In this study, the authors show that the MRTF–SRF transcriptional pathway modulates sustained plasma membrane blebbing and entotic cell-in-cell invasion via regulation of the expression of the ERM protein Ezrin.
Abstract
Entosis is a nonapoptotic form of cell death initiated by actomyosin-dependent homotypic cell-in-cell invasion that can be observed in malignant exudates during tumor progression. We previously demonstrated formin-mediated actin dynamics at the rear of the invading cell as well as nonapoptotic plasma membrane (PM) blebbing in this cellular motile process. Although the contractile actin cortex involved in bleb-driven motility is well characterized, a role for transcriptional regulation in this process has not been studied. Here, we explore the impact of the actin-controlled MRTF–SRF (myocardin-related transcription factor–serum response factor) pathway for sustained PM blebbing and entotic invasion. We find that cortical blebbing is tightly coupled to MRTF nuclear shuttling to promote the SRF transcriptional activity required for entosis. Furthermore, PM blebbing triggered SRF-mediated up-regulation of the metastasis-associated ERM protein Ezrin. Notably, Ezrin is sufficient and important to sustain bleb dynamics for cell-in-cell invasion when SRF is suppressed. Our results highlight the critical role of the actin-regulated MRTF transcriptional pathway for bleb-associated invasive motility, such as during entosis.
Introduction
Cell motility is an essential process during development, epithelial differentiation, and immune responses, but also during tumor cell invasion. Cancer cells adopt different modes of invasive motility to adapt to tissue environments and matrix properties by changes in cytoskeletal organization where actin-based protrusions and membrane dynamics provide motile force as well as cell shape changes (Friedl and Wolf, 2010; Nürnberg et al., 2011; Charras and Sahai, 2014). One important mode of cancer cell invasion is represented by bleb-associated motility, which facilitates rapid single-cell locomotion (Sahai, 2005; Fackler and Grosse, 2008). We previously showed that a specialized form of cell-in-cell invasion during entosis involves G protein–coupled receptor signaling and highly dynamic and persistent plasma membrane (PM) blebbing over time periods of several hours (Purvanov et al., 2014).
Entosis is a nonapoptotic cell death process involving cell-in-cell formation and occurring in epithelial cells and human tumors under conditions of low integrin-based adhesions, such as in malignant exudates (Overholtzer et al., 2007; Florey et al., 2015). As a consequence, entosis can result in cancer cell aneuploidy, thereby promoting tumor progression (Krajcovic et al., 2011).
Entotic invasion of neighboring cells requires Rho-actin–dependent signaling of the invading cell involving Rho-associated protein kinase (ROCK) and myosin-based contractility as well as mDial-mediated polarized actin assembly (Overholtzer et al., 2007; Purvanov et al., 2014), which bear similarities regarding cytoskeletal regulation to rounded, bleb-associated cell invasion of cancer cells (Sahai and Marshall, 2003; Kitzing et al., 2007; Lorentzen et al., 2011). However, whether sustained nonapoptotic blebbing during invasive motility over many hours requires transcriptional input has not been addressed.
Here, we investigate the relationship and impact of the actin-binding serum response factor (SRF) transcriptional coregulator myocardin-related transcription factor (MRTF, also known as MAL or MKL1; Olson and Nordheim, 2010) on PM blebbing and entotic invasion. We demonstrate that bleb dynamics and entosis require MRTF–SRF transcriptional activity by up-regulation of Ezrin expression. In turn, cortical blebbing controls MRTF nuclear accumulation for SRF function, thus providing a feedback mechanism for bleb-associated invasive motility.
Results and discussion
SRF is required for dynamic PM blebbing
The transcription factor complex MRTF–SRF controls the expression of genes involved in actin cytoskeleton dynamics regulating cell adhesion and motility (Olson and Nordheim, 2010; Esnault et al., 2014). It has been previously demonstrated that the SRF coactivator MRTF-A plays a critical role in invasion and experimental metastasis (Brandt et al., 2009; Medjkane et al., 2009); however, whether MRTF–SRF transcriptional activity affects PM blebbing or bleb-associated motility is unknown.
To analyze continuous PM blebbing, which promotes events of entotic invasion, we cultured MCF10A cells on poly-HEMA (2-hydroxy-ethyl methacrylate)–coated surfaces, which were previously shown to induce blebbing and entotic invasion (Fig. 1 A; Purvanov et al., 2014), and investigated their behavior when SRF was silenced using siRNA (Fig. 1 B). We first assessed the dynamics of blebbing that is characterized by rapid bleb expansion and followed by a slower phase of bleb retraction involving Ezrin recruitment and actomyosin contractility (Charras et al., 2006; Fackler and Grosse, 2008; Fritzsche et al., 2014).
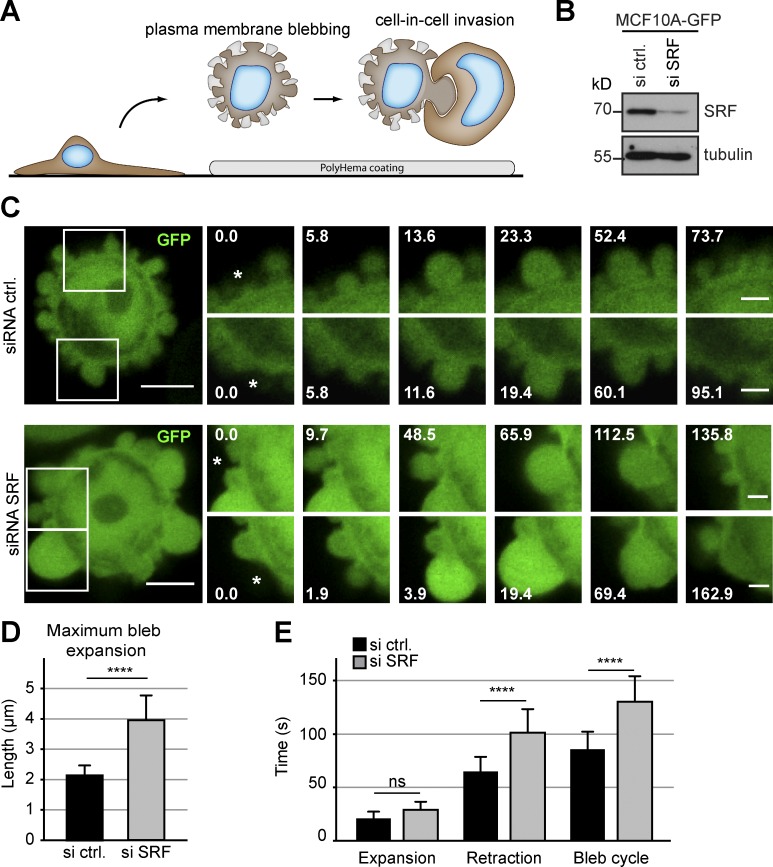
Figure 1.
Silencing of SRF affects PM blebbing. (A) Cartoon illustrating the induction of PM blebbing and entotic invasion by plating cells on poly-HEMA–coated surfaces to prevent cellular attachment. (B) Western blot confirming efficient siRNA-mediated knockdown of SRF in MCF10A cells stably expressing GFP. Tubulin served as a loading control. (C) Live MCF10A cells stably expressing GFP were plated on poly-HEMA and imaged over time to visualize the dynamics of PM blebbing. Cells were treated with either control or siRNA directed against SRF as indicated. Boxes indicate areas that are shown magnified over time to highlight outgrowth and retraction of individual blebs (asterisks). Time is indicated in seconds. Bars: (overview) 5 µm; (magnifications) 2 µm. (D) Quantification of maximum bleb expansion in cells stably expressing GFP. Cells were treated with the indicated siRNAs, and the maximum length reached by individual blebs was compared. (E) MCF10A cells stably expressing GFP were treated with siRNA as indicated to measure the time for expansion and retraction of individual blebs. Note a significant and specific difference in the bleb retraction times resulting in an overall prolonged bleb cycle in SRF-silenced cells. (D and E) ≥60 blebs and 15 cells per condition. Error bars indicate SD. Asterisks indicate statistical significance (****, P ≤ 0.0001). ns indicates no significance (P > 0.05).
RNA interference and live cell imaging of MCF10A cells stably expressing GFP revealed that suppression of SRF profoundly affected blebbing dynamics (Fig. 1 C and Video 1), with maximum bleb expansion size being increased in SRF-depleted cells (Fig. 1 D). Notably, this qualitative change in PM blebbing differs from our previous observations of overall reduced bleb activity in the absence of the actin nucleator mDia1 (Purvanov et al., 2014). Consistently, we did not observe altered expression of mDia1 in cells silenced for SRF (Fig. S1, A and B). Specifically, SRF silencing resulted in significantly prolonged bleb retraction, whereas the time for bleb expansion remained unaffected (Fig. 1 E), indicating a potential transcriptional impact of SRF on the reassembly of a contractile actin cortex during nonapoptotic PM blebbing.
Cortical contractility and blebbing control MRTF-A subcellular localization
Next, we assessed the importance of the SRF coactivator MRTF-A in PM blebbing. MRTF-A is a high-affinity actin-binding protein that requires G-actin interactions with its N-terminal, RPEL motif–containing domain for nuclear export (Vartiainen et al., 2007; Baarlink et al., 2013). In turn, MRTF-A nuclear localization is enhanced upon release of G-actin binding, resulting in SRF-dependent gene expression (Miralles et al., 2003; Mouilleron et al., 2011; Baarlink et al., 2013; Gualdrini et al., 2016). Bleb dynamics and subsequent retraction required actin polymerization at the bleb cortex (Charras et al., 2006), which may be sufficient to alter cellular G-actin levels.
To study directly the behavior of MRTF-A shuttling, we generated and imaged MCF10A cells stably expressing MRTF-A–GFP and coexpressing the actin marker LifeAct-mCherry during PM blebbing. We observed a striking correlation between cortical bleb activity and nuclear accumulation of MRTF-A, whereas in nonblebbing cells, MRTF-A showed mostly a predominant cytosolic localization (Fig. 2, A and B; and Video 2).
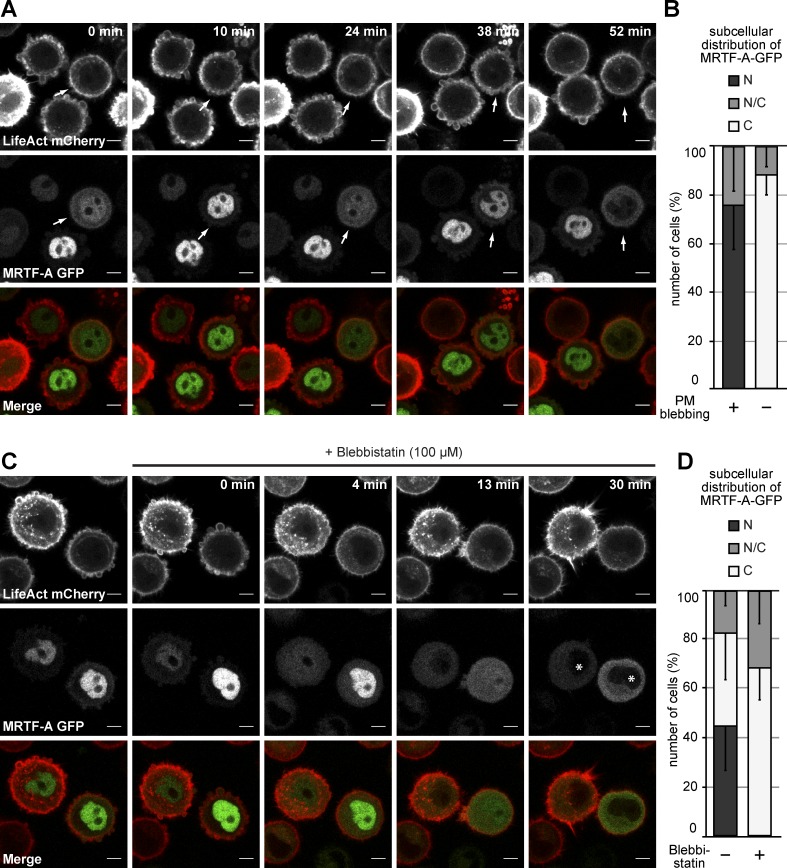
Figure 2.
PM blebbing triggers nuclear accumulation of MRTF-A. (A) Live MCF10A cells stably expressing MRTF-A–GFP together with LifeAct-mCherry were plated on poly-HEMA, and MRTF-A–GFP subcellular localization was monitored over time. LifeAct-mCherry is shown for each frame to visualize PM blebbing. Arrows highlight a cell showing dynamic oscillations of nuclear redistribution of MRTF-A–GFP in response to PM blebbing. Bars, 5 µm. (B) Quantification of MRTF-A–GFP subcellular localization in blebbing versus nonblebbing cells as in A. 180 cells from at least six experiments. (C) Live cells were imaged as in A. To interfere with cortical contractility and PM blebbing, cells were treated with 100 µM blebbistatin at 0 min, and MRTF-A–GFP subcellular localization was monitored over time. Coexpression of the actin marker LifeAct-mCherry reveals inhibition of membrane bleb activity. Asterisks indicate cytosolic redistribution of MRTF-A–GFP. Bars, 5 µm. (D) Quantification of MRTF-A–GFP subcellular localization in MCF10A cells treated with or without blebbistatin as indicated. (B and D) Localization was scored as predominantly cytoplasmic (C), pancellular (N/C), or predominantly nuclear (N). 200 cells from at least six experiments. Error bars indicate SD.
To further investigate a potential link between the contractile actin cortex involved in PM blebbing and MRTF-A subcellular localization, we treated cells with 100 µM blebbistatin, a myosin II inhibitor known to block blebbing (Cheung et al., 2002; Limouze et al., 2004; Charras et al., 2006; Norman et al., 2011). This resulted in efficient inhibition of blebbing within minutes accompanied by the complete redistribution of MRTF-A to the cytoplasm (Fig. 2, C and D; and Video 3), indicating that PM blebbing in conjunction with the contractile actin cortex represents a critical stimulus for MRTF-A nuclear accumulation.
PM blebbing triggers MRTF–SRF transcriptional activity and Ezrin up-regulation
SRF gene expression is under the control of its own transcriptional activity (Esnault et al., 2014). To understand whether MRTF–SRF activity is regulated by PM blebbing, we analyzed SRF mRNA induction in cells cultured on poly-HEMA to promote blebbing and associated cortical contractility. Under these conditions, SRF target gene expression was robustly induced (Fig. 3 A). A similar response could be observed in cells stably expressing a luciferase reporter under control of the MRTF–SRF pathway (Fig. S1, C and D). Interestingly, under these conditions we also observed a strong and significant bleb-associated induction of Ezrin mRNA expression that was fully SRF dependent as assessed by siRNA transfections (Fig. 3 B). Similar results were observed using combined siRNAs against MRTF-A and -B (Fig. 3 C) and could also be confirmed on the level of Ezrin protein (Fig. 3 D), demonstrating the specific requirement of MRTF-stimulated, SRF-dependent up-regulation of Ezrin during cellular blebbing.
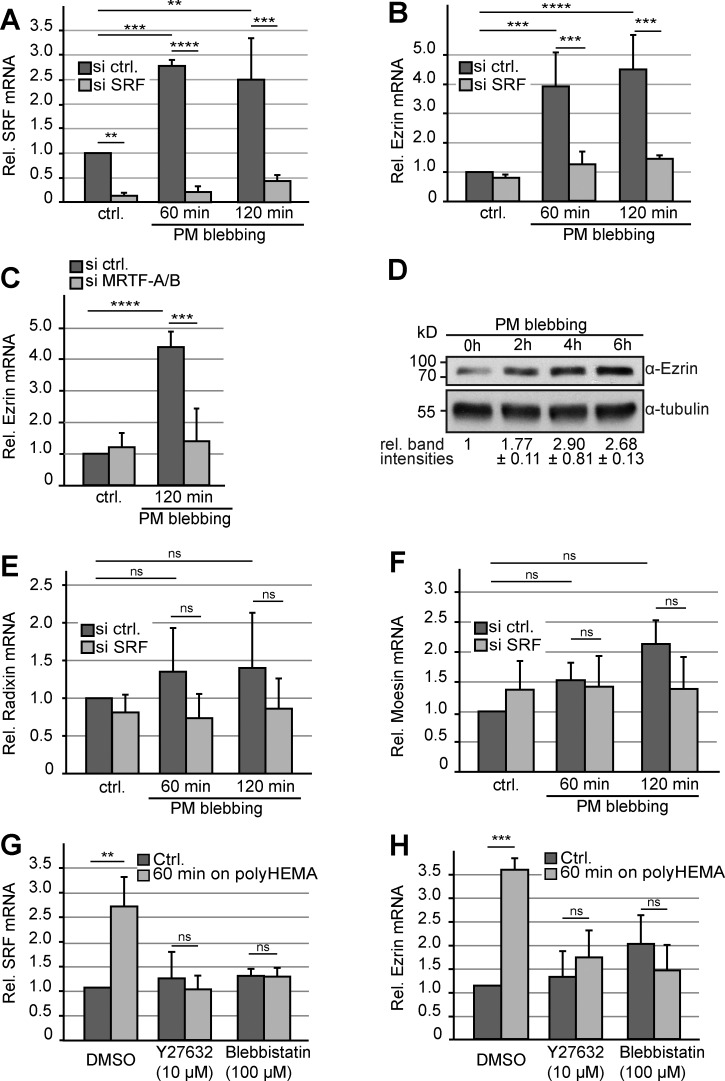
Figure 3.
PM blebbing induces MRTF–SRF-mediated up-regulation of Ezrin. (A–C, E, and F) Relative mRNA levels were assessed by quantitative RT-PCR in cells either attached (ctrl.) or plated on poly-HEMA to induce PM blebbing for the indicated periods of time. Results are shown as means from three independent experiments. Error bars indicate SD. Asterisks indicate statistical significance (**, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). ns indicates no significance (P > 0.05). (A) Relative levels of SRF mRNA were compared in cells treated with either control or siRNA directed against SRF. (B) Relative levels of Ezrin mRNA were compared in cells treated with either control or siRNA directed against SRF. (C) Relative levels of Ezrin mRNA were compared in cells treated with either control or combined siRNAs directed against MRTF-A and MRTF-B. (D) Western blot showing up-regulation of Ezrin upon induced PM blebbing for the indicated periods of time. Tubulin served as a loading control. Relative band intensities ± SD were quantified from two independent experiments. (E and F) Relative levels of Radixin mRNA (E) and Moesin mRNA (F) were compared in cells treated with either control or siRNA directed against SRF. (G) Relative levels of SRF mRNA were assessed by quantitative RT-PCR in cells either attached (ctrl.) or plated on poly-HEMA as indicated. 15 min before and during the time on poly-HEMA, cells were treated with either DMSO, 10 µM ROCK inhibitor Y27632, or 100 µM blebbistatin. Results are shown as means from three independent experiments. Error bars indicate SD. ns indicates no significance (P > 0.05). (H) Relative levels of Ezrin were assessed as in G.
Notably, this regulation was specific for Ezrin, as no significant differences were observed for the two closely related ERM proteins Radixin and Moesin (Fig. 3, E and F). Importantly, and consistent with our findings on MRTF subcellular localization, treatment of cells with the ROCK inhibitor Y27632 or blebbistatin abrogated blebbing-induced SRF and Ezrin mRNA expression (Fig. 3, G and H), suggesting that cortical contractility during PM blebbing represents the critical trigger to induce SRF-dependent gene expression.
MRTF–SRF and Ezrin are required for cell-in-cell invasion
Given its known function in PM blebbing and the need of efficient cortical blebbing for cell-in-cell invasion, we next determined the subcellular distribution of Ezrin during entotic invasion. Live imaging of cells stably expressing Ezrin-GFP together with H2B-mCherry to visualize the nuclei revealed an enrichment of Ezrin throughout the cell cortex (Fig. 4 A). During advanced stages of cell-in-cell invasion, Ezrin-GFP accumulated at the rear of the invading cell, which also showed extensive PM blebbing. This was followed by its redistribution toward the entire PM upon completion of invasive migration once a cell-in-cell structure was formed (Fig. 4 A and Video 4). Thus, Ezrin localization undergoes a dynamic and polar PM redistribution in the actively invading cell, which could further be confirmed by the invasion of an Ezrin-mCherry–expressing cell into an Ezrin-GFP–expressing neighbor (Fig. 4 B). Consistent with a critical function of Ezrin during entotic invasion, immunofluorescence microscopy not only confirmed a similar distribution pattern for endogenous Ezrin, but also indicated a high proportion of Ezrin at the rear of the cell being in its activated, phosphorylated state (Fig. 4 C).
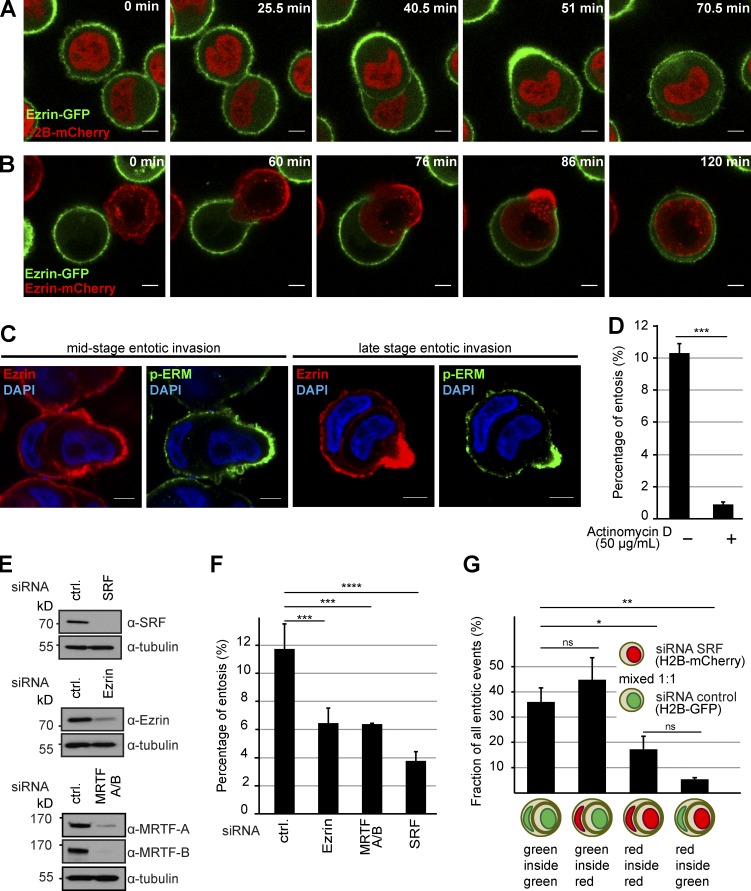
Figure 4.
Cell-in-cell invasion requires MRTF–SRF and Ezrin. (A) MCF10A cells stably expressing H2B-mCherry together with Ezrin-GFP were imaged during the process of entotic invasion. Note redistribution and enrichment of Ezrin-GFP at the rear of the invading cell during advanced stages of invasion. (B) MCF10A cells stably expressing either Ezrin-mCherry or Ezrin-GFP were mixed and imaged during the process of entotic invasion. Note that the redistribution and enrichment of Ezrin specifically originates from the invading cell. (C) Immunolabeling of endogenous Ezrin (red), phospho-Ezrin/Radixin/Moesin (green), and nuclei (DAPI) of MCF10A cells. Before fixation, cells were cultured for 4 h in suspension to promote entotic invasion. Examples illustrate the enrichment of Ezrin as well as p-ERM at the rear of the invading cell at different stages of the process. Bars, 5 µm. (D) Quantification of entotic invasion after 4 h of culture in suspension. To test for the effects of acute transcriptional inhibition, 50 µg/ml actinomycin D was added to the cells immediately after plating them on Ultra-Low attachment dishes. Entotic events were counted from three different experiments considering ≥2,000 cells for each condition. (E) Western blot confirming efficient siRNA-mediated knockdown of SRF, Ezrin, or MRTF-A together with MRTF-B. Tubulin served as a loading control. (F) Quantification of entotic invasion of cells treated with the indicated siRNAs for 72 h. Entotic events were counted from three independent experiments considering ≥800 cells for each condition. (G) Cells either expressing H2B-mCherry or H2B-GFP were treated with either control or siRNA directed against SRF as indicated, mixed, and allowed to undergo entotic invasion. Resulting entotic events were considered (100%) and scored according to the underlying color combinations as indicated. Note the significantly impaired invasion of SRF-silenced cells. ≥290 entotic events were scored from three independent experiments. Error bars indicate SD. Asterisks indicate statistical significance (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). ns indicates no significance (P > 0.05).
To test for an impact of transcription on entotic invasion, we blocked global transcription by the intercalating drug actinomycin D (Bensaude, 2011). Treatment of MCF10A cells with 50 µg/ml actinomycin D not only led to a rapid and almost complete block of serum-induced transcriptional stimulation (Fig. S2, A and B), but also resulted in a strongly reduced number of entotic events (Fig. 4 D). To examine whether Ezrin, as well as MRTF or SRF, is required for entosis, MCF10A cells were treated with the respective siRNAs, and successful protein depletion was assessed by Western blot analysis (Fig. 4 E). Interestingly, silencing Ezrin, MRTF-A and -B, or SRF all resulted in a significant and robust reduction of entotic invasion (Fig. 4 F).
To address whether the MRTF–SRF signaling axis is specifically required for the invading cell, we performed two-color entosis assays in which we mixed GFP-H2B–expressing control cells (green) together with SRF-silenced cells expressing mCherry-H2B (red). Quantification of entotic events according to the resulting combination of colors showed that cells silenced for SRF specifically failed to invade neighboring cells, but SRF depletion had no significant effect on the invaded host cells (Fig. 4 G). Consistently, predominantly nuclear MRTF-A was detected in a high percentage of cells that were about to invade into neighbors, whereas nuclear MRTF-A was mostly absent in invaded cells (Fig. S2, C and D). Furthermore, a high proportion of cells silenced for SRF failed to complete entotic invasion despite initiating the process (Fig. S2 E). Together, these data demonstrate that SRF functions critically in the actively invading cells during entosis.
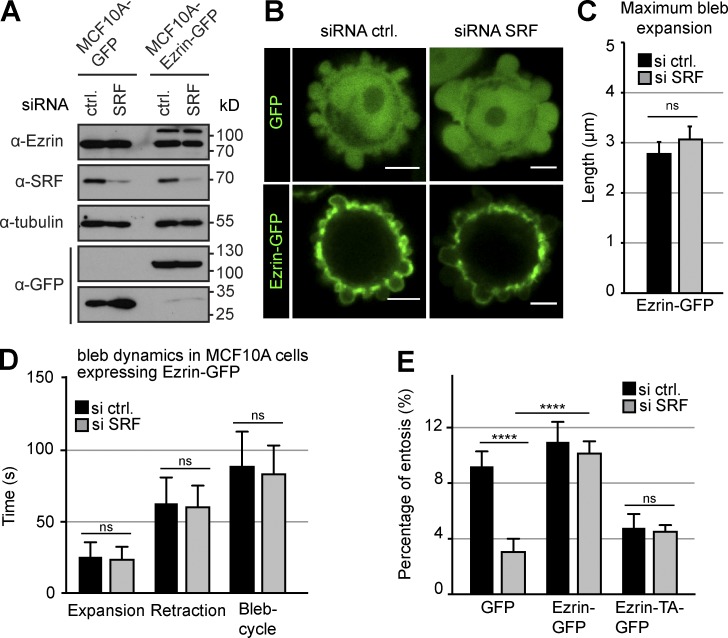
Ezrin expression is sufficient to rescue bleb dynamics and entotic invasion in SRF-depleted cells
To mimic Ezrin up-regulation during blebbing, we generated cells stably expressing Ezrin-GFP to compare bleb dynamics with GFP-expressing control cells in response to transient SRF depletion by siRNA (Fig. 5, A and B). Whereas SRF depletion consistently affected blebbing and induced larger blebs in control cells (Fig. 1, D and E), Ezrin-GFP–expressing cells remained unaffected (Fig. 5 C). Moreover, SRF suppression did not affect bleb retraction in Ezrin-GFP–expressing cells compared with the siRNA control (Fig. 5 D), demonstrating that increased Ezrin expression is sufficient to override a lack of SRF activity during blebbing.
Figure 5.
Ezrin expression restores membrane blebbing and entotic invasion in SRF-silenced cells. (A) Western blot confirming efficient siRNA-mediated knockdown of SRF in cells stably expressing either GFP or Ezrin-GFP. Tubulin served as a loading control. (B) Live MCF10A cells stably expressing either GFP or Ezrin-GFP were imaged over time to visualize PM blebbing. Bars, 5 µm. (C) Quantification of maximum bleb expansion in cells stably expressing Ezrin-GFP. Cells were treated with the indicated siRNAs, and the maximum length reached by individual blebs was compared. Note no significant differences between control and SRF-silenced cells in contrast to cells only expressing GFP (Fig. 1 D). (D) Cells stably expressing Ezrin-GFP were monitored to measure the time for expansion and retraction of individual blebs. Before imaging, cells were treated with siRNA as indicated. Note no significant differences between control and SRF-silenced cells in contrast to cells expressing GFP only (Fig. 1 E). (C and D) ≥60 blebs and 15 cells per condition. ns indicates no significance (P > 0.05). (E) Quantification of entotic invasion of cells stably expressing GFP, Ezrin-GFP, or Ezrin-T567A-GFP. Cells were transfected with either control or siRNA against SRF as indicated, and the number of entotic events was quantified from three independent experiments considering ≥800 cells as a percentage of the total number of cells. Error bars indicate SD. Asterisks indicate statistical significance (****, P ≤ 0.0001). ns indicates no significance (P > 0.05).
These data show that expression of Ezrin can rescue defective bleb dynamics in the absence of SRF and further corroborate Ezrin as an MRTF–SRF target (Fig. 3, B and C). Moreover and consistent with this notion, Ezrin expression restored entosis events to control levels in the absence of SRF (Fig. 5 E and Fig. S3), whereas the expression of the inactive mutant Ezrin-T567A (Gautreau et al., 2000) did not (Fig. 5 E). Thus, Ezrin activity is necessary and sufficient for bleb dynamics and entosis in SRF-depleted cells.
The actin-controlled MRTF–SRF transcriptional pathway regulates target genes involved in contractility, which is considered necessary for invasive cell migration (Gualdrini et al., 2016). During cancer invasion, cells often adopt a rounded bleb-associated form of rapid single-cell migration (Sahai and Marshall, 2003). Actomyosin contractility facilitates bleb retraction, which requires Ezrin to reassemble the contractile actin cortex (Charras et al., 2006). Consistent with this, Ezrin has been identified as a critical regulator of invasion and metastasis (Yu et al., 2004; Ren et al., 2009; Lorentzen et al., 2011). Our data identify a critical role for MRTF subcellular dynamics and transcriptional activity to regulate Ezrin during sustained blebbing and augmented cortical contraction, which in turn is necessary for bleb-associated cell-in-cell invasion. Thus, our findings implicate a novel mechanism in which cell-invasive bleb dynamics depend on an actin-controlled transcriptional feedback.
Materials and methods
Reagents, antibodies, and plasmids
Cell culture reagents were purchased from Invitrogen. Poly-HEMA was purchased from Polysciences. Antibodies used were purchased from BD (mouse anti–Ezrin 610602 and mouse anti–mDia1 610849), Santa Cruz Biotechnology (rabbit anti–SRF G20, mouse anti–GFP B20, and goat anti–MRTF-A C19), Cell Signaling Technology (rabbit anti-tubulin 2125S, rabbit anti–MRTF-B 14613, anti–goat HRP, and antiphospho–Ezrin/Radixin/Moesin 48G2 rabbit mAb), Bio-Rad Laboratories (anti–rabbit HRP), GE Healthcare (anti–mouse HRP), and Life Technologies (donkey anti–rabbit 488 and goat anti–mouse 555).
Cells were treated for the indicated times with the following inhibitors. 100 µM (S)-nitro-blebbistatin (Cayman Chemical) was used in live-cell imaging experiments (Fig. 2, C and D). Blebbistatin (Sigma-Aldrich) was used at 100 µM, Y-27632 (Sigma-Aldrich) was used at 10 µM, and actinomycin D (Santa Cruz Biotechnology) was used at 50 µg/ml.
pWPXL-based lentiviral expression vectors for Ezrin-GFP, Ezrin-T567A-GFP, Ezrin-mCherry, H2B-mCherry, H2B-GFP, LifeAct-mCherry, and the pIND20-based inducible lentiviral expression vector (Meerbrey et al., 2011) for MRTF-A–GFP were generated using standard PCR-based procedures. Lentiviral luciferase reporter constructs were generated using the FUGW plasmid. To obtain reporter gene constructs, the coding sequence of firefly luciferase was inserted into FUGW with either saving or deleting the hUbC promoter or replacing it with the MRTF–SRF-specific promoter 3Da.luc (Geneste et al., 2002). The FUGW lentiviral vector was a gift from D. Oliver (University of Marburg). In all reporter gene constructs, the luciferase gene was linked to GFP by a self-cleavable T2A peptide to allow for FACS-based cell sorting of successfully transduced cells.
Cell culture, transfection, and viral transduction
Human MCF10A cells were cultured in DMEM/F12 supplemented with 5% horse serum, 20 ng/ml epidermal growth factor, 10 µg/ml insulin, 0.5 µg/ml hydrocortisone, 100 ng/ml cholera toxin, 100 U/ml penicillin, and 100 g/ml streptomycin at 37°C in a CO2 atmosphere as described by Debnath et al. (2003). Human HEK293T cells were maintained in DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 g/ml streptomycin at 37°C in a CO2 atmosphere.
For gene silencing, MCF10A cells were transiently transfected with 30 nM siRNA oligonucleotides using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. After 72 h, knockdown was quantified by quantitative PCR or confirmed by Western blot analysis. The following FlexiTube siRNA sequences (QIAGEN) were used: Hs_SRF_5, 5′-CAAGATGGAGTTCATCGACAA-3′; Hs_MKL1_7, 5′-ATCACGTGTGATTGACATGTA-3′; Hs_MKL1_10, 5′-CCCGCCAAAGTCAGCAGGCGA-3′; Hs_VIL2_1, 5′-ACTAAGCTCTTATTAGCGCTC-3′; Hs_MKL2, 5′-AAGTAACAGTGGGAATTCAGC-3′; Hs_DIAPH1_1, 5′-AAGATATGAGAGTGCAACT-3′; and control siRNA, 5′-AATTCTCCGAACGTGTCACGT-3′.
HEK293T cells were transfected using the calcium phosphate method. For lentivirus production, HEK293T cells were cotransfected with the lentiviral packaging vectors psPAX and pMDG.2 together with the pInducer- or pWXPL-based plasmids of choice. The lentiviral packing plasmids and pWPXL were provided by J. Swiercz (Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany). After 48 h, supernatants containing viral particles were harvested, filtered, and used to transduce MCF10A cells. Transduced MCF10A cells were selected by FACS-based cell sorting. Expression of MRTF-A–GFP from pInducer20 was induced by 333 ng/ml doxycycline.
Microscopy, live cell imaging, and entosis assays
Microscopic imaging was performed using confocal laser-scanning microscopes (LSM 700 and LSM 800; Carl Zeiss) and a 63× 1.4 NA oil objective lens (Carl Zeiss). Time-lapse microscopy was performed in MCF10A medium at 37°C in a CO2-humidified incubation chamber (Pecon, CO2 module S1) using ZEN software (Carl Zeiss).
To induce PM blebbing and entotic invasion, cells were plated on 35-mm glass-bottom dishes (In Vitro Scientific) coated with 12% poly-HEMA wt/wt solution in ethanol to prevent cellular attachment as described previously by Overholtzer et al. (2007).
To quantify the number of entotic events, MCF10A cells were trypsinized and plated on Ultra Low cluster plates (3473; Costar) at densities of 300,000–400,000 cells per well. After 4 h, cells were fixed directly in suspension using 4% formaldehyde/PBS for 10 min. After washing, the cells were rehydrated in PBS and seeded onto 12-mm coverslips on a heating block at 60°C for 5 min. Fixed samples were washed in PBS and permeabilized by 0.05% PBS-Tween for 10 min. Nuclei were stained using DAPI (Sigma-Aldrich) at 1:10,000 for 20 min at RT, and F-actin was labeled by Alexa Fluor 555–phalloidin or Alexa Fluor 488–phalloidin (Invitrogen) at 1:1,000 overnight at 4°C.
Real-time RT-PCR
For total RNA extraction, TRIzol reagent (Peqlab) was used according to the manufacturer’s instructions. The reverse transcription was performed using RevertAid Reverse Transcriptase (Fermentas). Obtained cDNA was quantified using a SYBR green Master Mix (Bio-Rad Laboratories). The following primers were used: for human SRF, we used forward, 5′-CAGATCGGTATGGTGGTCGG-3′, and reverse, 5′-GTCAGCGTGGACAGCTCATA-3′. For human Ezrin, we used forward, 5′-TAAGGGTTCTGCTCTGACTCCA-3′, and reverse, 5′-GCTCTGCATCCATGGTGGTAA-3′. For human MRTF-A, we used forward, 5′-CATGAGTCCCAGGGTTCTGT-3′, and reverse, 5′-ACTTGGCAGTGGGGATAGTG-3′. For human MRTF-B, we used forward, 5′-ACATTCGCCCTTTCTTGCAGT-3′, and reverse, 5′-TCCGAGATTGCCATCTTATTGTC-3′. For human TATA-binding protein, we used forward, 5′-TGCACAGGAGCCAAGAGTGAA-3′, and reverse, 5′-CACATCACAGCTCCCCACCA-3′. For human Radixin, we used forward, 5′-CCATATTGCCGAGCTGTCTG-3′, and reverse, 5′-GGCAAATTCCAGCTCAGCAT-3′. For human Moesin, we used forward, 5′-ATCCAAGCCGTGTGTACTGC-3′, and reverse, 5′-AAATAGCTGCTTCCCGGTGG-3′. For human mDia1, we used forward, 5′-GTCAGGCTTGCGGGATATG-3′, and reverse, 5′-TTCAGCACCAAATGTTTGCAC-3′. For human Fos, we used forward, 5′-CTCTCTTACTACCACTCACCCGC-3′, and reverse, 5′-GGTCCGTGCAGAAGTCCTGCG-3′.
Relative mRNA levels were calculated using the comparative ΔΔCT model normalized to the abundance of TATA-binding protein cDNA.
Statistical analysis
Statistical analyses were done using Prism 6 (GraphPad Software). Data are expressed as mean ± SD. Statistical significance was evaluated with the ANOVA test (Fig. 1 E; Fig 3, A–C and E–H; Fig. 4, F and G; Fig 5, D and E; Fig. S1, B and D; and Fig S2 A) or the unpaired Student’s t test. Data distribution was assumed to be normal, but this was not formally tested (Fig. 1 D, Fig. 4 D, Fig. 5 C, and Fig. S2 E). Statistical differences were judged as significant at P ≤ 0.05: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001.
Online supplemental material
Fig. S1 shows that the SRF pathway does not affect expression of the formin mDia1 and confirms an induction of the MRTF–SRF pathway by PM blebbing using a luciferase reporter assay. Fig. S2 shows that entotic invasion requires active transcription and supports the notion that during entotic invasion, the MRTF–SRF pathway primarily functions in the actively invading cell. Fig. S3 illustrates entotic invasion of Ezrin-GFP–expressing cells silenced for SRF and compares the time necessary for entotic invasion of Ezrin-GFP–expressing cells, with or without silencing of SRF. Video 1 compares PM blebbing in MCF10A cells treated with either control or siRNA directed against SRF. Video 2 shows the strong correlation between PM blebbing and nuclear accumulation of MRTF-A. Video 3 shows the effects of blebbistatin treatment on the subcellular distribution of MRTF-A. Video 4 shows redistribution and accumulation of Ezrin at the rear of the invading cell during entotic invasion. Video 5 confirms that the enrichment of Ezrin during cell-in-cell invasion originates at the rear of the invading cell. Video 6 shows MRTF-A subcellular distribution before and during entotic cell-in-cell invasion.
Supplementary Material
Acknowledgments
We are grateful to H. Raifer from the Flow Cytometry Core Facility. We thank laboratory members for discussions.
This work was funded by grants to R. Grosse from the Deutsche Forschungsgemeinschaft (grant GRK 2213). M. Holst was supported by a Mildred-Scheel-Doktorandenprogramm fellowship from the Deutsche Krebshilfe (grant 110405).
The authors declare no competing financial interests.
Author contributions: L. Soto Hinojosa and M. Holst performed the experiments and analyzed the data. C. Baarlink and R. Grosse conceived the work and supervised and designed the experiments. R. Grosse wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
Abbreviations used:
- HEMA
- 2-hydroxy-ethyl methacrylate
- MRTF
- myocardin-related transcription factor
- PM
- plasma membrane
- SRF
- serum response factor
References
- Baarlink C., Wang H., and Grosse R.. 2013. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 340:864–867. 10.1126/science.1235038 [DOI] [PubMed] [Google Scholar]
- Bensaude O. 2011. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2:103–108. 10.4161/trns.2.3.16172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt D.T., Baarlink C., Kitzing T.M., Kremmer E., Ivaska J., Nollau P., and Grosse R.. 2009. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of β1-integrin. Nat. Cell Biol. 11:557–568. 10.1038/ncb1862 [DOI] [PubMed] [Google Scholar]
- Charras G., and Sahai E.. 2014. Physical influences of the extracellular environment on cell migration. Nat. Rev. Mol. Cell Biol. 15:813–824. [DOI] [PubMed] [Google Scholar]
- Charras G.T., Hu C.-K., Coughlin M., and Mitchison T.J.. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175:477–490. 10.1083/jcb.200602085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A., Dantzig J.A., Hollingworth S., Baylor S.M., Goldman Y.E., Mitchison T.J., and Straight A.F.. 2002. A small-molecule inhibitor of skeletal muscle myosin II. Nat. Cell Biol. 4:83–88. 10.1038/ncb734 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., and Brugge J.S.. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268. 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Esnault C., Stewart A., Gualdrini F., East P., Horswell S., Matthews N., and Treisman R.. 2014. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 28:943–958. 10.1101/gad.239327.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler O.T., and Grosse R.. 2008. Cell motility through plasma membrane blebbing. J. Cell Biol. 181:879–884. 10.1083/jcb.200802081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O., Kim S.E., and Overholtzer M.. 2015. Entosis: cell-in-cell formation that kills through entotic cell death. Curr. Mol. Med. 15:861–866. 10.2174/1566524015666151026100042 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Wolf K.. 2010. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188:11–19. 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche M., Thorogate R., and Charras G.. 2014. Quantitative analysis of ezrin turnover dynamics in the actin cortex. Biophys. J. 106:343–353. 10.1016/j.bpj.2013.11.4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A., Louvard D., and Arpin M.. 2000. Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J. Cell Biol. 150:193–203. 10.1083/jcb.150.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneste O., Copeland J.W., and Treisman R.. 2002. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell Biol. 157:831–838. 10.1083/jcb.200203126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdrini F., Esnault C., Horswell S., Stewart A., Matthews N., and Treisman R.. 2016. SRF co-factors control the balance between cell proliferation and contractility. Mol. Cell. 64:1048–1061. 10.1016/j.molcel.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing T.M., Sahadevan A.S., Brandt D.T., Knieling H., Hannemann S., Fackler O.T., Grosshans J., and Grosse R.. 2007. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 21:1478–1483. 10.1101/gad.424807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic M., Johnson N.B., Sun Q., Normand G., Hoover N., Yao E., Richardson A.L., King R.W., Cibas E.S., Schnitt S.J., et al. . 2011. A non-genetic route to aneuploidy in human cancers. Nat. Cell Biol. 13:324–330. 10.1038/ncb2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limouze J., Straight A.F., Mitchison T., and Sellers J.R.. 2004. Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 25:337–341. 10.1007/s10974-004-6060-7 [DOI] [PubMed] [Google Scholar]
- Lorentzen A., Bamber J., Sadok A., Elson-Schwab I., and Marshall C.J.. 2011. An ezrin-rich, rigid uropod-like structure directs movement of amoeboid blebbing cells. J. Cell Sci. 124:1256–1267. 10.1242/jcs.074849 [DOI] [PubMed] [Google Scholar]
- Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., and Treisman R.. 2009. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11:257–268. 10.1038/ncb1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., et al. . 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 108:3665–3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.-I., and Treisman R.. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 113:329–342. 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- Mouilleron S., Langer C.A., Guettler S., McDonald N.Q., and Treisman R.. 2011. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci. Signal. 4:ra40 10.1126/scisignal.2001750 [DOI] [PubMed] [Google Scholar]
- Norman L., Sengupta K., and Aranda-Espinoza H.. 2011. Blebbing dynamics during endothelial cell spreading. Eur. J. Cell Biol. 90:37–48. 10.1016/j.ejcb.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Nürnberg A., Kitzing T., and Grosse R.. 2011. Nucleating actin for invasion. Nat. Rev. Cancer. 11:177–187. 10.1038/nrc3003 [DOI] [PubMed] [Google Scholar]
- Olson E.N., and Nordheim A.. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353–365. 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M., Mailleux A.A., Mouneimne G., Normand G., Schnitt S.J., King R.W., Cibas E.S., and Brugge J.S.. 2007. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 131:966–979. 10.1016/j.cell.2007.10.040 [DOI] [PubMed] [Google Scholar]
- Purvanov V., Holst M., Khan J., Baarlink C., and Grosse R.. 2014. G-protein-coupled receptor signaling and polarized actin dynamics drive cell-in-cell invasion. eLife. 3 10.7554/eLife.02786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Hong S.H., Cassavaugh J., Osborne T., Chou A.J., Kim S.Y., Gorlick R., Hewitt S.M., and Khanna C.. 2009. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene. 28:792–802. 10.1038/onc.2008.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E. 2005. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 15:87–96. 10.1016/j.gde.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Sahai E., and Marshall C.J.. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719. 10.1038/ncb1019 [DOI] [PubMed] [Google Scholar]
- Vartiainen M.K., Guettler S., Larijani B., and Treisman R.. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 316:1749–1752. 10.1126/science.1141084 [DOI] [PubMed] [Google Scholar]
- Yu Y., Khan J., Khanna C., Helman L., Meltzer P.S., and Merlino G.. 2004. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat. Med. 10:175–181. 10.1038/nm966 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.