Bicaudal D2 (BICD2) is an adaptor protein that recruits and activates dynein–dynactin onto Rab6 membrane vesicles. Huynh and Vale reconstitute Rab6 regulation of BICD2-mediated dynein transport in vitro and show that disease-associated mutations in BICD2 cause an increase in retrograde transport.
Abstract
Bicaudal D2 (BICD2) joins dynein with dynactin into a ternary complex (termed DDB) capable of processive movement. Point mutations in the BICD2 gene have been identified in patients with a dominant form of spinal muscular atrophy, but how these mutations cause disease is unknown. To investigate this question, we have developed in vitro motility assays with purified DDB and BICD2’s membrane vesicle partner, the GTPase Rab6a. Rab6a–GTP, either in solution or bound to artificial liposomes, released BICD2 from an autoinhibited state and promoted robust dynein–dynactin transport. In these assays, BICD2 mutants showed an enhanced ability to form motile DDB complexes. Increased retrograde transport by BICD2 mutants also was observed in cells using an inducible organelle transport assay. When overexpressed in rat hippocampal neurons, the hyperactive BICD2 mutants decreased neurite growth. Our results reveal that dominant mutations in BICD2 hyperactivate DDB motility and suggest that an imbalance of minus versus plus end–directed microtubule motility in neurons may underlie spinal muscular atrophy.
Introduction
The retrograde motor cytoplasmic dynein is involved in a host of cellular activities, including the trafficking of diverse cargos such as organelles, vesicles, and mRNA as well as mitotic spindle alignment and chromosome positioning (Allan, 2011). In contrast to yeast dynein, which can achieve long-range transport on its own (Reck-Peterson et al., 2006), mammalian dynein must form a tripartite complex with the 1.2-MD dynactin complex and an adapter protein to move processively along microtubules (McKenney et al., 2014; Schlager et al., 2014a).
A number of different adaptor proteins that join dynein and dynactin into an active motile complex have been identified (McKenney et al., 2014; Cianfrocco et al., 2015). For example, bicaudal D2 (BICD2), one of the best-studied dynein adaptors, interacts with the small GTPase Rab6, which is found on early endosomes and ER–Golgi vesicles (Matanis et al., 2002). BICD2 is thought to be autoinhibited through an interaction between its C-terminal coiled coil domain with the N-terminal domain of the protein, which prevents it from associating with its other partners, including dynein (Matanis et al., 2002; Hoogenraad et al., 2003; Liu et al., 2013). Autoinhibition is thought to be relieved by binding to an effector, such as Rab6a in its GTP-bound form (Liu et al., 2013); however, evidence for this model is lacking.
BICD2 recently emerged as a genetic locus associated with dominant spinal muscular atrophy (SMA), a genetic disorder characterized by the degeneration of anterior horn cells and leading to eventual muscle atrophy and weakness. (Neveling et al., 2013; Oates et al., 2013; Peeters et al., 2013). The effects that these point mutations have on BICD2’s ability to function as a dynein adaptor are not known, although overexpression of these mutants has been reported to cause Golgi fragmentation and changes in BICD2 localization in cultured cells (Neveling et al., 2013; Peeters et al., 2013). Interestingly, point mutations in the tail region of heavy chain 1 of cytoplasmic dynein have been associated with another form of SMA, SMA-LED1. These mutations were reported to cause an increase in the affinity between the motor and BICD2 (Peeters et al., 2015) and a decrease in dynein–dynactin processivity (Hoang et al., 2017).
Here, using biochemical and single-molecule motility assays, we demonstrate that BICD2 disease mutations cause a gain of function in the motility of DDB complexes both in vitro and in cells. In addition, we show that the expression of BICD2 mutants in neurons leads to impaired neurite outgrowth. These results indicate that the SMA BICD2 mutants hyperactivate dynein motility, a gain of function consistent with a dominant genetic trait that could explain the gradual loss of motor neuron function in SMA.
Results and discussion
Activation of BICD2 via Rab6a–GTP increases dynein–dynactin binding and motility
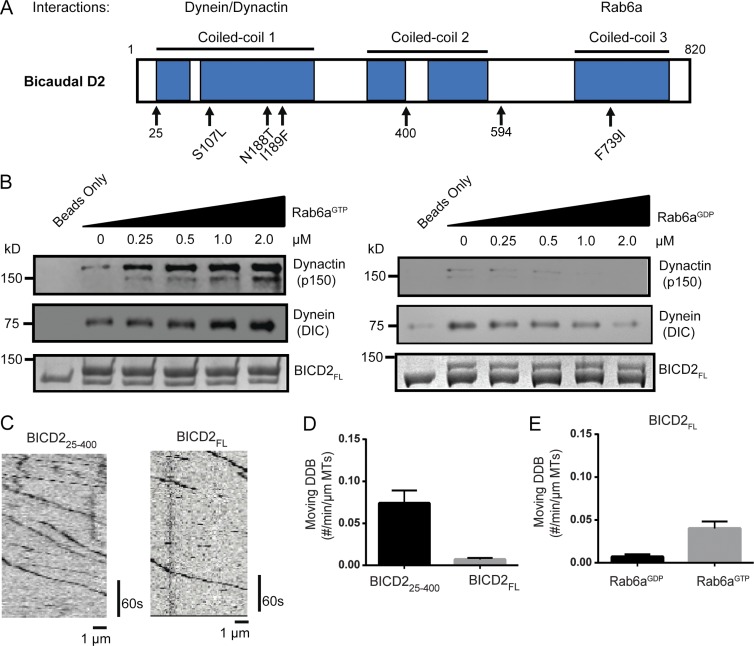
BICD2 is composed of five coiled coil domains that can be grouped into three distinct regions (Fig. 1 A). The C-terminal region of the protein is the most well-conserved and contains the binding site to different partners localized to different cargos, including the small GTPase Rab6a (Matanis et al., 2002). An N-terminal construct of the protein, which spans residues 25–400 (BICD225–400), was shown to be more effective in binding dynein–dynactin in cell lysate pull-down assays compared with autoinhibited full-length protein (BICD2FL; Splinter et al., 2012; McKenney et al., 2014). We replicated these findings; the difference in dynactin binding was especially striking, exhibiting about a 10-fold reduction for the BICD2FL pull-down compared with BICD225–400, whereas dynein was two- to threefold lower (Fig. S1, A and B).
Figure 1.
BICD2FL activation by Rab6aGTP. (A) Domain architecture of mammalian Bicaudal D2. N188T, I189F, and S107L are mutations associated with human SMA-LED, and F739I is the analogous mutation to the classical Drosophila mutation F684I. Amino acids 400 and 594 indicate the end of constructs used in this study. (B) Porcine brain lysate pull-down using full-length (FL) BICD2 as bait. Increasing amounts of recombinant Rab6aGTP (left) or Rab6aGDP (right) were added to the lysate, and the amount of endogenous dynein (dynein intermediate chain [DIC]) and dynactin (p150 subunit) bound was analyzed by immunoblot. (C) Sample kymographs of single-molecule DDB (aa 25–400 or FL BICD2) motility on microtubules. Fluorescence is from the N-terminal sfGFP tag on BICD2. (D) Quantification of the number of moving motors per micrometer of microtubule; mean and SEM from n = 3 independent experiments (each experiment measuring a minimum of 30 microtubules). (E) Quantification of number of moving motors per micrometer of microtubule of BICD2FL with either Rab6aGDP or Rab6aGTP added; mean and SEM from n = 3 independent experiments (each experiment measuring a minimum of 30 microtubules).
We next tested whether the autoinhibited state of BICD2FL could be activated by Rab6a. We added purified recombinant Rab6a–Q72L, which is locked in a GTP-bound state (hereafter referred to as Rab6aGTP), to porcine brain lysate and performed pull-downs using BICD2 as bait (Matanis et al., 2002). With increasing Rab6aGTP, there was a corresponding increase of dynein and dynactin in the BICD2FL pull-down. Conversely, the GDP-bound mutant of Rab6a (Rab6a–T27N, hereafter Rab6aGDP) appeared to somewhat reduce the pull-down of dynein and dynactin (Fig. 1 B). Rab6aGTP also was detected in the pellet fraction in these experiments, whereas Rab6aGDP was absent (Fig. S1 C). A densitometric analysis shows a 2.5-fold increase in dynein enrichment and a 10-fold increase in dynactin at the highest Rab6aGTP concentration used (Fig. S1 D). The greater pull-down of dynactin relative to dynein might reflect an interaction of the Rab6aGTP–BicD2 complex with dynactin alone. Together, these results indicate that Rab6aGTP is able to activate full-length BICD2 and enhance its ability to bind to dynein and dynactin.
To further examine whether Rab6aGTP can activate BICD2FL and enable dynein–dynactin motility, we used total internal reflection fluorescence microscopy to visualize DDB motility on microtubules. Purified dynein and dynactin from mammalian RPE-1 cell lysates were incubated with similar concentrations of either superfolder-GFP (sfGFP)–fused BICD225–400 or BICD2FL, and the numbers of moving motor complexes were analyzed via kymographs (Fig. 1 C). This single-molecule in vitro assay revealed that the truncated BICD225–400 produced more processive dynein–dynactin complexes when compared with BICD2FL, which is again consistent with the idea that BICD2FL is autoinhibited (Fig. 1, C and D; and Video 1). The addition of Rab6aGTP to BICD2FL and dynein–dynactin resulted in a three- to fourfold increase in the number of moving DDBFL complexes on microtubules compared with Rab6aGDP or the individual DDB components alone (Fig. 1 E). In contrast, Rab6aGTP neither increased the motility of DDB25–400 (Fig. S1 E) nor interacted with BICD225–400 (Fig. S1 F). Collectively, these results suggest that Rab6aGTP is capable of alleviating the autoinhibition of full-length BICD2 and activating its ability to form processive DDB complexes.
Disease-related point mutations in BICD2 increase binding to dynein/dynactin and motility
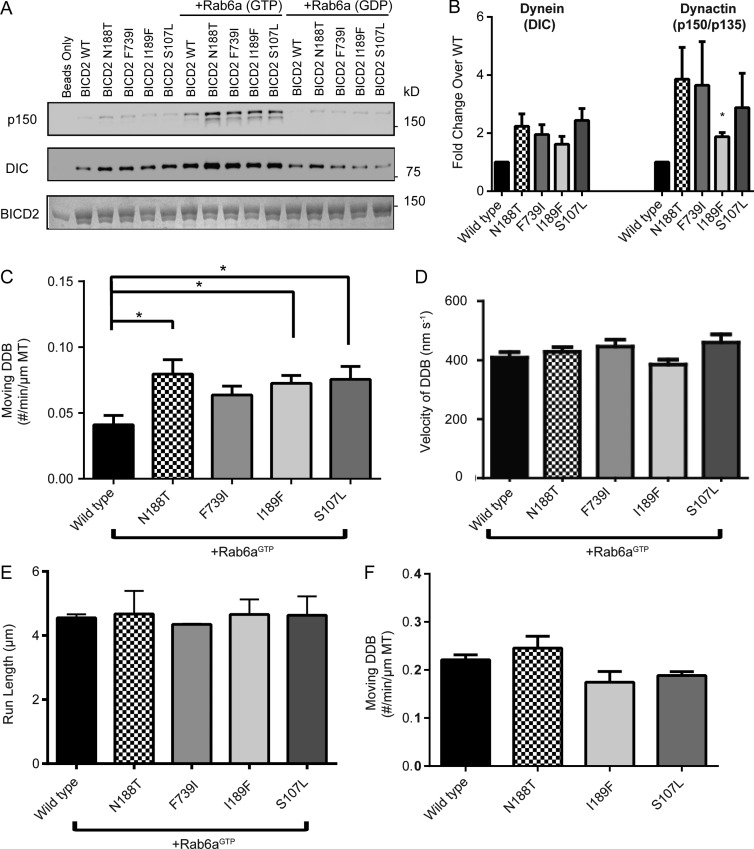
Next, using our assays, we sought to examine the effects of three BICD2FL mutants identified in patients afflicted with DC-SMA/SMA-LED2 (S107L, N188T, and I189F) and a mutant (F739I) that is equivalent to the Drosophila melanogaster F684I mutant that produces a bicaudal phenotype (Fig. 1 A; Mohler and Wieschaus, 1986; Wharton and Struhl, 1989). The latter mutation has been shown to have higher affinity toward dynein compared with WT and was proposed to be more readily activated upon binding to cargo (Liu et al., 2013). All four of the mutated residues are highly conserved in the BICD protein family.
In a brain lysate pull-down assay with Rab6aGTP, the four BICD2FL mutants recruited more dynein and dynactin compared with WT BICD2FL, with the greatest difference being observed in dynactin binding (Figs. 2 A and S2 B). Weak binding of dynein–dynactin also was observed with Rab6aGDP or without added Rab6a (Fig. 2, A and B; and Fig. S2, A andB). It is possible that endogenous Rab6a in the brain lysate might be responsible for this weak BICD2 mutant-specific pull-down or that other factors in the lysate allow for a loss of autoinhibition. In the single-molecule total internal reflection fluorescence assay in the presence of Rab6aGTP, three of the BICD2FL mutants (N188T, I189F, and S107L) showed a statistically significant 20–30% increase in the number of motile DDB complexes compared with WT BICD2FL (Fig. 2 C). Neither the velocity (Fig. 2 D) nor run-length (Fig. 2 E and Fig. S2 C) of DDB was different between WT and mutant BICD2FL. In the presence of Rab6aGDP, however, motility was much lower, and no statistical difference was observed between mutant and WT BICD2FL (Fig. S2 D). No significant difference also was observed in the number of motile DDB complexes formed by WT and mutant truncated BICD225–400 (Fig. 2 F). WT and mutant BICD225–400 also pulled down similar amounts of dynein and dynactin (Fig. S2 E). Collectively, these results indicate the point mutations in BICD2FL enhance Rab6aGTP-dependent activation from the autoinhibited state, producing more motile DDBFL complexes but not altering the measured parameters of DDBFL motility.
Figure 2.
BICD2FL mutants show increased binding to dynein/dynactin and single-molecule motility in vitro. (A) Porcine brain lysate pull-down using BICD2FL as bait. Rab6aGTP or Rab6aGDP was added to the lysate at a concentration of 1 µM. The amount of endogenous dynein (DIC) and dynactin (p150 subunit) bound was analyzed by immunoblot. The amount of BICD2 on beads was visualized using Coomassie stain. (B) The ratio of dynein or dynactin band intensity for mutants compared with WT BICD2 for the lysate only condition; mean and SEM from n = 3 independent experiments. Mean, SEM, and p-values for the mutants shown for DIC are as follows: N188T: 2.24 ± 0.42, P = 0.10; F739I: 1.95 ± 0.34, P = 0.11; I189F: 1.61 ± 0.27, P = 0.15; and S107L: 2.44 ± 0.41, P = 0.07. Mean, SEM, and p-values for p135/p150 are as follows: N188T: 3.86 ± 1.09, P = 0.12; F739I: 3.65 ± 1.5, P = 0.22; I189F: 1.88 ± 0.14, P = 0.027; and S107L: 2.88 ± 1.18, P = 0.25. Quantitation of the other conditions are shown in Fig. S2 (A and B). (C) Dynein and dynactin (purified from RPE-1 cells; see Materials and methods) were incubated with BICD2 and flowed into a motility chamber. The number of moving DDB motors, as visualized by sfGFP fluorescence on BICD2, was quantified per minute per micrometer length of microtubules in the assay; mean and SEM from n = 3 independent experiments (each experiment measuring a minimum of 30 microtubules); *, P ≤ 0.05. (D) Velocities of the moving motors from C is shown; mean and SEM from n = 3 independent protein preparations and experiments (each experiment measuring a minimum of 100 DDB complexes). (E) The run lengths of WT and mutant DDBFL; mean and SEM from n = 3 independent experiments (each experiment measuring a minimum of 130 DDB complexes). For processivity measurements, the NaCl concentration was increased from 50–65 mM to reduce the run length so that it could be more reliably measured (see Materials and methods). (F) Purified dynein and dynactin were incubated with truncated BICD225–400 (WT and mutant) and flowed into a motility chamber. The number of moving DDB complexes, as visualized by SNAP-TMR fluorescence on BICD2, was quantified per minute per micrometer of microtubule; mean and SEM from n = 3 independent protein preparations and experiments (each experiment measuring a minimum of 30 microtubules).
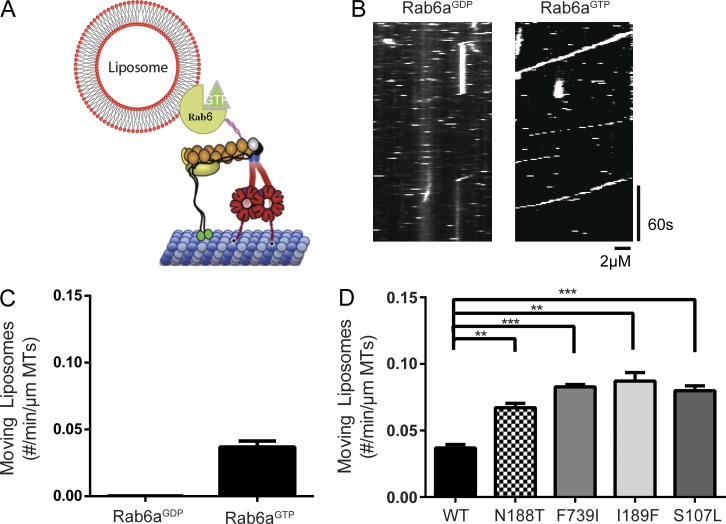
A liposome assay provides an in vitro system for cargo transport
The retrograde transport of native membranous cargos (Utskarpen et al., 2006; Grigoriev et al., 2007) involves prenylated, membrane-bound Rab6a interacting with BICD2. We sought to recapitulate this cargo motility in vitro using recombinant Rab6aGTP or Rab6aGDP covalently linked to large unilamellar vesicles via maleimide lipids (Fig. 3 A). With Rab6aGTP on the liposomes, we observed the frequent binding and movement of liposomes along microtubules in the presence of dynein, dynactin, and BICD2FL (Fig. 3, B and C). In contrast, motility events were only rarely observed with Rab6aGDP-coated liposomes (Fig. 3, B and C; and Video 2). Next, we examined the BICD2FL mutants in the Rab6aGTP liposome motility assay. All four mutants produced significantly more moving Rab6aGTP liposomes compared with WT BICD2FL (Fig. 3 D). The velocities of WT and mutant BICD2 were similar (500 nm/s; Fig. S2 F) and slightly higher than the single-molecule velocities. Thus, consistent with the single-molecule motility assays, the BICD2 mutants hyperactivated dynein–dynactin motility in an in vitro cargo transport assay.
Figure 3.
A motility assay using liposomes recapitulates single-molecule data. (A) A schematic of the liposome motility assay. Rab6a was conjugated to maleimide lipids incorporated into 200-nm-sized liposomes and incubated in solution with DDB. Liposomes were labeled with 0.1% rhodamine–PE. (B) Representative kymographs of Rab6aGDP versus Rab6aGTP liposomes moving along microtubule in the presence of DDBFL. Horizontal dashed lines represent liposomes transiently entering the field of focus. (C) Quantification of number of motile Rab6aGDP or Rab6aGTP liposomes when incubated with DDBFL; mean and SEM from n = 3 independent experiments (each experiment measuring a minimum of 30 microtubules; runs for Rab6aGDP occurred very infrequently). (D) Quantification of the number of motile Rab6aGTP liposomes moving on microtubules when incubated with DDBFL (WT vs. mutant); mean and SEM from n = 3 independent experiments (each experiment measuring a minimum of 30 microtubules); **, P ≤ 0.01; ***, P ≤ 0.001.
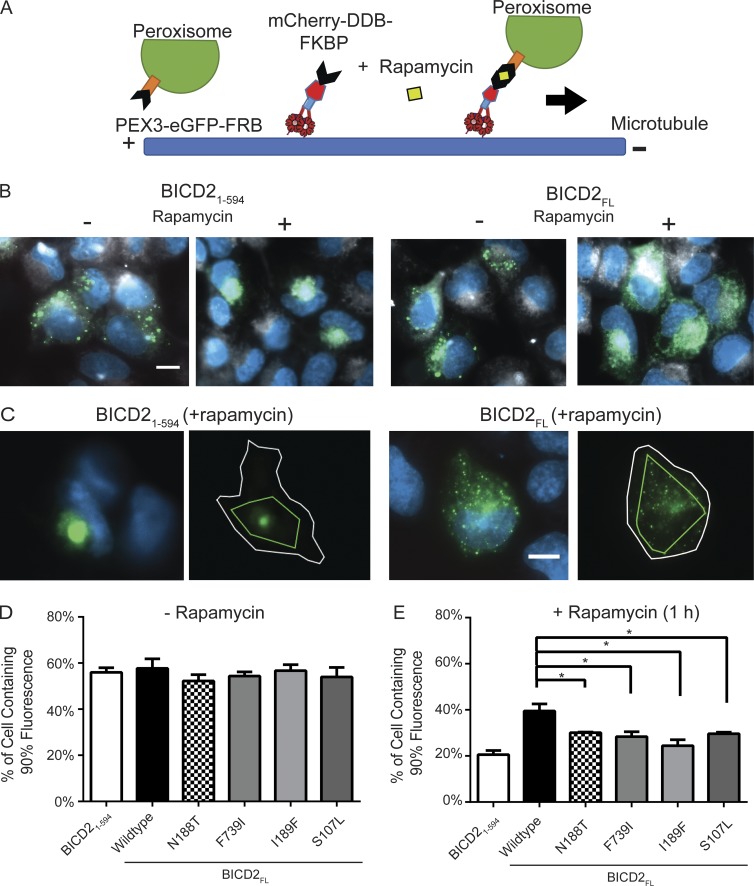
Enhanced retrograde motility induced by BICD2 mutants in a cell-based assay
To examine the behavior of WT and mutant BICD2 in cells, we turned to a previously established inducible cargo trafficking assay (Hoogenraad et al., 2003; Kapitein et al., 2010). In this assay, FKBP12-rapamycin binding domain (FRB) fused to GFP and a peroxisome localization sequence is coexpressed with BICD2 fused to FK506 binding protein (FKBP; Fig. 4 A). As shown previously, an N-terminal construct of BICD2 (BICD21–594-FKBP) in the presence of rapamycin increased retrograde transport of peroxisomes and produced significant clustering of the GFP–peroxisome signal around the perinuclear region (Hoogenraad et al., 2003; Fig. 4 B). This longer construct includes coiled coil 2 but still lacks the C-terminal region needed for autoinhibition. In contrast, the autoinhibited BICD2FL-FKBP construct displayed a more dispersed peroxisome signal (Fig. 4 B).
Figure 4.
Cellular dynein–dynactin motility induced by WT and mutant BICD2. (A) Schematic of the cell-based peroxisome motility assay. U2OS cells were cotransfected with a plasmid expressing GFP–FRB with a PEX3 localization sequence, targeting the protein to peroxisomes, and an mCherry-BICD2–FKBP construct. After addition of rapamycin, mCherry-BICD2–FKBP is recruited to peroxisomes. If in an active state, the BICD2 recruits dynein–dynactin, which transport the peroxisome along microtubules toward the centrosome. (B) Representative epifluorescence images of the assay for both BICD21–594 and BICD2FL. Perixosomes are shown in green, the white signal represents the far-red cell membrane marker, CellMask, and blue is DAPI. In the absence of rapamycin, peroxisomes decorated with GFP–FRB are distributed throughout the cytoplasm (columns 1 and 3). 1 h after rapamycin addition, peroxisomes in cells expressing BICD21–594 are heavily clustered at the cell center (column 2), whereas those expressing BICD2FL exhibit less clustering (column 4). Bar, 10 µm. (C) Example image of an analyzed cell for both BICD21–594 and BICD2FL. The white outline depicts the cell boundary; the inner green line indicates the area that contains 90% of fluorescence (see Materials and methods). Bar, 10 µm. (D and E) Quantification of the cell clustering data. The percentage of the cell area in which 90% of total fluorescence was contained is used as a measure for the degree of clustering (see Materials and methods). Lower values reflect a higher degree of clustering; mean and SD from n = 3 independent experiments (each experiment measuring a minimum of 25 cells for each construct); *, P ≤ 0.05.
We next tested the mutant BICD2FL constructs in this inducible peroxisome motility assay. To quantify the degree of peroxisome clustering due to minus end–directed motility, we used a previously created plugin for the image analysis software Icy that calculates the fraction of the cell area containing 90% of the total peroxisome GFP signal; increased transport to the centrosome results in a more clustered signal, and thus 90% of the signal occupies less area (de Chaumont et al., 2012; Mounier et al., 2012; Fig. 4 C). In the absence of rapamycin, the clustering values for the WT and mutants were similar (55%; Fig. 4 D). With the addition of rapamycin to cells expressing BICD21–594, the peroxisomes occupied 20% of the cell area compared with 40% expressing autoinhibited BICD2FL. The four mutants tested in the BICD2FL construct, while not achieving the same degree of peroxisome clustering observed for BICD21–594, showed an increase in the compaction of the peroxisome signal compared with WT BICD2FL (Fig. 4 D). Thus, similar to the in vitro data, the mutant BICD2FL proteins hyperactivated dynein–dynactin in a cellular context and more efficiently transported peroxisomes toward the centrosomes.
We also tested whether localizing Rab6aGTP containing a peroxisome-targeting sequence (Rab6aGTP–PEX) could increase the activity of FKBP–BICD2FL. However, we found that Rab6aGTP–PEX alone (without FKBP–BICD2FL coexpression) was sufficient to cluster peroxisomes, presumably through the recruitment of endogenous BICD2 and then dynein–dynactin (Fig. S3, A–C). Thus, targeting of Rab6aGTP on peroxisome membranes is sufficient for the recruitment and activation of endogenous BICD2 to produce dynein-mediated transport of peroxisomes.
Overexpression of BICD2 mutants in hippocampal neurons decreases neurite length
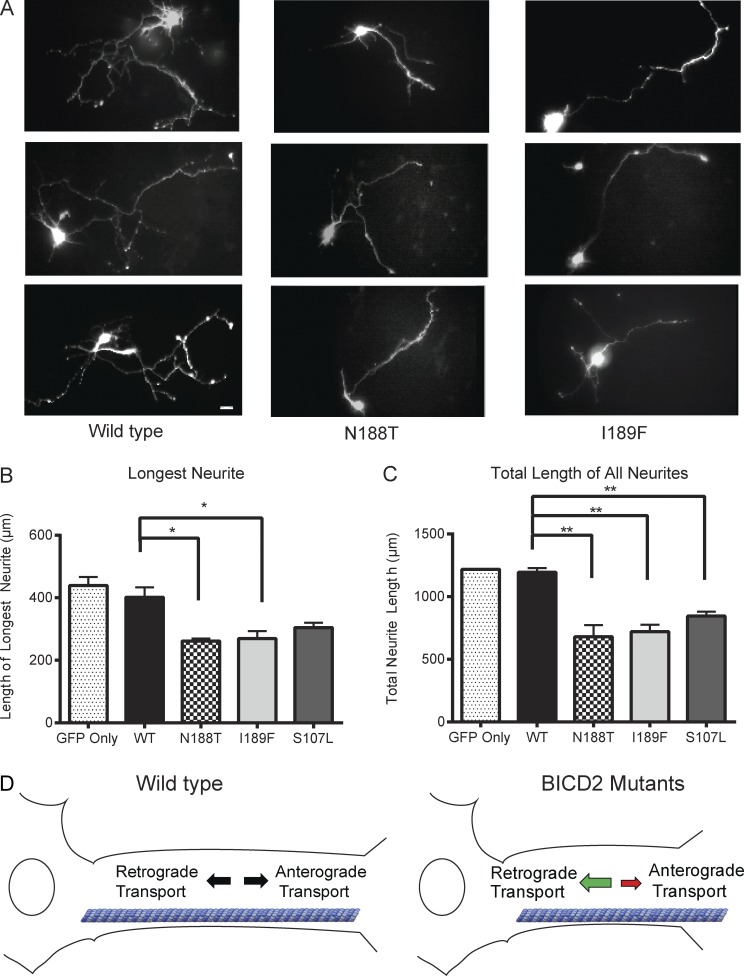
We next examined whether the hyperactivating BICD2FL mutants affected neurite outgrowth and neuronal morphology. Rat hippocampal neurons, dissected from embryonic day 18 tissue and grown in culture, were transfected with either WT or mutant mouse mCherry–BICD2FL constructs along with soluble GFP to visualize the cell body and neurites (Fig. 5 A). Three days later, dual mCherry- and GFP-positive neurons were then scored for the length of their longest process as well as the total length of all processes. Cells transfected with WT mCherry–BICD2FL and GFP showed similar neurite lengths compared with cells transfected with GFP alone (Fig. 5, B and C). This result is consistent with previous work showing that overexpression of WT BICD2 in either hippocampal or rat dorsal root ganglion neurons does not affect axon length or overall neurite length (Schlager et al., 2014b). However, cells transfected with the disease mutant BICD2FL constructs for 3 d showed a 40% decrease in both the length of the longest neurite as well as an overall decrease in total neurite length after 3 d (Fig. 5, B and C). This result is similar to what was reported when the BICD-related protein BICDR-1, which forms a higher velocity DDB complex, is overexpressed in neurons. (Schlager et al., 2010, 2014b). The number of long (>15-µm) processes emanating from the cell body was not affected by the BICD2 mutants (Fig. S3 D). The Drosophila mutant (F739I) also showed a significant reduction in total neurites, but not in the axon length (Fig. S3, E and F). These results demonstrate that mutations in BICD2 affect the process of neurite outgrowth of hippocampal neurons in culture.
Figure 5.
Overexpression of BICD2 mutants in rat hippocampal neurons results in a decrease in neurite length. (A) Representative images of hippocampal neurons overexpressing mCherry–BICD2FL (WT and mutants) or an empty plasmid control; neurons were also cotransfected with soluble GFP in order to mark the neurites. Bar, 10 µm. (B) Axon length after 3 d of overexpression of BICD2FL constructs; mean and SD from n = 3 independent experiments (each experiment measuring 15–20 neurons for each construct); *, P ≤ 0.05. (C) Total neurite length after 3 d of overexpression of BICD2FL constructs; mean and SD from n = 3 independent experiment (each experiment measuring 15–20 neurons for each construct); **, P ≤ 0.01. (D) Model of how a gain of function in dynein-based motility from BICD2 mutants might cause an imbalance in axonal transport and lead to SMA. See Discussion for more details.
In conclusion, our vitro and cell-based assays recapitulate Rab6aGTP-dependent activation of DDB motility and reveal that three BICD2 disease-associated mutations and one Drosophila mutant all display a similar phenotype of eliciting more dynein–dynactin motility. These effects were not evident with the Rab6a-independent, constitutively active BICD225–400 construct. These results suggest a scenario in which the mutants lower the energy barrier for Rab6aGTP to convert the autoinhibited BICD2FL into an active conformation. Future work can test this possibility by using assays that probe the conformational states of BICD2FL.
Previous cell-based models have suggested that BICD2 disease mutants may have a dominant-negative effect on dynein function, resulting in disrupted Golgi morphology (Burkhardt et al., 1997; Harada et al., 1998; Neveling et al., 2013). Although we cannot rule out dominant-negative effects that cannot be measured with our assays, our data suggest that the mutants produce a dominant-positive effect leading to enhanced dynein cargo transport. Although subtle (25% increase in motile DDB complexes), such effects may perturb transport in an axon (which can be up to 1 m long in a motor neuron) over time.
To explain the SMA-LED2 disease phenotype, we propose a model in which mutant BICD2 increases dynein activation and leads to an imbalance between anterograde and retrograde transport (Fig. 5 D). The shift toward dynein transport could produce a decrease in the delivery of cargo to the nerve terminal, which in turn could lead to a gradual decline in function of neurons with long axons. This model is congruous with experimental data from transgenic mouse models. BICD2-null mice possess a normal spinal cord and their motor neurons exhibit no evidence of abnormal retrograde transport (Jaarsma et al., 2014). These mice, which die at around p30, do not display a SMA phenotype, suggesting that the absence or inhibition of BICD2 activity cannot explain the disease etiology. In contrast, transgenic mice expressing the shorter, more active N-terminal construct of BICD2 were impaired in retrograde axonal transport and Golgi fragmentation, presumably through a dominant-negative effect of activating dynein and dynactin without cargo and causing its accumulation in the cell body (Teuling et al., 2008). Curiously, however, these mice, which live up to 2 yr, also did not develop any motor abnormalities. The phenotypic differences in mice expressing the N-terminal construct of BICD2 compared with haploid S107L, N188T, and I189F mutations suggests that the C terminus of the BICD2 protein that interacts with cargo is necessary for contributing to the SMA disease phenotype.
Recently, Hoang et al. (2017) showed that disease-related mutants in the human dynein heavy chain, including three mutations associated with SMA, exhibited normal DDB complex formation and velocity, but displayed a 60%–70% reduction in processivity. These results differ from those described for the SMA BICD2 mutants, which show no change in processivity and a gain-of-function increase in the amount of retrograde transport. Taken together, these studies on dynein and BICD2 mutations reveal multiple ways in which changes in motor activity can affect can affect the homeostasis and function of neurons in SMA.
Materials and methods
DNA constructs
The cDNAs for mouse BICD2 (GenBank accession no. AJ250106.1) and mouse Rab6a (GenBank accession no. BC019118.1) were obtained from the Thermo Fisher Scientific MGC collection. BICD2 N-terminal constructs were cloned into a pet28a vector containing an N-terminal 6xHis-strepII-sfGFP tag. Full-length BICD2 WT and mutant constructs were cloned into a pFastbac vector containing either an N-terminal StrepII-sfGFP or StrepII-SNAPf tag followed by a C-terminal 6xHis tag for tandem purification. Rab6a was cloned into a pGEX vector containing an N-terminal GST tag followed by a PreScission protease cleavage site. The Rab6a constructs span the GTPase domain of Rab6a (aa 8–195), followed by a short linker and either a KCK motif for maleimide labeling purposes (liposome experiment) or a His10 tag. Point mutations for both the BICD2 constructs and the Rab6a GDP/GTP (Q72L and T27N, respectively) constructs were created by the protocol of Phusion.
For the cell-based peroxisome assay, the sequence spanning aa 1–42 of human PEX3 was synthesized via a gBlock (IDT) and was cloned into the N-terminal of a modified eGFP vector, followed by a C-terminal FRB sequence. BICD2 constructs for this assay were cloned into a pHR vector containing an N-terminal mCherry sequence and C-terminal FKBPf36v sequence. The Rab6a–PEX construct was constructed by fusing the PEX3 sequence to the C terminus of Rab6a lacking the cysteines for lipidation (1–203 aa). An mTagBFP2 was fused to the N terminus of this construct as well. For neuron expression, BICD2 was cloned into an N-terminal pmCherry vector.
Protein purification
BICD2 N-terminal and Rab6a constructs were transformed into the Escherichia coli strain BL21 RIPL from Agilent. Cells were grown in LB at 37°C until growth reached 0.6 OD280. The temperature was then lowered to 18°C, and cells were induced overnight with 0.5 mM IPTG. Bacterial pellets were resuspended in either strep lysis buffer (30 mM Hepes, pH 7.4, 300 mM NaCl, 2 mM MgCl2, 5% glycerol, 5 mM DTT, and 1 mM PMSF) for BICD2 or PBS containing 5% glycerol, 5 mM DTT, and 1 mM PMSF for Rab6a. Cells were lysed using an Emulsiflex press (Avestin) and clarified at 40,000 g for 60 min. Lysates were bound to either Strep-Tactin Superflow Plus resin (QIAGEN) or GST Sepharose 4B (GE Lifesciences). Beads were then washed extensively, and the bound protein was eluted with 3 mM desthiobiotin in lysis buffer (BICD2) or via overnight cleavage with PreScission (Rab6a).
BICD2 full-length constructs were purified from SF9 cells and purified using tandem purification. Bacmids isolated from DH10Bac cells were transfected into SF9 cells. P2 virus was used to infect SF9 cells grown in shaker flasks to a density of 2 × 106 cells/ml. After 60 h of infection, cells were harvested, pelleted, and resuspended in NiNTA lysis buffer (30 mM Hepes, pH 7.4, 300 mM NaCl, 5% glycerol, 20 mM imidazole, 1 mM PMSF, and 5 mM βME) and lysed using an Emulsiflex. The lysate was clarified at 40,000 g for 1 h and bound to NiNTA resin from QIAGEN. After extensively washing with lysis buffer, cells were eluted with lysis buffer containing 500 mM imidazole. The eluted material was then diluted to lower the imidazole concentration to 100 mM, and bound to Strep-Tactin beads. The beads were then washed with lysis buffer and eluted with lysis buffer containing 3 mM desthio-biotin. After elution, proteins were concentrated and injected onto either a Superose 6 10/300GL column (BICD2 constructs) or a S200 10/300GL column (Rab6a) from GE Healthcare. Gel filtration buffer was composed of 30 mM Hepes, pH 7.4, 150 mM NaCl, 10% glycerol, and 2 mM tris(2-carboxyethyl)phosphine. Peak fractions were pooled and concentrated and then flash frozen. For SNAP-fusion constructs, purified proteins were bound to Strep-Tactin beads and incubated at a 2:1 ratio of SNAP-TMR Star:Protein on ice for 1 h. Beads were briefly washed in the gel filtration buffer, and the protein was eluted with 3 mM desthiobiotin in gel filtration buffer.
Dynein and dynactin were prepared from RPE-1 cells. Fifteen 15-cm dishes of RPE-1 cells were grown to 80%–900% confluency and then washed with PBS. The cells were then harvested using a cell scraper and pelleted at 500 g for 5 min. They were then resuspended in 2.5 ml buffer A (30 mM Hepes, pH 7.4, 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, and 10% glycerol) along with 5 mM DTT, 1 mM PMSF, and 1% Triton X-100 and incubated on ice for 10 min. The lysate was clarified at 266,000 g and diluted to a volume of 25 ml using buffer A containing 1 mM PMSF and 5 mM DTT to reach a final Triton X-100 concentration of 0.1%. Strep-Tactin Sepharose (GE Healthcare) bound to purified StrepII-SNAP-FIP3 was added to the lysate and incubated for 1 h. The beads were then washed four times with buffer A containing 5 mM DTT and 0.1% NP-40. Buffer A containing an additional 300 mM NaCl was then added to the beads to form a 50% slurry and incubated on ice for 10 min to elute the dynein and dynactin bound to FIP3. This slurry was then passed through a 0.22-µm filter to remove the beads. An equal volume of 50% Strep-Tactin Sepharose was added to this elution and allowed to gently mix for 30 min at 4°C to remove any remaining StrepII-SNAP-FIP3. The slurry was then filtered again and then diluted with buffer A to reach a final NaCl concentration of 200 mM. Sucrose was added to a final concentration of 6%, and the dynein and dynactin were then flash frozen as aliquots.
Porcine brain pull-downs
Pull-downs were performed as previously described (McKenney et al., 2014). 500 µl clarified porcine brain lysate in buffer A (30 mM Hepes, pH 7.4, 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, and 10% glycerol) was added to 80 µl of a 50% suspension of Strep-Tactin Sepharose beads (GE Healthcare) along with 100 nM of the BICD2 construct to be tested. To this, 0.1% of NP-40, 1 mM PMSF, and 5 mM DTT were added, and the mixture was incubated at 4°C for 1 h. The beads were then pelleted and washed four times with buffer A containing 0.1% NP-40 and 5 mM DTT. The proteins were then eluted using SDS loading buffer and resolved on an acrylamide gel (Invitrogen). For experiments in which Rab6a was supplemented, the appropriate amount of recombinant Rab6a protein was added before the 4°C incubation step and processed in the same fashion.
Immunoblot analysis
The SDS-PAGE gel was transferred onto a nitrocellulose membrane using the iBlot system from Invitrogen. Western blotting was performed using TBS buffer with 0.1% Tween-20 (TBS-T). Membranes were blotted for 1 h at RT with 5% milk in TBS-T, followed by a 1-h primary antibody incubation. Antibodies used were as follows: mouse anti-p150 (clone 12, 1:250; BD), mouse anti-dynein intermediate chain (clone 74.1, 1:1,000; EMD Millipore), mouse anti-6xHis (1:1,000; Roche), and mouse anti-Rab6a (ab55660, 1:500; Abcam). Membranes were washed three times with TBS-T and then incubated with a secondary antibody for 1 h at RT. Secondary antibodies were anti–mouse-800 or anti–rabbit-680 (1:10,000; Molecular Probes). Membranes were washed three times with TBS-T and then imaged using an Odyssey Clx Infrared Imaging System (LI-COR Biosciences). Quantification of band intensity was performed using ImageJ’s (National Institutes of Health) gel analysis feature. P-values were calculated using a one-sample t test with a hypothetical mean of 1.
Liposome preparation and Rab6a labeling
Liposomes were prepared from a mixture of lipids dissolved in chloroform at (82.4% POPC, 15% POPS, 2.5% 18:1 PE MCC, and 0.1% rhodamine PE; Avanti). After mixing, they were dried under a constant stream of argon and then desiccated in vacuum overnight. The lipid film was then resuspended with buffer A without glycerol and passed 25 times through an extruder containing a 200-nm pore size polycarbonate filter. A 3:1 ratio of Rab6a to maleimide lipid was then mixed together and incubated overnight. The next day, DTT was added to a final concentration of 10 mM to quench the reaction, and the liposomes were pelleted at 160,000 g for 20 min to remove unlabeled Rab6a. The pellet was then washed and resuspended with buffer A.
Single-molecule and liposome imaging
Preparation of microtubules
Microtubules were prepared as previously described (McKenney et al., 2014). Unlabeled tubulin was mixed with biotinylated tubulin and Alexa-640-labeled tubulin at a ratio of 10:2:2 in BRB80 (80 mM Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2). 5 mM GTP was added and the mixture was incubated in a 37°C water bath for 10 min before adding 20 µM Taxol and incubated for an additional 50 min at 37°C. Microtubules were spun over a 25% sucrose cushion at 160,000 g for 10 min in a tabletop centrifuge before use.
Preparation of DDB complexes and imaging
DDB complexes were formed in a 30-µl reaction volume containing ∼20 nM recombinant BICD2, ∼0.15 mg/ml of dynein/dynactin, and buffer A along with 0.1 mg/ml Biotin-BSA, 0.5% pluronic acid F-127, and 0.2 mg/ml κ-casein (buffer B). The proteins were mixed and incubated on ice for 20 min before use. In experiments in which Rab6a was also included, the respective construct was added to a final concentration of 1 µM. Flow chambers with attached microtubules were performed as described (Schroeder and Vale, 2016). A 1:4 or 1:5 dilution of the DDB complex in buffer B was then added in presence of 1 mM ATP and the Trolox/PCA/PCD oxygen scavenging system (Aitken et al., 2008). For processivity experiments, a SNAP-BICD2 construct labeled with the TMR-Star fluorophore from NEB was used. Because the run lengths using buffer B tended to span the entire lengths of our microtubules, we added 15 mM of KCl to buffer B (termed buffer C) for measurement of processivity. The increased amount of salt in buffer C reduced the run length for DDB (4 µm) so that it could be measured more accurately on microtubules (mean length of 16 µm used in these measurements).
For liposome assays, the same components as in the aforementioned DDB mixture were mixed in a tube, along with either Rab6aGTP or Rab6aGDP liposomes. The liposomes were diluted to a final concentration of 2.5 µM total lipid. After incubation, the solution was flowed in undiluted into an imaging chamber after addition of 1 mM ATP and Trolox/PCA/PCD. The amount of BICD2–GFP fluorescence on the liposomes was used to determine the mean number of molecules bound to each liposome by comparing it to that of single GFP molecules captured via biotin-anti-GFP stuck onto a coverslip coated with streptavidin. Fluorescence intensities were measured by integrating the average fluorescence intensity over five frames at 200-ms exposure and quantified with the FiJi plugin Spot Intensity Analysis (http://imagej.net/Spot_Intensity_Analysis). Liposome GFP fluorescence (bound to glass) was normalized to single GFP fluorescent yielded an average of 6.01 BICD2 molecules per Rab6aGTP liposomes (n = 6,000 intensity measurements of single GFP and n = 728 intensity measurement of BICD2–GFP on liposomes).
Total internal reflection fluorescence images were acquired on a Nikon Eclipse TE200-E microscope equipped with an Andor iXon electron-multiplying charge-coupled device camera, a 100× 1.49 NA objective, and MicroManager software (Edelstein et al., 2010). Exposures of 100 or 200 ms with a frame interval of 1 per second for 300 s was typically used for the acquisition. All assays were performed at RT.
Peroxisome assay in U2OS cells
U2OS cells from ATCC were cultured in DMEM containing 10% FCS and 1× penicillin/streptomycin/glutamine. Cells were seeded in glass-bottom 96-well plates and were at 40% confluence on the day of transfection. Cells were transfected with TransIT Lt1 as per the manufacturer’s protocol with a combination of the PEX–eGFP–FRB, mCherry–BICD2–FKBP, and PEX–Rab6a plasmids and incubated overnight. The next day, 1 µM rapamycin or DMSO was added to cells and allowed to incubate at 37°C for 1 h. 10 min before the end of this incubation, Cell Mask Far Red (Thermo Fisher Scientific) was added (0.5× of the manufacturer’s solution) to stain the plasma membrane. Cells were then washed with PBS and fixed with 4% PFA at RT, followed by extensive washing with PBS. Cells were imaged at RT using an inverted Nikon Eclipse Ti microscope using a 0.75 NA 40× air objective, Andor Xyla camera, and Micro-Manager software (Edelstein et al., 2010).
Primary hippocampal neuron cultures and transfection
Primary hippocampal neuron cultures were prepared from tissue from embryonic day 18 (E18) rat brains that were shipped overnight on ice from BrainBits. Cells were prepared according to provider’s instructions. Briefly, the tissue was incubated with a 2 mg/ml solution of papain in Hibernate E without calcium (BrainBits) for 10 min at 30°C. The tissue was then triturated using a fire-polished Pasteur pipette for 1 min, and then large tissue pieces were allowed to settle for 1 min. The supernatant was collected and centrifuged at 200 g for 1 min to collect cells. After removal of most of the supernatant, the pellet was resuspended with 1 ml NbActiv1 and the cells were counted after staining with Trypan Blue (1:5 dilution). 50,000 cells were seeded onto poly-d-lysine/laminin coated round coverslips (Corning BioCoat) placed into wells of a 24-well plate.
One day after plating, neurons were transfected using Lipofectamine 2000 (Invitrogen). Each transfection reaction contained 1 µg total plasmid DNA along with 4 µl Lipofectamine reagent incubated together for 20 min at RT. Media from the neurons was then removed and saved, and fresh NbActiv was added to the wells along with the transfection mixture. After 2.5 h at 37°C in 5% CO2, the neurons were washed with NbActiv, and the original conditioned media was added back. Cells were fixed 3 d after transfection using 4% PFA for 20 min at RT. After washing, the coverslips were mounted onto slides using VECTASHIELD (Vector Labs). Neurons were imaged for both soluble GFP and mCherry–BICD2 signal at RT using a Zeiss Axiovert 200M microscope with a 0.5 NA 20× air objective, a C4742-98 charge-coupled device camera (Hamamatsu), and Micro-Manager software (Edelstein et al., 2010).
Image analysis and quantification
Analysis of DDB and liposome motility and run length
Kymographs were created from the single-molecule or liposome movies using ImageJ. The number of runs per micrometer of MT per unit of time was obtained from these kymographs, with each run being scored if it was >1 µm in length. P-values were calculated using an unpaired t test. The cumulative frequency was used for analysis of run lengths, as previously described (McKenney et al., 2014). The one-cumulative frequency distribution was fitted with a one-phase exponential decay for run lengths of >1 µM, as runs shorter than this were undersampled or difficult to measure given our imaging conditions (Thorn et al., 2000).
Analysis of peroxisome clustering
The images acquired from fixed U2OS cells were processed and analyzed using the Compaction Profiler plugin previously developed for Icy software (Mounier et al., 2012). In brief, the fluorescent signal from the GFP channel corresponding to peroxisomes was detected after masking the shape of cell via the CellMask channel and quantified using the Spot Detector plugin in Icy. The detected fluorescence data were then passed along to the Compaction Profiler plugin, which then calculated the area of a region of interest inside the cell that represents 90% of the total fluorescence signal. A higher degree of clustering results in a smaller calculated area, whereas a more diffusive signal yields a larger area. P-values were calculated using an unpaired t test.
Analysis of primary hippocampal neurons
Neurite lengths were measured using the soluble GFP signal from the transfected plasmid. Acquired images were processed via ImageJ. The background was subtracted using the default setting, followed by a median filter with a value of 2.0. The images were adjusted for brightness and contrast and the ImageJ plugin NeuronJ was then used to trace all neurites, including branches from individual neurite. The longest neurite from a cell was also identified and measured. P-values were calculated using an unpaired t test.
Online supplemental material
Fig. S1 shows a comparison of a brain lysate pull-down between BICD225–400 and BICD2FL, a Rab6a blot for the titration experiment in Fig. 1, along with the quantification of dynein intermediate chain and p150 intensity for Rab6aGTP, single-molecule quantification of BICD225–400 with or without Rab6a, and a GST pull-down showing Rab6a binding to BICD2FL but not the truncated BICD2. Fig. S2 shows quantification of the pull-down experiment in Fig. 2, processivity and motility with Rab6aGDP between WT and mutants, a pull-down using the N-terminal construct of either WT and mutant, and velocity of liposomes from Fig. 3. Fig. S3 displays peroxisome experiments in which Rab6a–PEX is cotransfected as well as neuron data for the fly mutant and an analysis of the number of processes emanating from the neurons for both WT and mutants. Video 1 is a comparison of BICD225–400 versus BICD2FL DDB. Video 2 is a comparison of Rab6aGDP and Rab6aGTP liposome motility.
Supplementary Material
Acknowledgments
We thank N. Stuurman for help with microscopy; R. McKenney for both technical help and discussions; and C. Schroeder, Y. Wang, and S. Niekamp for discussions as well.
This work was supported by a Maximizing Investigators’ Research Award from the National Institute of General Medical Sciences (NIH 1R35 GM118106).
The authors declare no competing financial interests.
Author contributions: W. Huynh performed the experiments. W. Huynh and R.D. Vale designed the experiments and wrote the manuscript.
Footnotes
Abbreviations used:
- BICD2
- bicaudal D2
- FKBP
- FK506 binding protein
- FRB
- FKBP12-rapamycin binding domain
- sfGFP
- superfolder-GFP
- SMA
- spinal muscular atrophy
References
- Aitken C.E., Marshall R.A., and Puglisi J.D.. 2008. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94:1826–1835. 10.1529/biophysj.107.117689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V.J. 2011. Cytoplasmic dynein. Biochem. Soc. Trans. 39:1169–1178. 10.1042/BST0391169 [DOI] [PubMed] [Google Scholar]
- Burkhardt J.K., Echeverri C.J., Nilsson T., and Vallee R.B.. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469–484. 10.1083/jcb.139.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco M.A., Desantis M.E., Leschziner A.E., and Reck-peterson S.L.. 2015. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol. 31:83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chaumont F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y., et al. . 2012. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods. 9:690–696. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using manager. Curr. Protoc. Mol. Biol. Chapter 14:Unit 14.20 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., Splinter D., Keijzer N., Wulf P.S., Demmers J., Ohtsuka T., Modesti M., Maly I.V., Grosveld F., Hoogenraad C.C., and Akhmanova A.. 2007. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell. 13:305–314. 10.1016/j.devcel.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Harada A., Takei Y., Kanai Y., Tanaka Y., Nonaka S., and Hirokawa N.. 1998. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J. Cell Biol. 141:51–59. 10.1083/jcb.141.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang H.T., Schlager M.A., Carter A.P., and Bullock S.L.. 2017. DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein-dynactin-cargo adaptor complexes. Proc. Natl. Acad. Sci. USA. 114:E1597–E1606. 10.1073/pnas.1620141114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad C.C., Wulf P., Schiefermeier N., Stepanova T., Galjart N., Small J.V., Grosveld F., de Zeeuw C.I., and Akhmanova A.. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22:6004–6015. 10.1093/emboj/cdg592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D., van den Berg R., Wulf P.S., van Erp S., Keijzer N., Schlager M.A., de Graaff E., De Zeeuw C.I., Pasterkamp R.J., Akhmanova A., and Hoogenraad C.C.. 2014. A role for Bicaudal-D2 in radial cerebellar granule cell migration. Nat. Commun. 5:3411 10.1038/ncomms4411 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Schlager M.A., van der Zwan W.A., Wulf P.S., Keijzer N., and Hoogenraad C.C.. 2010. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J. 99:2143–2152. 10.1016/j.bpj.2010.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Salter H.K., Holding A.N., Johnson C.M., Stephens E., Lukavsky P.J., Walshaw J., and Bullock S.L.. 2013. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev. 27:1233–1246. 10.1101/gad.212381.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T., Akhmanova A., Wulf P., Del Nery E., Weide T., Stepanova T., Galjart N., Grosveld F., Goud B., De Zeeuw C.I., et al. . 2002. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4:986–992. 10.1038/ncb891 [DOI] [PubMed] [Google Scholar]
- McKenney R.J., Huynh W., Tanenbaum M.E., Bhabha G., and Vale R.D.. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 345:337–341. 10.1126/science.1254198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler J., and Wieschaus E.F.. 1986. Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics. 112:803–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J., Boncompain G., Senerovic L., Lagache T., Chrétien F., Perez F., Kolbe M., Olivo-Marin J.C., Sansonetti P.J., and Sauvonnet N.. 2012. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe. 12:381–389. 10.1016/j.chom.2012.07.010 [DOI] [PubMed] [Google Scholar]
- Neveling K., Martinez-Carrera L.A., Hölker I., Heister A., Verrips A., Hosseini-Barkooie S.M., Gilissen C., Vermeer S., Pennings M., Meijer R., et al. . 2013. Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am. J. Hum. Genet. 92:946–954. 10.1016/j.ajhg.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates E.C., Rossor A.M., Hafezparast M., Gonzalez M., Speziani F., MacArthur D.G., Lek M., Cottenie E., Scoto M., Foley A.R., et al. UK10K . 2013. Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am. J. Hum. Genet. 92:965–973. 10.1016/j.ajhg.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters K., Litvinenko I., Asselbergh B., Almeida-Souza L., Chamova T., Geuens T., Ydens E., Zimoń M., Irobi J., De Vriendt E., et al. . 2013. Molecular defects in the motor adaptor BICD2 cause proximal spinal muscular atrophy with autosomal-dominant inheritance. Am. J. Hum. Genet. 92:955–964. 10.1016/j.ajhg.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters K., Bervoets S., Chamova T., Litvinenko I., De Vriendt E., Bichev S., Kancheva D., Mitev V., Kennerson M., Timmerman V., et al. . 2015. Novel mutations in the DYNC1H1 tail domain refine the genetic and clinical spectrum of dyneinopathies. Hum. Mutat. 36:287–291. 10.1002/humu.22744 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S.L., Yildiz A., Carter A.P., Gennerich A., Zhang N., and Vale R.D.. 2006. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 126:335–348. 10.1016/j.cell.2006.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M.A., Kapitein L.C., Grigoriev I., Burzynski G.M., Wulf P.S., Keijzer N., de Graaff E., Fukuda M., Shepherd I.T., Akhmanova A., and Hoogenraad C.C.. 2010. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 29:1637–1651. 10.1038/emboj.2010.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M.A., Hoang H.T., Urnavicius L., Bullock S.L., and Carter A.P.. 2014a In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33:1855–1868. 10.15252/embj.201488792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M.A., Serra-Marques A., Grigoriev I., Gumy L.F., Esteves da Silva M., Wulf P.S., Akhmanova A., and Hoogenraad C.C.. 2014b Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell Reports. 8:1248–1256. 10.1016/j.celrep.2014.07.052 [DOI] [PubMed] [Google Scholar]
- Schroeder C.M., and Vale R.D.. 2016. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J. Cell Biol. 214:309–318. 10.1083/jcb.201604002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D., Razafsky D.S., Schlager M.A., Serra-Marques A., Grigoriev I., Demmers J., Keijzer N., Jiang K., Poser I., Hyman A.A., et al. . 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell. 23:4226–4241. 10.1091/mbc.E12-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuling E., van Dis V., Wulf P.S., Haasdijk E.D., Akhmanova A., Hoogenraad C.C., and Jaarsma D.. 2008. A novel mouse model with impaired dynein/dynactin function develops amyotrophic lateral sclerosis (ALS)-like features in motor neurons and improves lifespan in SOD1-ALS mice. Hum. Mol. Genet. 17:2849–2862. 10.1093/hmg/ddn182 [DOI] [PubMed] [Google Scholar]
- Thorn K.S., Ubersax J.A., and Vale R.D.. 2000. Engineering the processive run length of the kinesin motor. J. Cell Biol. 151:1093–1100. 10.1083/jcb.151.5.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utskarpen A., Slagsvold H.H., Iversen T.-G., Wälchli S., and Sandvig K.. 2006. Transport of ricin from endosomes to the Golgi apparatus is regulated by Rab6A and Rab6A′. Traffic. 7:663–672. 10.1111/j.1600-0854.2006.00418.x [DOI] [PubMed] [Google Scholar]
- Wharton R.P., and Struhl G.. 1989. Structure of the Drosophila BicaudalD protein and its role in localizing the posterior determinant nanos. Cell. 59:881–892. 10.1016/0092-8674(89)90611-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.