Cells can survive executioner caspase activation following transient apoptotic stimuli, a process called anastasis. Using whole-transcriptome RNA sequencing, Sun et al. show that anastasis is an active, two-stage program and characterize the cell behaviors and molecular signature involved in the process.
Abstract
During apoptosis, executioner caspase activity has been considered a point of no return. However, recent studies show that cells can survive caspase activation following transient apoptotic stimuli, a process called anastasis. To identify a molecular signature, we performed whole-transcriptome RNA sequencing of untreated, apoptotic, and recovering HeLa cells. We found that anastasis is an active, two-stage program. During the early stage, cells transition from growth-arrested to growing. In the late stage, HeLa cells change from proliferating to migratory. Recovering cells also exhibited prolonged elevation of proangiogenic factors. Strikingly, some early-recovery mRNAs, including Snail, were elevated first during apoptosis, implying that dying cells poise to recover, even while under apoptotic stress. Snail was also required for recovery. This study reveals similarities in the anastasis genes, pathways, and cell behaviors to those activated in wound healing and identifies a repertoire of potential targets for therapeutic manipulation.
Introduction
Apoptosis is a cell suicide program that is conserved in multicellular organisms and functions to remove excess or damaged cells during development, regulation of the immune system, and stress (Elmore, 2007; Fuchs and Steller, 2011). Excessive apoptosis contributes to degenerative diseases, whereas blocking apoptosis can cause (Favaloro et al., 2012) or treat (Chen and Han, 2015) cancer. Apoptotic cells exhibit distinctive morphological changes (Kerr et al., 1972) caused by activation of proteases called caspases (Martin and Green, 1995; Kumar, 2007). Activation of executioner caspases is a necessary step during apoptosis (Kumar, 2007) and until recently was considered a point of no return (Green and Kroemer, 1998).
However, executioner caspase activation is not always sufficient to kill cells under apoptotic stress. For example, caspase 3 activation in cells treated with sublethal doses of radiation or chemicals does not cause morphological changes or death but rather allows cells to survive with caspase-dependent DNA damage that can result in oncogenic transformation (Lovric and Hawkins, 2010; Ichim et al., 2015; Liu et al., 2015). In addition, transient treatment of cells with lethal doses of certain apoptosis inducers causes caspase 3 activation sufficient to cause apoptotic morphological changes, yet cells can survive after removing the toxin in a process called anastasis (Tang et al., 2012). Although most cells fully recover, a small fraction bear mutations and an even smaller fraction undergo oncogenic transformation. Cell survival after executioner caspase activation has also been reported in cardiac myocytes responding to transient ischemia, in neurons overexpressing Tau, and during normal Drosophila melanogaster development (de Calignon et al., 2010; Kenis et al., 2010; Ding et al., 2016; Levayer et al., 2016). Collectively, these studies suggest that cells can recover from the brink of apoptotic cell death and that this can salvage cells, limiting the permanent tissue damage that might otherwise be caused by a transient injury. However, the same process of anastasis in cancer cells might underlie recurrence after chemotherapy. Thus, defining the molecular changes occurring in cells undergoing this remarkable recovery from the brink of death is a critical step toward manipulating this survival mechanism for therapeutic benefit.
Results
Whole-transcriptome RNA sequencing (RNAseq) reveals that anastasis comprises two stages
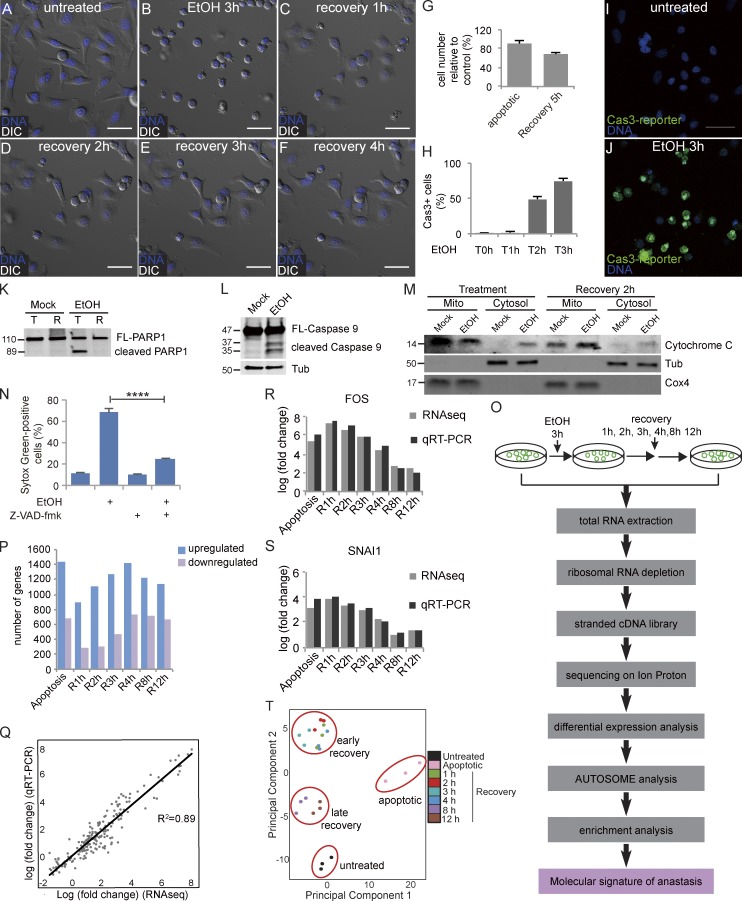
To initiate apoptosis, we exposed HeLa cells to a 3-h treatment with EtOH, which was sufficient to induce cell shrinkage and membrane blebbing (Fig. 1, A and B). Removal of the EtOH by washing allowed a striking recovery to take place over the course of several hours, during which time ∼70% of the cells reattached to the culture matrix and spread out again (Fig. 1, C–G; and Video 1; Tang et al., 2012). 3 h of EtOH treatment was sufficient to cause activation of a fluorescent reporter of caspase 3 activity in ∼75% of the cells (Fig. 1, H–J; and Video 2); cleavage of PARP1, which is a target of caspase 3/7 (Fig. 1 K); cleavage of caspase 9 (Fig. 1 L); and release of cytochrome c from mitochondria to the cytosol (Fig. 1 M). Therefore, EtOH activates the intrinsic apoptotic pathway. Inhibition of caspase activity blocked EtOH-induced cell death (Fig. 1 N).
Figure 1.
RNaseq defines anastasis as a two-stage, active process. (A–F) Time-lapse live imaging of HeLa cells before EtOH treatment (A), after 3 h of EtOH treatment (B), and after recovery for 1 h (C), 2 h (D), 3 h (E), and 4 h (F). (G) The ratio of the number of remaining cells immediately after washing away EtOH (apoptotic) or after 5 h of recovery to the number of cells after mock treatment (n = 3). (H) Quantification of the percentage of cells with active caspase 3 during EtOH treatment (n = 5). In G and H, error bars represent the standard error of the mean. (I and J) Caspase 3 activity (green fluorescence) in the same group of cells before (I) and after (J) 3 h of EtOH treatment. DAPI staining is shown in blue in A–F and I and J. Bars, 50 µm. (K) Western blots of full-length PARP1 (FL-PARP1) and cleaved PARP1 in cells after 3 h of mock or EtOH treatment (T) followed by 21 h of recovery (R). (L) Western blots of full-length caspase 9 (FL-Caspase 9) and cleaved caspase 9 in cells with or without EtOH treatment. (M) The level of cytochrome c in mitochondria (mito) and cytosol of cells with mock or EtOH treatment and of cells at 2 h of recovery after mock or EtOH treatment. Tubulin (Tub) was used as the control for the cytosol fraction, and Cox4 was used as the control for mitochondria. (N) The percentage of SYTOX Green–positive cells (dead cells) in cells with indicated treatment. Error bars represent the standard error of the mean. ****, P < 0.0001. (O) The workflow of RNAseq experiments. (P) Numbers of up-regulated and down-regulated genes in apoptotic cells and cells after 1, 2, 3, 4, 8, and 12 h of recovery compared with untreated cells (fold change > 1.5; FDR < 0.05). (Q) Correlation of RNAseq and qRT-PCR data for 27 genes. (R and S) Comparison of the changes in levels of FOS (R) and SNAI1 (S) mRNAs over time detected by RNAseq and qRT-PCR. In Q–S, fold change is compared with the expression level in untreated cells. (T) PCA of RNAseq data reveals four clusters: untreated cells, apoptotic cells, 1–4 h of recovery, and 8 and 12 h of recovery. Each color represents a different time point. Each time point was analyzed in triplicate.
To define anastasis at a molecular level, we performed RNAseq of untreated cells; apoptotic cells; and cells allowed to recover for 1, 2, 3, 4, 8, or 12 h (Fig. 1 O). These time points include and extend beyond the time needed for the major morphological changes, which appeared to be complete after 4 h (Fig. 1, A–F). Compared with untreated cells, 900–1,500 genes increased in abundance >1.5-fold at each time point, whereas 250–750 genes decreased >1.5-fold (false discovery rate [FDR] < 0.05; Fig. 1 P and Table S1). Well-characterized genes such as Fos, Jun, Klf4, and Snail were induced, as well as genes about which little is known, such as the long noncoding RNA LOC284454, a gene predicted to encode a deubiquitinating enzyme (OTUD1), a pseudokinase (TRIB1), and a phosphate carrier protein (SLC34A3).
We validated the expression patterns of 27 top-ranked differentially expressed (22 up-regulated and 5 down-regulated) genes using quantitative reverse transcription PCR (qRT-PCR). The results of RNAseq and qRT-PCR correlated well, with an R2 of 0.89 (Fig. 1, Q–S; and Fig. S1).
Principal component analysis (PCA) of the RNAseq data showed that cells undergoing anastasis clustered into two distinct groups: one group composed of cells allowed to recover for 1–4 h and a second containing cells that recovered for 8 or 12 h. Both groups were also clearly different from apoptotic cells and untreated cells (Fig. 1 T). We therefore defined the first 4 h of recovery as the early stage and 8–12 h as the late stage.
Distinct features of early and late recovery
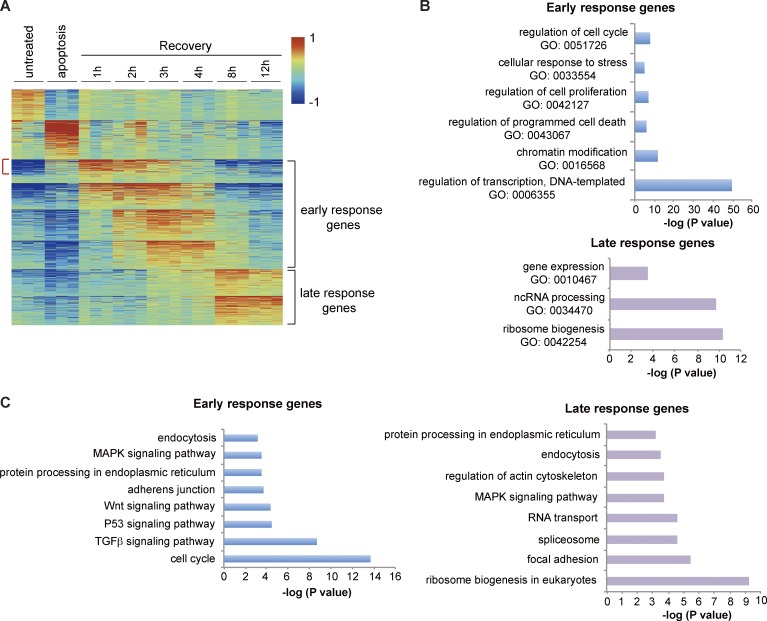
To compare the transcriptional profiles between early and late stages, we used the program AutoSOME (Newman and Cooper, 2010), which clusters genes according to similarities in their expression patterns over time (Table S2). This approach identified eight clusters containing a total of 1,172 genes up-regulated during early recovery and six clusters containing 759 genes up-regulated late (Table S3). We refer to these as early- and late-response genes, respectively (Fig. 2 A and Table S3). Gene ontology (GO) analysis revealed enrichment of expected categories such as “regulation of cell death” and “cellular response to stress” in the early response (Fig. 2 B). The GO term “transcription” was the most significantly enriched, indicating induction of transcription factors during initiation of anastasis (Fig. 2 B). The term “chromatin modification” was also enriched. Enrichment of early-response genes in “regulation of cell proliferation” and “regulation of cell cycle” terms suggested that removing apoptotic stress released cells from a growth-arrested state to reenter the cell cycle and proliferate (Fig. 2 B). Remarkably, the classes of early- and late-response genes were very different. The late response was enriched in posttranscriptional activities such as noncoding RNA processing and ribosome biogenesis (Fig. 2 B).
Figure 2.
AutoSOME and enrichment analyses of early- and late-response genes. (A) AutoSOME analysis. Red indicates increases and blue indicates decreases in mRNA abundance. Genes most highly up-regulated during 1–4 h of recovery are defined as early-response genes, and those that peak at 8 or 12 h are defined as late-response genes. Red bracket points out the genes up-regulated in both apoptosis and early recovery. (B) GO enrichment analysis of early-response genes and late-response genes. (C) KEGG pathway enrichment analysis of early- and late-response genes. In B and C, the p-value is the Bonferroni p-value.
To identify pathways that are overrepresented in the early- and late-induced genes relative to untreated controls, we performed a pathway analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Early-response genes were enriched in cell cycle and pro-survival pathways such as TGFβ, MAPK, and Wnt signaling (Fig. 2 C). The late response showed enrichment in general posttranscriptional pathways such as ribosome biogenesis, RNA transport, protein processing, and endocytosis, as well as specific pathways such as focal adhesion and regulation of actin cytoskeleton (Fig. 2 C).
HeLa cells transition from proliferation to migration during anastasis
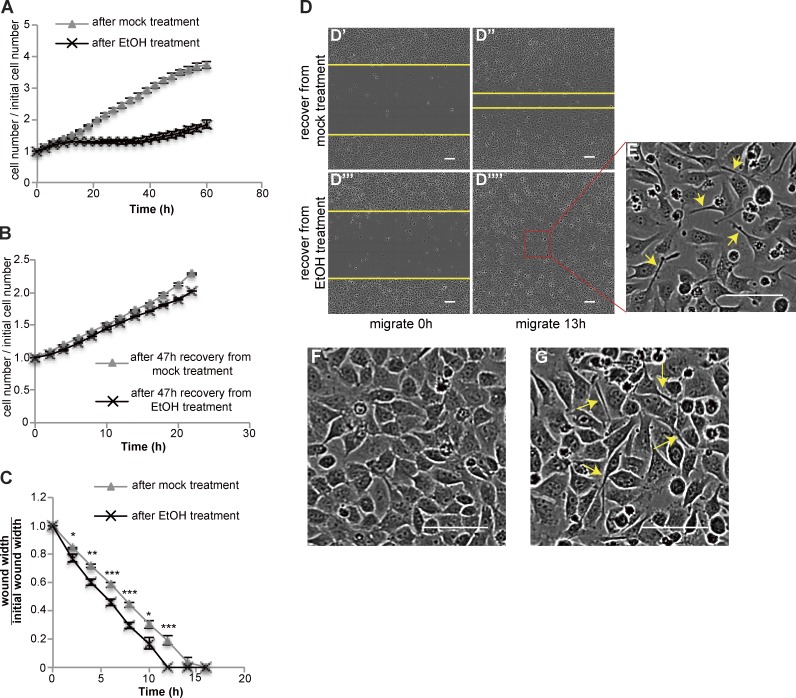
Consistent with enrichment of cell cycle and proliferation genes in the early response, cell numbers increased during the first 11 h of recovery (Fig. 3 A and Fig. S2), plateaued after 11 h, and began to increase again at ∼30 h. At even later time points (after replating), recovered cells exhibited a similar proliferation rate to control, mock-treated cells (Fig. 3 B).
Figure 3.
Cells transition from proliferation to migration during recovery. (A) Cell number during recovery after mock or EtOH treatment (n = 3). (B) Cells after 47 h of recovery from mock or EtOH treatment were trypsinized and replated at similar density. The curves show the change in the cell number over 22 h (n = 3). (C and D) Wound-healing assays. (C) Quantification of wound width over time (n = 8). The asterisks above each time point represent the statistical significance of the difference between cells after mock versus EtOH treatment. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Images of wounds made in cells recovering from mock treatment (D’ and D″) or EtOH treatment (D’’’ and D’’’’). The yellow lines mark the wound margins. (E) Magnified images of the outlined regions in D’’’’. (F and G) Images of confluent monolayers of cells recover from mock (F) or EtOH (G) treatment. In E and G, arrows point to elongated cells. In all plots, error bars represent the standard error of the mean. Bars, 100 µm.
Because of the enrichment of “focal adhesion” and “regulation of actin cytoskeleton” pathways in late-response gene clusters (Fig. 2 C), we hypothesized that cells might become migratory during the proliferation pause. To measure migration, we performed wound-healing assays. Scratch wounds made in monolayers of cells allowed to recover from EtOH treatment for 16 h closed faster than those in mock-treated monolayers (Fig. 3, C and D), even though they exhibited a lower cell number and a slightly slower proliferation rate (Fig. S3). In both mock-treated and EtOH-treated cells, those that migrated to fill the wound were more elongated than cells lagging behind (Fig. 3 E). A larger proportion of cells recovering from EtOH treatment showed this elongated morphology compared with mock-treated cells (Fig. 3, F and G), suggesting that this morphology might facilitate migration and wound closure.
Identification of the early-response genes common to multiple inducers and cell types
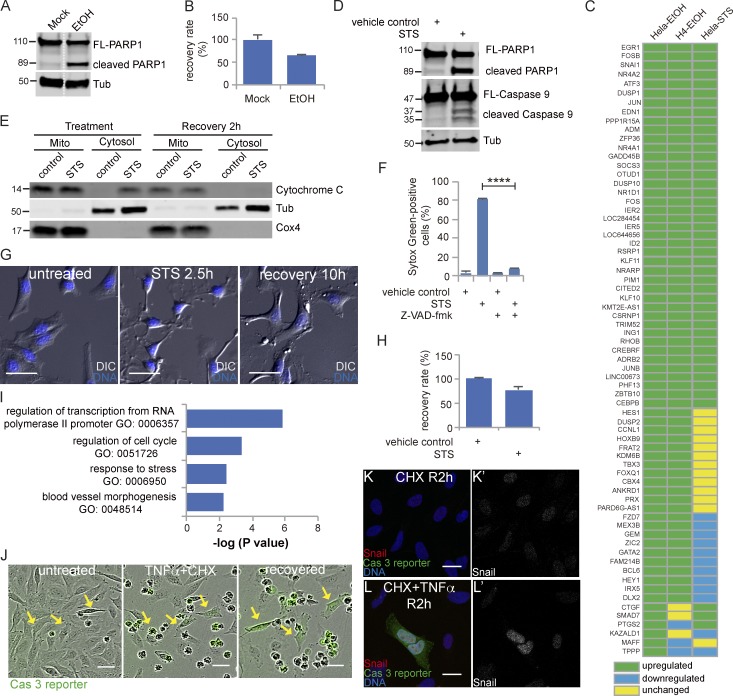
To identify which of the response genes induced during early recovery from EtOH treatment are common to recovery from other apoptotic inducers or in another cell line, we tested the expression of 69 top-ranked up-regulated early genes in H4 cells, a human glioma cell line, recovering from EtOH and in HeLa cells recovering from staurosporine (STS). Treatment of H4 cells with EtOH activated caspase 3, resulting in PARP1 cleavage (Fig. 4 A). Removal of EtOH after 4 h allowed 64% of the cells to recover (Fig. 4 B). 63 out of 69 genes were also up-regulated in H4 cells recovering from EtOH treatment (Fig. 4 C).
Figure 4.
Identification of the early-response genes common to multiple inducers and cell types. (A) Western blots of full-length PARP1 (FL-PARP1) and cleaved PARP1 in H4 cells treated with or without 6.5% EtOH for 4 h. The white dotted line divides the lanes that were cropped from the same blot. (B) The percentage of recovering H4 cells after washing away EtOH. (C) The expression of the 69 top-ranking early-recovery genes in HeLa cells recovering from EtOH, H4 cells recovering from EtOH, and HeLa cells recovering from STS. Green: up-regulated compared with control; blue: down-regulated compared with control; yellow: unchanged compared with control. (D) Western blots of full-length PARP1 (FL-PARP1), cleaved PARP1, full-length caspase 9 (FL-Caspase 9), and cleaved caspase 9 in HeLa cells treated with 250 nM STS or 0.1% DMSO (vehicle control) for 2.5 h. (E) The level of cytochrome c in mitochondria (mito) and cytosol of cells with control or STS treatment and of cells at 2 h of recovery after control or STS treatment. Tubulin (Tub) was used as the control for the cytosol fraction, and Cox4 was used as the control for mitochondria. (F) The percentage of SYTOX Green–positive (dead) cells in cells with indicated treatment. ****, P < 0.0001. (G) Time-lapse live imaging of HeLa cells before STS treatment (untreated), after STS treatment (STS 2.5 h), and after recovery for 10 h (recovery 10 h). DNA is stained in blue. Bars, 50 µm. (H) The percentage of recovering HeLa cells after washing away STS. In B, F, and H, error bars represent the standard error of the mean. (I) GO enrichment analysis of common genes. (J) Time-lapse live imaging of HeLa-GC3AI cells before (untreated) and after treatment with TNFα together with CHX (TNFα + CHX) and cells after 12 h of recovery (recovered). Green indicates caspase 3 activity. Arrows point to cells recovered from caspase 3 activation induced by TNFα together with CHX. Bars, 50 µm. (K and L) Snail expression in cells after 2 h of recovery from CHX alone (K) and from TNFα together with CHX (L). Snail is shown in red, caspase 3 reporter is shown in green, and DNA is shown in blue. K’ and L’ show Snail signal only. Bars, 25 µm.
STS is a protein kinase inhibitor and a classic apoptosis inducer that activates the intrinsic pathway, as shown by cytochrome c release from mitochondria to cytosol, caspase 9 activation, and PARP1 cleavage (Fig. 4, D and E). As expected, inhibition of caspase blocked STS-induced cell death (Fig. 4 F). 76% of STS-treated cells recovered after removal of the drug (Fig. 4, G and H; and Video 3). 44 of 69 genes tested were up-regulated in cells recovering from STS (Fig. 4 C). GO analysis showed that the 44 genes up-regulated in both cells recovering from EtOH and cells recovering from STS were enriched in “regulation of transcription from RNA polymerase II promoter,” “regulation of cell cycle,” “response to stress,” and “blood vessel morphogenesis” (Fig. 4 I), similar to what we found using the full list of early-response genes in cells recovering from EtOH treatment (Fig. 2 B).
Both STS and EtOH induce apoptosis through the intrinsic pathway. To test if cells can survive apoptosis induced by extrinsic inducers, we treated HeLa cells stably expressing a caspase 3 reporter (HeLa-GC3AI) with TNFα and cycloheximide (CHX). Active caspase 3 induces a conformational change of the reporter, switching it from dark to green fluorescent (Zhang et al., 2013). As reported previously, cotreatment of TNFα and CHX induced caspase 8 activation and PARP1 cleavage and turned on the reporter (Fig. 4 J and Fig. S4). After washing away the TNFα and CHX, 5.6% of the caspase 3–positive cells recovered, possibly because of the poor reversibility of TNFα binding to its receptor. We tested the protein expression of Snail, one of the early-response genes, and found that GFP-positive cells that recovered contained a higher level of Snail (Fig. 4, K and L).
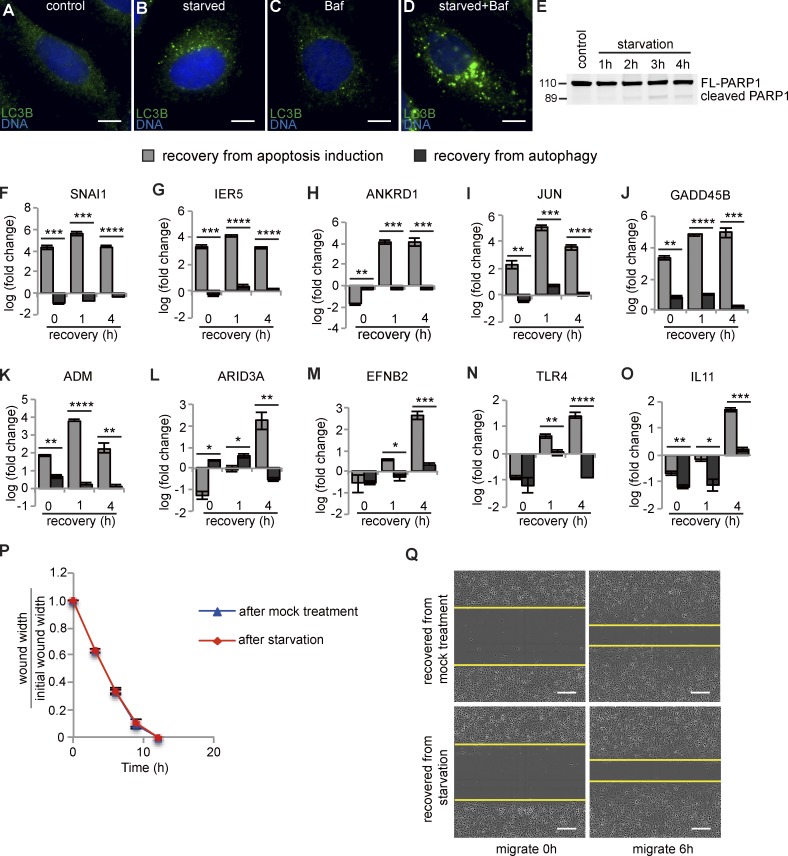
Recovery from apoptotic stress is distinct from recovery from autophagy
The observed enrichment of cell cycle components in the early response suggested that one facet of anastasis is reentry into the cell cycle after growth arrest during apoptosis. To distinguish which molecular features of anastasis were common to another type of growth arrest and recovery and to identify those more likely to be specific to anastasis, we evaluated the expression of the top-ranked, differentially expressed anastasis genes in cells undergoing recovery from nutrient deprivation. Nutrient deprivation induces growth arrest and autophagy, a process that can promote survival (Nikoletopoulou et al., 2013; Mariño et al., 2014). Autophagy results in degradation of cytoplasmic components in autophagosomes, which are double membrane–bound vesicles that sequester cytoplasm and fuse with lysosomes (Mariño et al., 2014). However, expression of autophagy genes was not induced during anastasis, suggesting that the two survival mechanisms differ. A time course showed that amino acid starvation for 2 h induced autophagy in HeLa cells, as shown by increased LC3 staining (Fig. 5, A–D), which is a marker for autophagosomes. LC3 staining is typically further augmented by blocking fusion between autophagsomes and lysosomes with bafilomycin A1 (Mizushima et al., 2010), and this was also true for nutrient-deprived HeLa cells (Fig. 5, A–D). 2 h of starvation did not induce caspase 3 activation (Fig. 5 E), although longer treatments did. Of the 24 genes up-regulated during anastasis that we tested, 10 were down-regulated or only slightly up-regulated during recovery from autophagy (Fig. 5, F–O). Thus, elevated transcription of these 10 genes distinguishes cells in early anastasis from those recovering from autophagy. Furthermore, cells recovering from autophagy showed no measurable difference in the rate of wound closure compared with that of mock-treated cells (Fig. 5, P and Q). Thus, cells recovering from transient apoptotic stress exhibit both molecular and behavioral hallmarks that distinguish anastasis from recovery from other types of stress that induce growth arrest.
Figure 5.
Comparison between anastasis and recovery from autophagy. (A–D) LC3B autophagosome marker staining (green) in cells incubated with growth medium containing 1% DMSO (control; A), HBSS containing 1% DMSO (B), growth medium containing 100 nM Bafilomycin A1 (C), or HBSS containing Bafilomycin A1 (D) for 2 h. Bars, 25 µm. (E) Western blot for PARP1 showing little cleavage during amino acid starvation. The 2-h time point was chosen for further studies. (F–O) Comparison between the mRNA levels of indicated genes after 0, 1, and 4 h of recovery from apoptosis induction (gray bars) or from autophagy (black bars). “Fold change” is compared with expression level of mock-treated cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n = 3. (P) Relative wound width over time in wound-healing assay (n = 5). (Q) Images of wounds made in cells recovering from mock treatment (upper two panels) or amino acid starvation (lower two panels). Yellow lines mark the margin of the wound. Bars, 200 µm. In all graphs, error bars represent the standard error of the mean.
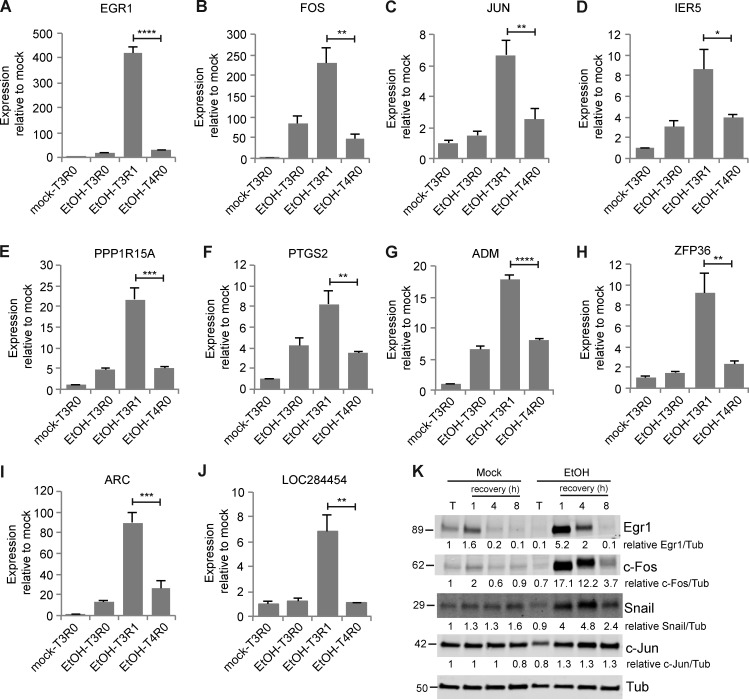
Cells poise for recovery during apoptosis
The transcriptional profile of cells undergoing anastasis revealed an unknown feature of apoptotic cells that appears to contribute to the rapid transition to recovery. We noticed that transcripts corresponding to a subset of early-response genes that were induced during the first hour of anastasis were already elevated in abundance in apoptotic cells relative to untreated cells (Fig. 1, R and S; Fig. 2 A; and Fig. S1, A–G). One possible explanation is that these are genes that drive apoptosis and that apoptosis had not completely stopped 1 h after removal of the chemical stress. Alternatively, these could be genes encoding proteins that contribute to recovery, and cells prepare for the possibility of recovery even while caspase is activated. To distinguish between these opposing possibilities, we compared the levels of expression of 10 such early genes at 1 h of recovery after 3 h of EtOH treatment to the levels in cells left in EtOH for 4 h. The mRNA levels after 4 h of EtOH treatment were significantly lower than those at 1 h of recovery, indicating that accumulation of these mRNAs was associated with the survival response (Fig. 6, A–J). We analyzed the corresponding protein levels for five of the early-response targets for which antibodies were available. The protein levels remained unchanged or were slightly reduced during apoptosis (Fig. 6 K), suggesting that although the mRNAs accumulated, their translation was inhibited, consistent with prior observations of down-regulated protein synthesis in apoptotic cells (Liwak et al., 2012). This intriguing finding supports the idea that even during apoptosis, cells actually poise for recovery by synthesizing, or protecting from degradation, specific mRNAs encoding survival proteins, which are, however, not translated. If apoptotic stress persists, the mRNAs are degraded and the cells die. However, if the apoptotic stress disappears, cells are prepared to rapidly synthesize survival proteins. This “poised for recovery” state may help to explain the rapid recovery after stress removal.
Figure 6.
Apoptotic cells were poised for recovery. (A–J) mRNA levels of the indicated genes relative to mock-treated cells after 3 h of mock treatment (mock-T3R0), after 3 h of EtOH treatment (EtOH-T3R0), after 1 h of recovery from EtOH treatment (EtOH-T3R1), and after 4 h of EtOH treatment (EtOH-T4R0; n = 3). Error bars represent the standard error of the mean. Asterisks show statistical significance between EtOH-T3R1 and EtOH-T4R0. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (K) The protein levels of Egr1, c-Fos, c-Jun, and Snail of cells with mock or EtOH treatment (T) and of cells during recovery from mock treatment and EtOH treatment. The numbers below each blot are the normalized intensity of the bands.
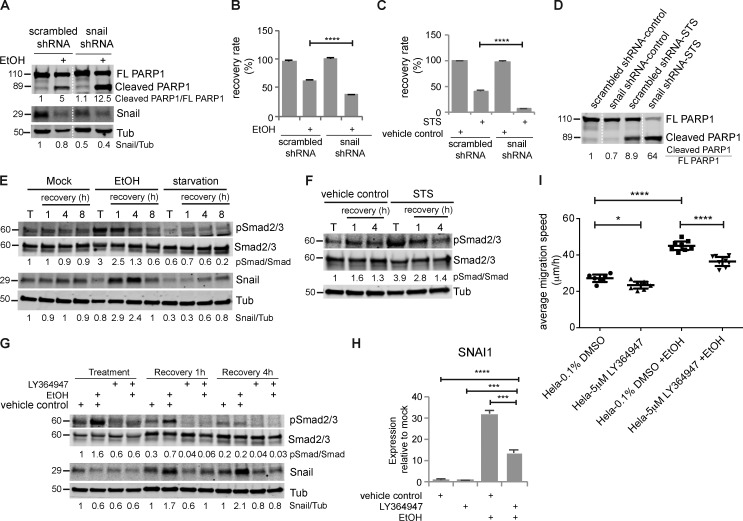
Snail knockdown impairs recovery
Snail is one of the mRNAs enriched in apoptotic cells and then highly induced in early recovery and has been reported to protect cells from apoptosis (Inukai et al., 1999; Metzstein and Horvitz, 1999; Franco et al., 2010; Wan et al., 2015). We found that Snail protein levels increased during recovery (Fig. 6 K). In addition, knocking down Snail expression by stably expressing shRNA reduced the endogenous Snail protein level (Fig. 7 A) and suppressed recovery from EtOH or STS treatment (Fig. 7, B and C). We also found increased PARP1 cleavage in Snail-depleted cells after EtOH or STS treatment (Fig. 7, A and D). Therefore, the poor recovery after Snail knockdown may result from enhanced caspase activation during treatment, impaired anastasis, or both.
Figure 7.
Transient activation of TGFβ signaling induced Snail up-regulation, which was required for anastasis, and caused increased migration in the late stage. (A) Cleavage of PARP1 and Snail protein in HeLa cells stably expressing scrambled shRNA or Snail shRNA treated with or without EtOH for 3 h. The white dotted line divides the lanes that were cropped from the same blot. (B) Recovery rate of HeLa-scrambled shRNA and HeLa-Snail shRNA cells after mock treatment or EtOH treatment (n = 3). (C) Recovery rate of HeLa-scrambled shRNA and HeLa-Snail shRNA cells after control treatment or STS treatment (n = 3). In all bar graphs, error bars represent the standard error of the mean. (D) Western blots of full-length PARP1 (FL PARP1) and cleaved PARP1 in HeLa-scrambled shRNA and HeLa-Snail shRNA cells treated with STS or control. The white dotted line divides the lanes that were cropped from the same blot. (E) Western blots of phosphor-Smad2/3 (pSmad2/3), total Smad2/3, Snail in cells after mock treatment, EtOH treatment, or starvation (T) and in cells recovering from these treatments (recovery). (F) The level of pSmad2/3 and Smad2/3 in HeLa cells treated with STS or vehicle control (T) and in cells recovering from STS or control treatment (recovery). (G) The level of pSmad2/3, Smad2/3, and Snail in apoptotic cells (treatment); cells after 1 h of recovery (recovery 1 h); and cells after 4 h of recovery (recovery 4 h). The addition of LY364497 and EtOH is indicated. The numbers under the blots are the intensity of the bands or the indicated ratio relative to the mock-treated sample. (H) The mRNA expression of Snail (SNAI1) after 1 h of recovery. The addition of LY364947 and EtOH is indicated. (I) Mean migration speed of the indicated group of cells during wound-healing assay (n = 8). Before wound-healing assay, cells were treated with or without EtOH together with 0.1% DMSO or 5 µM LY364947 for 3 h, followed by 4 h of recovery with 0.1% DMSO or 5 µM LY364947 and an additional 16 h of recovery without any inhibitor. Error bars represent 95% confidence interval. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Activation of TGFβ signaling contributes to Snail up-regulation and migration
One important upstream regulator of Snail is TGFβ signaling (Peinado et al., 2003), and this pathway was enriched in the early-recovery gene set. TGFβ signaling regulates transcription through phosphorylation and activation of the downstream transcription factors Smad2 and Smad3 (Massagué, 1998). Phosphorylation of Smad2/3 increased during apoptosis and in the first hour of recovery from EtOH treatment, then diminished after 4 h of recovery, indicating transient activation of TGFβ signaling (Fig. 7 E), which was also observed in HeLa cells recovering from STS treatment (Fig. 7 F).
To determine when and to what extent Snail induction depends on TGFβ signaling, we treated cells with the TGFβ receptor I–specific inhibitor LY364947. LY364947 did not affect basal cell survival or proliferation (Fig. S5 A). LY364947 also did not measurably affect Snail expression during EtOH treatment (Fig. 7 G). At 1 h of recovery, LY364947 treatment prevented induction of Snail mRNA and protein, reducing them to ∼40% of the control, and at 4 h, LY364947 eliminated Snail induction (Fig. 7, G and H). Whereas direct knockdown of Snail by shRNA decreased survival (Fig. 7, B and C), LY364947 treatment did not (Fig. S5, B and C), suggesting that the “poised” Snail is required at the earliest time points, before further induction by TGFβ. This is also consistent with the observation that dramatic transcriptional and morphological changes occur already within the first hour of recovery (Fig. 1, B–F and T).
We then tested whether the effect of TGFβ inhibition might occur later. TGFβ signaling activation can promote epithelial-to-mesenchymal transition (EMT) and cell migration (Xu et al., 2009). To determine if the transient activation of TGFβ signaling was responsible for the increased migration later, we inhibited TGFβ signaling during apoptosis and the first 4 h of recovery and then tested cell migration using the wound-healing assay. LY364947 treatment reduced the mean migration speed of EtOH-treated cells from 45 to 37 μm/h while reducing the mean migration speed of mock-treated cells from 27 to 23 μm/h, suggesting that TGFβ signaling contributes to both basal motility and anastasis-induced migration in HeLa cells (Fig. 7 I). Interestingly, TGFβ signaling, Snail mRNA, and Snail protein were all down-regulated during autophagy and recovery (Fig. 5 F and Fig. 7 E). Recovery from autophagy did not stimulate cell migration (Fig. 5, P and Q). Thus, in HeLa cells, activation of TGFβ signaling, induction of Snail, and increased migration characterize the recovery from the brink of apoptotic cell death but not recovery from a nonapoptotic stress.
Induction of angiogenesis-related genes throughout recovery
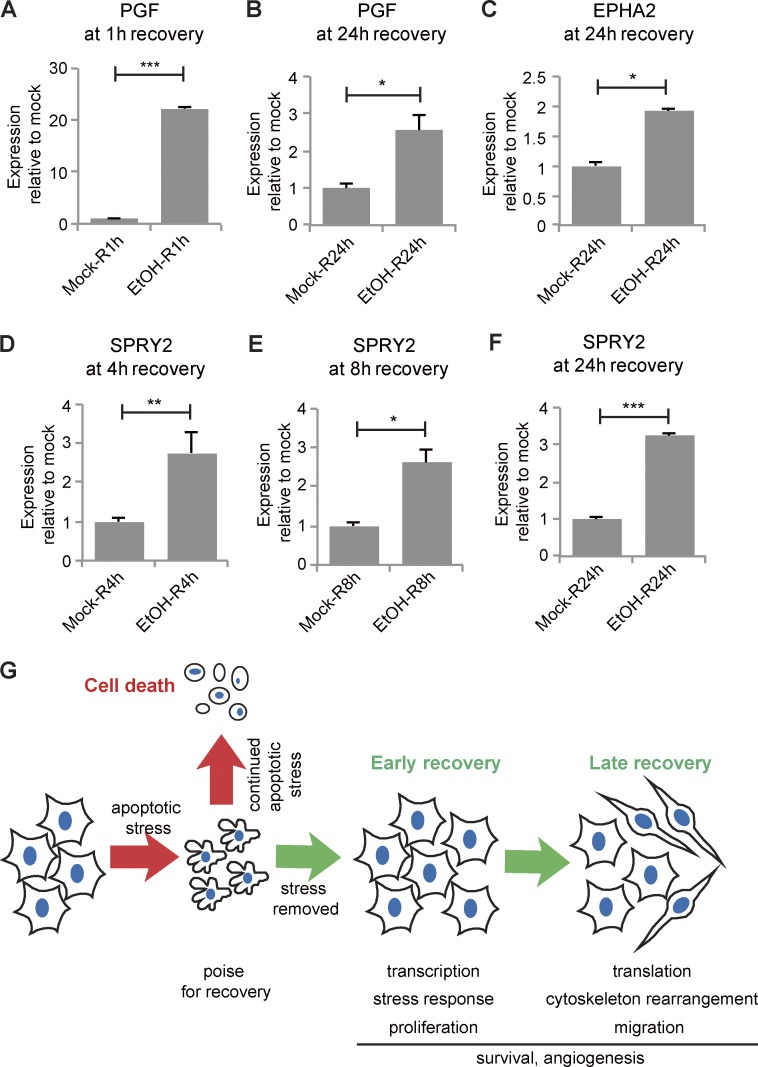
Although TGFβ signaling and Snail expression were transiently elevated during early recovery, some angiogenesis-related genes were persistently elevated throughout the 12 h examined. Placenta growth factor (PGF) binds VEGF receptor (VEGFR) and stimulates endothelial cell proliferation and migration (De Falco, 2012). PGF was among the top 10 up-regulated genes at every time point during apoptosis and recovery (Table S1). PGF mRNA increased ∼22-fold at 1 h of recovery, and even after 24 h of recovery, PGF mRNA was threefold higher in EtOH-treated cells compared with mock-treated cells (Fig. 8, A and B).
Figure 8.
Angiogenesis-related genes were persistently up-regulated during recovery and schematic of events in anastasis. mRNA expression of PGF (A and B), EPHA2 (C), and SPRY2 (D–F) in cells recovering from mock or EtOH treatment for the indicated time (R). Error bars represent the standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G) Summary of events during anastasis. During apoptosis, cells poise for recovery. If the stress persists, cells die. If the stress is removed, cells undergo two stages of recovery.
Ephrin and Ephrin receptor (EphR) signaling are also important in blood vessel development and angiogenesis (Salvucci and Tosato, 2012; Barquilla and Pasquale, 2015). Several EphRs (EPHA2, EPHB2, and EPHB4) and Ephrins (EFNB1, EFNB2) were up-regulated throughout recovery (Table S1). For example, expression of EFNB2 in the first hour of recovery was ∼1.6-fold that of mock-treated cells and elevated approximately two- to threefold during 3–12 h of recovery (Fig. S1K). EPHA2 was significantly up-regulated after 24 h of recovery (Fig. 8 C). Sprouty 2 (SPRY2) is a common transcriptional target of VEGFR and EphR signaling (Cabrita and Christofori, 2008). SPRY2 expression was up-regulated from 4 to 24 h of recovery (Fig. 8, D–F), suggesting activation of VEGFR and EphR signaling during recovery.
Discussion
The ability of cells to survive caspase 3 activity has implications for normal development, cancer, and degenerative and ischemic diseases. Herein, we discuss the molecular characterization of cells recovering from the brink of apoptotic cell death. The data show that anastasis proceeds in two clearly defined stages that are characterized by distinct repertoires of genes. In the early stage, cells transcribe mRNAs encoding many transcription factors and reenter the cell cycle. In the late stage, cells pause in proliferation while increasing migration. Whereas the proliferation and migration responses were transient, others were longer lasting. For example, we found that cells that have undergone anastasis elevate expression of angiogenesis-related genes for 24 h. In vivo, these factors would be expected to exert a nonautonomous effect of stimulating blood vessel growth. Collectively, the results presented herein demonstrate that cells recovering from the brink of apoptotic cell death express factors that promote proliferation, survival, migration, and angiogenesis (Fig. 8 G). The cell biological processes involved in anastasis are thus reminiscent of wound-healing responses (Gurtner et al., 2008), consistent with the idea that cells evolved this capacity to limit permanent tissue damage after a transient injury.
Many of the same molecular pathways are up-regulated during wound healing and in cells undergoing anastasis, including TGFβ, receptor tyrosine kinase, MAPK signaling, and angiogenesis-promoting pathways (Gurtner et al., 2008). TGFβ signaling and Snail expression are thought to promote EMT and chemotherapy resistance during tumor progression (Kalluri and Weinberg, 2009; Kurrey et al., 2009; Brunen et al., 2013). HeLa cells undergoing anastasis activate TGFβ signaling, activate Snail expression, and become migratory—all features of EMT. Two recent studies reported that EMT, although dispensable for tumor metastasis, is required for tumor recurrence after chemotherapy (Fischer et al., 2015; Zheng et al., 2015), suggesting that EMT is a survival strategy for tumor cells under stress in vivo. Our results suggest a possible relationship among tumor recurrence, EMT, and anastasis. If cancer cells exposed to radiation or chemotherapy during treatment escape death via anastasis, TGFβ signaling and Snail expression would be induced, and these critical regulators of EMT (Kalluri and Weinberg, 2009) would confer resistance to further chemotherapy and radiation (Kurrey et al., 2009). Therefore, anastasis could, in principle, drive EMT-dependent tumor recurrence.
In vivo, fast-growing tumors require formation of new blood vessels to supply nutrients and provide a route to metastasis (Nishida et al., 2006). Common cancer treatments, such as tumor resection, can induce extensive angiogenesis, which may promote tumor recurrence (Kong et al., 2010). In fact, elevated expression of angiogenic factors and/or increased blood vessel formation have been found in recurrent craniopharyngioma, bladder cancer, and squamous cell carcinoma after surgery or irradiation (Dinh et al., 1996; Sun et al., 2010; Agrawal et al., 2011). Our results imply that if tumor cells survive apoptosis triggered by chemotherapy, irradiation, or surgery, these survivors may up-regulate production of proangiogenic factors to promote angiogenesis and tumor recurrence.
In addition to providing insight into the molecular nature of anastasis, the work presented herein uncovers an unanticipated aspect of apoptosis. Even during apoptosis, cells poise for recovery by accumulating mRNAs encoding survival proteins. We propose that this facilitates rapid recovery upon removal of the apoptotic stimulus.
Apoptosis and caspase 3 have been linked to tumor recurrence. In cancer patients, the rate of recurrence is positively correlated with the amount of activated caspase 3 in tumor tissues (Huang et al., 2011; Flanagan et al., 2016). One explanation that has been offered for this somewhat paradoxical result is that after radiotherapy or chemotherapy, activated caspase 3 promotes production of progrowth signals that are released from dying cells to stimulate proliferation of living tumor cells, leading to tumor recurrence (Huang et al., 2011; Donato et al., 2014; Cheng et al., 2015). Dying cells can also secrete VEGF in a caspase-dependent way to promote angiogenesis after irradiation (Feng et al., 2015). These previous studies proposed that apoptotic cells induce nonautonomous compensatory proliferation in neighboring cells. The model is that cells that activate caspase 3 die and stimulate cells without activated caspase to proliferate and grow. However, our study suggests another possible mechanism underlying tumor recurrence: that tumor cells with activated caspase 3 themselves may eventually survive, proliferate, migrate, and trigger angiogenesis, contributing to tumor repopulation.
In addition to characterizing the process of anastasis using EtOH-treated HeLa cells, we found 41 genes up-regulated in early recovery in STS-treated HeLa and EtOH-treated H4 glioma cells. The GO analysis of these 41 genes indicates that the biological processes involved in early recovery, like transcription regulation, cell cycle regulation, stress response, and angiogenesis, are common to anastasis in different cell lines and after treatment with different apoptotic inducers (Fig. 4 I). These 41 genes also serve as candidates for regulators of anastasis. Among them, we found that knocking down Snail impaired survival, whereas knocking down TGFβ impaired migration. Unfortunately, H4 cells were not amenable to the migration assay, so we were not able to test the effect of anastasis on motility in this additional cell type.
When facing tissue injury, anastasis could, in principle, facilitate repair and regeneration and limit the permanent damage that might otherwise occur in response to a powerful but temporary insult. However, anastasis would be detrimental if adopted by cancer cells in response to chemotherapy or radiation therapy, thus potentially promoting recurrence. Thus, the mechanisms described herein fit into the general idea that cancers mimic and co-opt wound-healing behaviors (Dvorak, 2015). Enhancing anastasis would be expected to be beneficial in the context of degenerative or ischemic disease, whereas inhibiting anastasis would be expected to be beneficial in cancer treatment. Further study will be required to determine which of the molecular markers and cell biological features described herein are general to many cells undergoing anastasis and in response to various inducers. Although some might be specific to particular cell types, both general and specific molecules and behaviors are of interest. General features may reveal a conserved molecular pathway. However, specific features may provide important therapeutic targets—if, for example, increased motility or a particular molecular pathway turns out to be characteristic of cancer cells but not normal cells or vice versa.
Materials and methods
Cell culture
Human cervical cancer HeLa cells (cell line CCL-2; ATCC) were grown in MEM supplemented with GlutaMAX (Thermo Fisher Scientific), 10% FBS (Sigma), and 100 U/ml penicillin–streptomycin (Thermo Fisher Scientific). Human neuroglioma H4 cells (cell line HTB-148; ATCC) were grown in high-glucose DMEM supplemented with GlutaMAX (Thermo Fisher Scientific), 10% FBS, and 100 U/ml penicillin–streptomycin. All cells were maintained at 37°C with 5% CO2 and 90% humidity. Cells were tested for Mycoplasma contamination.
For RNAseq, 1.2 × 106 cells were seeded in each 100-mm dish and cultured overnight. The next day, cells were treated with either fresh growth medium or fresh growth medium with 4.3% EtOH (Thermo Fisher Scientific) for 3 h. Samples from untreated and apoptotic cells were collected at this moment. For recovery, medium was carefully removed and fresh growth medium was added. For each time point, three biological replicates were included.
For qRT-PCR, Western blotting, and immunofluorescent antibody staining, 2 × 105 cells were seeded in a 35-mm dish or six-well plate and cultured overnight. The next day, cells were treated with different chemicals. The concentration and time for different treatments were as follows: 4.3% EtOH on HeLa for 3 h, 250 nM STS (Santa Cruz Biotechnology) on HeLa for 2.5 h, 6.5% EtOH on H4 for 4 h, and HBSS (Thermo Fisher Scientific) on HeLa for 2 h. The precise concentrations and time points were chosen based on titration studies to achieve the highest possible percentage of apoptotic cells that could recover. For recovery, medium was carefully removed and fresh growth medium was added for the indicated period of time.
For TGFβ signaling inhibition, 5 µM LY364947 (Sigma) was added to cells together with mock or EtOH treatment.
To test if cell death induced by STS or EtOH is caspase dependent, caspase inhibitor Z-VAD-fmk (10 µM) was applied to cells together with STS or EtOH. SYTOX Green was used to label permeabilized cells that entered secondary necrosis.
To activate the extrinsic apoptosis pathway, 100 µg/ml TNFα (Abcam) was cotreated with 10 µg/ml cycloheximide (Santa Cruz Biotechnology) for 6 h.
RNA extraction
For RNA sequencing, total RNA was extracted using a mirVana miRNA isolation kit (Thermo Fisher Scientific) and then treated with TURBO DNase (Thermo Fisher Scientific) to get rid of the genomic DNA. Ribosomal RNA (rRNA) was removed using RiboMinus Eukaryote System v2 (Thermo Fisher Scientific). The quality of RNA was examined using a fragment analyzer (Advanced Analytical).
For qRT-PCR, RNA was extracted using RNeasy Mini kit (QIAGEN) and treated with TURBO DNase to remove the genomic DNA.
RNA sequencing and data analysis
cDNA libraries used for sequencing were made from rRNA-depleted RNA using Ion Total RNA-seq kit v2 (Thermo Fisher Scientific) and sequenced on an Ion Torrent Proton sequencer (Thermo Fisher Scientific). Strand-specific, single-end reads were generated from sequencing with mean read lengths of 75 bp. Reads were mapped to University of California, Santa Cruz Human Reference Genome (hg19) using Tophat (v2.0.13; Trapnell et al., 2010). Reads covering gene coding regions were counted using htseq (v0.6.1; Anders et al., 2015), and the resulting count data were used for downstream analysis. Count data were first filtered by removing genes with low expression or genes with <50 reads in more than two replicates per sample. The remaining count data were normalized using the trimmed mean of the M-values method using edgeR (v3.14.0; Robinson et al., 2010). Normalized count distributions were fit to a generalized linear model to test for differential expression of genes (P < 0.05) among multiple samples. The differential expression test was corrected for multiple testing by applying the Benjamini–Hochberg method on p-values to control the FDR. AutoSOME (Newman and Cooper, 2010) was used for identification of gene clusters with similar expression patterns on counts per million (CPM) and log2-transformed count data.
GO enrichment analysis was performed using DAVID (https://david.ncifcrf.gov/) and PANTHER (http://pantherdb.org). Only common GO terms with Bonferroni p-values <0.001 and FDRs <0.001 were considered to be significantly enriched. KEGG pathway enrichment analysis was performed using WebGestalt (http://www.webgestalt.org) to identify the enriched pathways in the gene sets based on the KEGG database. The pathways with adjusted p-values <0.001 and FDRs <0.01 were considered to be significantly enriched.
qRT-PCR
RNA samples were reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific), and qPCR was performed on the QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific) with the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) or on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) with the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories). The primers used in qPCR are listed in Table S4.
Live imaging
2 × 105 cells were seeded in each glass-bottom, 35-mm dish (MatTek Corporation). Cells were incubated with Hoechst 33342 (Molecular Probes) for 20 min and imaged on a Zeiss LSM 780 or Leica DMi8 microscope with temperature and CO2 control. Images were taken every 10 or 5 min as indicated in the figure legends. Medium change for treatment and recovery was performed between scans. To live-monitor caspase 3 activation during EtOH treatment, NucView 488 (Biotium) was added. NucView 488 binds irreversibly to DNA and thus inhibits anastasis.
Mitochondria isolation
After treatment, 1.5 × 106 HeLa cells were scraped and pelleted. The cell pellets were resuspended in 300 µl of ice-cold fractionation buffer (250 mM sucrose, 20 mM Hepes, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT) supplemented with the protease inhibitor cocktail (Roche), 10 µM NaF, and 1 mM PMSF. The resuspended cells were placed on ice for 20 min and passed through a 26G needle 10–15 times to shear the cells. The needle-treated cell suspension was cleared from both nuclear fraction and nonsheared cells by centrifugation at 3,000 rpm for 5 min at 4°C. 200 µl of the supernatant (which represents the combined cytosolic and mitochondrial fractions) was transferred to new, prechilled tubes for further processing. To separate the mitochondrial fraction, the supernatant after nuclear fractionation was spun down for an additional 10 min at 10,000 rpm at 4°C. The supernatant was the cytosol fraction, and the pellet was the mitochondrial fraction. The pellet was resuspended in 50 µl of ice-cold fractionation buffer supplemented with inhibitors.
Western blotting
Cells were lysed in Laemmli sample buffer and run in 4–20% Mini-PROTEAN TGX precast protein gels (Bio-Rad Laboratories). The primary antibodies used were rabbit anti-Egr1 (no. 4154; Cell Signaling), rabbit anti–c-Fos (no. 2250; Cell Signaling), rabbit anti–c-Jun (no. 9165; Cell Signaling), mouse anti-Snail (no. 3895; Cell Signaling), rabbit anti-PARP1 (no. 9532; Cell Signaling), rabbit anti-Smad2/3 (no. 8685; Cell Signaling), rabbit anti-pSmad2/3 (no. 8828; Cell Signaling), mouse anti–cytochrome c (no. sc-13560; Santa Cruz Biotechnology), mouse anti-Cox4 (no. 11967; Cell Signaling), rabbit anti–caspase 9 (no. 9502; Cell Signaling), mouse anti–caspase 8 (no. 9746; Cell Signaling), rabbit anti-Tubulin (no. 2128; Cell Signaling), and mouse anti–α-Tubulin (T6199; Sigma). The secondary antibodies used were IRDye 800CW donkey anti–rabbit IgG (H+L), IRDye 680LT donkey anti–mouse IgG (H+L), and IRDye 800CW donkey anti–mouse IgG (H+L; LI-COR Biosciences). The blots were scanned on an Odyssey imaging system (LI-COR Biosciences). Cell treatment, sample collection, and Western blotting were repeated at least three times, and the representative blots are shown in the figures.
Immunofluorescent staining
Cells were seeded in six-well plates with a coverslip. After treatment, cells were washed once with PBS and fixed with cold MeOH for 5 min. Cells were then rinsed with PBS twice and washed with PBS containing 0.2% Triton X-100 (Thermo Fisher Scientific). Thereafter, cells were blocked with PBS containing 0.2% Triton X-100 and 5% goat serum (Sigma). The primary and secondary antibodies used were rabbit anti-LC3B (no. 2775; Cell Signaling), rabbit anti-Snail (no. 3879; Cell Signaling), and Alexa Fluor 488–conjugated goat anti–rabbit IgG (H+L) secondary antibody (Thermo Fisher Scientific). The images were acquired on an DMi8 microscope (Leica). Cell treatment and staining were repeated three times, and the representative images are shown in the figures.
shRNA construct, transfection, and stable cell line
shRNA constructs were made in the pLVX vector. The sequences of Snail shRNA and scrambled shRNA were 5′-GGATCTCCAGGCTCGAAAGTCAAGAGCTTTCGAGCCTGGAGATCCTTTTTT-3′ (Lee et al., 2012) and 5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTT-3′ (no. 1864; Addgene). The constructs were transfected into HeLa cells using TurboFect transfection reagent (Thermo Fisher Scientific) and selected using 2 µg/ml puromycin (Thermo Fisher Scientific) to obtain stable cell lines.
The GC3AI caspase 3 reporter plasmid was obtained from B. Li (Tianjin Medical University, Tianjin, China). The plasmid was transfected into HeLa cells and selected using puromycin (Thermo Fisher Scientific) to obtain stable cell lines.
Proliferation assay
HeLa NucLight Red cells (Essen BioScience) were seeded in six-well plates and cultured overnight. After EtOH treatment, cells were cultured in growth medium and imaged using IncuCyte Zoom (Essen BioScience) every hour. Nine fields of view were taken per well, and the number of red fluorescent nuclei was counted using IncuCyte Zoom.
Wound-healing assay
HeLa NucLight Red cells were seeded in 100-mm dishes and cultured overnight. After treatment and 16 h of recovery, cells were trypsinized and seeded in Matrigel-coated 96-well ImageLock plates (Essen BioScience) at 4 × 104 cells per well. After 4 h, a wound was made in each well using WoundMaker (Essen BioScience). The wound closure process was imaged in IncuCyte Zoom every hour.
Recovery rate quantification
Before treatment, cells seeded in six-well plates were incubated with growth medium with DRAQ5 (Thermo Fisher Scientific) for 10 min at 37°C and imaged in IncuCyte Zoom to quantify the original cell number. After 4 h of recovery, cells were washed twice with PBS to remove floating dead cells and stained with DRAQ5 again. The cell number was quantified again as the number of survivors. The recovery rate was calculated as the ratio between the number of survivors and the original cell number.
Statistical analyses
The statistical analysis used in RNAseq data analysis is described in the RNA sequencing and data analysis section. For other experiments, statistical significance was determined using unpaired, two-tailed t tests with Welch’s correction for comparison between two samples and one-way ANOVA to compare more than two samples, with P < 0.05 set as the criteria for significance. The Tukey test was used to derive the adjusted p-values for pairwise comparison among multiple samples. Sample size was not predetermined.
Data availability
The RNAseq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE86480.
Online supplemental material
Fig. S1 shows qRT-PCR validation of differentially expressed genes. Fig. S2 shows examples of cell division in early recovery stage. Fig. S3 shows change of cell number during wound-healing assay. Fig. S4 shows that treatment of TNFα together with CHX induces apoptosis through the extrinsic pathway. Fig. S5 shows inhibition of TGFβ signaling does not affect cell proliferation and recovery from EtOH or STS treatment. Video 1 shows live imaging of HeLa cells undergoing EtOH-induced apoptosis and recovery. Video 2 shows live imaging of caspase 3 activation in HeLa cells treated with EtOH. Video 3 shows live imaging of HeLa cells undergoing STS-induced apoptosis and recovery. Table S1 is a list of differentially expressed genes. Table S2 shows the results of AutoSOME gene clustering. Table S3 is a list of early- and late-response genes. Table S4 is a list of qPCR primers.
Supplementary Material
Acknowledgments
The authors thank Ugochukwu Ihenacho, Jing Xiong, and Rebecca Cheng for technical assistance.
This work was supported by National Institutes of Health grants DP1OD019313-01 and 5DP1CA195760-02 to D.J. Montell.
The authors declare no competing financial interests.
Author contributions: E. Guzman participated in preparing libraries for RNAseq and carried out bioinformatics analyses of the data. H.R. Zhou carried out the mapping of RNAseq reads. V. Balasanyan tested the cytochrome c level in cells with different treatments. C.M. Conner imaged HeLa cells recovering from STS treatment. K. Wong performed qPCR and Western blotting. K.S. Kosik provided advice on the design, execution, and interpretation of the RNAseq analysis. G. Sun carried out the majority of experiments presented, participated in preparing libraries for RNAseq, and carried out bioinformatics analyses. G. Sun also prepared the figures and wrote the first draft of the manuscript. D.J. Montell conceived and coordinated the study, advised G. Sun in all aspects of the work, contributed to the experimental design and interpretation of the results, and revised the manuscript.
Footnotes
Abbreviations used:
- EMT
- epithelial-to-mesenchymal transition
- FDR
- false discovery rate
- GO
- gene ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- PGF
- placenta growth factor
- qRT-PCR
- quantitative RT-PCR
- STS
- staurosporine
References
- Agrawal U., Mishra A.K., Salgia P., Verma S., Mohanty N.K., and Saxena S.. 2011. Role of tumor suppressor and angiogenesis markers in prediction of recurrence of non muscle invasive bladder cancer. Pathol. Oncol. Res. 17:91–101. 10.1007/s12253-010-9287-1 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A., and Pasquale E.B.. 2015. Eph receptors and ephrins: Therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 55:465–487. 10.1146/annurev-pharmtox-011112-140226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunen D., Willems S.M., Kellner U., Midgley R., Simon I., and Bernards R.. 2013. TGF-β: An emerging player in drug resistance. Cell Cycle. 12:2960–2968. 10.4161/cc.26034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita M.A., and Christofori G.. 2008. Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis. 11:53–62. 10.1007/s10456-008-9089-1 [DOI] [PubMed] [Google Scholar]
- Chen L., and Han X.. 2015. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Invest. 125:3384–3391. 10.1172/JCI80011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Tian L., Ma J., Gong Y., Zhang Z., Chen Z., Xu B., Xiong H., Li C., and Huang Q.. 2015. Dying tumor cells stimulate proliferation of living tumor cells via caspase-dependent protein kinase Cδ activation in pancreatic ductal adenocarcinoma. Mol. Oncol. 9:105–114. 10.1016/j.molonc.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Calignon A., Fox L.M., Pitstick R., Carlson G.A., Bacskai B.J., Spires-Jones T.L., and Hyman B.T.. 2010. Caspase activation precedes and leads to tangles. Nature. 464:1201–1204. 10.1038/nature08890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco S. 2012. The discovery of placenta growth factor and its biological activity. Exp. Mol. Med. 44:1–9. 10.3858/emm.2012.44.1.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding A.X., Sun G., Argaw Y.G., Wong J.O., Easwaran S., and Montell D.J.. 2016. CasExpress reveals widespread and diverse patterns of cell survival of caspase-3 activation during development in vivo. eLife. 5:e10936 10.7554/eLife.10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T.V., Hannigan E.V., Smith E.R., Hove M.J., Chopra V., and To T.. 1996. Tumor angiogenesis as a predictor of recurrence in stage Ib squamous cell carcinoma of the cervix. Obstet. Gynecol. 87:751–754. 10.1016/0029-7844(96)00039-7 [DOI] [PubMed] [Google Scholar]
- Donato A.L., Huang Q., Liu X., Li F., Zimmerman M.A., and Li C.-Y.. 2014. Caspase 3 promotes surviving melanoma tumor cell growth after cytotoxic therapy. J. Invest. Dermatol. 134:1686–1692. 10.1038/jid.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak H.F. 2015. Tumors: Wounds that do not heal—Redux. Cancer Immunol. Res. 3:1–11. 10.1158/2326-6066.CIR-14-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. 2007. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 35:495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro B., Allocati N., Graziano V., Di Ilio C., and De Laurenzi V.. 2012. Role of apoptosis in disease. Aging (Albany NY). 4:330–349. 10.18632/aging.100459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Tian L., Zhang Z., Yu Y., Cheng J., Gong Y., Li C.-Y., and Huang Q.. 2015. Caspase 3 in dying tumor cells mediates post-irradiation angiogenesis. Oncotarget. 6:32353–32367. 10.18632/oncotarget.5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T.C., Choi H., El Rayes T., Ryu S., Troeger J., et al. . 2015. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 527:472–476. 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan L., Meyer M., Fay J., Curry S., Bacon O., Duessmann H., John K., Boland K.C., McNamara D.A., Kay E.W., et al. . 2016. Low levels of Caspase-3 predict favourable response to 5FU-based chemotherapy in advanced colorectal cancer: Caspase-3 inhibition as a therapeutic approach. Cell Death Dis. 7:e2087 10.1038/cddis.2016.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco D.L., Mainez J., Vega S., Sancho P., Murillo M.M., de Frutos C.A., Del Castillo G., López-Blau C., Fabregat I., and Nieto M.A.. 2010. Snail1 suppresses TGF-β-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J. Cell Sci. 123:3467–3477. 10.1242/jcs.068692 [DOI] [PubMed] [Google Scholar]
- Fuchs Y., and Steller H.. 2011. Programmed cell death in animal development and disease. Cell. 147:742–758. 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D., and Kroemer G.. 1998. The central executioners of apoptosis: Caspases or mitochondria? Trends Cell Biol. 8:267–271. 10.1016/S0962-8924(98)01273-2 [DOI] [PubMed] [Google Scholar]
- Gurtner G.C., Werner S., Barrandon Y., and Longaker M.T.. 2008. Wound repair and regeneration. Nature. 453:314–321. 10.1038/nature07039 [DOI] [PubMed] [Google Scholar]
- Huang Q., Li F., Liu X., Li W., Shi W., Liu F.-F., O’Sullivan B., He Z., Peng Y., Tan A.-C., et al. . 2011. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 17:860–866. 10.1038/nm.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichim G., Lopez J., Ahmed S.U., Muthalagu N., Giampazolias E., Delgado M.E., Haller M., Riley J.S., Mason S.M., Athineos D., et al. . 2015. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol. Cell. 57:860–872. (published erratum appears in Mol. Cell 2015. 58:900) 10.1016/j.molcel.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai T., Inoue A., Kurosawa H., Goi K., Shinjyo T., Ozawa K., Mao M., Inaba T., and Look A.T.. 1999. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol. Cell. 4:343–352. 10.1016/S1097-2765(00)80336-6 [DOI] [PubMed] [Google Scholar]
- Kalluri R., and Weinberg R.A.. 2009. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119:1420–1428. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenis H., Zandbergen H.R., Hofstra L., Petrov A.D., Dumont E.A., Blankenberg F.D., Haider N., Bitsch N., Gijbels M., Verjans J.W.H., et al. . 2010. Annexin A5 uptake in ischemic myocardium: Demonstration of reversible phosphatidylserine externalization and feasibility of radionuclide imaging. J. Nucl. Med. 51:259–267. 10.2967/jnumed.109.068429 [DOI] [PubMed] [Google Scholar]
- Kerr J.F., Wyllie A.H., and Currie A.R.. 1972. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 26:239–257. 10.1038/bjc.1972.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B., Michalski C.W., Friess H., and Kleeff J.. 2010. Surgical procedure as an inducer of tumor angiogenesis. Exp. Oncol. 32:186–189. [PubMed] [Google Scholar]
- Kumar S. 2007. Caspase function in programmed cell death. Cell Death Differ. 14:32–43. 10.1038/sj.cdd.4402060 [DOI] [PubMed] [Google Scholar]
- Kurrey N.K., Jalgaonkar S.P., Joglekar A.V., Ghanate A.D., Chaskar P.D., Doiphode R.Y., and Bapat S.A.. 2009. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 27:2059–2068. 10.1002/stem.154 [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Jeon H.M., Ju M.K., Kim C.H., Yoon G., Han S.I., Park H.G., and Kang H.S.. 2012. Wnt/Snail signaling regulates cytochrome c oxidase and glucose metabolism. Cancer Res. 72:3607–3617. 10.1158/0008-5472.CAN-12-0006 [DOI] [PubMed] [Google Scholar]
- Levayer R., Dupont C., and Moreno E.. 2016. Tissue crowding induces caspase-dependent competition for space. Curr. Biol. 26:670–677. 10.1016/j.cub.2015.12.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., He Y., Li F., Huang Q., Kato T.A., Hall R.P., and Li C.-Y.. 2015. Caspase-3 promotes genetic instability and carcinogenesis. Mol. Cell. 58:284–296. 10.1016/j.molcel.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liwak U., Faye M.D., and Holcik M.. 2012. Translation control in apoptosis. Exp. Oncol. 34:218–230. [PubMed] [Google Scholar]
- Lovric M.M., and Hawkins C.J.. 2010. TRAIL treatment provokes mutations in surviving cells. Oncogene. 29:5048–5060. 10.1038/onc.2010.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G., Niso-Santano M., Baehrecke E.H., and Kroemer G.. 2014. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 15:81–94. 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.J., and Green D.R.. 1995. Protease activation during apoptosis: Death by a thousand cuts? Cell. 82:349–352. 10.1016/0092-8674(95)90422-0 [DOI] [PubMed] [Google Scholar]
- Massagué J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67:753–791. 10.1146/annurev.biochem.67.1.753 [DOI] [PubMed] [Google Scholar]
- Metzstein M.M., and Horvitz H.R.. 1999. The C. elegans cell death specification gene ces-1 encodes a Snail family zinc finger protein. Mol. Cell. 4:309–319. 10.1016/S1097-2765(00)80333-0 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Levine B.. 2010. Methods in mammalian autophagy research. Cell. 140:313–326. 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., and Cooper J.B.. 2010. AutoSOME: A clustering method for identifying gene expression modules without prior knowledge of cluster number. BMC Bioinformatics. 11:117 10.1186/1471-2105-11-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V., Markaki M., Palikaras K., and Tavernarakis N.. 2013. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta. 1833:3448–3459. 10.1016/j.bbamcr.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Nishida N., Yano H., Nishida T., Kamura T., and Kojiro M.. 2006. Angiogenesis in cancer. Vasc. Health Risk Manag. 2:213–219. 10.2147/vhrm.2006.2.3.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Quintanilla M., and Cano A.. 2003. Transforming growth factor β-1 induces Snail transcription factor in epithelial cell lines: Mechanisms for epithelial mesenchymal transitions. J. Biol. Chem. 278:21113–21123. 10.1074/jbc.M211304200 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci O., and Tosato G.. 2012. Essential roles of EphB receptors and EphrinB ligands in endothelial cell function and angiogenesis. Adv. Cancer Res. 114:21–57. 10.1016/B978-0-12-386503-8.00002-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.I., Akgun E., Bicer A., Ozkan A., Bozkurt S.U., Kurtkaya O., Koc D.Y., Pamir M.N., and Kilic T.. 2010. Expression of angiogenic factors in craniopharyngiomas: Implications for tumor recurrence. Neurosurgery. 66:744–750. 10.1227/01.NEU.0000367553.65099.14 [DOI] [PubMed] [Google Scholar]
- Tang H.L., Tang H.M., Mak K.H., Hu S., Wang S.S., Wong K.M., Wong C.S.T., Wu H.Y., Law H.T., Liu K., et al. . 2012. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell. 23:2240–2252. 10.1091/mbc.E11-11-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z., Pan H., Liu S., Zhu J., Qi W., Fu K., Zhao T., and Liang J.. 2015. Downregulation of SNAIL sensitizes hepatocellular carcinoma cells to TRAIL-induced apoptosis by regulating the NF-κB pathway. Oncol. Rep. 33:1560–1566. [DOI] [PubMed] [Google Scholar]
- Xu J., Lamouille S., and Derynck R.. 2009. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 19:156–172. 10.1038/cr.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang X., Cui W., Wang W., Zhang H., Liu L., Zhang Z., Li Z., Ying G., Zhang N., and Li B.. 2013. Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage. Nat. Commun. 4:2157 10.1038/ncomms3157 [DOI] [PubMed] [Google Scholar]
- Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.-C., LeBleu V.S., and Kalluri R.. 2015. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 527:525–530. 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE86480.