Dasso discusses work from Beaven et al. on the regulation of Ncd in the meiotic spindle by 14-3-3 proteins.
Abstract
During Drosophila melanogaster oogenesis, spindle assembly occurs without centrosomes and relies on signals from chromosomes. Beaven et al. (2017. J. Cell. Biol. https://doi.org/10.1083/jcb.201704120) show that 14-3-3 proteins bind and inhibit a key microtubule motor, Ncd, during oogenesis, but Aurora B releases Ncd inhibition near chromosomes, allowing Ncd to work in the right time and place.
Meiosis is the process of specialized divisions that produce haploid gametes from diploid cells. Meiotic chromosome distribution is a critical event in sexual reproduction. Failure of chromosome segregation during meiosis results in progeny that are aneuploid, possessing chromosomes above or below the 2N number of the diploid genome. Such failures have many important consequences, not least of which is the high rate of aneuploid oocyte production in mammals, a central cause of human infertility, miscarriages, and birth defects (Bennabi et al., 2016). Beaven et al. examined the role of Drosophila melanogaster 14-3-3 proteins in meiotic chromosome segregation and found that they promote accurate assembly of meiotic spindles through spatial regulation of claret nondisjunctional (Ncd), a minus end–directed kinesin-14 family motor protein that mediates sliding of antiparallel microtubules and cross-links parallel microtubules (Fink et al., 2009). This inhibition is relieved locally through phosphorylation by Aurora B, the kinase subunit of the chromosomal passenger complex (CPC)—a key regulator of spatial organization within meiotic and mitotic spindles (Weaver and Walczak, 2015; Radford et al., 2017b). Collectively, these findings reveal an intricate mechanism for organizing microtubules with the meiotic spindle to achieve accurate chromosome segregation.
Female meiosis presents at least two sets of important challenges for accurate chromosome segregation (Bennabi et al., 2016). First, oocytes arrest in late prophase for extended periods of time, which can last for weeks to months in mice and up to several decades in humans. This arrest requires prolonged stability of both meiotic spindle structures and cell cycle regulatory states. Second, female meiotic spindles in many species, including humans, mice, frogs, and flies, lack centrosomes to direct their microtubule assembly and instead rely on cues from the chromosomes to guide spindle organization. Moreover, oocytes are larger than most somatic cells, and they divide asymmetrically, partitioning resources preferentially into the fertilized egg; this asymmetry necessitates precise formation of meiotic spindles adjacent and perpendicular to the cell cortex.
Two pathways have been implicated as major contributors to chromatin-based signaling during acentrosomal spindle formation: the Ran pathway and CPC-mediated phosphorylation (Weaver and Walczak, 2015; Radford et al., 2017b). Although acentrosomal spindle assembly in different situations generally relies on one or both of these pathways, their relative importance can vary greatly between different organisms and situations (Radford et al., 2017b). Ran is a small GTPase whose nucleotide-binding state is controlled by a chromatin-bound exchange factor (RCC1) and a GTPase-activating protein (RanGAP1) that is cytoplasmic during interphase and dispersed or spindle associated during cell division. The distribution of these regulators results in higher levels of Ran-GTP within interphase nuclei and near chromosomes of dividing cells as well as lower levels in interphase cytoplasm and distal to condensed chromosomes during cell division. Ran controls the association of nuclear transport substrates to a family of nuclear transport receptors (called karyopherins) that also bind Ran-GTP. During interphase, this pathway drives a large fraction of nuclear–cytoplasmic protein trafficking. After nuclear envelope breakdown, the same interactions regulate numerous spindle assembly factors through karyopherin binding in a manner that is responsive to Ran-GTP levels and thus dictated by proximity to RCC1 localized on chromosomes.
The CPC consists of four subunits: inner centromere protein (INCENP), Survivin, Borealin/Dasra, and Aurora B kinase. The CPC complex concentrates on centromeres during early mitosis (Weaver and Walczak, 2015). The CPC is pivotal for the fidelity of chromosome segregation because it controls kinetochore structure, kinetochore–microtubule attachments, and signal transduction pathways that specify the timing of anaphase onset. The CPC localizes within a ring encircling the chromosomes during the first meiotic division (meiosis I), where it is essential for microtubule polymerization and spindle organization (Radford et al., 2012). During meiosis I, the CPC is also essential for centromere biorientation, i.e., the correct attachment of homologous chromosomes to microtubules emanating from opposite poles; notably, Drosophila meiotic biorientation is similarly dependent on Ncd function (Radford et al., 2012).
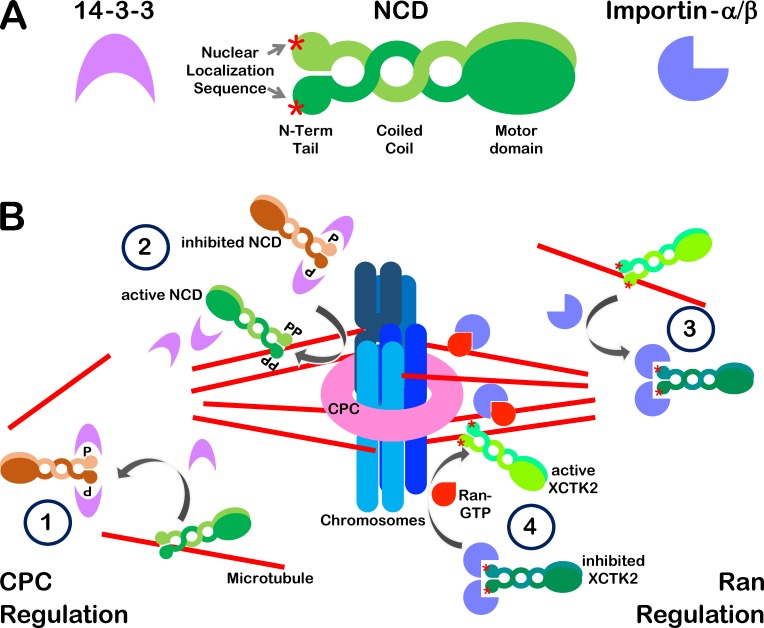
Ncd modulates spindle length and morphology as well as pole organization in acentrosomal spindles (Radford et al., 2017b; She and Yang, 2017). Like other kinesin-14 proteins, Ncd consists of three functional domains: a C-terminal ATPase motor domain, a central coiled-coil domain, and an N-terminal microtubule-binding tail domain (Fig. 1 A; She and Yang, 2017). Many kinesin-14 proteins including Ncd as well as human HSET and Xenopus laevis XCTK2 possess nuclear import signals (NLSs) within their N-terminal domains that can be recognized by Importin-α/β karyopherin, conferring nuclear localization during interphase through the Ran nuclear transport pathway (She and Yang, 2017). The capacity of Ncd to interact with microtubules through both its motor and tail domains allows it to bridge microtubules, both sliding antiparallel microtubules and statically cross-linking parallel microtubules (Fink et al., 2009). Within spindles, kinesin-14 proteins generate inward pulling forces that are antagonized by outward forces generated by kinesin-5 family members (She and Yang, 2017); this in the case of Drosophila, where Ncd antagonizes the KLP61F protein, a kinesin-5 (Radford et al., 2017a). To avoid spindle collapse, it is vital to keep the inward and outward forces generated by these motors in balance. Xenopus eggs control this balance via the Ran pathway: Importin-α/β binds the NLS within the XCKT2 tail domain, competitively inhibiting tail binding to microtubules and thereby increasing XCKT2 turnover rates and reducing the microtubule anchoring of XCKT2 (Weaver et al., 2015). Variation in local Ran-GTP concentrations provides a dynamic and spatially controlled mechanism to release this inhibition, modulating XCKT2 activity relative to its proximity to chromosomes, with its highest activity in the vicinity of chromosomes (Fig. 1 B, right side).
Figure 1.
Regulated deposition of kinesin-14 on spindle microtubules. (A) Schematic representations of 14-3-3, Ncd, and Importin-α/β. (B) Alternative pathways for kinesin-14 regulation in flies and frogs. (Left) Ncd is regulated by the CPC during Drosophila meiosis: (1) The tail microtubule-binding domain of Ncd is bound by 14-3-3 proteins, releasing its interactions with microtubules distal to chromosomes. (2) Near chromosomes, phosphorylation by Aurora B releases 14-3-3 and promotes Ncd tail–microtubule association. (Right) XCTK2 is regulated by the Ran pathway in Xenopus egg extracts: (3) The tail microtubule-binding domain of XCTK2 is bound by Importin-α/β, releasing its interactions with microtubules distal to chromosomes. (4) Near chromosomes, elevated levels of Ran-GTP bind Importin-α/β and promote XCTK2 tail–microtubule association.
In contrast, Beaven et al. (2017) find that flies use a mechanism based on similar catch-and-release principles, albeit responsive to spatial signals from CPC (Fig. 1 B, left side). This mechanism was discovered through examination of the meiotic role of 14-3-3 proteins, a family of proteins that bind specifically to phosphorylated targets, changing the localization, activity, or protein–protein interactions of their binding partners (Darling et al., 2005). Drosophila has two 14-3-3 isoforms, ε and ζ; expression of shRNAs in the female germline to deplete these proteins produced abnormal meiotic spindle formation in the absence of 14-3-3ε that was further aggravated by 14-3-3ζ depletion (Beaven et al., 2017). 14-3-3ε–depleted oocytes showed disorganized and unstable spindles that mislocalized minispindles (Msps). Msps is a homologue of vertebrate XMAP215/TOG, a microtubule-associated protein that enhances microtubule assembly. Mass spectrometry showed that 14-3-3ε binds to Ncd within oocyte extracts, and Ncd mutants had been previously reported to have similar spindle defects to those observed in the absence of 14-3-3 proteins. These observations prompted Beaven et al. (2017) to further examine the relationship between these proteins, leading them to find that 14-3-3ε binds to a site within the N-terminal tail domain of Ncd (phosphoserine-96).
Beaven et al. (2017) compared the microtubule binding of an unphosphorylated Ncd tail fragment (Ncd58–192) to one that was fully phosphorylated on S96 in vitro by protein kinase D2 (PKD2). In the absence of 14-3-3ε, phosphorylation did not substantially alter microtubule binding. However, in the presence of 14-3-3ε, binding was reduced, suggesting that phosphorylation-dependent association of Ncd with 14-3-3ε suppresses Ncd tail domain interactions with microtubules. This inhibition could be relieved through a second phosphorylation of Ncd on an adjacent residue (serine-94) by Aurora B while at the same time releasing 14-3-3ε association with Ncd58–192. Mutants lacking serine-94 (NcdS94A) do not localize on meiotic spindles or rescue spindle assembly in Ncd− oocytes. Beaven et al. (2017) interpret these findings to collectively support a model (Fig. 1 B, left side) wherein 14-3-3 binding to Ncd prevents its association to microtubules distal to the meiotic spindle within the large volume of oocyte cytosol. In the vicinity of the meiotic chromosomes, elevated levels of CPC activity cause Ncd serine-94 phosphorylation and 14-3-3 release, directing Ncd to bind specifically to spindle microtubules.
Xenopus eggs and Drosophila oocytes both control the activity of kinesin-14 family members during the process of chromosome-dependent spindle assembly through capture mechanisms that preclude N-terminal tail binding to nonspindle microtubules, with the release of these restrictions near chromosomes. It is striking, however, that these mechanisms respond to separate chromatin-based signaling pathways. It is interesting to speculate whether these mechanisms arose separately to meet similar needs in the different systems for careful regulation of kinesin-14 activity. Alternatively, one of them may have supplanted the other; for example, 14-3-3 binding may have replaced karyopherin binding in flies as CPC became the dominant signaling pathway in this system as Ran became less central. Moreover, many of the major components of both organizational pathways (e.g., karyopherins, Ran, 14-3-3, and CPC) are well conserved and present across species. It will be fascinating to examine in the future whether each organism adheres to only one mechanism for kinesin-14 control, or if, for example, there are situations in which Ran becomes important for Ncd regulation in flies. In either case, several practical questions remain to be experimentally addressed: for example, what kinase phosphorylates Ncd on serine-96 in vivo, and whether this modification is regulated to alter Ncd activity during mitosis or other parts of the cell cycle.
In summary, Beaven et al. (2017) have demonstrated a novel mechanism through which chromosomes regulate acentrosomal spindle assembly via a series of phosphorylation events. These events control the binding and sequential displacement of 14-3-3 proteins, thereby regulating the accessibility of microtubule-binding sites in the N-terminal tail of Ncd. This mechanism is not only interesting in itself but also suggests a remarkable plasticity in the way that similar regulatory events are controlled in different organisms.
Acknowledgments
M. Dasso is supported by the National Institute for Child Health and Human Development intramural project Z01 HD008954.
The author declares no competing financial interests.
References
- Beaven R., Bastos R.N., Spanos C., Romé P., Cullen C.F., Rappsilber J., Giet R., Goshima G., and Ohkura H.. 2017. 14-3-3 regulation of Ncd reveals a new mechanism for targeting proteins to the spindle in oocytes. J. Cell Biol. 10.1083/jcb.201704120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennabi I., Terret M.E., and Verlhac M.H.. 2016. Meiotic spindle assembly and chromosome segregation in oocytes. J. Cell Biol. 215:611–619. 10.1083/jcb.201607062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling D.L., Yingling J., and Wynshaw-Boris A.. 2005. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68:281–315. 10.1016/S0070-2153(05)68010-6 [DOI] [PubMed] [Google Scholar]
- Fink G., Hajdo L., Skowronek K.J., Reuther C., Kasprzak A.A., and Diez S.. 2009. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 11:717–723. 10.1038/ncb1877 [DOI] [PubMed] [Google Scholar]
- Radford S.J., Jang J.K., and McKim K.S.. 2012. The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics. 192:417–429. 10.1534/genetics.112.143495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S.J., Go A.M., and McKim K.S.. 2017a Cooperation Between Kinesin Motors Promotes Spindle Symmetry and Chromosome Organization in Oocytes. Genetics. 205:517–527. 10.1534/genetics.116.194647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S.J., Nguyen A.L., Schindler K., and McKim K.S.. 2017b The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes. Chromosoma. 126:351–364. 10.1007/s00412-016-0618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Z.Y., and Yang W.X.. 2017. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J. Cell Sci. 130:2097–2110. 10.1242/jcs.200261 [DOI] [PubMed] [Google Scholar]
- Weaver L.N., and Walczak C.E.. 2015. Spatial gradients controlling spindle assembly. Biochem. Soc. Trans. 43:7–12. 10.1042/BST20140243 [DOI] [PubMed] [Google Scholar]
- Weaver L.N., Ems-McClung S.C., Chen S.H., Yang G., Shaw S.L., and Walczak C.E.. 2015. The Ran-GTP gradient spatially regulates XCTK2 in the spindle. Curr. Biol. 25:1509–1514. 10.1016/j.cub.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]