WEE1 inhibition eliminated the Y15 phosphorylation in RAD18-depleted cells (Fig. 6 A).
Figure 6.
Polκ is specifically required to prevent PCC and confer viability after WEE1 inhibition. (A) U2OS cells were transfected with siRAD18, siWEE1, or nontargeting control RNA. 48 h later, cells were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (B and C) Replicate cultures of isogenic Polk+/+ and Polk−/− MEFs were treated with 2.5 µM MK-1775 (or vehicle) for 7 h. Metaphase spreads from the resulting cells were analyzed by light microscopy and scored for aberrant nuclei undergoing mitotic catastrophe. Molecular masses are given in kilodaltons. Panel B shows representative examples of normal metaphase spreads and of pulverized nuclei displaying hallmarks of PCC. Bars, 10 µm. The histogram in C shows the effect of Polk and Rad18 on the relative incidence of PCC in control and MK-1775–treated cells. The results shown in the histogram are derived from three independent experiments in which at least 200 nuclei were scored for every experimental condition. The error bars represent SEM. Statistical analyses using ANOVA and Tukey’s multiple comparison of means test show significant differences in the percentage of nuclei undergoing PCC when comparing Rad18+/+ versus Rad18−/− MEFs (P < 0.01) and also when comparing Polk+/+ and Polk−/− cells (P < 0.01). There was no significant difference between Rad18−/− and Polk−/− cells basally or after MK-1775 treatment (P > 0.05), indicating that Polk−/− cells recapitulate the PCC phenotype of Rad18-null MEFs. (D–G) The indicated pairs of isogenic cell lines with mutations in Rad18, Polk, and POLH were treated with different doses of MK-1775, and sensitivity to Wee1/WEE1 inhibition was evaluated by clonogenic survival assays. For each experiment, the number of surviving colonies from MK-1775–treated cultures was expressed as a percentage of colony numbers from cells that received vehicle (DMSO) for control. On the survival curves, each data point represents the mean of triplicate determinations, and error bars represent the range. For each experiment, we performed a Student’s t test with the data obtained using 0.25 µM MK-1775. When comparing Rad18+/+ versus Rad18−/− cells or Polk+/+ versus Polk−/− cells, p was <0.01 (indicating that Rad18−/− and Polk−/− MEFs are MK-1775 sensitive when compared with their respective WT control cell lines). When comparing XP30RO and XP30RO + Polη cells, p was <0.05, indicating that Polη-complemented XPV cells are MK-1775 sensitive when compared with Polη-deficient XPV patient cells. When comparing “knock-in” MEFs expressing RAD18 WT and RAD18 DC2, p was <0.01, indicating that MEFs expressing the RAD18 DC2 mutant cells are more MK-1775 tolerant than WT RAD18-expressing cells. All data shown are from representative experiments that were repeated at least three times and yielded similar results on each occasion. (H) Primary untransformed Rad18+/+ or Rad18−/− MEFs were cultured in 3% oxygen and infected with pMXs-derived retrovirus encoding HRASV12 or with empty retroviral vector for control. Retrovirus-infected cells were seeded in medium containing puromycin (1 µg/ml), and puromycin-resistant cells were enumerated every day for 1 wk. Data points represent means of triplicate determinations with error bars representing SEM. Using the data from the day 4 post-infection time point, we performed ANOVA between groups followed by Tukey’s multiple comparison of means test to evaluate statistical significance of differences in growth of various genotypes. Results of the Tukey test were as follows: Rad18+/+ (empty vector) versus Rad18−/− (empty vector), P > 0.05 (indicating no significant difference in proliferation of Rad18+/+ and Rad18−/− in the absence of oncogenic RAS); Rad18+/+ (empty vector) versus Rad18+/+ (RASV12), P < 0.01 (indicating significant stimulation of proliferation by oncogenic RAS in Rad18-replete cells); Rad18−/− (empty vector) versus Rad18−/− (RASV12), P < 0.01 (indicating significant inhibition of proliferation by RASV12 in Rad18-deficient cells). The cell proliferation data are from a representative experiment that yielded similar results on three separate occasions. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We hypothesized that the persistent ssDNA induced by MK-1775 in TLS-deficient cells resulted from aberrant activation of CDK2 (which deregulates DNA synthesis) and CDK1 (which abrogates the G2/M checkpoint). To test whether the G2/M checkpoint is a failsafe when TLS is compromised, we used ectopically overexpressed CDC25C (a Y15-directed CDK1 phosphatase) as an MK-1775–independent strategy to bypass the G2/M checkpoint. As expected, overexpressed CDC25C caused a reduction in levels of Y15-phosphorylated CDK1 (Fig. S3 A). We determined the effects of CDC25C on mitotic ssDNA in RAS-transformed BJ cells (which have high CDK2 activity as shown in Fig. 1 C) and parental BJ fibroblasts. As shown in Fig. S3 C, CDC25C overexpression led to an ∼20-fold increase in mitotic ssDNA levels in RAS-transformed cells but not in parental BJ fibroblasts. Interestingly, in RAD18-depleted BJ-RAS cells, levels of mitotic ssDNA were further increased to ∼29-fold above control (Fig. S3 C). In three separate experiments, RAD18 depletion increased the levels of RAS-induced ssDNA by 1.4-fold ± 0.16 (P = 0.028). Collectively, the results of Fig. S3 indicate that in the absence of TLS, CDK2-induced ssDNA persists into mitosis when the replication checkpoint is bypassed by CDK1.
Forced bypass of the replication checkpoint can elicit premature chromatin condensation (PCC), abortive mitoses (termed “mitotic catastrophe”), and cell death (Nghiem et al., 2001). To determine whether TLS averts mitotic catastrophe, we analyzed metaphase spreads from Rad18−/− and Polk−/− MEFs and isogenic WT cells after MK-1775 treatment and scored metaphase chromosomes for PCC.
Images of representative metaphase spreads illustrating normal mitotic chromosomes and pulverized chromosomes caused by PCC are shown in Fig. 6 B. In control (MK-1775 untreated) MEFs, the incidence of spontaneous PCC was negligible (<1%) in WT, Rad18−/−, and Polk−/− cultures, indicating minimal mitotic defects when TLS alone is compromised. After MK-1775 treatment, the incidence of PCC increased to 14% and 8% in Rad18+/+ and Polk+/+ MEFs, respectively (Fig. 6 C). Remarkably, the numbers of nuclei undergoing PCC were 3.1- and 4.4-fold higher in Rad18−/− and Polk−/− MEFs, respectively, when compared with isogenic WT control cells (Fig. 6 C). In clonogenic survival assays, both Rad18−/− and Polk−/− MEFs were sensitive to Wee1 inhibition when compared with WT cell lines (Fig. 6, D and E). We conclude that Rad18 and Polκ are important for preventing PCC and maintaining viability when Wee1 is inhibited.
To test whether Polκ is specifically required to tolerate aberrant CDK activity, we also determined the MK-1775 sensitivity of an XP-variant (XPV) cell line (XP30RO), which harbors an inactivating mutation in Polη (Volpe and Cleaver, 1995). In contrast with Polk-null MEFs, Polη-deficient XPV fibroblasts were not MK-1775 sensitive when compared with isogenic Polη-complemented (XP30RO + Polη) cells. Surprisingly, Polη-deficient XP30RO cells were significantly more tolerant of WEE1 inhibition when compared with the corrected XP30RO + Polη line (Fig. 6 E). To further test the role of Polη on MK-1775 tolerance, we compared the MK-1775 sensitivity of MEFs derived from transgenic knock-in mice expressing WT RAD18 or a Polη interaction-deficient RAD18 DC2 mutant (Yang et al., 2016). Cells expressing RAD18 DC2 are compromised for Polη activation and have modest UV sensitivity (Watanabe et al., 2004; Day et al., 2010). Interestingly, similar to Polη-deficient XPV cells, RAD18 DC2 MEFs were MK-1775 resistant when compared with WT RAD18-expressing MEFs (Fig. 6 G). Therefore, Polκ is specifically required to tolerate excessive CDK activity, and Polη inhibits tolerance of CDK2-induced replication stress.
To extend our analysis of Rad18 functions in tolerance of excess CDK2 activity, we also determined the role of Rad18 in cellular response to oncogenic RAS. Rad18+/+ and Rad18−/− MEFs were transduced with retroviruses encoding oncogenic RAS or with an empty vector. As shown in Fig. 6 H, growth curves of empty vector–transduced Rad18+/+ and Rad18−/− cells were identical. However, RAS expression stimulated proliferation of Rad18+/+ MEFs but was inhibitory for Rad18−/− cells. These results further demonstrate that Rad18 facilitates tolerance of oncogenic stress.
Rad18 deficiency sensitizes cancer cells and Brca1-deficient cells to Wee1 inhibition
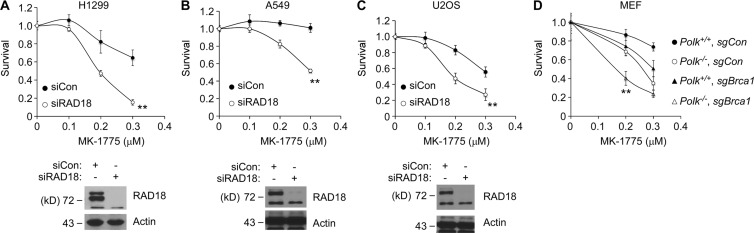
Because MK-1775 is being considered as a therapeutic agent, we determined the extent to which RAD18 signaling allows cancer cell lines to tolerate WEE1 inhibition. As shown in Fig. 7 (A–C), three representative and commonly studied cancer cell lines, H1299, A549, and U2OS, were MK-1775 sensitive after RAD18 depletion.
Figure 7.
RAD18 confers MK-1775 tolerance in multiple cancer cells and cooperates with DSB repair genes to promote cell viability. (A–C) The indicated cancer cell lines were transfected with siRAD18 or control nontargeting siRNAs. 48 h later, cells were treated with the indicated doses of MK-1775, and drug sensitivities were determined using clonogenic survival assays. For each experiment, the number of surviving colonies from MK-1775–treated cultures was expressed as a percentage of colony number from cells that received vehicle (DMSO) for control. On the survival curves, each data point represents the mean of triplicate determinations, and error bars represent the range. For each experiment, we performed a Student’s t test with data obtained using the 0.3-µM MK-1775 concentration. When comparing siCon and siRAD18, p was <0.01 for all three cell lines, indicating that these cancer cells depend on RAD18 for MK-1775 tolerance. All data shown are from representative experiments that were repeated at least three times and yielded similar results on each occasion. (D) Cultures of isogenic Polk+/+ and Polk−/− MEFs were infected with lentiviruses encoding nontargeting control guide RNA or sgRNA targeting the murine Brca1 gene. After lentiviral transduction, pools of puromycin-resistant cells were treated with different concentrations of MK-1775, and sensitivity to Wee1 inhibition was evaluated by clonogenic survival assays. For each experiment, the number of surviving colonies from MK-1775–treated cultures was expressed as a percentage of colony number from cells that received vehicle (DMSO) for control. On the survival curves, each data point represents the mean of triplicate determinations, and error bars represent the range. For each dose of MK-1775, we performed ANOVA between groups followed by Tukey’s multiple comparison of means test. From results obtained with 0.2 µM MK-1775, either Polk or Brca1 deficiency led to MK-1775 sensitivity (P = 0.006 for both genotypes when compared with Polk- and Brca1-replete controls), but there was no significant difference in the MK-1775 sensitivity of Polk- and Brca1-deficient cells (P = 0.06). Combined loss of Polk and Brca1 led to increased MK-1775 sensitivity when compared with individual deficiencies in Polk−/− or sgBrca1 cells (P = 0.001), indicating that Brca1 and Polk are nonepistatic and have redundant roles in MK-1775 tolerance. The graph shown is from a single experiment that was repeated three times with similar results on each occasion. **, P < 0.01.
Sensitivity to replicative stress is likely determined by the repertoire of available DNA damage tolerance pathways. Therefore, we sought to identify the DNA repair mechanisms that cooperate with RAD18 to confer tolerance of MK-1775–induced stress. TLS deficiency predisposes to DSB formation (Fig. 5 D). Therefore, we determined the contributions of the two major S phase–coupled DSB repair processes (homologous recombination [HR] and θ-mediated end joining [TMEJ]) to MK-1775 tolerance in Polk-deficient cells. We used lentivirally expressed single guide RNAs (sgRNAs) to functionally inactivate Brca1 or Polq genes (encoding essential mediators of HR and TMEJ, respectively) in Polk+/+ or Polk−/− MEFs. As shown in Fig. 7 D, disruption of Brca1 led to increased MK-1775 sensitivity of both Polk+/+ and Polk−/− MEFs, indicating that the HR and TLS jointly (and independently) facilitate tolerance MK-1775–induced DNA damage. As shown in Fig. S4, Polq-deficient MEFs were MK-1775 sensitive, also indicating a role for TMEJ in tolerance of CDK2-induced DSBs. However, inactivating the Polq gene did not increase the MK-1775 sensitivity of Rad18−/− or Polk−/− MEFs (Fig. S4). Collectively, these results indicate that there is partial redundancy between TLS and HR (but not between TLS and TMEJ) for MK-1775 tolerance. These findings validate RAD18-Polκ as a therapeutic target pathway for augmenting MK-1775–induced lethality in cancer cells, particularly those with HR defects.
Discussion
It is widely accepted that diverse oncogenes induce DNA replicative stresses that in turn activate canonical DNA damage signaling and repair pathways (Halazonetis et al., 2008). Many bona fide oncogenes including RAS induce CDK2 activity (Fikaris et al., 2006). The high-level CDK2 activity of oncogene-expressing cells is a likely source of DNA replication stress. We show in this study that three independent CDK2-activating stimuli (oncogenic RAS, Cyclin E, and WEE1 inhibition) induce a DNA damage response that involves TLS pathway activation. Interestingly, Soria et al. (2006) previously showed that p21–CDK2 interactions suppress PCNA monoubiquitination and speculated that CDK activity may promote TLS—a prediction borne out by our results.
The precise nature of the aberrant DNA replication structures induced by oncogenic stimuli or excess CDK activity is not well understood. Rereplication resulting in initiation of DNA synthesis more than once per S phase may be a major mechanism of oncogene-induced DNA replication stress and DNA damage (Bartkova et al., 2006; Di Micco et al., 2006). Rereplication induces ssDNA gaps and replication fork reversal (Neelsen et al., 2013b) that eventually lead to DSB and checkpoint activation (Vaziri et al., 2003). The ssDNA generated by rereplicating DNA provides a potential mechanism for activation of both CHK1 and RAD18 pathways. However, CDT1 overexpression or geminin depletion—treatments that induce massive relicensing and rereplication (Vaziri et al., 2003)—triggered modest PCNA ubiquitination when compared with Cyclin E overexpression or MK-1775 treatment. Therefore, rereplication intermediates may not be the sole mediators of CDK2-induced TLS pathway activation.
Regardless of the proximal DNA replication structures that give rise to TLS pathway activation, ssDNA is clearly generated in response to aberrant CDK2 activity (Neelsen et al., 2013a) and most likely initiates TLS pathway activation. In many of our experiments, CDK2 led to increased chromatin loading of PCNA (in addition to PCNA monoubiquitination). Similarly, both PCNA and TLS polymerases are recruited to oxidative damage-induced ssDNA (Yang et al., 2013). It is established that TLS can remediate post-replicative ssDNA gaps including ssDNA arising outside of S phase (Daigaku et al., 2010; Zlatanou et al., 2011; Yang et al., 2013). Genomes containing persistent ssDNA are vulnerable to nucleolytic attack and are likely to generate lethal DSBs. Collectively, our results indicate that RAD18-mediated repair synthesis prevents accumulation of CDK2-induced ssDNA gaps in the genome.
Of the Y family DNA polymerases, Polη is the most versatile enzyme and is probably the default TLS polymerase recruited to most stalled DNA replication forks (Watanabe et al., 2004). It is very clear that Polη resides constitutively in replication factories of unperturbed cells and facilitates tolerance of intrinsic replication stresses at fragile sites, telomeres, and probably elsewhere in the genome (Bétous et al., 2009; Bergoglio et al., 2013; Garcia-Exposito et al., 2016). It is unexpected therefore that Polκ but not Polη is specifically required for tolerance of CDK-induced replication stress. Moreover, it is surprising that Polη inhibits MK-1775 tolerance. When compared with other TLS polymerases, Polη has a high affinity for PCNA conferred by a PLTH motif immediately flanking the C-terminal PCNA-interacting peptide box motif (Hishiki et al., 2009; Durando et al., 2013; Despras et al., 2016). We speculate that the increased MK-1775 tolerance of Polη-compromised cells is caused by relief of competition with Polκ for access to replicating DNA. Regardless of whether Polη competes with Polκ for CDK2-induced replication intermediates, our results suggest a novel tumor-suppressive role for Polη in sensitizing cells to oncogenic DNA replication stress. Conversely, Polκ supports viability in the face of oncogenic stress and may facilitate tumorigenesis.
What, then, is the basis for Polκ dependency of MK-1775 tolerance, and what are the putative DNA substrates that are preferentially replicated by Polκ? Persistent ssDNA is known to generate G4 structures (Cea et al., 2015), and TLS polymerases are implicated in the bypass of quadruplex DNA (Bétous et al., 2009). Polκ preferentially binds G4 when compared with non-G4 DNA, and Polκ activity is enhanced when the enzyme is within two to three nucleotides of a G4 motif (Eddy et al., 2016). However, Polk−/− MEFs did not show increased sensitivity to the G4 stabilizer pyridostatin (PDS) when compared with Polk+/+ cells (Fig. S5). Therefore, reduced tolerance of quadruplex DNA cannot explain the MK-1775 sensitivity of Polk−/− cells. Interestingly, Rad18−/− cells were highly PDS sensitive, indicating that a Rad18 effector other than Polκ is necessary for G4 tolerance. Collectively, our results along with several recent studies (Despras et al., 2016; Garcia-Exposito et al., 2016) show that the different TLS pols have separable roles in processing distinct forms of intrinsically arising DNA replication stress.
Whether TLS polymerases evolved solely to cope with bulky chemically induced DNA lesions has been a subject of extensive debate. In this regard, we note that the TLS-defective (Rad18 and Polk mutant) cell lines used in this study are only modestly sensitive to UV light, and other bulky lesion-inducing agents yet show remarkable hypersensitivity to MK-1775. Our results are consistent with the view that TLS serves primarily to respond to replicative stresses such as those induced by CDK2.
CDK2-induced oncogenic stress is often interchangeably modeled in cultured cells using overexpressed Cyclin E or MK-1775 treatment (Sogo et al., 2002; Beck et al., 2010, 2012; Neelsen et al., 2013a). However, these experimental approaches differ considerably in that MK-1775 de-represses both CDK2 and CDK1 (which promote S phase and G2/M progression, respectively), whereas Cyclin E activates only CDK2. Our studies with WEE1 inhibition and CDC25C overexpression reveal a requirement for an intact G2/M restriction point to repress phenotypic defects (such as persistent ssDNA in mitosis and PCC) that arise if TLS fails. Historically, the UV sensitivities of some XPV cell lines were only evident after caffeine treatment (Maher et al., 1976). Based on our results, the requirement for caffeine to reveal DNA damage sensitivity is most likely caused by inhibition of the ATR–CHK1 pathway and override of the G2/M replication checkpoint. Our results reveal extensive interactions between TLS and multiple cell cycle and genome maintenance pathways: We demonstrate that p53 enforces G1 arrest and suppresses TLS activation in cells experiencing CDK2-induced replication stress. The G2/M replication checkpoint represses mitotic entry of TLS-defective cells containing underreplicated DNA (Fig. 8). Finally, our analysis of interactions between Rad18/Polk and DSB repair genes (Polq and Brca1) indicates that there is partial redundancy between TLS and HR for tolerance of CDK2 activity. Thus, TLS is a single component of a genome maintenance network comprising multiple pathways that collectively determine oncogenic stress tolerance.
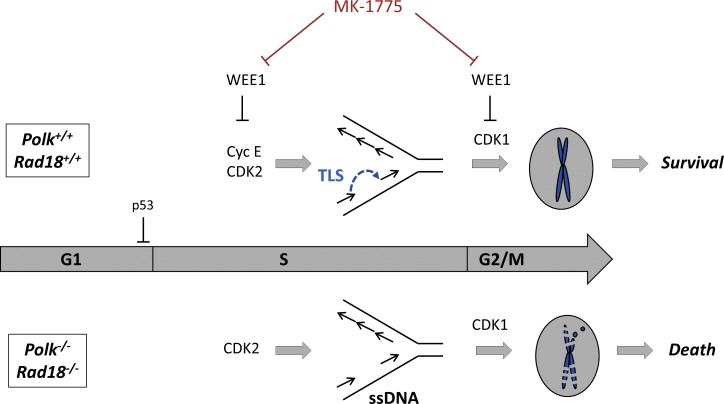
Figure 8.
Roles of Rad18 and Polκ in tolerance of CDK2-induced DNA replication stress. MK-1775 treatment de-represses CDK2 and CDK1 activities, which induce DNA replication stress and aberrant G2/M progression, respectively. Rad18 and Polκ prevent accumulation of post-replicative ssDNA gaps arising from aberrant CDK2 activity. In the absence of Rad18 or Polκ, CDK2-induced ssDNA persists into mitosis (because of aberrant CDK1 activity) and triggers PCC.
Current paradigms suggest that diverse oncogenic stresses must be tolerated for neoplastic cells to bypass tumor-suppressive barriers (Halazonetis et al., 2008). Our results suggest that RAD18/Polκ-mediated TLS could endow neoplastic cells with two important tumorigenic phenotypes: DNA damage/replication stress tolerance and error-prone (mutagenic) DNA synthesis. Therefore, tumorigenesis may impose selective pressure for TLS proficiency. It is reported that Polκ is up-regulated in tumors (Bavoux et al., 2005), and our previous identification of a cancer cell–specific RAD18 activation mechanism (Gao et al., 2016a,b) is also consistent with a role for TLS as a carcinogenic driver. Other DNA repair pathways are also elevated in some cancers. For example, POLQ (which we show in this study can confer tolerance of excess CDK2 activity) is up-regulated in BRCA1 mutant breast and ovarian cancers (Ceccaldi et al., 2015). It will eventually be important to systematically define the roles of Rad18, Polk, and Polq in carcinogenesis in response to defined oncogenic drivers using mouse models.
An important byproduct of the selective pressure for DNA damage tolerance during tumorigenesis is the emergence of chemoresistant cancer cells. If cancer cells are commonly dependent on pathological DNA repair (via up-regulated TLS, TMEJ, or other mechanisms), those genome maintenance mechanisms represent molecular vulnerabilities that could be harnessed to sensitize cells to intrinsic or therapy-induced DNA damage. WEE1 inhibition is currently being considered as a therapeutic strategy in cancer (Sakurikar et al., 2016). Our data show that the anticancer activity of MK-1775 and other WEE1 inhibitors could be improved with concurrent inhibition of the TLS pathway. In particular, our results suggest that BRCA1-deficient cells will be highly MK-1775 sensitive when TLS is inhibited.
Materials and methods
Cell culture and transfection
hTERT-expressing NHFs were provided by W. Kaufmann (University of North Carolina at Chapel Hill, Chapel Hill, NC). hTERT-expressing NHFs are diploid untransformed cells with no known mutations in oncogenes or tumor suppressors. Primary MEFs were derived from E13.5 embryos of WT, Rad18−/−, Polk−/−, HA-RAD18-WT knock-in, and HA-RAD18 (DC2) knock-in C57/BL6 mice as previously described (Bi et al., 2005; Yang et al., 2016). Polq−/− and isogenic Polq+/+ MEFs were provided by R. Wood (The University of Texas, Smithville, TX). With the exception of the engineered mutations in DNA repair genes, the MEFs used in this study are not known to harbor additional genetic lesions. Cancer cell lines H1299, A549, U2OS, and 293T were purchased from ATCC and used for the described experiments without further authentication. H1299 is a nonsmall cell lung carcinoma–adherent epithelial cell line that lacks p53 protein. A549 is a nonsmall cell lung carcinoma–adherent epithelial cell line which expresses WT p53 protein. U2OS is an osteosarcoma cell line with adherent epithelial morphology that is intact for p53 and RB signaling. The 293T cell line was derived from the embryonic kidney and was transformed by sequential stable expression of the adenovirus E1A and E1B genes and SV40 large T antigen. NHFs harboring a doxycycline-inducible human Cyclin E cDNA were generated using the pINDUCER20 lentiviral vector (Meerbrey et al., 2011). XP30RO XPV and XP30RO + Polη fibroblast cells were provided by G. Stewart (University of Birmingham, Birmingham, England, UK). XP30RO cells are UV sensitive, owing to homozygous deletion of the 5′ region of the POLH gene, leading to expression of truncated and inactive Polη protein (Volpe and Cleaver, 1995). With the exception of mutations in the POLH gene, XP30RO cells are not known to harbor any genetic lesions. The XP30RO+ Polη cell line is a corrected derivative of XP03O cells that stably expresses WT Polh and displays normal UV sensitivity (Kannouche et al., 2001). All mouse and human fibroblasts and cancer cell lines were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. XP30RO and XP30RO+ Polη cells were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin. All cell lines tested negative for mycoplasma contamination as determined using the Universal Mycoplasma Detection kit (301012K; ATCC). Plasmid DNA and siRNA oligonucleotides were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions except that plasmid DNA and Lipofectamine 2000 concentrations used in each transfection reaction were decreased by 50% to reduce toxicity.
Adenovirus construction and infection
The Tet-regulated Cyclin B adenovirus was constructed as previously described by Jin et al. (1998). All other adenoviruses were constructed and purified as described previously (Vaziri et al., 2003). In brief, cDNAs encoding various cell cycle regulators (including cyclins, oncogenes, and DNA replication and repair factors) were subcloned into the pACCMV shuttle vector. The resulting shuttle vectors were cotransfected with the pJM17 adenovirus plasmid into 293T cells. Recombinant adenovirus clones were isolated by plaque purification and verified by restriction analysis and Southern blotting. The empty vector AdCon (used to control for adenovirus infections) was derived similarly but by cotransfection of the parental pACCMV shuttle vector with pJM17. Adenovirus particles were purified from 293T cell lysates by polyethylene glycol precipitation, CsCl gradient centrifugation, and gel filtration column chromatography. Adenovirus preparations were quantified by A260 measurements. Cells were typically infected with 0.1−1.0 × 1010 pfu/ml (as indicated in Figs. 1, 2, 3, and 4) by direct addition of purified virus to the culture medium.
RNA interference
For cancer cell lines, siRNAs were reverse transfected using Lipofectamine 2000. In brief, siRNAs were incubated with Lipofectamine 2000 and serum-free OptiMEM for 15 min at room temperature in the dark. Cells were then trypsinized and resuspended in 1 ml of OptiMEM and added directly into the siRNA/OptiMEM/Lipofectamine solution to give a plating density of 50%, and then they were incubated for 48 h. For NHFs and mouse fibroblasts, siRNAs were transfected using electroporation with a GenePulser Xcell (Bio-Rad Laboratories). Electroporation was performed according to the manufacturer’s instructions. 200 µl PBS containing 107 cells and 5 µM siRNA was electroporated in a 0.2-cm cuvette using a 150 V, 10 ms, and 1 pulse for NHFs and using the preset 3T3 program (160 V and 500 µF) for mouse fibroblasts. Sequences of custom siRNA oligonucleotides used in this study are as follows: control nontargeting siRNA, 5′-UAGCGACUAAACACAUCAA-3′; RAD18, 5′-GAGCAUGGAUUAUCUAUUCAAUU-3′; RNF8, 5′-GAGAAGCUUACAGAUGUUU-3′; RPA32, 5′-GGCTCCAACCAACATTGTT-3′; NBS1, 5′-GUACGUUGUUGGAAGGAAA-3′; Chk1, 5′-GCGUGCCGUAGACUGUCCA-3′; Polκ, 5′-GUAAAGAGGUUAAGGAAA-3′; Polη, 5′-GCAGAAAGGCAGAAAGUUA-3′; Cdc7, 5′-GCUCAGCAGGAAAGGAGUUdTdT-3′; and Cdc6, 5′-ACAAUUAAGUCUCCUAGCA-3′. siCDK1 was the ON-TARGETplus SMARTpool siRNA from GE Healthcare.
IP and immunoblotting
To prepare extracts containing soluble and chromatin-associated proteins, monolayers of cultured cells typically in 10-cm plates were washed twice in ice-cold PBS and lysed in 500 µl of ice-cold cytoskeleton buffer (CSK buffer; 10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF, and 0.1% Triton X-100) freshly supplemented with protease inhibitor cocktail and PhosSTOP (Roche). Lysates were centrifuged at 4,000 rpm for 4 min to remove the CSK-insoluble nuclei. The detergent-insoluble nuclear fractions were washed once with 1 ml of CSK buffer and then resuspended in a minimal volume of CSK before analysis by SDS-PAGE and immunoblotting. For all IP experiments, input samples were sonicated and normalized for protein concentration. Magnetic beads containing covalently conjugated antibodies against the HA tag (Cell Signaling Technology) were added to the extracts, and incubations were performed for 30 min at 4°C using rotating racks. Immune complexes were recovered using magnetic stands. The beads were washed five times with 1 ml CSK (1 min per wash) to remove nonspecifically associated proteins. The washed immune complexes were boiled in protein loading buffer for 10 min to release and denature for SDS-PAGE.
For immunoblotting, cell extracts or immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose membranes, and incubated overnight with the following primary antibodies: PCNA (sc-56), Chk1 (sc-7898), β-actin (sc-130656), cyclin E (sc-198), GAPDH (sc-32233), Rnf8 (SC-134492), Cdc6 (SC-9964), and GFP (SC-9996) from Santa Cruz Biotechnology, Inc.; Cdt1 (A300-786A), pRPA32 S4/8 (A300-245A), Polη (A301-231A), Polι (A301-304A), Polκ (A301-977A), and RAD18 (A301-340A) from Bethyl Laboratories, Inc.; p42 MAPK (9107), p-Cdc2 Y15 (9111), p-Chk1 S317 (2344), and p-Chk2 (2661) antibodies were purchased from Cell Signaling Technology; γH2AX (05-636) and RPA34/RPA32 (NA19L) were from EMD Millipore; CDC7 (DCS-342) was from MEDICAL & BIOLOGICAL LABORATORIES CO.; and Cdc45 rat monoclonal antibody was as previously described (Liu et al., 2006). Antibody dilutions used for immunoblotting were 1:1,000 with exceptions for the following antibodies: PCNA (1:500), β-actin (1:5,000), GAPDH (1:5,000), and γH2AX (1:10,000).
CDK2 activity assays
CDK2 activities were determined by IP–kinase assays as described by Rosenblatt et al. (1992). In brief, cells were lysed in CSK as described in the IP and immunoblotting section. Lysates were clarified by centrifugation (10,000 g for 10 min), normalized for protein content (typically ∼500 µg protein in 0.5 ml), and immunoprecipitated with anti-CDK2 antibody (sc-163) or control IgG using magnetic beads as described in Fig. 1 C. After the final CSK wash, beads were washed in 50 mM Hepes, pH 7.4, and 1 mM dithiothreitol and then combined with a 30-µl reaction mix containing 10 µg of histone H1 (Sigma-Aldrich), 50 µM ATP, 10 mM MgCl2, and 2.5 µCi of [γ-32P]ATP (3,000 Ci/mmol; NEN). After a 15-min incubation at room temperature, reaction mixtures were boiled in Laemmli sample buffer and then resolved by SDS-PAGE using 12% gels. After electrophoresis, gels were fixed in 40% methanol/10% acetic acid containing 0.05% Coomassie blue dye and then were washed extensively in 40% methanol/10% acetic acid to remove unincorporated radioisotope. Phosphorylated histone bands were visualized by autoradiography of the dried gels.
COMET assay
Relative levels of DNA strand breaks were measured by a single-cell gel electrophoresis assay (Olive et al., 1990) using a commercially available COMET Assay kit (Trevigen). For COMET assays, NHF cells were plated in six-well dishes at a density of 0.5 × 105 cells per well. Oncogenes (Ha-RAS, MYC, Cyclin E, and CDT1) were delivered to cells by infection with 1010 pfu/ml of purified adenoviral vectors. After 24 h, cells were embedded in agarose and subjected to electrophoresis according to the manufacturer’s instructions. Cells were visualized using an IX61 inverted microscope (Olympus), and images of cells were acquired as TIFF files. For each experimental condition, “tail moments” (defined as the product of tail length and the fraction of total DNA in the tail) were determined for 50 nuclei using ImageJ software (National Institutes of Health) with the COMET assay plugins (original macro from Herbert M. Geller, added by Robert Bagnell). To compare tail moments between experimental groups, we performed ANOVAs followed by Tukey’s tests to correct for experiment-wide error rates between multiple comparisons.
Analysis of DNA replication dynamics using DNA fiber assays
Growing cells were pulse labeled for 20 min with 25 µM of the thymidine analogue chlorodeoxyuridine (CIdU; C6891; Sigma-Aldrich). At the end of the CldU-labeling period, cells were washed twice with a warm medium and immediately pulse labeled for 20 min with 250 µM of idodeoxyuridine (IdU; I7125; Sigma-Aldrich). Labeled cells were harvested and DNA fiber spreads were prepared and stained exactly as described previously by Jones et al. (2013). Images of stained DNA fibers were acquired on a 710 confocal microscope (UNC Microscopy Services Laboratory; ZEISS) using a 20× lens. The lengths of CldU (AF 555; red)- and IdU (AF 488; green)-labeled fibers were measured using the ImageJ software, and micrometer values were converted into kilobases using the conversion factor 1 µM = 2.94 kb (one bp corresponds to ∼340 pm). 50 representative DNA fibers were measured for each experimental condition. Prism 6 (GraphPad Software) was used for variation analysis and to generate dot plots. Fiber distribution figures were generated using the RColorBrewer package in R.
Clonogenic survival assays
For experiments in MEFs, cells were seeded at a density of 1,000 cells/well in triplicate in six-well dishes. The WEE1 inhibitor MK-1775 was diluted in growth medium and added directly to the cells for 24 h before switching to fresh medium. Growth medium was replenished every 3 d. Colonies containing ∼50 surviving cells were stained with 0.05% crystal violet in 1× PBS containing 1% methanol and 1% formaldehyde. The ImageJ plugin ColonyArea (Guzmán et al., 2014) was used to automatically quantify stained colonies. In some experiments, lentivirally expressed sgRNAs were used to functionally inactivate PolQ and Brca1 genes exactly as described previously (Wyatt et al., 2016). In brief, MEFs were infected with lentivirus encoding Cas9 and either control nontargeting guides or sgRNAs targeting mouse PolQ and Brca1 genes. Lentivirus-infected cells were selected in puromycin-containing medium for 3 d. Puromycin-resistant cells were seeded at a density of 1,000 cells/well in triplicate six-well dishes, treated with MK-1775, and analyzed for clonogenic survival exactly as described in Figs. 7 D and S4. The PolQ sgRNA was validated as previously described (Wyatt et al., 2016). To validate the gene editing experiments, quantitative PCR was used to measure Brca1 mRNA levels in puromycin-selected cells. The quantitative PCR primer sequences used in these experiments were 5′-CTGCCGTCCAAATTCAAGAAGT-3′ and 5′-CTTGTGCTTCCCTGTAGGCT-3′ (corresponding with Brca1 forward and reverse primers, respectively). As a positive control for the quantitative PCR reactions, β-actin mRNA was amplified using the primers 5′-TTCTTTGCAGCTCCTTCGTTGCCG-3′ (forward) and 5′-TGGATGGCTACGTACATGGCTGGG-3′ (reverse). In three independent experiments, Brca1 mRNA was reduced by 54 ± 4% in sgBrca1-transduced MEFs when compared with controls. After lentiviral transduction, pools of puromycin-resistant cells were seeded in six-well plates and treated with different concentrations of MK-1775. Cell sensitivity to Wee1 inhibition was evaluated by clonogenic survival assay. For experiments in the cancer cell lines, siRNA transfections were performed as described in the RNA interference section. 24 h after transfection, cells were seeded at a density of 1,000 cells/well in triplicate six-well dishes. MK-1775 treatments and colony-forming assays were performed exactly as described above.
Flow cytometry
To detect ssDNA, growing cells were first cultured for 24 h in medium containing 10 µM BrdU to label genomic DNA. Cell monolayers were washed to remove unincorporated BrdU, placed in fresh BrdU-free medium, and returned to the incubator. After treatment with genotoxins or oncogenes as described in all figure legends, cells were harvested by trypsinization, washed in PBS, resuspended in 65% PBS with 35% ethanol, and fixed by overnight incubation at 4°C. Fixed cells were stained with fluorescent anti-BrdU antibodies (FITC mouse anti-BrdU kit; 556028; BD) without prior acid treatment to detect only ssDNA. To confirm that the experimental cells had incorporated BrdU, aliquots of all nuclei were also denatured using HCl and then neutralized with borax before staining with anti-BrdU antibody. For cell cycle analysis, nuclei were incubated in 1× PBS containing 10 µg/ml of propidium iodide and 1 µg/ml of RNaseA to stain total DNA. Stained cells were analyzed by flow cytometry on an Accuri C6 flow cytometer (BD) using the manufacturer’s software.
Fluorescence microscopy
To visualize nuclear foci containing TLS proteins, U2OS cells were grown to ∼50% confluency on glass-bottomed plates (MatTek Corporation) then infected with AdYFP-Polη (5 × 108 pfu/ml), AdGFP-Polκ (109 pfu/ml), AdCFP-RAD18 (4 × 109) pfu/ml, or equivalent concentrations of an “empty” control adenovirus (AdCon). To induce replication stress by Cyclin E overexpression, cells were coinfected with 109 pfu/ml of AdCyclin E adenovirus and fixed 24 h after infection. To induce replication stress with WEE1 inhibition, cells were treated with 0.25 µM MK-1775 18 h after viral infection and then fixed 24 h after infection. At the time of harvesting, cells were washed three times with PBS and then extracted for 5 min in cold CSK buffer and fixed for 10 min in 2% PFA in PBS. For PCNA staining, cells were fixed with cold methanol for 20 min at −20°C instead of PFA. For experiments not requiring antibody staining, cells were covered with VECTASHIELD Antifade Mounting Medium containing DAPI (Vector Laboratories) and imaged within 2 h. For antibody staining, cells were blocked in 3% BSA + 5% normal donkey serum in PBS for 1 h and then incubated in primary antibody for 1 h at room temperature. Cells were washed three times with PBS and then incubated with secondary antibody (1:300) for 1 h at room temperature. After three washes with PBS, cells were covered with Vectashield Solution and imaged within 2 h. Antibodies used for fluorescence microscopy in this study were: mouse-RPA32 (1:200; ab2), mouse-G4 (1:200; 1H6), and rabbit anti–phosphor-H3 (1:400; 06-570) from EMD Millipore, and rabbit-53BP1 (1:300; sc-22760) and mouse PCNA (1:500; sc-56) from Santa Cruz Biotechnology, Inc. Fixed-cell imaging was performed in the University of North Carolina Microscopy Services Laboratory core facility using an LSM700 confocal laser scanning microscope (ZEISS) and the following objective lenses: Plan Apochromat 20×/0.80 differential interference contrast II/0.8 NA, Plan-NEOFLUAR 40×/1.30 oil/1.3 NA, and Plan Apochromat 63×/1.4 oil differential interference contrast/1.4 NA. Imaging was performed at 25°C. Flourochromes used were DAPI, Alexa Fluor 488, and Alexa Fluor 647. Images were acquired directly using ZEN 2011 Acquisition software (ZEISS). Images were analyzed using ImageJ and saved as TIFF files with no further adjustment.
Mitotic spreads
Mitotic spreads were prepared and analyzed for PCC assays as described previously (Nghiem et al., 2001). After a 6-h incubation with 0.25 µM MK-1775 (or DMSO for controls), cells were harvested with trypsin and collected by centrifugation at 300 g for 10 min. Cell pellets were resuspended by gentle pipetting in 10 ml of 75 mM KCl, and the resulting suspensions were incubated for 10 min at room temperature. Cells were then collected by centrifugation (300 g for 10 min) and resuspended in 5 ml of freshly prepared Carnoy’s fixative (three parts methanol and one part glacial acetic acid) and incubated for 10 min at room temperature. Fixed cells were collected by centrifugation (300 g and 10 min) and resuspended in 200 µl of Carnoy’s fixative. 10 µl of each cell suspension was dropped from a height of 10 cm onto a glass slide and allowed to dry. 15 µl of DAPI solution (Vectashield antifade mounting medium with DAPI fluorochrome) was spotted onto each slide. Coverslips were gently placed above the DAPI droplet, and edges were sealed with clear nail polish. A BX61 upright widefield fluorescence microscope (Olympus) was used to identify and count mitotic cells that had characteristic features of either a normal mitosis or PCC. Imaging was performed at 25°C using a Plan Apochromat N 60×/1.42 oil UIS 2 BFP1 objective lens with an NA of 1.42. Images were acquired using an ORCA RC camera (Hamamatsu Photonics) and Improvision’s Velocity software. Subsequent to acquisition, images were analyzed with ImageJ and saved as TIFF files with no additional adjustment. 200 metaphase cells were scored for each independent experiment, and percentages of PCC were calculated using compiled data from three separate experiments.
Statistics
Statistical analyses were performed using Prism 6. The unpaired two-tailed Student’s t test was used to compare two datasets in various experiments as indicated in the figure legends. In some experiments in which more than two datasets were compared, we performed ANOVA with post hoc Tukey’s honest significance difference test. On graphs and charts, asterisks indicate p-values: *, P < 0.05; **, P < 0.01; ***, P < 0.001. P > 0.05 indicates a difference that is not significant.
Online supplemental material
Fig. S1 is related to Fig. 1 and tests the differential effects of various CDK2 and CDK1 activators (including Cyclins E, A, B, and CDC25C) on PCNA monoubiquitination. Fig. S2 shows validation of the flow cytometry–based ssDNA assays used in Figs. 3 and 5. Fig. S3 shows that bypass of the G2/M checkpoint by overexpressed CDC25C promotes the accumulation of mitotic cells harboring persistent ssDNA. Fig. S4 shows that Polq-deficient cells lacking Polθ (the key mediator of the alternative end-joining DSB repair pathway) are sensitive to MK-1775. Fig. S5 shows that Rad18 and Polkκ have separable roles in tolerance of the G4 quadruplex DNA-stabilizing agent PDS.
Supplementary Material
Acknowledgments
We thank Dr. Tovah Day for helpful discussions and advice throughout the study. We are grateful to Dr. Wen Huang for providing the R script used to generate the DNA fiber plots. We thank Dr. Rick Wood for providing Polq+/+ and Polq−/− MEFs.
This study was supported by the National Institutes of Health grant R01 ES09558 to C. Vaziri, a University of North Carolina Institute of Clinical and Translational Sciences Clinical and Translational Science Award UL1TR001111 to Y. Yang, and a University of North Carolina Lineberger Comprehensive Cancer Center postdoctoral award to Y. Gao. Microscopy was performed at the Microscopy Services Laboratory at the University of North Carolina, Chapel Hill, which is supported in part by the Cancer Center Core Support grant P30 CA016086 to the UNC Lineberger Comprehensive Cancer Center.
The authors declare no competing financial interests.
Author contributions: all of the authors designed and discussed experiments. Y. Yang, Y. Gao, M. Durando, W. Ding, and D. Wyatt conducted the experimental work. C. Vaziri conceived the project, supervised experiments, provided funding, and wrote the manuscript. All authors helped with editing.
Footnotes
Abbreviations used:
- DSB
- double-strand break
- HR
- homologous recombination
- IP
- immunoprecipitation
- MEF
- mouse embryonic fibroblast
- NHF
- normal human fibroblast
- OIS
- oncogene-induced senescence
- PCC
- premature chromatin condensation
- PCNA
- proliferating cell nuclear antigen
- PDS
- pyridostatin
- ROS
- reactive oxygen species
- sgRNA
- single guide RNA
- ssDNA
- single-stranded DNA
- TLS
- trans-lesion synthesis
- TMEJ
- θ-mediated end joining
- XPV
- XP-variant
References
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637. 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Bavoux C., Leopoldino A.M., Bergoglio V., O-Wang J., Ogi T., Bieth A., Judde J.-G., Pena S.D.J., Poupon M.-F., Helleday T., et al. 2005. Up-regulation of the error-prone DNA polymerase κ promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res. 65:325–330. [PubMed] [Google Scholar]
- Beck H., Nähse V., Larsen M.S., Groth P., Clancy T., Lees M., Jørgensen M., Helleday T., Syljuåsen R.G., and Sørensen C.S.. 2010. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J. Cell Biol. 188:629–638. 10.1083/jcb.200905059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H., Nähse-Kumpf V., Larsen M.S., O’Hanlon K.A., Patzke S., Holmberg C., Mejlvang J., Groth A., Nielsen O., Syljuåsen R.G., and Sørensen C.S.. 2012. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32:4226–4236. 10.1128/MCB.00412-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergoglio V., Boyer A.S., Walsh E., Naim V., Legube G., Lee M.Y., Rey L., Rosselli F., Cazaux C., Eckert K.A., and Hoffmann J.S.. 2013. DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J. Cell Biol. 201:395–408. 10.1083/jcb.201207066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., and Kerem B.. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 145:435–446. 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Rey L., Wang G., Pillaire M.J., Puget N., Selves J., Biard D.S., Shin-ya K., Vasquez K.M., Cazaux C., and Hoffmann J.S.. 2009. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol. Carcinog. 48:369–378. 10.1002/mc.20509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Slater D.M., Ohmori H., and Vaziri C.. 2005. DNA polymerase κ is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 280:22343–22355. 10.1074/jbc.M501562200 [DOI] [PubMed] [Google Scholar]
- Bi X., Barkley L.R., Slater D.M., Tateishi S., Yamaizumi M., Ohmori H., and Vaziri C.. 2006. Rad18 regulates DNA polymerase κ and is required for recovery from S-phase checkpoint-mediated arrest. Mol. Cell. Biol. 26:3527–3540. 10.1128/MCB.26.9.3527-3540.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 310:1821–1824. 10.1126/science.1120615 [DOI] [PubMed] [Google Scholar]
- Canman C.E. 2001. Replication checkpoint: Preventing mitotic catastrophe. Curr. Biol. 11:R121–R124. 10.1016/S0960-9822(01)00057-4 [DOI] [PubMed] [Google Scholar]
- Cea V., Cipolla L., and Sabbioneda S.. 2015. Replication of structured DNA and its implication in epigenetic stability. Front. Genet. 6:209 10.3389/fgene.2015.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Liu J.C., Amunugama R., Hajdu I., Primack B., Petalcorin M.I., O’Connor K.W., Konstantinopoulos P.A., Elledge S.J., Boulton S.J., et al. 2015. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 518:258–262. 10.1038/nature14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L., Sotiriou S.K., Rantala J.K., Magin S., Mladenov E., Helleday T., Haber J.E., Iliakis G., Kallioniemi O.P., and Halazonetis T.D.. 2014. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 343:88–91. 10.1126/science.1243211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y., Davies A.A., and Ulrich H.D.. 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 465:951–955. 10.1038/nature09097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I.F., Li A., and Blow J.J.. 2006. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol. Cell. 24:433–443. 10.1016/j.molcel.2006.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A.A., Huttner D., Daigaku Y., Chen S., and Ulrich H.D.. 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell. 29:625–636. 10.1016/j.molcel.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T.A., Palle K., Barkley L.R., Kakusho N., Zou Y., Tateishi S., Verreault A., Masai H., and Vaziri C.. 2010. Phosphorylated Rad18 directs DNA polymerase η to sites of stalled replication. J. Cell Biol. 191:953–966. 10.1083/jcb.201006043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. 2011. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 475:106–109. 10.1038/nature10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despras E., Sittewelle M., Pouvelle C., Delrieu N., Cordonnier A.M., and Kannouche P.L.. 2016. Rad18-dependent SUMOylation of human specialized DNA polymerase eta is required to prevent under-replicated DNA. Nat. Commun. 7:13326 10.1038/ncomms13326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant N., Hendel A., Vered I., Carell T., Reissner T., de Wind N., Geacinov N., and Livneh Z.. 2012. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 40:170–180. 10.1093/nar/gkr596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P.G., Bensimon A., et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 444:638–642. 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Durando M., Tateishi S., and Vaziri C.. 2013. A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 41:3079–3093. 10.1093/nar/gkt016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S., Tillman M., Maddukuri L., Ketkar A., Zafar M.K., and Eoff R.L.. 2016. Human translesion polymerase κ exhibits enhanced activity and reduced fidelity two nucleotides from G-quadruplex DNA. Biochemistry. 55:5218–5229. 10.1021/acs.biochem.6b00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikaris A.J., Lewis A.E., Abulaiti A., Tsygankova O.M., and Meinkoth J.L.. 2006. Ras triggers ataxia-telangiectasia-mutated and Rad-3-related activation and apoptosis through sustained mitogenic signaling. J. Biol. Chem. 281:34759–34767. 10.1074/jbc.M606737200 [DOI] [PubMed] [Google Scholar]
- Gao Y., Mutter-Rottmayer E., Greenwalt A.M., Goldfarb D., Yan F., Yang Y., Martinez-Chacin R.C., Pearce K.H., Tateishi S., Major M.B., and Vaziri C.. 2016a A neomorphic cancer cell-specific role of MAGE-A4 in trans-lesion synthesis. Nat. Commun. 7:12105 10.1038/ncomms12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Tateishi S., and Vaziri C.. 2016b Pathological trans-lesion synthesis in cancer. Cell Cycle. 15:3005–3006. 10.1080/15384101.2016.1214045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Exposito L., Bournique E., Bergoglio V., Bose A., Barroso-Gonzalez J., Zhang S., Roncaioli J.L., Lee M., Wallace C.T., Watkins S.C., et al. 2016. Proteomic profiling reveals a specific role for translesion DNA polymerase η in the alternative lengthening of telomeres. Cell Reports. 17:1858–1871. 10.1016/j.celrep.2016.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán C., Bagga M., Kaur A., Westermarck J., and Abankwa D.. 2014. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 9:e92444 10.1371/journal.pone.0092444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn W.C., Counter C.M., Lundberg A.S., Beijersbergen R.L., Brooks M.W., and Weinberg R.A.. 1999. Creation of human tumour cells with defined genetic elements. Nature. 400:464–468. 10.1038/22780 [DOI] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., and Bartek J.. 2008. An oncogene-induced DNA damage model for cancer development. Science. 319:1352–1355. 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Hedglin M., and Benkovic S.J.. 2015. Regulation of Rad6/Rad18 activity during DNA damage tolerance. Annu. Rev. Biophys. 44:207–228. 10.1146/annurev-biophys-060414-033841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki A., Hashimoto H., Hanafusa T., Kamei K., Ohashi E., Shimizu T., Ohmori H., and Sato M.. 2009. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 284:10552–10560. 10.1074/jbc.M809745200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huen M.S., Kim H., Leung C.C., Glover J.N., Yu X., and Chen J.. 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11:592–603. 10.1038/ncb1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Hardy S., and Morgan D.O.. 1998. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J. Cell Biol. 141:875–885. 10.1083/jcb.141.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.M., Mortusewicz O., Afzal I., Lorvellec M., García P., Helleday T., and Petermann E.. 2013. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 32:3744–3753. 10.1038/onc.2012.387 [DOI] [PubMed] [Google Scholar]
- Kannouche P., Broughton B.C., Volker M., Hanaoka F., Mullenders L.H., and Lehmann A.R.. 2001. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 15:158–172. 10.1101/gad.187501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.R., Zeman M.K., Chen J.Y., Yee M.C., and Cimprich K.A.. 2011. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol. Cell. 42:237–249. 10.1016/j.molcel.2011.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liontos M., Koutsami M., Sideridou M., Evangelou K., Kletsas D., Levy B., Kotsinas A., Nahum O., Zoumpourlis V., Kouloukoussa M., et al. 2007. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 67:10899–10909. 10.1158/0008-5472.CAN-07-2837 [DOI] [PubMed] [Google Scholar]
- Liu P., Barkley L.R., Day T., Bi X., Slater D.M., Alexandrow M.G., Nasheuer H.P., and Vaziri C.. 2006. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J. Biol. Chem. 281:30631–30644. 10.1074/jbc.M602982200 [DOI] [PubMed] [Google Scholar]
- Lopes M., Foiani M., and Sogo J.M.. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 21:15–27. 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Maher V.M., Ouellette L.M., Curren R.D., and McCormick J.J.. 1976. Caffeine enhancement of the cytotoxic and mutagenic effect of ultraviolet irradiation in a xeroderma pigmentosum variant strain of human cells. Biochem. Biophys. Res. Commun. 71:228–234. 10.1016/0006-291X(76)90272-2 [DOI] [PubMed] [Google Scholar]
- Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., et al. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 108:3665–3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K.J., Zanini I.M., Herrador R., and Lopes M.. 2013a Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 200:699–708. 10.1083/jcb.201212058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K.J., Zanini I.M., Mijic S., Herrador R., Zellweger R., Ray Chaudhuri A., Creavin K.D., Blow J.J., and Lopes M.. 2013b Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 27:2537–2542. 10.1101/gad.226373.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem P., Park P.K., Kim Y., Vaziri C., and Schreiber S.L.. 2001. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. USA. 98:9092–9097. 10.1073/pnas.161281798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive P.L., Banáth J.P., and Durand R.E.. 1990. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat. Res. 122:86–94. 10.2307/3577587 [DOI] [PubMed] [Google Scholar]
- Palle K., and Vaziri C.. 2011. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle. 10:1625–1638. 10.4161/cc.10.10.15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., and Prakash L.. 2005. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 74:317–353. 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Raderschall E., Golub E.I., and Haaf T.. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA. 96:1921–1926. 10.1073/pnas.96.5.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M., Smeenk G., Hansen R.K., Temu T., Oka Y., Hein M.Y., Nagaraj N., Long D.T., Walter J.C., Hofmann K., et al. 2015. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 348:1253671 10.1126/science.1253671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regairaz M., Zhang Y.W., Fu H., Agama K.K., Tata N., Agrawal S., Aladjem M.I., and Pommier Y.. 2011. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195:739–749. 10.1083/jcb.201104003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J., Gu Y., and Morgan D.O.. 1992. Human cyclin-dependent kinase 2 is activated during the S and G2 phases of the cell cycle and associates with cyclin A. Proc. Natl. Acad. Sci. USA. 89:2824–2828. 10.1073/pnas.89.7.2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurikar N., Thompson R., Montano R., and Eastman A.. 2016. A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK-8776 as monotherapy due to CDK2 activation in S phase. Oncotarget. 7:1380–1394. 10.18632/oncotarget.6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppy D.W., Ragland R.L., Gilad O., Shastri N., Peters A.A., Murga M., Fernandez-Capetillo O., Diehl J.A., and Brown E.J.. 2012. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J. Clin. Invest. 122:241–252. 10.1172/JCI58928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., and Foiani M.. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602. 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Sørensen C.S., and Syljuåsen R.G.. 2012. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 40:477–486. 10.1093/nar/gkr697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G., Podhajcer O., Prives C., and Gottifredi V.. 2006. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 25:2829–2838. 10.1038/sj.onc.1209315 [DOI] [PubMed] [Google Scholar]
- Srinivasan S.V., Dominguez-Sola D., Wang L.C., Hyrien O., and Gautier J.. 2013. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Reports. 3:1629–1639. 10.1016/j.celrep.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K., Abbas T., Jazaeri A.A., and Dutta A.. 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell. 37:143–149. 10.1016/j.molcel.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y., Watanabe K., Araki K., Shinohara M., Yamagata Y., Tsurimoto T., Hanaoka F., Yamamura K., Yamaizumi M., and Tateishi S.. 2008. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 13:343–354. 10.1111/j.1365-2443.2008.01176.x [DOI] [PubMed] [Google Scholar]
- Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D.S., and Dutta A.. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell. 11:997–1008. 10.1016/S1097-2765(03)00099-6 [DOI] [PubMed] [Google Scholar]
- Volpe J.P., and Cleaver J.E.. 1995. Xeroderma pigmentosum variant cells are resistant to immortalization. Mutat. Res. 337:111–117. 10.1016/0921-8777(95)00015-C [DOI] [PubMed] [Google Scholar]
- Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., and Yamaizumi M.. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886–3896. 10.1038/sj.emboj.7600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.A., Longerich S., Sung P., Vaziri C., and Kupfer G.M.. 2011. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood. 117:5078–5087. 10.1182/blood-2010-10-311761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., Wood R.D., Gupta G.P., and Ramsden D.A.. 2016. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell. 63:662–673. 10.1016/j.molcel.2016.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara H., Kobayashi J., Tateishi S., Kato A., Matsuura S., Tauchi H., Yamada K., Takezawa J., Sugasawa K., Masutani C., et al. 2011. NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol η-dependent translesion DNA synthesis. Mol. Cell. 43:788–797. 10.1016/j.molcel.2011.07.026 [DOI] [PubMed] [Google Scholar]
- Yang Y., Durando M., Smith-Roe S.L., Sproul C., Greenwalt A.M., Kaufmann W., Oh S., Hendrickson E.A., and Vaziri C.. 2013. Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 41:2296–2312. 10.1093/nar/gks1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Poe J.C., Yang L., Fedoriw A., Desai S., Magnuson T., Li Z., Fedoriw Y., Araki K., Gao Y., et al. 2016. Rad18 confers hematopoietic progenitor cell DNA damage tolerance independently of the Fanconi Anemia pathway in vivo. Nucleic Acids Res. 44:4174–4188. 10.1093/nar/gkw072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanou A., Despras E., Braz-Petta T., Boubakour-Azzouz I., Pouvelle C., Stewart G.S., Nakajima S., Yasui A., Ishchenko A.A., and Kannouche P.L.. 2011. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol η in response to oxidative DNA damage in human cells. Mol. Cell. 43:649–662. 10.1016/j.molcel.2011.06.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.