Filopodia, actin-rich finger-like protrusions, contribute to many pathophysiological conditions. Jacquemet et al. developed FiloQuant, a versatile open-source tool to quantify filopodia imaged using different techniques, including intravital microscopy, and reveal enhanced filopodia dynamics during local breast cancer invasion.
Abstract
Defective filopodia formation is linked to pathologies such as cancer, wherein actively protruding filopodia, at the invasive front, accompany cancer cell dissemination. Despite wide biological significance, delineating filopodia function in complex systems remains challenging and is particularly hindered by lack of compatible methods to quantify filopodia properties. Here, we present FiloQuant, a freely available ImageJ plugin, to detect filopodia-like protrusions in both fixed- and live-cell microscopy data. We demonstrate that FiloQuant can extract quantifiable information, including protrusion dynamics, density, and length, from multiple cell types and in a range of microenvironments. In cellular models of breast ductal carcinoma in situ, we reveal a link between filopodia formation at the cell–matrix interface, in collectively invading cells and 3D tumor spheroids, and the in vitro invasive capacity of the carcinoma. Finally, using intravital microscopy, we observe that tumor spheroids display filopodia in vivo, supporting a potential role for these protrusions during tumorigenesis.
Introduction
The extension of membrane protrusions is a prominent morphological feature during many cellular processes and serves as an important mechanism to probe the ECM and to ascertain the appropriate cellular response. Cellular protrusions are broadly classified in function of membrane shape and/or size and primarily include lamellipodia, membrane blebs, filopodia, and filopodia-like protrusions (Chhabra and Higgs, 2007; Petrie and Yamada, 2012). Filopodia are thin, finger-like projections exploited widely by different cell types, including neurons, endothelial cells, epithelial cells, fibroblasts, and immune cells (Mattila and Lappalainen, 2008; Heckman and Plummer, 2013; Jacquemet et al., 2015), wherein they contribute to cellular communication (Sagar et al., 2015), directional cell migration (Jacquemet et al., 2015), and the establishment of cell–cell junctions (Biswas and Zaidel-Bar, 2017). In vivo, filopodia have been reported to contribute to processes such as endothelial sprouting and angiogenesis (Phng et al., 2013; Wakayama et al., 2015), ECM deposition and remodeling (Sato et al., 2017), epithelial sheet migration during wound healing and dorsal closure (Wood et al., 2002; Millard and Martin, 2008), and embryonic development (Fierro-González et al., 2013).
Filopodia may also contribute to pathological conditions, including cancer and brain disorders (Jacquemet et al., 2015; Kanjhan et al., 2016). We and others have reported that filopodia and filopodia-like protrusions are extensively used by cancer cells to support directional single-cell migration and invasion as well as survival at distant metastatic sites (Shibue et al., 2012, 2013; Jacquemet et al., 2013a, 2016; Arjonen et al., 2014; Paul et al., 2015). In addition, the expression of several filopodia regulatory proteins has been shown to correlate with poor patient survival in multiple cancer types, the down-regulation of which impedes cancer metastasis in animal models (Yap et al., 2009; Arjonen et al., 2014; Li et al., 2014). Therefore, targeting filopodia formation could prove a viable strategy to impair cancer cell metastasis (Jacquemet et al., 2016). However, cancer cell dissemination is an intricate multistep process (Gupta and Massagué, 2006), and the significance of filopodia at every stage of the metastatic cascade is not clear.
In spite of their wide biological importance, filopodia remain poorly studied, primarily because of technical difficulties. In particular, filopodia are difficult to observe, especially in vivo, owing to their small size, the absence of specific markers, and an often labile nature, which is particularly affected by fixation protocols (Wood and Martin, 2002; Sato et al., 2017). In addition, automatic quantification of filopodia properties remains a challenge, despite the availability of dedicated tools, and therefore, filopodia features are often described using manual analyses. To our knowledge, currently available tools to quantify filopodia include FiloDetect (Nilufar et al., 2013), CellGeo (Tsygankov et al., 2014), and ADAPT (Barry et al., 2015), each with unique strengths and shortcomings (Table 1). Limitations of these tools include requirement for proprietary software (i.e., MATLAB and MATLAB Image Processing Toolbox), lack of customizable options to improve filopodia detection, selective dedication to live-cell data or to fixed samples only, designation for single cells only, quantification of filopodia numbers, but not density, and the usage of an unmodifiable and/or complex code source that precludes addition of extra functionalities by nonexperts.
Table 1. Comparison of FiloQuant with previously described filopodia analysis software.
| FiloDetect (Nilufar et al., 2013) | CellGeo (Tsygankov et al., 2014) | ADAPT (Barry et al., 2015) | FiloQuant (this study) | |
|---|---|---|---|---|
| Requirement for proprietary software | Yes (MATLAB) | Yes (MATLAB) | No (ImageJ) | No (ImageJ) |
| Analysis of single images | Yes | No | No | Yes |
| Analysis of movies/filopodia tracking | No | Yes | Yes | Yes in combination with a freely available ImageJ plugin |
| Optimized for single cells | Yes | Yes | Yes | Yes |
| Optimized for cell sheets | No | No | No | Yes |
| Quantification of filopodia length and number | Yes | Yes | Yes | Yes |
| Quantification of edge length to measure filopodia density | No | Yes (indirectly) | No | Yes |
| Options to improve faint filopodia detection | No | No | No | Yes |
| Option to filter filopodia in function of size | No | Yes | Yes | Yes |
| Option to filter filopodia in function of proximity to the cell or colony edge | No | No | No | Yes |
| Batch mode available | No | Yes | Yes | Yes |
| Customizable code (easily) | No | No | No | Yes |
| Easy access to update | No | No | Yes | Yes |
| Analysis of lamellipodia and/or blebs | No | Yes | Yes | No |
In the process of addressing these limitations, we developed FiloQuant to detect filopodia and filopodia-like protrusions (finger-like) and to extract quantifiable data on protrusion dynamics, density, and length from both fixed- and live-cell microscopy images. Here, we provide three versions of FiloQuant (software 1: single image analysis; software 2: semiautomated; software 3: fully automated for large analyses and/or live-cell imaging) on the freely available ImageJ platform, each designed with a different purpose in mind and with alternative levels of user control over the analysis. Using FiloQuant, we demonstrate that filopodia can be successfully detected in different cells and in a range of microenvironments, including during collective or single cancer cell migration in 2D and 3D, in fixed neuronal cultures, and in sprouting endothelial cells in vivo, regardless of the imaging modality. Using FiloQuant, we report that in a cellular model of breast cancer progression filopodia density and length, during collective invasion and in 3D tumor spheroids, appears to correlate with the previously described capacity of these breast cancer cells to invade locally in vivo (Miller et al., 2000; Behbod et al., 2009; Lodillinsky et al., 2016). Finally, using intravital microscopy, we report that tumor spheroids in vivo display a high number of filopodia.
Results
FiloQuant, an ImageJ tool to rapidly quantify filopodia length and density under different cellular contexts
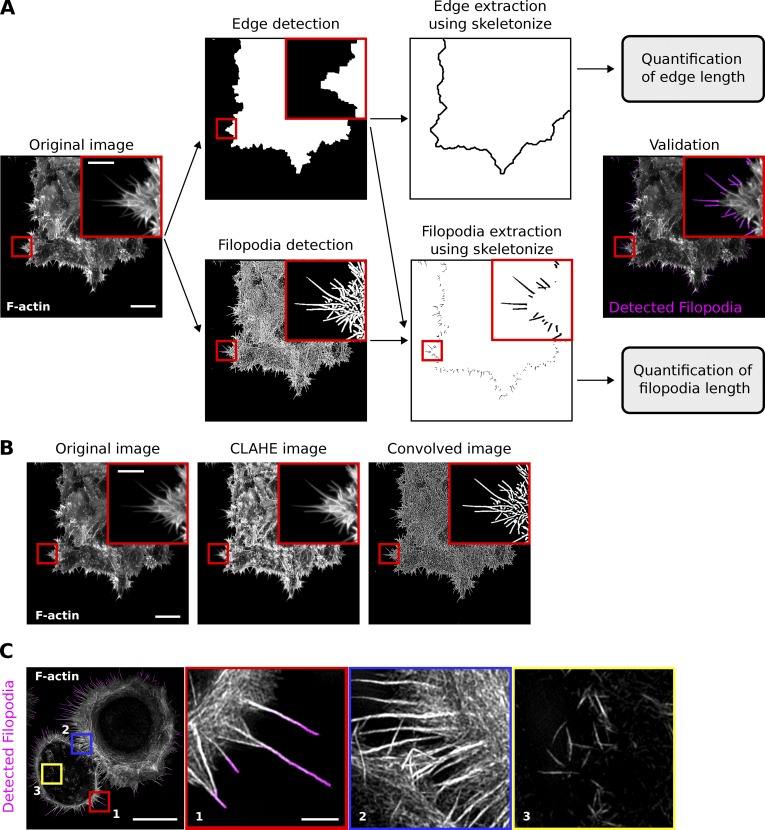
We developed FiloQuant as a plugin for the ImageJ software with inter operating systems compatibility (Schindelin et al., 2012). In brief, FiloQuant works by first defining the cell/colony edge in an input image after intensity based thresholding and removal of long thin protrusions such as filopodia from the plasma membrane (Fig. 1 A). In parallel, the same input image is separately enhanced (Fig. 1 B) to optimize filopodia detection and is then superimposed on the filopodia-erased cell-edge image to specifically isolate filopodia at the cell boundary (Fig. 1 A). The number and length of these cell-edge filopodia are then automatically analyzed by FiloQuant using the skeletonize and AnalyzeSkeleton algorithms (Arganda-Carreras et al., 2010). Filopodia density can also be determined by calculating the ratio of filopodia number to edge length (extracted by FiloQuant from the edge detection image; Fig. 1 A).
Figure 1.
FiloQuant, an ImageJ tool to rapidly quantify filopodia length and density. (A) Workflow depicting FiloQuant analysis of filopodia density and length. Representative images obtained at the different stages of analysis are displayed. In brief, the original image (input) undergoes two parallel processing steps. In the top panel, the cell edge is defined and detected by intensity based thresholding and by ”erasing” the filopodia (edge detection). In the bottom panel, the image is enhanced to optimize detection of faint filopodia without introducing noise (filopodia detection). The resulting images are subtracted to isolate edge filopodia (filopodia extraction), and filopodia number and length are automatically analyzed using the Skeletonize3D and AnalyzeSkeleton algorithms. Detected filopodia are highlighted in magenta in the final image. Filopodia density can be also quantified by determining the ratio of filopodia number to edge length (extracted from the edge detection image). The original image shows MCF10A ductal carcinoma in situ (DCIS.COM) cells invading collectively through a fibrillar collagen gel (circular invasion assay), stained for actin and imaged using a spinning disk confocal (SDC) microscope (100× objective, CMOS camera). Bars: (main) 20 µm; (inset) 5 µm. (B) Images illustrate how two of the settings (CLAHE and Convolve) available in FiloQuant can help to improve the detection of faint filopodia. Bars: (main) 20 µm; (inset) 5 µm. (C) Image illustrating that FiloQuant detects and quantifies only the filopodia present at the cell edge (inset 1) and not the filopodia present at cell-cell junctions (inset 2) or ventral (inset 3) and dorsal filopodia. The image shows a DCIS.COM cell plated on fibronectin for 2 h, stained for actin, and imaged using an SIM. Bars: (main) 20 µm; (inset) 2 µm.
To make this software as easy to use as possible, FiloQuant (single image analysis version; software 1) contains step-by-step user validation of the various processing stages to help users achieve optimal settings for filopodia detection. This is especially important, as efficient detection of filopodia can vary from image to image, even when acquired under similar settings, mainly because the small size of these structures results in weak signals. Several enhancements and filtering options are available in FiloQuant to improve filopodia detection, and we provide here a detailed manual explaining how to use FiloQuant (see supplemental zip file). In particular, among the enhancement strategies tested, a fairly conservative convolution kernel (Fig. 1 B) was found to be very effective in improving detection of faint filopodia. Importantly, FiloQuant is currently limited to the detection and quantification of filopodia and other finger-like protrusions that extend out from the cell edge and is not applicable to filopodia localized at cell–cell junctions or to filopodia that protrude dorsally on top of the cell body or ventrally underneath the cell (Fig. 1 C).
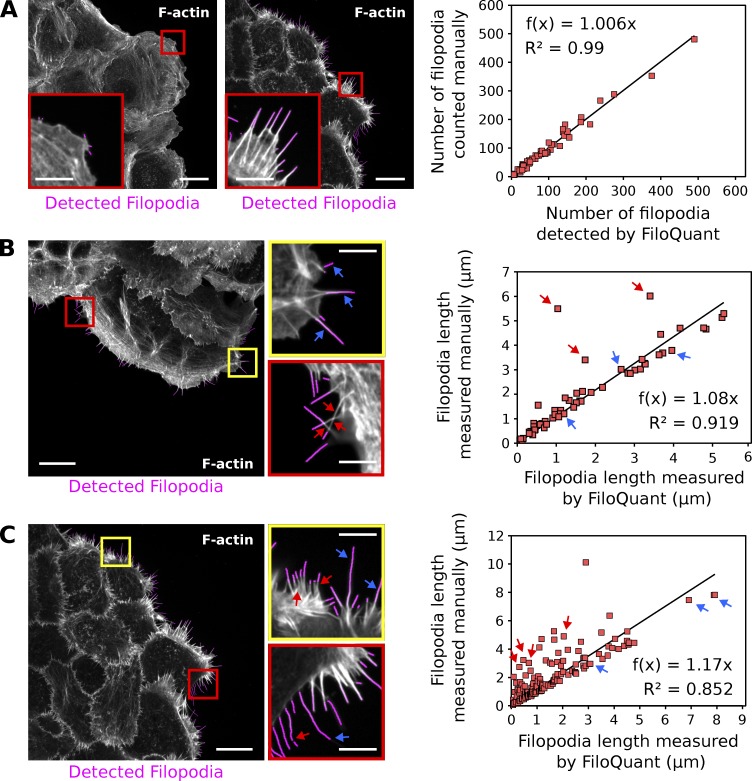
To assess the reliability of FiloQuant in scoring filopodia, results obtained using this software were compared with manual analyses of 59 images displaying a broad range of filopodia densities (Fig. 2 A). Importantly, FiloQuant readouts of filopodia number in this dataset correlated well with manual counting (Fig. 2 A). Moreover, filopodia length measurements obtained with FiloQuant were, overall, very comparable to manual analyses (Fig. 2, B and C). In some cases, FiloQuant measurements did underestimate filopodium length, including when two or more filopodia were found to intersect each other (Fig. 2 B; for more information, see the FiloQuant manual provided in the supplemental zip file), when the filopodia density was very high and the cell edge was difficult to resolve (Fig. 2 C), or when the labeling intensity of a filopodium was extremely weak and resulted in difficulties to threshold the full length of the protrusion (Fig. 2 C). However, as the coordinates of each filopodia measured are also provided by the software (see the FiloQuant manual provided in the supplemental zip file), it is possible to reevaluate the data and to surmise if a filopodium has been incorrectly measured. Together, these analyses demonstrate that FiloQuant can successfully extract filopodia parameters from microscopy images.
Figure 2.
FiloQuant readouts in comparison to manual analyses. (A) FiloQuant readouts of filopodia number were compared with manual analyses (only the filopodia at the colony edge were considered) in a total of 59 random images of MCF10A and DCIS.COM colonies. Images show examples of MCF10A (left image; no overlay; low filopodia density) and DCIS.COM (right image; fibrillar collagen overlay; high filopodia density) cells migrating collectively in circular invasion assays (imaged using an SDC microscope; 100×, CMOS camera). Insets denote magnified areas, and magenta highlights the filopodial protrusions detected by FiloQuant. Correlation between FiloQuant and manual analyses is displayed on the right. (B and C) FiloQuant readouts of filopodia length were compared with manual analyses in two images of DCIS.COM cells migrating collectively in circular invasion assays (B, no overlay; C, fibrillar collagen overlay). Cells were stained for actin and imaged using an SDC microscope (100×, CMOS camera). Red and yellow insets denote magnified areas, and magenta highlights the filopodial protrusions detected by FiloQuant. Blue arrows point to examples of filopodia that were accurately measured using FiloQuant. Red arrows highlight filopodia that were assigned a shorter length by FiloQuant compared with manual analyses. Discrepancies between FiloQuant readouts and manual measurements are primarily caused by intersecting filopodia (B, red insert), high filopodia density (C, yellow inset), or broken filopodia (C, red inset). Correlation between FiloQuant and manual analyses are displayed on the right, where the blue and red arrows represent the values for the same filopodia indicated in the corresponding images. Bars: (main) 20 µm; (inset) 5 µm.
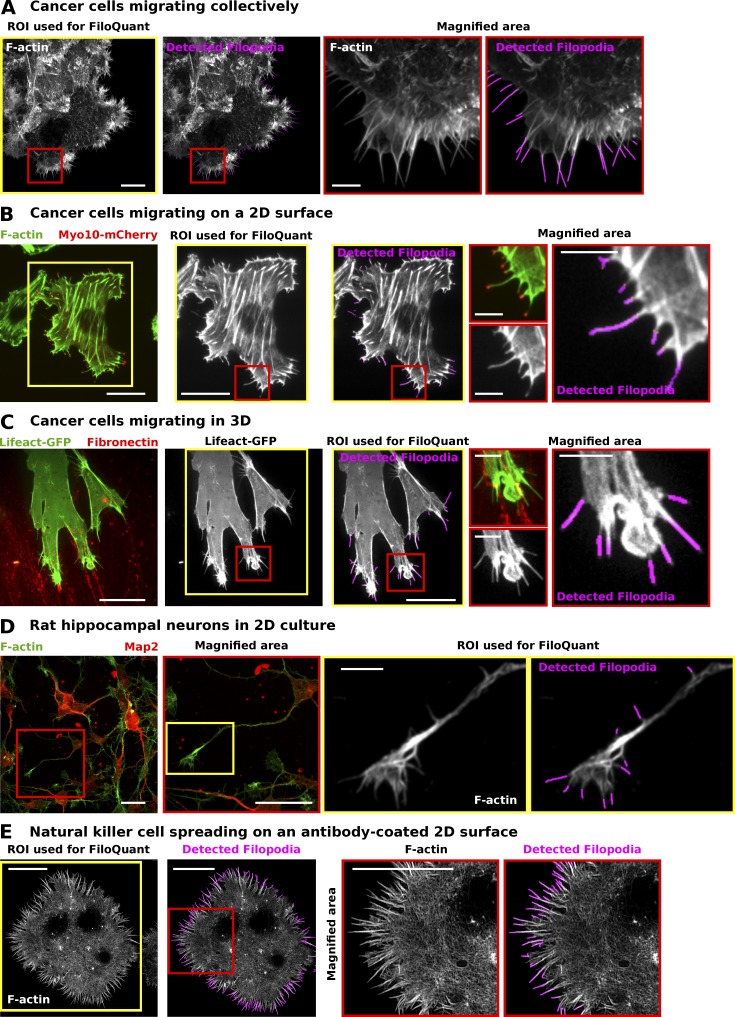
We further demonstrate that FiloQuant can be broadly applied to detect filopodia and filopodia-like protrusions from an assortment of cell images acquired on different microscopes. These include cells migrating collectively (Fig. 3 A) or as single cells (Fig. 3, B and C) in various environments such as on 2D fibronectin (FN; Fig. 3 B) or on 3D cell-derived matrices (Fig. 3 C). In addition, we show that FiloQuant can detect filopodia in neurons, which have a more complex morphology (Fig. 3 D), and can distinguish filopodia-like protrusions in activated natural killer cells (Fig. 3 E).
Figure 3.
FiloQuant can be used to detect filopodia under different cellular contexts and imaging modalities. (A–E) FiloQuant (single image analysis version; software 1) was used to detect filopodia in images acquired from different cell types and using different imaging modalities. (A) DCIS.COM cells migrating collectively were stained for actin and imaged using an SDC microscope (100×, CMOS camera). Bars: (main) 20 µm; (inset) 5 µm. (B) MDA-MB-231 cells transiently expressing mCherry-Myo10 (to visualize filopodia tips) were plated for 2 h on fibronectin (FN), stained for actin, and imaged using a total internal reflection fluorescence (TIRF) microscope. Bars: (main) 20 µm; (inset) 5 µm. (C) A2780 cells transiently expressing mEmerald-LifeAct and migrating on cell-derived matrices in the presence of exogenous FN (labeled with Alexa Fluor 568) were imaged live on an SDC microscope (63× oil objective, EMCCD camera). Bars: (main) 20 µm; (inset) 5 µm. (D) Primary rat hippocampal neurons plated on laminin were fixed, stained for actin and MAP2 (a neuronal marker), and imaged using an SDC microscope (100× objective, CMOS camera). Bars: (main) 20 µm; (inset) 5 µm. (E) NK-92 natural killer cells were seeded on antibody-coated glass (anti-CD18 and anti-NKp30) for 20 min before being fixed, stained for actin, and imaged by TIRF-SIM. The actin-rich filopodia-like protrusions were then detected using FiloQuant (bars, 10 µm). For each panel, FiloQuant analysis was performed on the region of interest defined by the yellow inset, red insets denote magnified areas, and magenta highlights the filopodial protrusions detected by FiloQuant. ROI, region of interest.
Detection of filopodia in vivo during angiogenesis using intravital imaging and FiloQuant
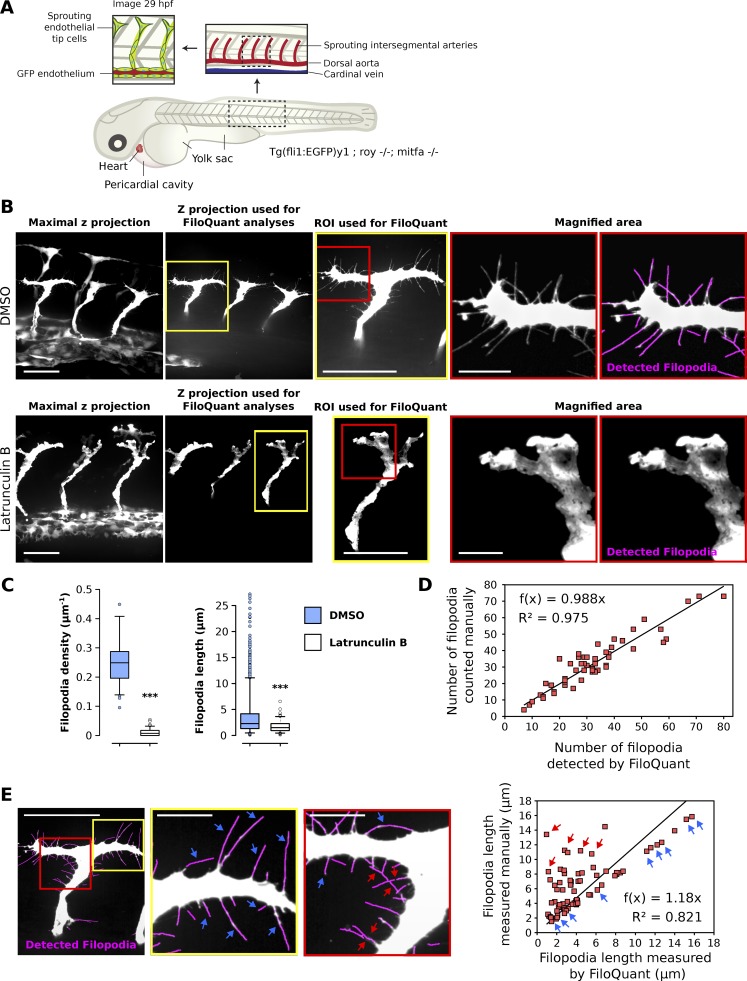
To test further the flexibility of FiloQuant, we analyzed filopodia properties in images acquired using high-resolution intravital microscopy (Fig. 4). In living organisms, filopodia are used by multiple cell types (Jacquemet et al., 2015), including endothelial tip cells, which generate long filopodial protrusions during the process of angiogenesis (Wakayama et al., 2015). To visualize angiogenesis in vivo, we imaged genetically modified zebrafish embryos expressing GFP in the endothelium (Tg(fli1:EGFP)y1; roy−/−; mitfa−/−; Fig. 4 A). To disrupt filopodia formation in this system, zebrafish embryos were treated with a low concentration of latrunculin B previously reported to inhibit filopodia formation in endothelial tip cells (Phng et al., 2013). The sprouting segmental arteries of the embryos were then imaged live on a spinning disk confocal (SDC) microscope, and z projections were created for analysis (Fig. 4 B).
Figure 4.
Filopodia can be detected and quantified in sprouting endothelia in the developing zebrafish embryo using intravital imaging and FiloQuant. (A) Simplified cartoon of a zebrafish embryo. Insets represent magnified regions of interest (ROIs). In particular, endothelial tip cells in the sprouting intersegmental arteries are highlighted to indicate the region being imaged in 29 h postfertilization (hpf) transgenic embryos expressing GFP in the endothelium (Tg(fli1:EGFP)y1; roy−/−; mitfa−/−). (B and C) Transgenic zebrafish embryos (Tg(fli1:EGFP)y1; roy−/−; mitfa−/−) were treated with 1% DMSO or150 ng/ml latrunculin B from 25 hpf to 29 hpf. The embryos were then anesthetized and mounted in low-melting point agarose on glass-bottom dishes. Z stack images of the sprouting segmental arteries were obtained live using an SDC microscope (long working distance 63× water objective, CMOS camera). Representative maximal z projection and the z projection used for FiloQuant analyses are shown (B). In addition, ROIs (yellow square) used for FiloQuant analyses, magnified area (red squares), and filopodia detected using FiloQuant (magenta) are displayed. Bars: (main) 20 µm; (inset) 5 µm. Quantification of filopodia density and filopodia length using the semiautomated version of FiloQuant are displayed as Tukey box plots; C; DMSO, 15 embryos imaged; latrunculin B, 17 embryos imaged; filopodia density: DMSO, 60 ROIs analyzed; latrunculin B, 71 ROIs analyzed; filopodia length: DMSO, 2,232 filopodia measured; latrunculin B, 138 filopodia measured; ***, P < 5.49 × 10−25). The Tukey box plots represent the median and the 25th and 75th percentiles (interquartile range); points are displayed as outliers if 1.5 times above or below the interquartile range; outliers are represented by dots. Statistical analysis: Student’s t test (unpaired, two tailed, unequal variance). (D) FiloQuant (semiautomated; software 2) readouts of filopodia number were compared with manual analyses from a total of 54 images of sprouting endothelial tip cells from DMSO-treated embryos (related to Fig. 4, A–C). (E) FiloQuant readouts of filopodia length were compared with manual analyses in one image of a sprouting endothelial tip cell from a DMSO-treated embryo. Bars: (main) 20 µm; (inset) 5 µm. Red and yellow insets denote magnified areas and magenta highlights the filopodial protrusions detected by FiloQuant. Blue arrows point to examples of filopodia that were accurately measured using FiloQuant. Red arrows highlight filopodia that were assigned a shorter length by FiloQuant compared with manual analyses. Correlation between FiloQuant and manual analyses are displayed on the right where the blue and red arrows represent the same filopodia indicated in the corresponding images (75 filopodia measured).
To analyze larger datasets while retaining control over the settings used to analyze each image, a second version of FiloQuant was created (semiautomated; software 2). Using the semiautomated version of FiloQuant, we were able to rapidly and efficiently detect and analyze filopodia properties in sprouting endothelial cells, and, as previously reported (Phng et al., 2013), demonstrated a significant inhibition in filopodia density and length after latrunculin B treatment (Fig. 4, B and C). To assess reliability of FiloQuant in scoring filopodia in vivo, we compared FiloQuant results, obtained from images of sprouting endothelial tip cells in DMSO-treated zebrafish embryos, to manual analyses (Fig. 4, D and E). Similar to our previous validation of the software in vitro (Fig. 2), FiloQuant readouts of filopodia number (Fig. 4 D) and filopodia length (Fig. 4 E) in vivo correlated well with manual measurements. However, FiloQuant did underestimate filopodia length in some cases, in particular when two or more filopodia were found to intersect.
This example, in addition to those presented in Figs. 1, 2, and 3, clearly demonstrates that FiloQuant is a powerful tool to detect and quantify filopodial features under different cellular contexts and imaging modalities.
Filopodia density and length correlate with increased invasiveness in vitro
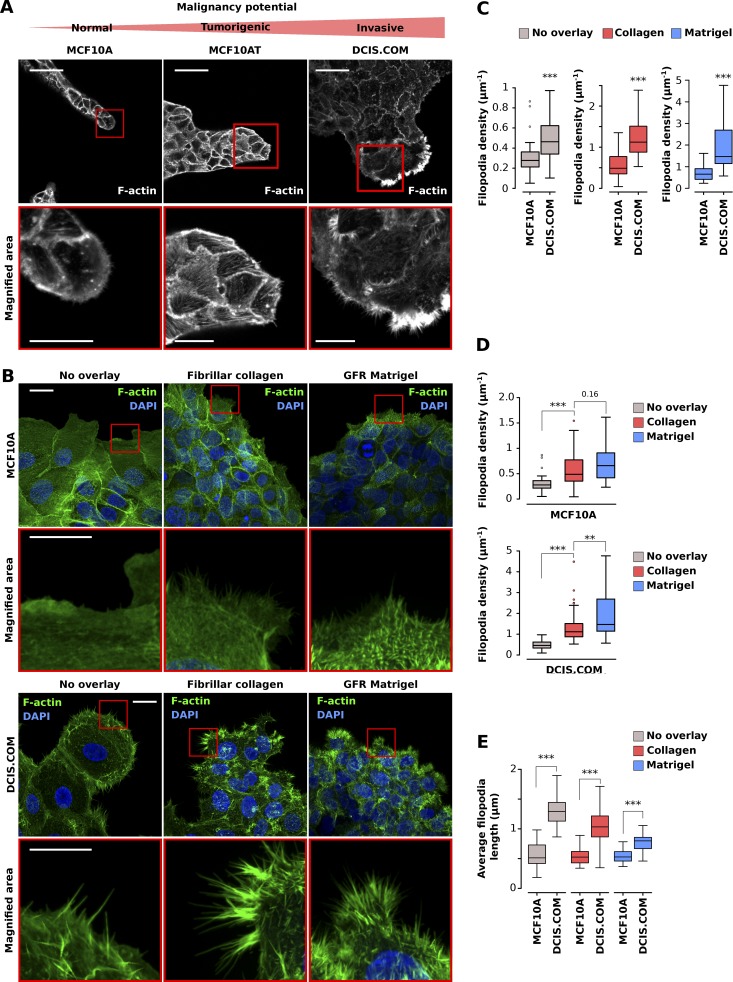
Filopodia and filopodia-like protrusions are prominent features of migrating/invading cancer cells in vitro (Petrie and Yamada, 2012; Jacquemet et al., 2013a; Paul et al., 2015). However, the significance of filopodia at the different stages of the metastatic cascade remains unclear. In breast cancer, metastasis is initiated by cells breaking through a basement membrane to escape the tumor in situ and to invade locally in the surrounding stroma. To study local invasion, a cellular model of breast cancer that recapitulates the different stages of ductal carcinoma in situ (DCIS) progression in animal models and, importantly, mimics the human disease (Miller et al., 2000; Behbod et al., 2009; Lodillinsky et al., 2016) was used. This model is composed of three cell lines: (1) normal immortalized breast epithelial cells (MCF10A), (2) premalignant H-Ras transformed MCF10A variants (MCF10AT cell line) that are tumorigenic as xenografts, and (3) tumorigenic and invasive MCF10A variants (MCF10DCIS.COM cell line; Dawson et al., 1996; Miller et al., 2000). When growing MCF10A, MCF10AT, and MCF10DCIS.COM (DCIS.COM) cells under an overlay of growth factor–reduced (GFR) Matrigel, the actin protrusions formed at the invading edge of cell colonies were strikingly different (Fig. 5 A). In particular, the highly invasive DCIS.COM cells exhibited substantially more filopodia than MCF10A and MCF10AT cells (Fig. 5 A). To further validate this observation, MCF10A and DCIS.COM cells were plated in cell culture inserts and left to invade for 3 d through an overlay of fibrillar collagen or GFR Matrigel or left in media only (Figs. S1 and 5 B). Filopodia density and length at the invasive edge were then quantified using FiloQuant (Fig. 5, C–E; and Fig. S2). Importantly, DCIS.COM colonies displayed higher filopodia density and longer filopodia than MCF10A colonies regardless of the composition of the microenvironment (Fig. 5, B, C, and E; and Fig. S2). Manual counting of filopodia also provided a similar pattern of filopodia number in MCF10A and DCIS.COM cells compared with FiloQuant (Fig. S2). Surprisingly, the composition of the microenvironment did impact filopodia density within each cell population (Fig. 5, B, D, and E). In particular, in DCIS.COM cells, invasion into GFR Matrigel induced very dense arrays of short filopodia compared with media alone or fibrillar collagen. Invasion into fibrillar collagen, in turn, triggered higher filopodia density than media alone, without affecting filopodia length. Similarly, in MCF10A cells, invasion through GFR Matrigel and fibrillar collagen increased filopodia density, but not filopodia length, compared with media alone (Fig. 5, B, D, and E). Collectively, these data are indicative of a prominent role for the cell microenvironment in regulating filopodia formation, which was previously believed to be dictated primarily by cancer cell–intrinsic properties such as oncogenes.
Figure 5.
Filopodia density and length in circular invasion assays correlate with reported degree of cancer cell malignancy. (A) MCF10A, MCF10AT, and DCIS.COM cells were left to migrate into GFR Matrigel for 14 d, fixed, stained, and imaged using a confocal microscope. Bars: (main) 50 µm; (inset) 20 µm. (B–E) MCF10A and DCIS.COM cells were plated in circular invasion assays and left to invade through GFR Matrigel, fibrillar collagen I, or media (no overlay) for 3 d. Cells were then fixed, stained for actin and DAPI, and imaged using an SDC microscope (100× objective, CMOS camera). For each condition, representative maximal z projection and magnified area (squares) are displayed. Bars: (main) 20 µm; (inset) 5 µm. Filopodia density was compared between cells (C; ***, P < 1.54 × 10−4) and in the same cells in different overlay conditions (D; **, P = 0.002; ***, P < 9.99 × 10−05). Mean filopodia length was also calculated in each cell line (E; ***, P < 3.46 × 10−10). Results from three independent experiments are displayed as Tukey box plots (condition, fields of view analyzed; MCF10A no overlay, 43; MCF10A fibrillar collagen I, 31; MCF10A GFR Matrigel, 37; DCIS.COM no overlay, 30; DCIS.COM fibrillar collagen I, 73; DCIS.COM GFR Matrigel, 37). The Tukey box plots represent the median and the 25th and 75th percentiles (interquartile range); points are displayed as outliers (represented by dots) if 1.5 times above or below the interquartile range (represented by whiskers). Statistical analysis: Student’s t test (unpaired, two tailed, unequal variance). The filopodia detection images generated by FiloQuant are displayed in Fig. S2.
Analysis of filopodia dynamics using FiloQuant
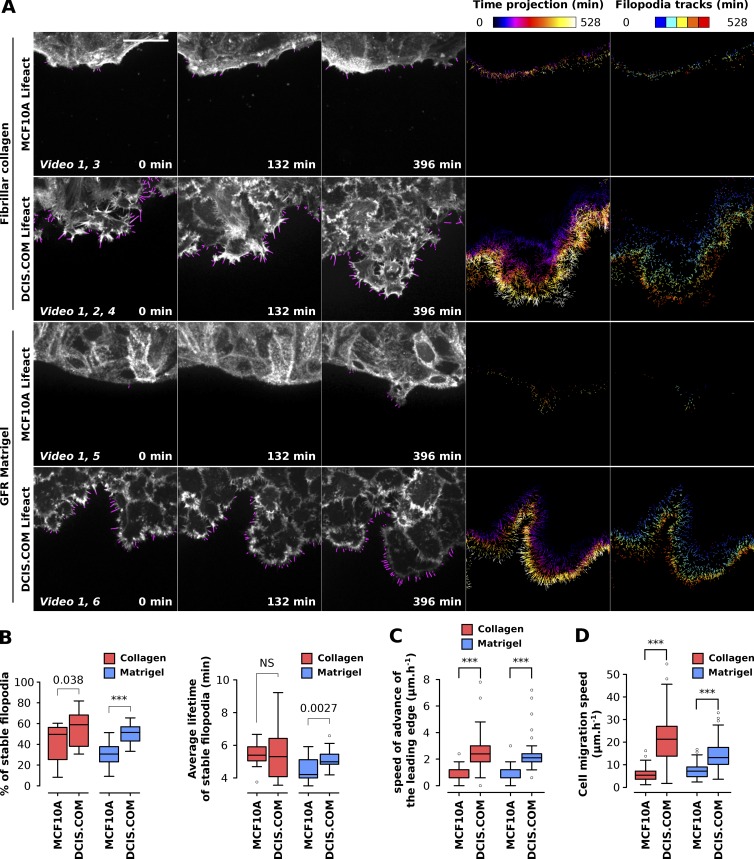
As filopodia formation appeared to be different in MCF10A and DCIS.COM cells (Fig. 5), we set out to further characterize these structures using live-cell imaging. To visualize actin dynamics in live cells, MCF10A and DCIS.COM cell lines expressing LifeAct-RFP (MCF10A LifeAct and DCIS.COM LifeAct, respectively) were generated using lentivirus (see Materials and methods). Importantly, constitutive expression of LifeAct-RFP did not perturb filopodia density, filopodia length, or proliferation in DCIS.COM cells (Fig. S3). To study filopodia dynamics, MCF10A LifeAct and DCIS.COM LifeAct cells were plated in circular invasion assays (Fig. S1) and left to invade for 3 d through fibrillar collagen or GFR Matrigel before being imaged live using an SDC microscope (Fig. 6 and Video 1). Notably, DCIS.COM cells were able to invade collectively and efficiently through fibrillar collagen and GFR Matrigel, whereas MCF10A cells migrated mostly within the cell sheet and did not invade (Fig. 6 A and Video 1).
Figure 6.
Filopodia dynamics can be analyzed using FiloQuant in circular invasion assays. (A and B) MCF10A and DCIS.COM cells stably expressing LifeAct-mRFP were plated in circular invasion assays and left to invade through fibrillar collagen I or GFR Matrigel for 3 d before being imaged live using an SDC microscope (100× objective, EMCCD camera) for over 9 h (one picture every 3 min). Filopodia were then automatically detected using the automated version of FiloQuant (software 3) and tracked using TrackMate (ImageJ-based tracking tool). For each condition, stills of single z planes with filopodia detected by FiloQuant (magenta) are displayed (bar, 25 µm) at different time points. In addition, time projections of detected filopodia as well as filopodia tracks are shown. The time projections were directly generated by FiloQuant and are color coded as a function of time. For filopodia tracking, tracking files, generated by FiloQuant, were entered into TrackMate for automated tracking. Filopodia tracks generated by TrackMate are color coded as a function of the track starting time (track index; A). For each condition, the percentage of stable filopodia (lifetime > 3 min) and the mean lifetime of stable filopodia were quantified from two independent experiments and displayed as Tukey box plots (B; ***, P < 2.79 × 10−04). The Tukey box plots represent the median and the 25th and 75th percentiles (interquartile range); points are displayed as outliers (represented by dots) if 1.5 times above or below the interquartile range (represented by whiskers). (C and D) For the same experiment as in A, the speed of advance of the leading edge (invasion speed through the indicated overlay; C; ***, P < 2.01 × 10−10) and the speed of individual cell migration (D; 100 cells tracked per conditions; ***, P < 3.49 × 10−23) were measured using manual tracking (see Materials and methods for details). n numbers for A to D: condition, movies analyzed; MCF10A LifeAct fibrillar collagen I, 14; MCF10A LifeAct GFR Matrigel, 19; DCIS.COM LifeAct fibrillar collagen I, 23; DCIS.COM LifeAct GFR Matrigel, 30. Statistical analysis: Student’s t test (unpaired, two tailed, unequal variance).
For analysis of large datasets, where the same settings can be applied to many images at once at the beginning of the analysis, a third version of FiloQuant was created (fully automated; software 3). This is especially useful for high-throughput assays and/or to analyze filopodia properties and dynamics from live-cell imaging data. Movies acquired from MCF10A LifeAct and DCIS.COM LifeAct cells were analyzed using this automated version of FiloQuant (software 3) and the “stack analysis” option (see the FiloQuant manual) for sequential processing of images. In particular, this mode enables the analysis of filopodia properties per frame, creates a time projection of all detected filopodia (Fig. 6 A and Videos 2, 3, 4, 5, and 6) and provides a tracking file (Videos 2 and 7), which can then be used to automatically track filopodia dynamics using freely available ImageJ tools such as TrackMate (Tinevez et al., 2017). Using FiloQuant and TrackMate, we found that DCIS.COM cells generate a higher proportion of stable filopodia (>3 min lifetime) compared with MCF10A cells regardless of the composition of the microenvironment (Fig. 6 B). DCIS.COM cells are able to break through basement membranes and invade locally in vivo (Miller et al., 2000; Behbod et al., 2009; Lodillinsky et al., 2016). We demonstrate that the higher density of long filopodia displayed by DCIS.COM cells appears to correlate with enhanced ability to invade through Matrigel and collagen (Fig. 6 C) and with increased migration speed of single cells within the monolayer (Fig. 6 D). As filopodia formation (density and length) are particularly enhanced in DCIS.COM cells compared with normal MCF10A breast epithelial cells (Figs. 5 and 6), and as filopodia are known to play a role in cell invasion and migration (Jacquemet et al., 2015), it is tempting to speculate that filopodia may contribute to DCIS.COM cell invasion through the microenvironment; however, this hypothesis remains to be formally tested.
Filopodia density in 3D spheroids
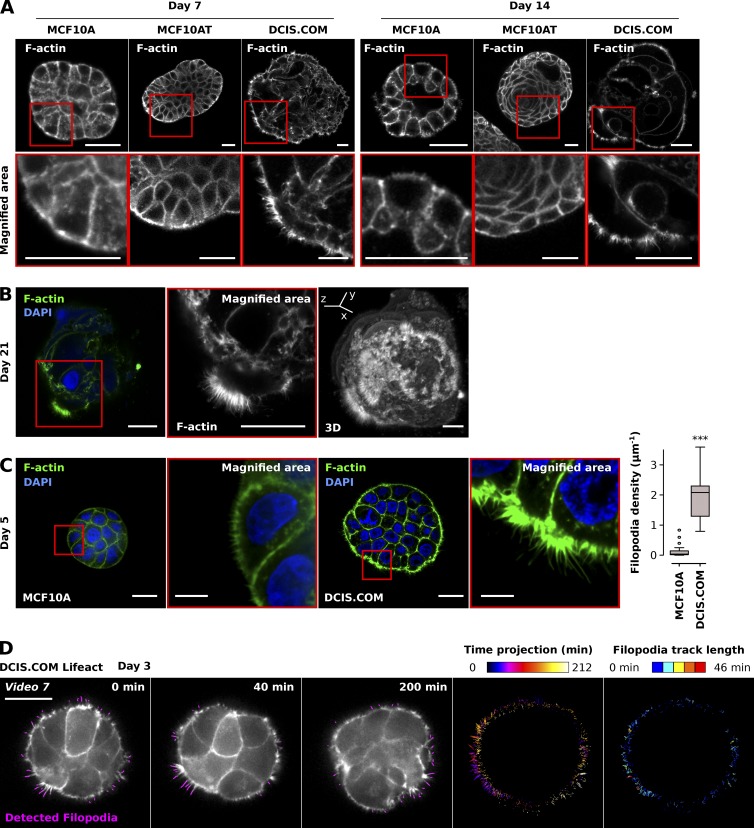
Within a 3D environment, MCF10A and MCF10A-derived cell lines form spheroids encapsulated within a basement membrane that is believed to better recapitulate the in vivo situation. To investigate whether these MCF10A cell line–derived spheroids also exhibit distinct filopodia similar to those observed in 2D (Figs. 5 and 6), we plated single MCF10A, MCF10AT, and DCIS.COM cells on GFR Matrigel and monitored filopodia formation in spheroids at different time points after plating using a confocal microscope (Fig. 7 A). Strikingly, DCIS.COM spheroids displayed very prominent edge filopodia at days 7 and 14 after plating compared with MCF10A or MCF10AT spheroids (Fig. 7 A). These dense arrays of filopodia persisted in the larger DCIS.COM spheroids at day 21 after plating (Fig. 7 B). We analyzed filopodia density in MCF10A and DCIS.COM spheroids at day 5 (using single z planes; see Fig. S4 A for details on the imaging plane), when the entire spheroid was still relatively small and could therefore be accommodated within the imaging window at high magnification and resolution (requirement for filopodia detection), using an SDC microscope (Fig. 7 C). Quantification of filopodia density using FiloQuant clearly demonstrated extensive filopodia assembly in DCIS.COM cells at the spheroid edge, whereas in MCF10A spheroids, filopodia were largely absent from the borders (Figs. 7 C and S4 B).
Figure 7.
Analysis of filopodia density in 3D tumor spheroids using FiloQuant. (A) MCF10A, MCF10AT, and DCIS.COM cells were seeded as single cells on GFR Matrigel and left to form spheroids over 7 or 14 d, fixed, stained, and imaged using a confocal microscope. For each condition, representative single z planes (see Fig. S4 A for cartoon depicting imaging plane) as well as magnified areas (red squares) are displayed. Bars, 25 µm. (B) DCIS.COM cells were seeded as in A and left to form spheroids for 21 d, fixed, stained and imaged using a confocal microscope. A representative single z plane, magnified region (red square) and an Imaris-generated 3D reconstruction are displayed. Bars, 25 µm. (C) MCF10A and DCIS.COM cells were seeded as in A and allowed to form spheroids for 5 d, fixed, stained, and imaged using an SDC microscope (100× objective, CMOS camera). Representative single z planes and magnified areas (red square) are displayed. Bars: (main) 20 µm; (inset), 5 µm. Filopodia density at spheroid borders was quantified using FiloQuant. Results from two independent experiments are displayed as Tukey box plots (condition, fields of view analyzed: MCF10A, 18; DCIS.COM, 11; ***, P = 3.06 × 10−05). The Tukey box plots represent the median and the 25th and 75th percentiles (interquartile range); points are displayed as outliers (represented by dots) if 1.5 times above or below the interquartile range (represented by whiskers). The filopodia detection images generated by FiloQuant are displayed in Fig. S4. (D) DCIS.COM LifeAct cells were seeded as in A and allowed to form spheroids for 3 d before being imaged live using an SDC microscope (100× objective, EMCCD camera) for over 3 h (one picture every 2 min). Filopodia were then automatically detected using FiloQuant and tracked using TrackMate as in Fig. 6 B. Stills of single z planes with filopodia detected by FiloQuant (magenta) and a time projection of the detected filopodia and filopodia tracks are displayed. Bar, 25 µm. The time projections are color coded as a function of time. Filopodia tracks are color coded as a function of the track length to highlight filopodia stability.
To investigate filopodia dynamics in 3D, DCIS.COM LifeAct cells were plated as single cells on GFR Matrigel and allowed to form spheroids for 3 d before being imaged live using an SDC microscope (Video 7). Filopodia were then automatically detected and tracked using FiloQuant and TrackMate (Fig. 7 D and Video 7). These analyses revealed that, in 3D, DCIS.COM cells can produce extremely stable filopodia with a long lifetime approaching 40–50 min (Fig. 7 D).
Tumor spheroids generate filopodia in vivo
We next sought to investigate whether DCIS.COM filopodia observed in vitro are present in vivo using intravital microscopy. Specifically, DCIS.COM LifeAct cells were injected as single cells into the pericardial cavity of zebrafish embryos and imaged live 24 h after injection using an SDC microscope (Fig. 8 A). DCIS.COM cells were able to survive in the pericardial cavity and to form 3D spheroids similar to those observed in 3D GFR Matrigel (Fig. 8, B and C). High-resolution intravital imaging of these tumors revealed the presence of dense filopodial networks at the spheroid border (Fig. 8, C–E; and Videos 8 and 9), thus confirming that filopodia formation is not limited to in vitro cultures and that filopodia are generated in vivo by the invasive DCIS.COM breast cancer cells.
Figure 8.
Tumor spheroids generate filopodia in vivo. (A–C) DCIS.COM LifeAct cells were injected as single cells into the pericardial cavity of zebrafish embryos (A and B) and imaged live using an SDC microscope 24 h postinjection. Before imaging, the embryos were anesthetized and mounted in low-melting-point agarose on glass-bottom dishes. A representative image of a DCIS.COM LifeAct spheroid growing in the zebrafish embryo pericardial cavity (20× objective, CMOS camera; bar, 100 µm; B) and a single z plane (63× water objective, CMOS camera; bars: [main] 20 µm; [inset] 5 µm) and magnification (C) are shown. (D and E) Live imaging of the DCIS.COM LifeAct spheroid, shown in B, using an SDC microscope (63× water objective, EMCCD camera, one frame per 10 s). Single z planes (D; bars: [main] 20 µm; [inset] 5 µm) and Imaris-generated 3D reconstructions (E; bars: [main] 20 µm; [inset] 5 µm) are displayed. Red insets denote magnified areas.
Altogether, we developed FiloQuant, a novel software that can be successfully applied in different settings to detect and quantify properties of filopodia and filopodia-like protrusions. Using FiloQuant, we focused on investigating the presence and dynamics of filopodia in cell models reflecting different stages of breast cancer progression; however, we expect that FiloQuant would also be useful in quantifying filopodia in other contexts and/or to quantify other finger-like protrusions such as retraction fibers or nanotubes. In addition, despite the broad functions offered by the software, we appreciate that other users may require extra functionalities or the ability to include FiloQuant within larger analysis routines. To this end, and to facilitate easy modification, the simple ImageJ macro language used to generate FiloQuant has been fully annotated (software 1–3) and can therefore be edited with limited coding knowledge. For easy installation and rapid distribution, we have also deposited FiloQuant on an ImageJ Update site (see Materials and methods).
Discussion
The extensive employment of filopodia in vivo by different cell types, as well as the mounting in vitro and clinical evidence linking filopodia and/or filopodia regulatory pathways to disease (Jacquemet et al., 2015), emphasizes the need for more comprehensive analyses of filopodia function. Here, we developed FiloQuant, an ImageJ-based computational tool, to simplify quantification of filopodia, and we applied this tool to investigate filopodial properties in cell models of cancer progression.
In cancer, filopodium-like protrusions have been demonstrated to enable the outgrowth of macrometastases and cancer survival at distal sites (Shibue et al., 2012, 2013). Moreover, we and others have shown a dense arrangement of actin-rich filopodial structures at the invasive front of cancer cells in vitro that is required for single-cell invasion in a 3D setting (Jacquemet et al., 2013a, 2016; Arjonen et al., 2014; Paul et al., 2015). Here, using FiloQuant and a cellular model of breast cancer that recapitulates the different stages of DCIS (Dawson et al., 1996; Miller et al., 2000; Behbod et al., 2009), we identified an association between the formation of stable and dense arrays of filopodia, in tumor spheroids and during collective invasion, with the reported ability of these cells to invade in vivo. These data suggest that filopodia may also be important for collective local invasion and the initiation of metastasis in vivo; however, filopodia requirement for collective cell invasion requires further investigation. Intriguingly, in our study, breast cancer cell invasion through a gel composed of basement membrane constituents induced the highest filopodia density. This observation supports the notion that filopodia formation is not merely a cancer cell–intrinsic property but is strongly influenced by the surrounding matrix, potentially via integrin receptor signaling (Jacquemet et al., 2016).
Filopodia are important for many other pathophysiological processes in addition to cancer, and we show that FiloQuant can be applied to quantify filopodia properties (length, density, and dynamics) across different cell types, microenvironments, and image acquisition techniques. Importantly, we showed that FiloQuant readouts performed well compared with manual analyses. FiloQuant presents several advantages over previously described filopodia analysis tools (Nilufar et al., 2013; Tsygankov et al., 2014; Barry et al., 2015; Table 1): (1) to the best of our knowledge, this is the only currently available software capable of extracting filopodia properties from either single images (individual or in batch) or live-cell imaging data; (2) FiloQuant can be applied to detect edge protrusions in both single cells and in cell colonies; (3) FiloQuant analyses include quantification of filopodia density; and (4) FiloQuant contains modifiable parameters, in addition to filtering options based on filopodia size and proximity to the cell or colony edge, that improve the detection of faint filopodia. The only systems requirement, the installation of the ImageJ/Fiji (Schindelin et al., 2012; Schneider et al., 2012) platform (with the addition of several plugins already packaged in Fiji), enables easy and free dissemination of the software throughout the cell biology community. Importantly, FiloQuant’s basic ImageJ macro language can be easily modified by nonexperts to add extra functionalities and/or as a means to incorporate FiloQuant in larger analysis routines. Extra functionalities could include quantification of protein recruitment to filopodia or evaluation of other filopodia and/or cell edge parameters such as filopodia angles or straightness and cell shape. In addition, FiloQuant outputs can be effortlessly coupled to existing ImageJ plugins. As an example we connected FiloQuant to TrackMate (Tinevez et al., 2017) to automatically track filopodia dynamics. A detailed user manual to FiloQuant is provided in the supplemental zip file. Furthermore, to allow customization to address specific needs, the original FiloQuant source code has been fully annotated and deposited in an open online repository (ImageJ update site).
Despite its numerous advantages, FiloQuant has some limitations. (1) Similarly to the other filopodia quantification software, FiloQuant is currently limited to the detection and quantification of filopodia and other finger-like protrusions that extend out from the cell edge. (2) FiloQuant currently works only on 2D images and does not yet support 3D analyses. We anticipate that the filopodia analysis pipeline used here could be translated to work with reconstructed 3D images. The main limitation in studying filopodia in 3D stems from the lack of imaging strategies with sufficient axial resolution to enable successful reconstruction and separation of individual filopodia. Furthermore, FiloQuant is unable to measure filopodia dipping between z-planes in a 3D z projection, resulting in underestimation of filopodia length in such instances. (3) FiloQuant will not work well on noisy images or images taken with insufficient resolution to separate individual filopodia (see FiloQuant manual for advice on sample preparation and imaging strategies to optimize filopodia detection and segmentation). For instance, FiloQuant did not successfully identify the filopodia in the images presented in Figs. 5 A, 7 B, and Fig. 8. (4) Finally, FiloQuant can underestimate true filopodia length in some cases, such as when filopodia originate from the cell body (only the part extending out from the edge is measured), when filopodia intersect each other, when filopodia density is very high, or when the filopodium intensity is extremely weak. Future work will aim at refining filopodia tracing to improve the robustness of filopodia length quantification. Problems associated with high filopodia density could be solved by improving the image resolution using superresolution microscopy and by image processing (Gustafsson et al., 2016).
FiloQuant was designed specifically to assess filopodia properties; however, we anticipate a broader application for FiloQuant in the analysis of other finger-like protrusions such as microvilli, retraction fibers, or nanotubes in different biological settings. As an example, FiloQuant successfully detected filopodia-like structures in activated natural killer cells. Although the role of these structures remains poorly documented in natural killer cells, similar cytoplasmic extensions have been described in T cells and suggested to contribute to T cell activation (Jung et al., 2016).
Materials and methods
FiloQuant installation
The files necessary to run FiloQuant in Fiji (https://fiji.sc/) are provided as software together with test images in the supplemental zip file. Alternatively, FiloQuant installation in Fiji can be easily achieved through the ImageJ update site (see supplemental FiloQuant manual). In brief, in Fiji, click on “Help → Update,” then “Manage update sites,” and “add my site.” In the field “ImageJ Wiki account,” input “FiloQuant,” then click “OK.” Close the “Manage update sites” window and, in the ImageJ Updater window, click on “Apply changes.” FiloQuant can then be found under “plugin → FiloQuant.” To run FiloQuant in ImageJ, users need to install the following dependencies: Enhanced Local Contrast (CLAHE.class; http://imagej.net/Enhance_Local_Contrast_(CLAHE)), Skeletonize3D.jar (http://imagej.net/Skeletonize3D), AnalyzeSkeleton.jar (http://imagej.net/AnalyzeSkeleton; Arganda-Carreras et al., 2010), and Temporal-Color Code (http://imagej.net/Temporal-Color_Code).
Cell culture and transient transfection
Immortalized normal breast epithelial cells (MCF10A), T24 c-Ha-ras oncogene-transfected MCF10A cells (MCF10AT), and invasive variant MCF10 DCIS.COM (DCIS.COM) cells were cultured in a 1:1 mix of DMEM (Sigma-Aldrich) and F12 (Sigma-Aldrich) supplemented with 5% horse serum (16050-122; GIBCO BRL), 20 ng/ml human EGF (E9644; Sigma-Aldrich), 0.5 mg/ml hydrocortisone (H0888-1G; Sigma-Aldrich), 100 ng/ml cholera toxin (C8052-1MG; Sigma-Aldrich), 10 µg/ml insulin (I9278-5ML; Sigma-Aldrich), and 1% (vol/vol) penicillin/streptomycin (P0781-100ML; Sigma-Aldrich). DCIS.COM cells were cloned from a cell culture initiated from a xenograft obtained after two trocar passages of a lesion formed by MCF10AT cells (Miller et al., 2000). MCF10A LifeAct and DCIS.COM LifeAct cells were generated by lentiviral transduction (see Virus production section). A2780 (ovarian carcinoma) cells were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FCS. MDA-MB-231 (triple-negative human breast adenocarcinoma) cancer cells and telomerase immortalized human fibroblasts (TIFs) were grown in DMEM supplemented with 10% FCS. The 293FT packaging cell line was grown in high-glucose DMEM supplemented with 10% FCS, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 6 mM l-glutamine, 1% (vol/vol) penicillin/streptomycin, and 0.5 mg/ml geneticin (all from Thermo Fisher Scientific). All cells were maintained at 37°C and 5% CO2. MDA-MB-231 and MCF10A were provided by the ATCC. A2780 cells were a gift from P. Caswell (University of Manchester, Manchester, England, UK). TIF cells were donated by J. Norman (CRUK Beatson Institute, Glasgow, Scotland, UK). MCF10AT and DCIS.COM were provided by J.F. Marshall (Barts Cancer Institute, Queen Mary University of London, London, England, UK).
Primary hippocampal neurons were isolated from embryonic day 20 rat embryos. In brief, embryonic brain tissue was dissected, and neurons were recovered by enzymatic digestion with trypsin and mechanical dissociation. Cells were maintained in neurobasal medium supplemented with 2% B27 supplement, 0.5 mM l-glutamine, 0.1 mg/ml primocin, and 25 µM glutamate (all from Invitrogen).
NK-92 cells were maintained in α MEM complemented with 0.2 mM myoinositol, 0.1 mM β-mercaptoethanol, 0.02 mM folic acid, 12.5% heat inactivated horse serum, 12.5% heat-inactivated FCS (all from Sigma-Aldrich), 2 mM l-glutamine, and 1× nonessential amino acids (Gibco). The growth medium was replaced every 2 d and supplemented with 100 U/ml human recombinant interleukin-2 (Roche).
All cells were tested for mycoplasma contamination. Plasmids of interest were transfected using Lipofectamine 3000 and the P3000TM Enhancer Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Reagents, antibodies, plasmids, and compounds
The anti-MAP2 antibody was acquired from Antibodies Online (ABIN372661; used at 1:1,000 for immunofluorescence). The Alexa Fluor 488 Phalloidin (A12379), used to stain filamentous actin, and DAPI (D1306) were purchased from Life Technologies. Bovine plasma FN was purchased from Merck (341631). DMSO and latrunculin B (L5288-1MG) were obtained from Sigma-Aldrich. GFR Matrigel was bought from BD Biosciences (354230). PureCol EZ Gel (fibrillar collagen I, concentration 5 mg/ml) was provided by Advanced BioMatrix. DQ collagen (type I collagen from bovine skin, fluorescein conjugate; D12060) was provided by Thermo Fisher Scientific. mEmerald-Lifeact-7 was a gift from M. Davidson (plasmid 54148; Addgene). psPAX2 and pMD2.G were gifts from D. Trono (École polytechnique fédérale de Lausanne, Lausanne, Switzerland; plasmids 12260 and 12259; Addgene). pCDH-LifeAct-mRFP was a gift from P. Caswell. Full-length bovine FN was labeled with Alexa Fluor 568 using an Alexa Fluor 568 Protein Labeling kit (A10238; Thermo Fisher Scientific) according to the manufacturer’s instructions.
Virus production
LifeAct mRFP lentiviral particles were generated in the 293FT packaging cell line after transient cotransfection of pCDH-LifeAct-mRFP, psPAX2, and pMD2.G constructs, in a 7:2:1 ratio, using the calcium-phosphate precipitation method (Graham and van der Eb, 1973). Virus-containing medium was collected 72 h after transfection, concentrated for 2 h at 25,000 rpm, resuspended in residual medium, and flash frozen in liquid nitrogen. Functional titer was evaluated in 293FT cells by FACS (BD LSRFortessa; Becton Dickinson). To obtain stable LifeAct expression, DCIS.COM cells were transduced with MOI 1 (viral particle to cell number ratio of 1:1) and MOI 4 (viral particle to cell number ratio of 4:1), and MCF10A cells were transduced with MOI 4 and MOI 10 of viral stock. Cells exposed to different MOIs were then pooled 3 d after transduction and sorted using a BD FACSaria II cell sorter (Becton Dickinson) with a gating strategy to obtain medium expression.
Production of cell-derived matrices to monitor cell migration
Cell-derived matrices were generated as previously described (Jacquemet et al., 2013b). In brief, TIFs were seeded at a density of 50,000 cells/ml in a 24-well plate. When confluent, cells were cultured for a further 10 d, with medium being changed every 48 h to complete medium supplemented with 50 µg/ml ascorbic acid (Sigma-Aldrich) to ensure collagen cross-linking. Mature matrices were then denuded of cells using lysis buffer (PBS containing 20 mM NH4OH and 0.5% [vol/vol] Triton X-100). After PBS washes, matrices were incubated with 10 µg/ml DNase I (Roche) at 37°C for 30 min. Matrices were then stored in PBS containing 1% (vol/vol) penicillin/streptomycin at 4°C before use.
Circular invasion assay
A cartoon of the circular invasion assay protocol can be found in Fig. S1. In brief, 5 × 104 DCIS.COM or MCF10A cells were plated in one well of a culture-insert 2 well (ibidi) preinserted within a well of a µ-Slide 8 well (ibidi). After 24 h, the culture-insert 2 well was removed, and a gel of GFR Matrigel or fibrillar collagen (PureCol EZ Gel) was casted. The gels were allowed to polymerize for 30 min at 37°C before normal media was added on top. Cells were left to invade for 3 d before fixation or live imaging (over 9 h).
Proliferation assay
To monitor cell proliferation, cells were plated at low density in a well of a six-well plate and imaged using a live-cell microscopy incubator (IncuCyte ZOOM). Growth rates were calculated using the confluency method within the IncuCyte ZOOM software.
3D spheroid formation assay
To form spheroids in 3D Matrigel, cells were seeded as single cells, in normal growth media, at very low density (∼3,000 cells per well) on GFR Matrigel–coated glass-bottom dishes (coverslip No. 0; MatTek). After 12 h, the medium was replaced by normal growth medium supplemented with 2% (vol/vol) GFR Matrigel. The GFR Matrigel medium was then changed every other day until the completion of the experiment.
Zebrafish work
Zebrafish maintenance
Zebrafish (Danio rerio) housing and experimentation was performed under license no. MMM/465/712-93 according to the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes and the Statutes 1076/85 and 62/2006 of The Animal Protection Law in Finland and EU Directive 86/609. Zebrafish were maintained and mated using standard procedures (Westerfield, 2000; Nüsslein-Volhard and Dahm, 2002).
Zebrafish intersegmental vessel sprouting assay
Transgenic zebrafish embryos expressing GFP in the endothelium (genotype Tg(fli1:EGFP)y1; roy−/−; mitfa−/−; Lawson and Weinstein, 2002; White et al., 2008) were cultured at 28.5°C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) before treatment with 150 ng/ml latrunculin B or 1% DMSO from 25 h postfertilization (hpf) to 29 hpf. For live imaging of the sprouting segmental arteries, the embryos were dechorionated with forceps, anesthetized, and mounted in 0.7% low-melting point agarose on glass-bottom dishes. Agarose was overlaid with E3 medium supplemented with 160 mg/l tricaine (Sigma-Aldrich) and 150 ng/ml latrunculin B (Sigma-Aldrich) or 1% DMSO (Sigma-Aldrich). Imaging was performed at 28.5°C using a 3i SDC microscope equipped with a 63× (NA 1.15) long-distance water-immersion objective. Long-term treatment (up to 17 h) of embryos with low concentration of 150 ng/ml latrunculin B has been previously reported to have no adverse effects on embryo viability (Phng et al., 2013). Consistently, no reduction in embryo viability during our short-term experiments (3-h treatment) was observed.
Zebrafish embryo xenograft assay
Zebrafish embryos of the pigment-free casper strain (roy−/−; mitfa−/−) were used in the experiments. One 10-cm plate of MCF10 DCIS.COM cells stably expressing LifeAct mRFP were trypsinized, washed twice in PBS, and resuspended in 30 µl of 2% polyvinylpyrrolidone (Sigma-Aldrich) diluted in PBS for injection. Before injections, 24 hpf embryos were dechorionated, anesthetized (160 mg/l tricaine; Sigma-Aldrich), and immobilized with 0.7% low-melting point agarose (Sigma-Aldrich). Tumor cells were microinjected as a suspension of single cells, using glass microinjection capillaries (TransferTip; Eppendorf), into the pericardial cavity of 24-hpf zebrafish embryos using Celltram vario microinjector (Eppendorf) and Injectman (Eppendorf) micromanipulator mounted on a SteroLumar V12 stereomicroscope (Zeiss). After injection, the embryos were released from the agarose with forceps, washed with E3 medium, and cultured at 34°C in E3 medium. For imaging, the embryos were anesthetized and mounted in low-melting point agarose on glass-bottom dishes.
Microscopy setup
The confocal microscope used was a laser scanning confocal microscope LSM780 (Zeiss) with a 63× (NA 1.2 water) objective controlled by ZEN software (2010).
The SDC microscope used was a Marianas spinning disk imaging system with a Yokogawa CSU-W1 scanning unit on an inverted Zeiss Axio Observer Z1 microscope controlled by SlideBook 6 (Intelligent Imaging Innovations, Inc.). Objectives used were a 20× (NA 0.8 air, Plan Apochromat, DIC) objective (Zeiss), a 63× oil (NA 1.4 oil, Plan-Apochromat, M27 with DIC III Prism) objective (Zeiss), a 63× water (NA 1.2 water C Apo, Korr C Apochromat UV-VIS-IR, DIC) objective (Zeiss), a long-working-distance 63× water (NA 1.15 water, LD C-Apochromat, M27) objective or a 100× (NA 1.4 oil, Plan-Apochromat, M27) objective. Images were acquired using either an Orca Flash 4 sCMOS camera (chip size 2,048 × 2,048; Hamamatsu Photonics) or an Evolve 512 EMCCD camera (chip size 512 × 512; Photometrics).
The total internal reflection fluorescence (TIRF) microscope used was a Zeiss Laser-TIRF 3 Imaging System equipped with a 100× (NA 1.46 oil, α Plan-Apochromat, DIC) objective. Images were acquired on an EMCCD camera (ImageEM C9100-13; chip size 512 × 512; Hamamatsu Photonics) controlled by Zen software (Zen 2012 Blue Edition Systems; Zeiss).
The structured illumination microscope (SIM) used was DeltaVision OMX v4 (GE Healthcare Life Sciences) fitted with a 60× Plan-Apochromat objective lens, 1.42 NA (immersion oil RI of 1.516) used in SIM illumination mode (five phases × three rotations). Emitted light was collected on a front illuminated pco.edge sCMOS (pixel size 6.5 µm, readout speed 95 MHz; PCO AG) controlled by SoftWorx.
The TIRF SIM used was an OMX SR (GE Healthcare Life Sciences) fitted with a 60× Plan-Apochromat objective lens, 1.42 NA (immersion oil RI of 1.516) used in 2D-SIM-TIRF illumination mode (three phases × three rotations within the TIRF plane per final image). Emitted light was collected on a front illuminated pco.edge sCMOS (pixel size 6.5 µm, readout speed 286 MHz; PCO AG) controlled by SoftWorx.
Sample preparation for light microscopy
For TIRF microscopy experiments (related to Fig. 3 B), cells transiently expressing bovine Myo10-mCherry were plated for 2 h on glass-bottom dishes (MatTek Corporation) precoated with 10 µg/ml bovine plasma FN overnight at 4°C.
If not stated otherwise, all samples were fixed in 4% (wt/vol) PFA for 10 min, washed with PBS, and permeabilized with PBS containing 0.5% (vol/vol) Triton X-100 for 3 min. Cells were then washed with PBS, blocked using a solution of 1 M glycine for 30 min, and incubated overnight at 4°C with Alexa Fluor 488 Phalloidin (1/100 in PBS) and, when indicated, with 1 µg/ml (in PBS) DAPI. After washing, samples were stored in PBS in the dark at 4°C before analysis. NK-92 natural killer cells were plated for 20 min on similar dishes precoated with 5 µg/ml anti-CD18 antibody (clone IB4, produced in-house) and 5 µg/ml anti-NKp30 (anti-human CD337, clone PG30.15; BioLegend).
If not stated otherwise, all live-cell imaging experiments were performed in normal growth media supplemented with 50 mM Hepes at 37°C and in the presence of 5% CO2.
FiloQuant and TrackMate analysis of filopodia dynamics
To analyze filopodia dynamics in the circular invasion or 3D spheroid assays, filopodia were first detected and analyzed using the automated version of FiloQuant and the “stack analysis” option. In addition, filopodia further than 40 pixels away from the detected cell edge were excluded using the “maximal distance from cell edges” option. The tracking file generated by FiloQuant was then used as an input for TrackMate, an automated tracking software freely available within ImageJ (Tinevez et al., 2017). TrackMate was chosen over other available ImageJ tracking plugins because of its user-friendly interface and high flexibility. In TrackMate, the LoG detector (estimated bob diameter = 0.1 µm; threshold = 10; subpixel localization enabled) and the simple LAP tracker (linking max distance = 1 µm; gap-closing max distance = 1 µm; gap-closing max frame gap = 1) were used.
Manual tracking analyses
The manual tracking analyses (related to Fig. 6, C and D) were performed in ImageJ using the manual tracking and chemotaxis tool plugins. To measure the speed of the advancing leading edge, three separate points were tracked in each field of view. To measure the migration speed of individual cells within the monolayer, cells at the leading edge and cells behind the edges were tracked.
Statistical analysis
The Tukey box plots represent the median and the 25th and 75th percentiles (interquartile range); points are displayed as outliers (represented by dots) if 1.5 times above or below the interquartile range (represented by whiskers). Box plots were generated using the online tool BoxPlotR (http://shiny.chemgrid.org/boxplotr/). Statistical analyses were performed when appropriate, and p-values are indicated in the figure legends. Unless otherwise indicated, the Student’s t test was used (unpaired, two tailed, and unequal variance, performed within LibreOffice Calc).
Data availability and software updates
The data supporting the findings of this study are available within the article and from the authors on request. FiloQuant code is available as supplementary files associated with this article. Updates of FiloQuant will be released through the FiloQuant ImageJ update site. Update of the FiloQuant manual will be released on the ImageJ website (http://imagej.net/FiloQuant).
Online supplemental material
Fig. S1 illustrates the principle of circular invasion assay. Fig. S2 shows FiloQuant outputs of the images displayed in Fig. 5. Fig. S3 shows that overexpression of LifeAct-RFP does not affect filopodia formation or proliferation of DCIS.COM cells. Fig. S4 shows FiloQuant outputs of the images displayed in Fig. 7. Video 1 displays MCF10A and DCIS.COM cells invading through fibrillar collagen I or GFR Matrigel. Video 2 demonstrates the full FiloQuant analysis of a movie of DCIS.COM cells invading through fibrillar collagen. Video 3 shows the FiloQuant analysis of a movie of MCF10A cells invading through fibrillar collagen. Video 4 shows the FiloQuant analysis of a movie of DCIS.COM cells invading through fibrillar collagen. Video 5 shows the FiloQuant analysis of a movie of MCF10A cells invading through GFR Matrigel. Video 6 shows the FiloQuant analysis of a movie of DCIS.COM cells invading through GFR Matrigel. Video 7 shows the full FiloQuant analysis of a movie monitoring a single DCIS.COM spheroid in 3D GFR Matrigel. Videos 8 and 9 show the actin dynamics of a DCIS.COM spheroid growing in the pericardial cavity of a zebrafish embryo. Software 1 is a version of FiloQuant designed to analyze a single image already opened in ImageJ. Software 2 is a version of FiloQuant designed to analyze images within a specified folder. Software 3 is a version of FiloQuant designed to automatically analyze images within a specified folder by using the same settings for all images (batch analysis). The FiloQuant manual contains detailed instructions on how to install and use FiloQuant as well as troubleshooting advice. Test images are also provided as supplemental files.
Supplementary Material
Acknowledgments
We thank Hans-Juergen Kreienkamp (University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for providing the primary hippocampal neuron samples. We thank Camilo Guzmán for his input in the software and the manuscript. We are grateful to Aki Stubb and Johanna Lilja for testing the FiloQuant software. We thank Patrick Caswell, Jim Norman, and John F. Marshall for providing reagents. We thank Adam Hurlstone for providing the zebrafish lines. The Cell Imaging Core and Zebrafish Core Facility (Turku Centre for Biotechnology, University of Turku and Åbo Akademi University), as well as the Live Cell Imaging Core Facility at the University of Texas Southwestern Medical Center, are acknowledged for services, instrumentation, and expertise.
This study was supported by the Academy of Finland (J. Ivaska), the European Research Council (consolidator grant 615258; J. Ivaska), the Sigrid Juselius Foundation (J. Ivaska), the Finnish Cancer Organization (J. Ivaska), and the National Institutes of Health (grant R01AI067946-11; J.S. Orange). G. Jacquemet was supported by a European Molecular Biology Organization long-term fellowship.
The authors declare no competing financial interests.
Author contributions: Conceptualization, G. Jacquemet and J. Ivaska; Methodology, G. Jacquemet, I. Paatero, A.F. Carisey, A. Padzik, and J. Ivaska; Formal Analysis, G. Jacquemet; Investigation, G. Jacquemet, I. Paatero, A.F. Carisey, and A. Padzik; Resources, J. Ivaska; Writing – Original Draft, G. Jacquemet and H. Hamidi; Writing – Review and Editing, G. Jacquemet, H. Hamidi, I. Paatero, A.F. Carisey and J. Ivaska; Visualization, G. Jacquemet; Supervision, G. Jacquemet and J. Ivaska; Funding Acquisition, J. Ivaska and J.S. Orange.
Footnotes
Abbreviations used:
- DCIS
- ductal carcinoma in situ
- FN
- fibronectin
- GFR
- growth factor–reduced
- hpf
- hours postfertilization
- SDC
- spinning disk confocal
- SIM
- structured illumination microscope
- TIF
- telomerase immortalized human fibroblast
- TIRF
- total internal reflection fluorescence
References
- Arganda-Carreras I., Fernández-González R., Muñoz-Barrutia A., and Ortiz-De-Solorzano C.. 2010. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc. Res. Tech. 73:1019–1029. 10.1002/jemt.20829 [DOI] [PubMed] [Google Scholar]
- Arjonen A., Kaukonen R., Mattila E., Rouhi P., Högnäs G., Sihto H., Miller B.W., Morton J.P., Bucher E., Taimen P., et al. 2014. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J. Clin. Invest. 124:1069–1082. 10.1172/JCI67280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry D.J., Durkin C.H., Abella J.V., and Way M.. 2015. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J. Cell Biol. 209:163–180. 10.1083/jcb.201501081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbod F., Kittrell F.S., LaMarca H., Edwards D., Kerbawy S., Heestand J.C., Young E., Mukhopadhyay P., Yeh H.-W., Allred D.C., et al. 2009. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 11:R66 10.1186/bcr2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K.H., and Zaidel-Bar R.. 2017. Early events in the assembly of E-cadherin adhesions. Exp. Cell Res.:S0014-4827(17)30090-3. [DOI] [PubMed] [Google Scholar]
- Chhabra E.S., and Higgs H.N.. 2007. The many faces of actin: Matching assembly factors with cellular structures. Nat. Cell Biol. 9:1110–1121. 10.1038/ncb1007-1110 [DOI] [PubMed] [Google Scholar]
- Dawson P.J., Wolman S.R., Tait L., Heppner G.H., and Miller F.R.. 1996. MCF10AT: A model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 148:313–319. [PMC free article] [PubMed] [Google Scholar]
- Fierro-González J.C., White M.D., Silva J.C., and Plachta N.. 2013. Cadherin-dependent filopodia control preimplantation embryo compaction. Nat. Cell Biol. 15:1424–1433. 10.1038/ncb2875 [DOI] [PubMed] [Google Scholar]
- Graham F.L., and van der Eb A.J.. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 52:456–467. 10.1016/0042-6822(73)90341-3 [DOI] [PubMed] [Google Scholar]
- Gupta G.P., and Massagué J.. 2006. Cancer metastasis: Building a framework. Cell. 127:679–695. 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Gustafsson N., Culley S., Ashdown G., Owen D.M., Pereira P.M., and Henriques R.. 2016. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 7:12471 10.1038/ncomms12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman C.A., and Plummer H.K. III. 2013. Filopodia as sensors. Cell. Signal. 25:2298–2311. 10.1016/j.cellsig.2013.07.006 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Green D.M., Bridgewater R.E., von Kriegsheim A., Humphries M.J., Norman J.C., and Caswell P.T.. 2013a RCP-driven α5β1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J. Cell Biol. 202:917–935. 10.1083/jcb.201302041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Morgan M.R., Byron A., Humphries J.D., Choi C.K., Chen C.S., Caswell P.T., and Humphries M.J.. 2013b Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J. Cell Sci. 126:4121–4135. 10.1242/jcs.121988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Hamidi H., and Ivaska J.. 2015. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 36:23–31. 10.1016/j.ceb.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Baghirov H., Georgiadou M., Sihto H., Peuhu E., Cettour-Janet P., He T., Perälä M., Kronqvist P., Joensuu H., and Ivaska J.. 2016. L-type calcium channels regulate filopodia stability and cancer cell invasion downstream of integrin signalling. Nat. Commun. 7:13297 10.1038/ncomms13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Riven I., Feigelson S.W., Kartvelishvily E., Tohya K., Miyasaka M., Alon R., and Haran G.. 2016. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl. Acad. Sci. USA. 113:E5916–E5924. 10.1073/pnas.1605399113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjhan R., Noakes P.G., and Bellingham M.C.. 2016. Emerging roles of filopodia and dendritic spines in motoneuron plasticity during development and disease. Neural Plast. 2016:3423267 10.1155/2016/3423267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N.D., and Weinstein B.M.. 2002. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248:307–318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Li A., Morton J.P., Ma Y., Karim S.A., Zhou Y., Faller W.J., Woodham E.F., Morris H.T., Stevenson R.P., Juin A., et al. 2014. Fascin is regulated by slug, promotes progression of pancreatic cancer in mice, and is associated with patient outcomes. Gastroenterology. 146:1386–1396. 10.1053/j.gastro.2014.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodillinsky C., Infante E., Guichard A., Chaligné R., Fuhrmann L., Cyrta J., Irondelle M., Lagoutte E., Vacher S., Bonsang-Kitzis H., et al. 2016. p63/MT1-MMP axis is required for in situ to invasive transition in basal-like breast cancer. Oncogene. 35:344–357. 10.1038/onc.2015.87 [DOI] [PubMed] [Google Scholar]
- Mattila P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454. 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- Millard T.H., and Martin P.. 2008. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development. 135:621–626. 10.1242/dev.014001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F.R., Santner S.J., Tait L., and Dawson P.J.. 2000. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J. Natl. Cancer Inst. 92:1185–1186. 10.1093/jnci/92.14.1185a [DOI] [PubMed] [Google Scholar]
- Nilufar S., Morrow A.A., Lee J.M., and Perkins T.J.. 2013. FiloDetect: automatic detection of filopodia from fluorescence microscopy images. BMC Syst. Biol. 7:66 10.1186/1752-0509-7-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Dahm R.. 2002. Zebrafish: A Practical Approach. Oxford University Press, New York. 303 pp. [Google Scholar]
- Paul N.R., Allen J.L., Chapman A., Morlan-Mairal M., Zindy E., Jacquemet G., Fernandez del Ama L., Ferizovic N., Green D.M., Howe J.D., et al. 2015. α5β1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J. Cell Biol. 210:1013–1031. 10.1083/jcb.201502040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., and Yamada K.M.. 2012. At the leading edge of three-dimensional cell migration. J. Cell Sci. 125:5917–5926. 10.1242/jcs.093732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phng L.-K., Stanchi F., and Gerhardt H.. 2013. Filopodia are dispensable for endothelial tip cell guidance. Development. 140:4031–4040. 10.1242/dev.097352 [DOI] [PubMed] [Google Scholar]
- Sagar A., Pröls F., Wiegreffe C., and Scaal M.. 2015. Communication between distant epithelial cells by filopodia-like protrusions during embryonic development. Development. 142:665–671. 10.1242/dev.115964 [DOI] [PubMed] [Google Scholar]
- Sato Y., Nagatoshi K., Hamano A., Imamura Y., Huss D., Uchida S., and Lansford R.. 2017. Basal filopodia and vascular mechanical stress organize fibronectin into pillars bridging the mesoderm-endoderm gap. Development. 144:281–291. 10.1242/dev.141259 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T., Brooks M.W., Inan M.F., Reinhardt F., and Weinberg R.A.. 2012. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2:706–721. 10.1158/2159-8290.CD-11-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T., Brooks M.W., and Weinberg R.A.. 2013. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell. 24:481–498. 10.1016/j.ccr.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., and Elston T.C.. 2014. CellGeo: A computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204:443–460. 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama Y., Fukuhara S., Ando K., Matsuda M., and Mochizuki N.. 2015. Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebrafish. Dev. Cell. 32:109–122. 10.1016/j.devcel.2014.11.024 [DOI] [PubMed] [Google Scholar]
- Westerfield M. 2000. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., and Zon L.I.. 2008. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2:183–189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., and Martin P.. 2002. Structures in focus--filopodia. Int. J. Biochem. Cell Biol. 34:726–730. 10.1016/S1357-2725(01)00172-8 [DOI] [PubMed] [Google Scholar]
- Wood W., Jacinto A., Grose R., Woolner S., Gale J., Wilson C., and Martin P.. 2002. Wound healing recapitulates morphogenesis in Drosophila embryos. Nat. Cell Biol. 4:907–912. 10.1038/ncb875 [DOI] [PubMed] [Google Scholar]
- Yap L.F., Jenei V., Robinson C.M., Moutasim K., Benn T.M., Threadgold S.P., Lopes V., Wei W., Thomas G.J., and Paterson I.C.. 2009. Upregulation of Eps8 in oral squamous cell carcinoma promotes cell migration and invasion through integrin-dependent Rac1 activation. Oncogene. 28:2524–2534. 10.1038/onc.2009.105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and from the authors on request. FiloQuant code is available as supplementary files associated with this article. Updates of FiloQuant will be released through the FiloQuant ImageJ update site. Update of the FiloQuant manual will be released on the ImageJ website (http://imagej.net/FiloQuant).