Abstract
Purpose
Patients with anaplastic thyroid cancer have a very high death rate. In contrast, deaths from non-anaplastic thyroid cancer are much less common. The genetic alterations in fatal non-anaplastic thyroid cancers have not been reported.
Experimental Design
We performed next-generation sequencing of 410 cancer genes from 57 fatal non-anaplastic thyroid primary cancers. Results were compared to The Cancer Genome Atlas study (TCGA study) of papillary thyroid cancers (PTC) and to the genomic changes reported in anaplastic thyroid cancer (ATC).
Results
There was a very high prevalence of TERT promoter mutations, comparable to that of anaplastic thyroid cancer, and these co-occurred with BRAF and RAS mutations. A high incidence of chromosome 1q gain was seen highlighting its importance in tumor aggressiveness. Two novel fusion genes DLG5-RET and OSBPL1A-BRAF were identified. There was a high frequency of mutations in MED12 and these were mutually exclusive to TERT promoter mutations and also to BRAF and RAS mutations. In addition, a high frequency of mutations in RBM10 were identified and these co-occurred with RAS mutations and PIK3CA mutations. Compared to the PTCs in TCGA, there were higher frequencies of mutations in TP53, POLE, PI3K/AKT/mTOR pathway effectors, SWI/SNF subunits, and histone methyltransferases.
Conclusions
These data support a model whereby fatal non-anaplastic thyroid cancers arise from well-differentiated tumors through the accumulation of key additional genetic abnormalities. The high rate of TERT promoter mutations, MED12 mutations, RBM10 mutations and chromosome 1q gain highlight their likely association with tumor virulence.
Keywords: Non-anaplastic, Thyroid cancer, Next-generation sequencing, MED12, RBM10
Introduction
Patients diagnosed with anaplastic thyroid cancer (ATC) have a very high death rate. Such patients have a mean survival after diagnosis of only 6 months. We have recently reported the genomic hallmarks of ATC showing a very high incidence of TERT promoter mutations in 73% of cases with a co-occurrence with either BRAF or RAS mutations (1). We also identified a high rate of mutations in TP53 (73%) as well as a higher frequency of alterations in genes such as EIF1AX (9%), PIK3CA (18%), and ATM (9%) as compared to those reported by The Cancer Genome Atlas Network (TCGA Network) study of well-differentiated papillary thyroid cancer, which showed a low frequency of somatic alterations (2).
Non-anaplastic thyroid cancers (NAT) derived from thyroid follicular cells comprise well differentiated (WDTC) and poorly differentiated thyroid cancer (PDTC). Deaths from WDTC are extremely rare occurring in 1–2%, however because these tumors are the most common form of the disease, this small percentage represents a significant fraction of patient dying of thyroid cancer. Deaths from PDTC are more common, occurring in 30% of patients (3). The genomic characteristics of fatal cases of non-anaplastic thyroid cancers (FNAT) has not been reported before. We hypothesized that such cancers may harbor genetic similarities to ATC. The objective of our study was to report the genetic alterations in fatal cases of non-anaplastic thyroid cancer and compare their molecular profile to that of ATC and to the TCGA landscape of PTCs. Fatal cases of non-anaplastic thyroid cancer are invariably refractory to radioiodine therapy, and traditional chemotherapy and radiotherapy are of marginal benefit (4). Two multikinase inhibitors, sorafenib and lenvatinib, have been approved for treatment of radioiodine refractory non-anaplastic thyroid cancer. Other drugs are currently being tested in early human clinical trials (5) but these efforts are hindered by the lack of knowledge on the genomics of fatal cases of non-anaplastic thyroid cancer. The identification of the genetic alterations in these cancers will therefore have major implications both in our ability to identify those rare patients at high risk of death and also to develop novel drugs which target the pathways responsible for their poor prognosis.
Methods
Patients and tumor samples
After IRB approval, patients with fatal non-anaplastic thyroid cancer (FNAT) were identified from a database of 3774 patients who had primary surgery treatment at Memorial Sloan Kettering Cancer Center from 1985 to 2010. 86 (2.3%) patients were identified who had either died of disease or died with disease. Of these, paraffin embedded tissue blocks from primary tumors were available on 57 (66%) patients. Following Institutional Review Board (IRB) approval, tumor and matched normal (non-neoplastic normal tissue) specimens were obtained and then hematoxylin and eosin stained tumor sections were independently re-evaluated by head and neck pathologists (R.A.G; D.L.C; BX). Tumors were then classified into poorly differentiated thyroid cancer (PDTC), as defined by histological and/or immunohistochemical evidence of follicular cell differentiation and presence of tumor necrosis and/or ≥ 5 mitoses per 10 high-power fields (×400) (6) and into well differentiated thyroid cancer (WDTC). Patients with well differentiated thyroid cancer were further classified into follicular carcinoma, Hurthle cell carcinoma and the different histological subtypes of papillary thyroid carcinoma such as classical, follicular variant and tall cell variant. Patient demographics, tumor histology, treatment and outcomes were determined by retrospective review of patient charts. Tumors were staged according to the 7th edition of the AJCC staging manual.
Sequencing platform and variant calling
The dataset comprised 35 PDTC and 22 WDTC tumor samples. All 57 tumors were sequenced using the MSK-IMPACT platform, a deep-coverage, targeted next-generation sequencing (NGS) assay encompassing 410 cancer-related genes and approved for clinical use by the NY State Department of Health (7). Of the 35 PDTC, 20 had previously been sequenced using an earlier iteration of MSK-IMPACT comprising 341 genes and reported as part of a cohort of 84 PDTC (1). The MSK-IMPACT (Memorial Sloan Kettering Integrated Mutation Profiling of Actionable Cancer Targets) assay is a next-generation sequencing (NGS) assay approved for clinical use through CLIA (Clinical Laboratory Improvement Amendments) by the Centers for Medicare and Medicaid Services (8). MSK-IMPACT is optimized for DNA extracted from low-input formalin-fixed, paraffin embedded (FFPE) samples. The assay is designed to detect single nucleotide variants (SNVs), indels, copy number variants (CNVs) and structural variants in genes that are functionally relevant to cancer and/or clinically actionable targets. The current assay uses hybrid capture technology (NimbleGen SeqCap EZ library custom oligo) to perform deep (>200×) sequencing (Illumina HiSeq 2500) of all 5781 exons and selected introns of 410 cancer genes, including canonical and selected non-canonical transcripts, the TERT promoter region, and 33 introns of 14 rearranged genes (Supplementary Table 1). The panel includes 1042 tiling probes covering single nucleotide polymorphisms (SNPs), allowing genotyping to ensure tumor-normal matching, identify contaminating DNA, and serve as a low-density SNP array for CNV analysis. MSK-IMPACT has been extensively validated.
Copy number aberrations were identified by comparing sequence coverage of targeted regions in a tumor sample relative to a standard diploid normal sample (7). To call allele-specific somatic DNA copy number we also applied an integrated pipeline called FACETS (9) to Tumor/Normal pairs of bam files according to authors’ recommendations. The bam files were processed to generate a read count matrix for all the potentially polymorphic sites from dbSNP/1000-genome database as well as pseudo SNPs to account for regions that have large gaps between consecutive SNPs. The read counts are then used to compute the GC-corrected normalized log-ratio of tumor to normal read depths for total copy number and log odds ratio from cross tabulating the tumor and normal reads into ref and alt alleles for loci that are heterozygous in the germline. These are then segmented jointly to obtain the regions of constant allele specific copy numbers and the segmented data used for allele specific integer copy number calls as well as cellular fractions.
Results
Patient, tumor, treatment and outcome characteristics (Table 1)
Table 1.
Patient, tumor, treatment and outcome characteristics of 57 patients with fatal non anaplastic thyroid cancer
| N | % | ||
|---|---|---|---|
| Age | < 45 years | 5 | 9% |
| ≥ 45 years | 52 | 91% | |
| Sex | Female | 32 | 56% |
| Male | 25 | 44% | |
| pT size | ≤ 4 cm | 22 | 39% |
| > 4 cm | 32 | 56% | |
| Unknown | 3 | 5% | |
| pT stage | T1 | 2 | 4% |
| T2 | 0 | 0% | |
| T3 | 19 | 32% | |
| T4 | 34 | 60% | |
| TX | 2 | 4% | |
| ETE | No | 9 | 15% |
| Yes | 46 | 81% | |
| microscopic | 12 | 21% | |
| gross | 34 | 60% | |
| Unknown | 2 | 4% | |
| Margins | Negative | 25 | 44% |
| Positive/Close | 30 | 52% | |
| Unknown | 2 | 4% | |
| pN stage | N0/Nx | 25 | 44% |
| N1a | 8 | 14% | |
| N1b | 23 | 40% | |
| Unknown | 1 | 2% | |
| M stage | M0 | 27 | 47% |
| M1 | 30 | 53% | |
| Stage | 1 | 3 | 5% |
| 2 | 0 | 0% | |
| 3 | 5 | 9% | |
| 4 | 47 | 82% | |
| Unknown | 2 | 4% | |
| Tm grade | PDTC | 35 | 61% |
| WDTC | 22 | 39% |
ETE extrathyroid extension
pNx clinically negative neck
Of 57 patients, 52 (91%) patients were over 45 years of age and 32 (56%) were female. The majority of patients had advanced stage disease; 53 (92%) had pT3 or T4 tumors, 34 (60%) gross extra thyroidal extension and 31 (54%) had central or lateral neck metastases. 30 (53%) patients presented with distant metastatic disease. 35 patients had poorly differentiated thyroid cancer and 22 had well differentiated thyroid cancer of whom 2 were classical papillary thyroid cancer (PTC), 2 follicular variant of PTC, 1 micro PTC, 1 PTC with tall cell features, 12 tall cell variant of PTC and 4 Hurthle cell carcinoma. 52 patients were treated by total thyroidectomy and 5 had less than total thyroidectomy (4 lobectomy and 1 subtotal thyroidectomy). The cause of death was distant metastatic disease in 51 patients, loco regional and distant disease in 3 patients, and loco regional disease in 3 patients. The median time to death was 52 months.
Somatic mutations
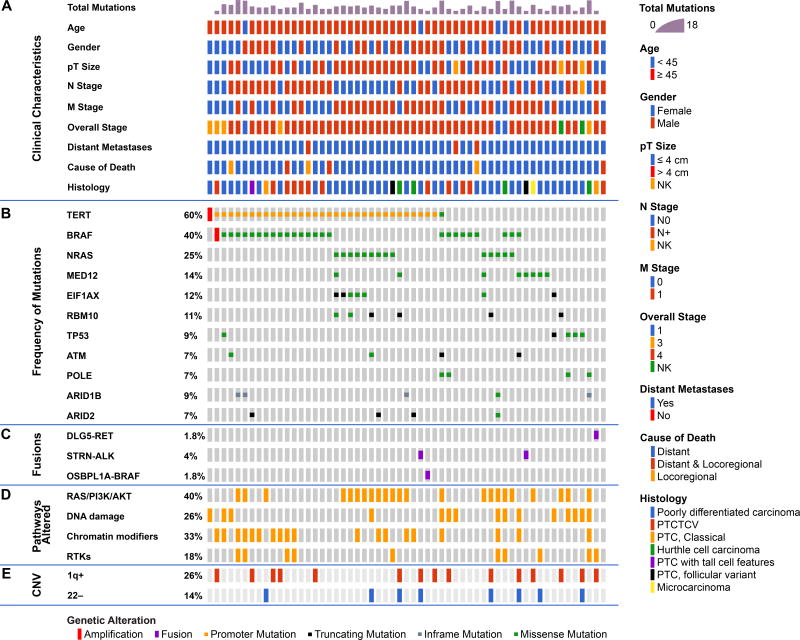
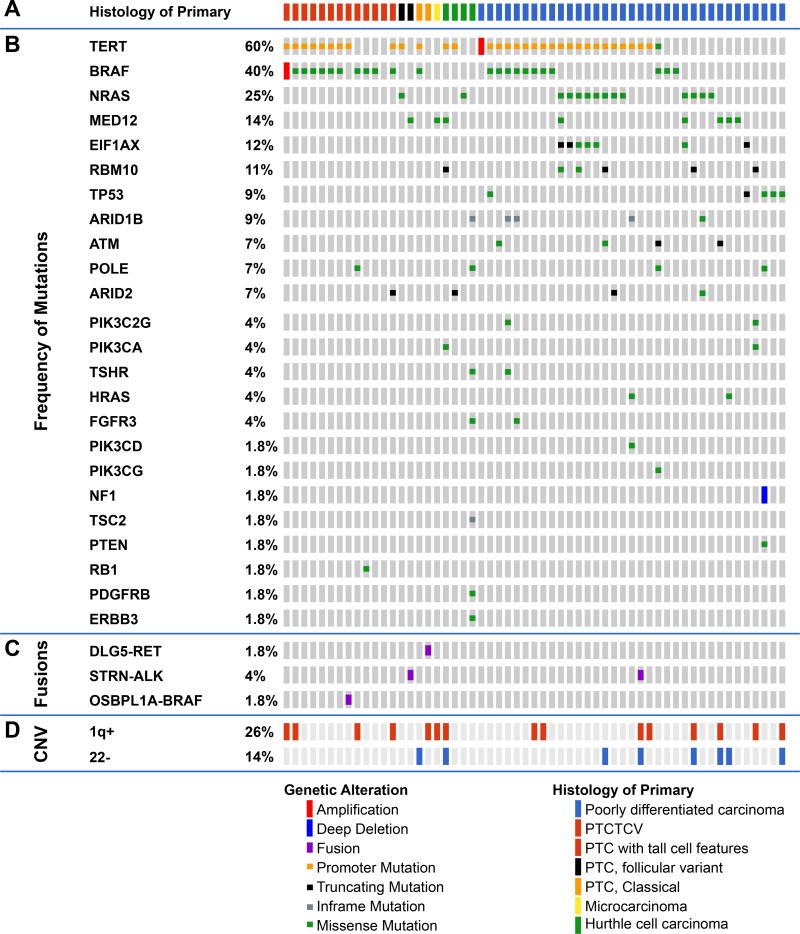
Frequently altered genes (Figure 1)
Figure 1.
Genomic landscape of fatal non-anaplastic thyroid cancer found in 410 genes in MSKCC IMPACT. Clinicopathological characteristics (A) included age, gender, tumor size, N stage, M stage, overall stage, distant metastases and histology. The most frequent genes mutated are shown in panel (B) with fusions shown in panel (C). The percentage of tumors with alterations in different pathways is shown in panel (D); RAS/PI3K/AKT pathway includes NRAS, HRAS, PIK3CA, ,PTEN, PIK3C2G, PIK3CG, AKT1, AKT3, TSC2, and MTOR),Chromatin modifier genes including KMT2A, KMT2C, KMT2D, KDM6A, PRDM1, BCOR, NCOA3, HIST1H3H, HIST1H1C, ARID1B, ARID2, SMARCB1, PBRM1, ATRX, CREBBP. Alterations in DNA damage control included mutations in TP53, RB1, MSK2, CHEK2, and POLE. Alterations in RTKs included mutations in PDGFRA, PDGFRB, FGFR3, ERBB3, MET, EGFR, TSHR, FGF3, TGFBR1, IFNGR1. The percentage of tumors with gain of chromosome 1q and loss of chromosome 22 are shown in panel (E).
There was a high prevalence of telomerase reverse transcriptase (TERT) promoter mutations occurring in 60% of patients (PDTC 60%, WDTC 60%). This high prevalence is comparable to 73% of ATC (1) and far higher than the 9% of PTCs from TCGA (2). TERT mutations co-occurred with BRAF mutations (18/26 p<0.001). They also showed a trend to co-occurrence with RAS mutations (10/16) and EIF1AX mutations (5/7). TERT mutations were mutually exclusive with TP53 mutations (p=0.08) and with MED12 mutations (p=0.04), consistent with alternate pathways toward progression to FNAT.
BRAFV600E mutations were present in 40% and mutations in NRAS and HRAS occurred in 25% and 4% respectively. RAS mutations were mutually exclusive with BRAF and gene fusions (Figure 1). Mutations in the eukaryotic translation initiation factor EIF1AX, reported in 1% of PTCs (2), occurred in 12% of FNAT (Figure 1 and Figure S1A), and were strongly associated with RAS (p<0.001, Figure S1B). EIF1AX mutations clustered in two regions: the N-terminal domain, as also observed in uveal melanomas (10), or at a unique splice acceptor site between exons 5 and 6 (p.A113splice). This splice site is unique to thyroid cancer and also occurs with high frequency in anaplastic thyroid cancer. It results in a 12–amino acid in-frame deletion (1).
Novel genes altered in fatal nonanaplastic thyroid cancer
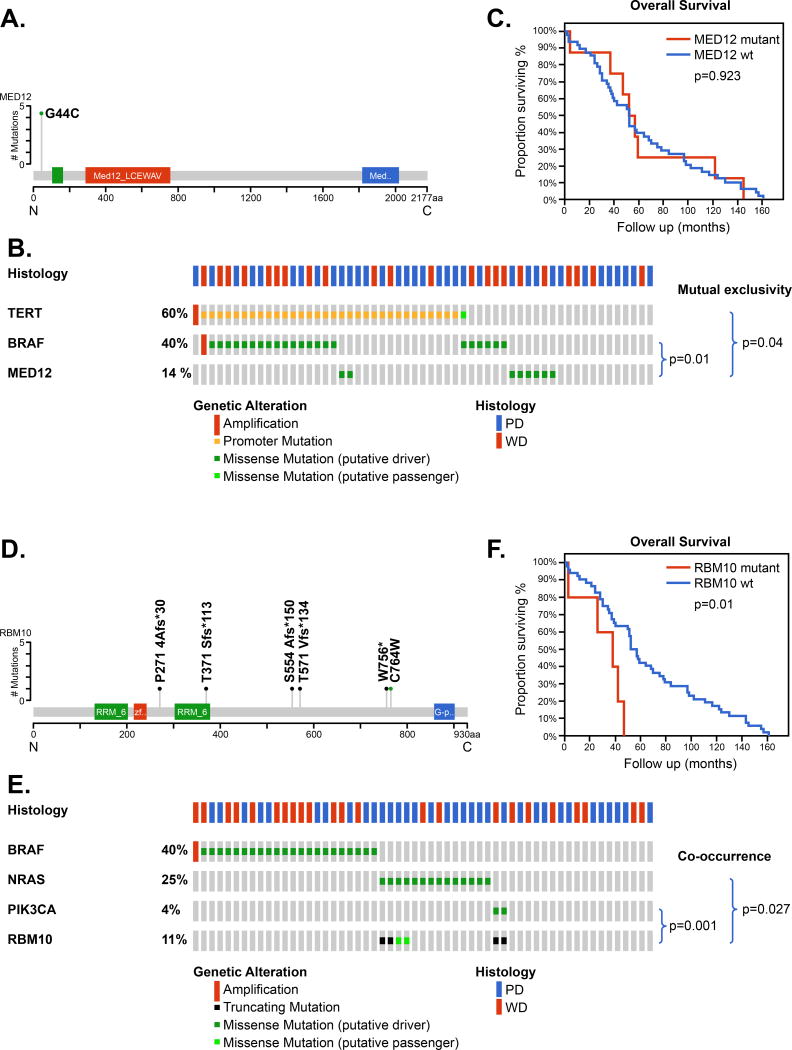
Mutations of the gene MED12 were seen in 14% of patients and all MED12 mutations were at the same site, resulting in a MED12-G44C substitution, consistent with a gain- or change-of-function (Figure 2A). These were mutually exclusive to tumors with a TERT promoter mutation (p=0.04) and were also mutually exclusive to BRAF (p=0.011) (Figure 2B). The overall survival of patients with MED12 mutations were similar to those with wild type MED12 (Figure 2C).
Figure 2.
Mutations in MED12 and RBM10 in fatal non-anaplastic thyroid cancer
Mutations of the gene RBM10 were seen in 11% of patients (Figure 2D). RBM10 mutations were of 2 types: 4 truncating mutations and 2 missense mutations. These were mutually exclusive to tumors with a BRAF mutation (p=0.037) but showed co-occurrence with NRAS (p=0.027) and PIK3CA ( p=0.001) (Figure 2E). The overall survival of patients with RBM10 mutations was significantly poorer to those with wild type RBM10 (p=0.01, Figure 2F).
TP53, ATM, RB1, POLE mutations
We found mutations in TP53 (9%), ATM (7%) and RB1 (1.8%) at a higher prevalence than for PTCs in the TCGA (2) (Figure S2A). Mutations in the POLE gene were seen in 4/57 (7%) of patients.
PI3K/AKT/mTOR pathway alterations
Mutations of genes encoding members of the PI3K/AKT/mTOR pathway were seen in 2557 (44%) of patients. Mutations occurred in PIK3CA (4%), PTEN (1.8%), PIK3C2G (5%), PIK3CD (1.8%), PIK3CG (1.8%), PIK3R2 (1.8%), AKT3 (1.8%), TSC2 (1.8%) and RPS6KA4 (1.8%). These mutations tend to be mutually exclusive to one another. (Figure S2B).
Epigenetic gene alterations
Genes encoding components of the SWI/SNF chromatin remodeling complex were mutated in 9/57 (16%) of patients (Figure S2C). Mutations in ARID1B (9%), ARID2 (7%), SMARCB1 (4%) and PBRM1 (1.8%) genes were identified. These mutations tended to be mutually exclusive indicating that alteration in only 1 of these genes is sufficient to alter function. It was reported previously that individual SWI/SNF chromatin remodeling complexes can contain e.g. either ARID1A or ARID1B but not both. Namely, the combined absence of ARID1A and ARID1B destabilizes SWI/SNF complexes and results in dissociation of subunits which eventually leads to synthetic lethality (11).
Mutations of the histone methyltransferases (HMTs) KMT2A (1.8%), KMT2C (4%- 1 amplification, 1 mutation), KMT2D (4%), KDM6A (1.8%) and PRDM1 (7%) ( 1 mutation and 3 amplifications) were found in 10/57 (18%) of tumors (Figure S2C). Additional mutations in other chromatin remodeling and epigenetic regulators were also seen, including histone acetyltransferase CREBBP (1.8%) and BCOR (4%).
Other gene alterations
Other genes were mutated in a small number of patients. Mutations of other receptor tyrosine kinases (RTKs) such as PDGFRA (1.8%), PDGFRB (1.8%), FGFR3 (4%), ERBB3 (1.8%) and MET (1.8%) were identified (Figure S2D). Mutations in NOTCH2 (5%) and NOTCH3 (1.8%) occurred in 4/57 ( 7%) of patients. There were infrequent mutations in FLT3 (VEGFR3) (1.8%), GNAQ (1.8%), GNAS (4%), KDR (1.8%), ASXL1 (1.8%), DNMT1 (1.8%), DNMT3A (4%).
Gene Fusions (Figure 1)
Of the 57 patients, 4 had a gene fusion identified; 1 in a patient with PDTC and 3 in patients with WDTC.
DLG5-RET
(DLG5: 10q23;RET: 10q11.2) was identified in 1 WDTC patient with classical papillary thyroid cancer. This is a balanced rearrangement involving DLG5 exons 1–13, including the N terminal coiled-coil domains, and RET exons 12 and the rest of the downstream exons which involves the entire RET kinase domain. This patient had no other mutations.
STRN-ALK
(STRN: 2p22.2;ALK: 2p23) was identified in 1 WDTC (follicular variant of PTC) and 1 PDTC patient. This is an 8 Mb deletion between STRN and ALK leading to a fusion between STRN exons 1–3 and ALK exons 20–29 which involves the entire ALK kinase domain. One patient also had a TERT promoter mutation and the other patient had a MED12 mutation.
OSBPL1A-BRAF
(OSBPL1A: 18q11.1 ;BRAF: 7q34 ) was identified in 1 WDTC patient with tall cell variant of papillary thyroid cancer. This is a balanced rearrangement t(7;18) (q34;q18) involving OSBPL1A exons 1–16 and BRAF exons 10 and the rest of the downstream exons including the entire BRAF kinase domain (AA 457–714). This patient also had a TERT promoter mutation.
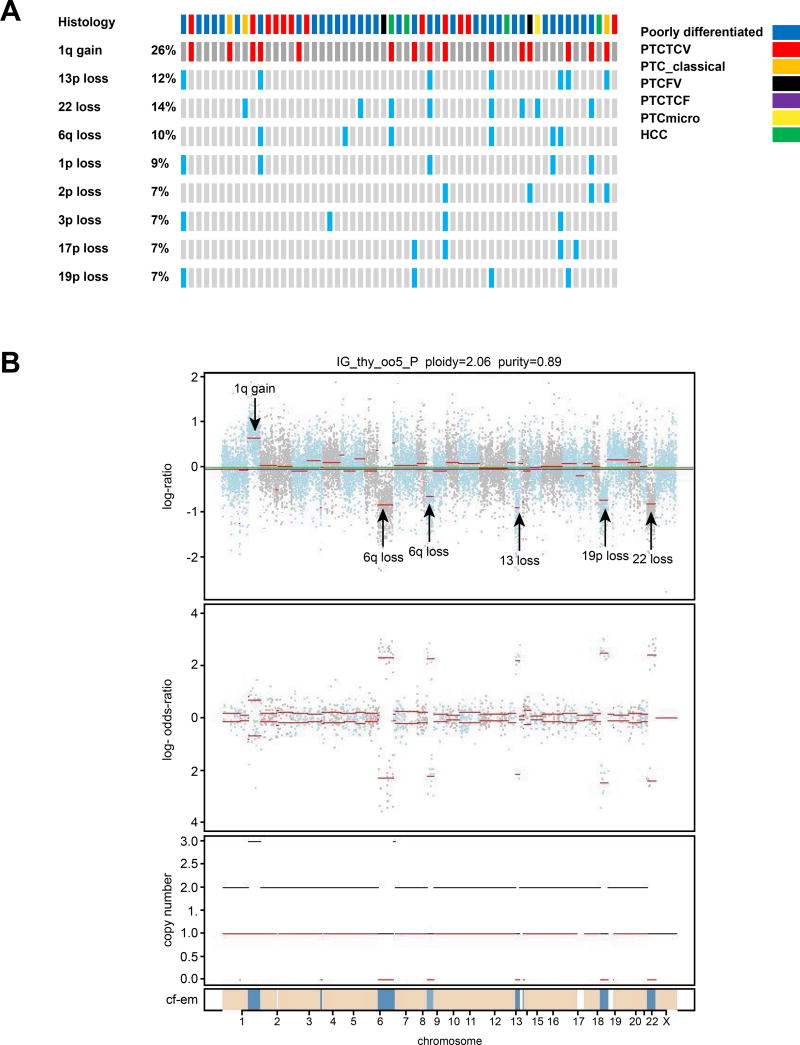
Copy number alterations (Figure 1, Figure 3)
Figure 3.
Most frequent copy number alterations for each patient are shown in A. Figure 3B shows an example of copy number alterations using FACETS in tumor IG_thy_005 which has 1q gain, 6q loss, 9p loss, partial 13 loss, 19p loss and 22 loss.
Of the 57 tumors, there were several arm level alterations identified. The most frequent copy number alterations are shown in Figure 3A. Arm level gains were identified in chromosome 1q in 15 (26%) patients. Arm level losses were identified in chromosomes 22 (14%), 13p (12%), 6q (10%), 1p (9%), 2p (7%), 3p (7%), 17p (7%) and 19p (7%). Example of copy number alterations in 1 tumor are shown in Figure 3B; tumor IG_thy_005 has 1q gain, 6q loss, 9p loss, partial 13 loss, 19p loss and 22 loss. The different components of the FACETS plot are: the top figure shows the GC corrected normalized log-ratio of tumor to normal read depths at a set of SNP loci; the second figure is the log odds ratio from cross tabulating the tumor and normal reads into ref and alt alleles for loci that are heterozygous in the germline; the third figure is the total (black) and minor (red) integer copy number assignment for the segments and the final band shows the cellular fractions where dark blue represents 1 with lighter shades representing lower numbers and beige represents no copy number change.
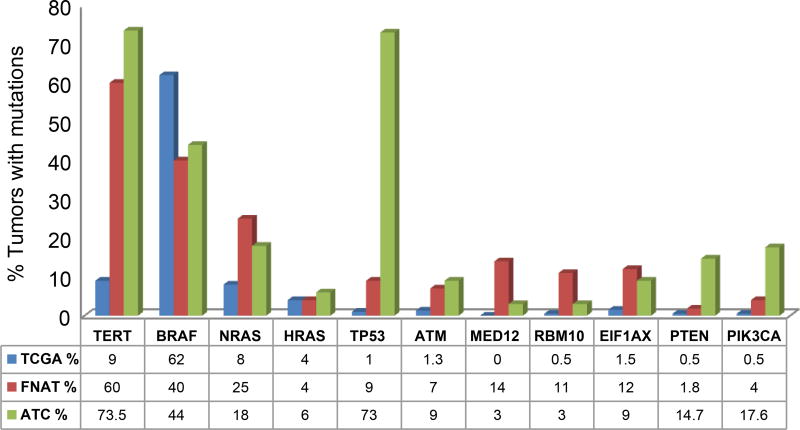
Pathways altered and mutational comparison to ATC and PTC (Figure 1, Figure 4)
Figure 4.
Comparison of the most commonly mutated genes in well differentiated thyroid cancer (TCGA), fatal cases of nonanaplastic thyroid cancer and anaplastic thyroid cancer.
Overall, pathways altered in FNAT included the RAS/PI3K/AKT/MTOR pathway in 40% of patients, DNA damage pathway in 26% patients, chromatin modifying pathways in 33% and alterations in RTKs in 18% of patients (Figure 1). Figure 4 shows the most commonly mutated genes in FNAT compared to well differentiated papillary thyroid cancers (TCGA) and ATC. Compared to WDTC reported in the TCGA, the prevalence of mutations of TERT (60% versus 9%), MED12 (14% versus 0%) and RBM10 (11% versus 0.5%), are higher in FNAT indicating the importance of these genes in tumor virulence. Compared to ATC, FNAT showed a similar MAPK alteration profile with similar frequencies of mutations in BRAF (40%), NRAS (25%) and HRAS (4%) genes (1). Mutations in thyroid-stimulating hormone receptor gene (TSHR) were also comparable to 6% of ATC (1). Mutations in the eukaryotic translation initiation factor 1A, X–linked (EIF1AX), occurred in 12% of our FNAT which was comparable to 9% in our report on ATC (1) and higher than the 1% observed in PTCs (2). We have already reported before a significant association between EIF1AX and RAS mutation, suggesting that this may predict for more aggressive behavior (1). In contrast, mutations of PI3K/AKT/mTOR signaling were uncommon, occurring at a frequency more comparable to PTC (2). Mutations in tumor suppressor genes TP53 and RB1 were much less common than in ATC. However, mutated ATM showed a similar rate of mutation in more aggressive tumors: 1.3% PTC (2) vs. 7% FNAT vs. 9% ATC (1). Mutations of the POLE gene were seen in 7% (4/57) of FNAT patients. DNA polymerase epsilon catalytic subunit (POLE) gene mutations affect the active site of the exonuclease domain of DNA polymerase. This mutation has been described in familial forms of colorectal adenomas and cancers of the colon, pancreas, ovaries and small intestine (c.1373A>T) (12) and in familial cutaneous melanoma (c.1041G>T) (13).
FNAT comprose both WDTC and PDTC.. A comparison of fatal WDTC to nonfatal WDTC using the TCGA cohort is shown in Figure S3A. From this we can conclude that the prevalence of mutations of TERT ( 60% versus 9%), MED12 (13% versus 0%), RBM10 (4% versus 0.5%) and PIK3CA (4% versus 0.5%) are higher in fatal forms of WDTC. We have also shown the comparison of fatal PDTC to nonfatal PDTC using nonfatal PDTC identified from the cohort in Landa et al (1). This is shown in Figure S3B. From this we can conclude that the prevalence of mutations of TERT ( 60% versus 21%), MED12 (15% versus 0%), RBM10 (12% versus 0%) ) are also higher in fatal forms of PDTC. In addition, prevalence of other genes is higher in fatal forms of PDTC. These included BRAF( 29% versus 4%), HRAS (6% versus 1.8%),TP53(15% versus 9%), ATM (12% versus 0%) and EIF1AX( 21% versus 4%).
Molecular profile of FNAT carcinomas according to their various histotypes
The molecular alterations categorized by histology are shown in Figure 5. Of the 13 patients with tall cell variant or tall cell features, 9 had both a TERT promoter mutations and BRAF mutation indicating the importance of this combination in this histology. Of 2 patients with PTC follicular variant, 1 patient had a TERT promoter mutation with NRAS mutation and the other patient had a MED12 mutation alone. Of 2 patients with PTC classical type, 1 patient had a TERT promoter mutation with a BRAF mutation and the other patient harbored the DLG5-RET fusion gene. There was 1 patient with a PTC micro carcinoma and this patient had a MED12 mutation. There were 4 patients with Hurthle cell cancer; 1 patient had TERT, MED12, RBM10 and PIK3CA mutations, 1 patient had TERT and ARID2 mutations, 1 patient had NRAS mutation and 1 patient had ARID1B,POLE, TSHR, FGFR3, TSC2, PDGFRB and ERBB3 mutations.
Figure 5.
Molecular alterations in FNAT stratified by histology
Of patients with PDTC, 60% had a TERT promoter mutation either with a BRAF mutation or a NRAS mutation. 5 patients with TERT/NRAS mutations also harbored an EIF1AX mutation. All EIF1AX mutations occurred in the PDTC patients usually in combination with NRAS mutations. Of the patients who did not have a TERT promoter mutation, mutations were observed in MED12, RBM10, TP53, and ATM amongst others.
Discussion
In this study we report a mutational assessment and clinicopathological features of 57 fatal cases of non-anaplastic thyroid (FNAT) cancer and we examine our results in the context of PTC TCGA study (2) and in the context of deep sequencing studies of ATC we reported previously (1).
Telomerase reverse transcriptase (TERT) promoter mutations showed the highest prevalence of mutation occurring in 60% of FNAT patients. These occurred with equivalent frequencies in both PDTC (60%) and WDTC (60%) patients. Promoter mutations occurred in the two usual hotspot positions (14). The 60% mutation rate in FNAT is far higher than the 9% of PTCs from TCGA (2) and comparable to 73% of ATC (1). This stepwise increase in the frequency of TERT promoter mutations as thyroid differentiation decreases is consistent with our previous reports (1,15) and reports from other studies (16). TERT mutations co-occurred with BRAF or RAS, which enhance the negative prognostic impact of TERT promoter mutations (17). In contrast, TERT promoter mutations in FNAT were mutually exclusive with TP53 mutations (p=0.08) and with MED12 mutations (p=0.04), consistent with alternate pathways toward progression to FNAT.
Compared to ATC, FNAT also showed a similar MAPK alteration profile (NRAS, HRAS, BRAF) and a similar incidence of mutations in the eukaryotic translation initiation factor 1A, X–linked (EIF1AX) (18). In contrast, mutations of PI3K/AKT/mTOR signaling were uncommon, occurring at a frequency more comparable to PTC (2). In addition, mutations in tumor suppressor genes TP53 and RB1 were much less common than in ATC, supporting the notion that mutations in TP53 are infrequent in all histologic types of thyroid cancer with the exception of ATC (1).
We identified chromosomal rearrangements in only a small percentage of FNAT (4/57; 7%) and all involved the entire kinase domain of the fusion partner. Importantly, 2 of the 3 fusion genes identified have never been reported before (DLG5-RET and OSBPL1A-BRAF). DLG5-RET fusion involved the entire RET kinase domain and therefore DLG5 may lead to constitutive activation of RET kinase. Disc large homolog 5 (DLG5) gene is located in a region that undergoes substantial recombination and has a possible role in inflammatory bowel and Crohns disease (19), in cell division, proliferation, cell migration and invasion (20), however it has not been reported as a RET partner in chromosomal rearrangements. As with other RET fusion partners, the DLG5 gene has a coiled-coil domain which acts as the dimerisation domain for the RET gene. OSBPL1A-BRAF is a balanced rearrangement that involves the entire BRAF kinase domain. In this novel fusion, OSBPL1A may lead to constitutive activation of BRAF kinase. OSBPL1A (Oxysterol-binding protein-related protein 1, which acts as an intracellular lipid receptor which is a member of the oxysterol-binding protein family) was shown to have differential expression of isoforms in several cancer types as a result of alternative transcription start site (in colorectal, lung, bladder, liver, prostate, gastric, and brain cancer) (21). The other fusion identified was STRN-ALK. This involves the entire ALK kinase domain, which leads to constitutive activation of ALK kinase via dimerization mediated by the coiled-coil domain of STRN (22). This rare rearrangement has been reported in thyroid cancer before (2) as well as renal cell carcinoma (23) and colorectal adenocarcinoma (24). Patients with this fusion have shown significant initial clinical response to the ALK inhibitors crizotinib and TAE864.
Several copy number alterations (CNA) were identified with the most common being gain of chromosome 1q and also loss of chromosome 22. Chromosome 1q gain was present in 15 FNAT patients (26%), which is higher than 14.8% (2) and 16% (25) reported in PTC. In PTC, 1q gain has been reported in more aggressive tumors (26) and associated with significantly higher MACIS scores, risk profiles and PTC tumor stage (2) as well as distant metastases (25). In PDTC, 1q gains were among the most common arm level CNA (1) and PDTC patients with 1q gains had worse survival rates (1). The high incidence of 1q gain that we observe is in keeping with these findings. Arm level losses in chromosome 22 were present in 8 FNAT patients (14%). This corresponds with previous reports on PTC and PDTC where 22q loss was reported (1,2,25). Loss of 22q region includes tumor suppressor genes NF2 and CHEK2 (25). NF2 loss promotes RAS induced tumorigenesis (27) which is consistent with strong association between 22q loss and RAS-mutated PDTC (1). Therefore our report of 1q gain and 22 loss in FNAT is consistent with previous reports of these CNA in aggressive thyroid tumors.
Our study identified a remarkably high prevalence of mutations in 2 genes, MED12 and RBM10, suggesting a role of these genes in tumor virulence. When we carried out a comparison of fatal forms of WDTC and PDTC to nonfatal forms of WDTC and PDTC, these 2 genes had a higher mutation prevalence indicating their importance in both WDTC and PDTC tumor virulence. MED12 (Mediator of RNA polymerase II transcription subunit 12 homolog) is located on X chromosome and encodes for a subunit of the macromolecular complex known as Mediator. Mediator complex consists of the core Mediator and the kinase module and initiates DNA transcription by interacting with RNA polymerase II (RNA Pol II) (28). Since MED12 plays an essential role in the assembly and activation of the kinase module (29,30) mutations in MED12 can lead to loss or gain of kinase activity. The latter can act as a promoter or suppressor of tumorigenesis, depending on biologic function that the kinase module carries out in the particular tissue (31,32).
MED12 has recently been included as a cancer driver gene in a recent large scale genomic analyses (33,34), reflecting its growing importance. In our study of FNAT, MED12 mutation clustered in a hotspot region in the N terminal region of exon 2 and this clustering suggests a specific change of function. This is consistent with the vast majority of reports where missense mutations of MED12 clustered in a hotspot region within exon 2 (31). Mutation of MED12 has been reported to alter highly conserved amino acids residues (L36, Q43 and G44) in exon 2 (35) which points to a possible gain or change of function. Exon 2 mutations were initially found in uterine leiomyomas (UL) (35) and were the first MED12 mutations reported in human tumors which implicated the role for disrupted Mediator-associated CDK8 kinase activity in tumorigenesis. Since then, MED12 mutations have been reported in typical UL (up to 86%), breast fibroadenomas (59–67 %) and phyllodes tumors (80–88%) (31). Comparative expression profiling and gene set enrichment analyses from mutant and wt MED12 reveal TGF-β signaling and Wnt/β-catenin signaling in mutant UL (31). Further research is needed to determine the impact of MED12 exon 2 mutations on the estrogen signaling pathway and its possible dysregulation during tumorigenesis. Exon 2 mutations are recurrent albeit less frequent in malignant uterine leiomyosarcomas (4–30%), chronic lymphocytic leukemias (5%) and colorectal cancers (0.5%) (31). MED12 mutations have also been reported outside of exon 2 in 5% of prostate cancers and may act through disruption of CDK8 kinase with subsequent transcriptional dysregulation of p53 and androgen signaling (36,37). In our study, all MED12 mutations were recurrent mutation in a single codon resulting in a MED12-G44C substitution. We anticipate that the MED12 mutations in our patients may represent gain or change of function. However, given the complexity of the Mediator complex and the variability in either gain or loss of function depending on tumor type, more research is required to properly explore the true function of the MED12 mutation that we have identified.
In addition to MED12 mutations, we also found mutations in the RNA Binding Motif Protein 10 (RBM10) in 11% of FNAT patients. RBM10 is an RNA binding protein which participates in alternative pre-mRNA splicing. Splicing has a direct role in regulation of gene expression and maintaining the homeostasis of cellular processes. Indeed there has been growing evidence of the involvement of mutated splicing factors in tumor progression (38). Mutation of splicing factors can impair expression of genes crucial for maintaining homeostasis of cell growth and therefore represents a novel mechanism which may promote growth advantage and tumorigenesis of select clonal populations. Mutations of genes encoding splicing factors have been most commonly reported in hematologic malignancies (myelodysplastic syndromes (MDS), acute myeloid leukemia, and chronic lymphocytic leukemia), and less frequently in several solid tumors (38). Most frequent mutations occur in SF3B1, U2AF1, SRSF2 and ZRSR2 and are generally mutually exclusive (38). With regards to RBM10, mutations have been reported in lung adenocarcinomas (39,40), where it acts as an alternative splicing regulator (41) modulating the product of NUMB, a NOTCH pathway regulator gene (42) critical for progression of lung adenocarcinomas. Studies point to loss of tumor suppressor properties of wild type RBM10 and oncogenic function of mutated RBM10 as possible mechanisms for causing uncontrolled growth (40). RBM10 knockdown (RBM10KD) in human cancer cells enhanced tumor growth of xenografts in nude mice with similar results in lung adenocarcinoma cells expressing an RBM10 valine to glutamic acid (V354E) substitution (40). In addition to missense mutations, truncation mutations also occur in RBM10. RBM10 truncated mutants lacking the C-terminal Zn-finger and glycin patch are basically non-functional; moreover the shortest variants appear to exert a dominant-negative effect (40). In our study, RBM10 mutations were of 2 types: 4 truncating mutations and 2 missense mutations. We anticipate that the RBM10 mutations in our patients may represent loss of tumor suppressor function. Furthermore, our study showed statistically significant co occurrence of mutation in RBM10 with NRAS and PI3KCA and also a mutual exclusivity with BRAF mutations. This suggests mutual independence in oncogenic potential of RBM10 regulated proteins and BRAF signaling. Importantly, patients who had RBM10 mutations had a significantly poorer survival compared to patients who did not have these mutations. This suggests tumors harboring RBM10 mutations are biologically more virulent.
Our study has identified several genetic alterations which may have therapeutic implication. There is a great interest to specifically target mutated TERT promoter and our findings suggest aggressive thyroid cancer with these mutations would be an ideal cancer to treat. In addition, new insights into TERT genetics and biology may also offer a potential for personalized immunotherapy (43). The rare STRN-ALK rearrangement can be targeted with ALK inhibitors, crizotinib and TAE864, as previously mentioned. The central role of MED12 in the proper function & assembly of the Mediator kinase module makes MED12 an attractive therapeutic target. So far efforts in targeted therapy have mostly been directed towards CDK8 kinase; e.g. Sorafenib as a multi tyrosine kinase inhibitor/CDK8 inhibitor and Senexin A as a novel CDK8/19 inhibitor (31). An important obstacle in targeting Mediator kinase module may be its versatile role in both repression and activation of transcription depending on the context (31,44) and consequent impact on oncogenic or tumor suppressor signaling. Mutated RBM10 may also represent a novel therapeutic strategy at the level of transcription. Splicing factor inhibitors have already been tested in clinical trials but not in patients with splicing factor mutations (38). A phase I trial of E7107, a spliceosome inhibitor, have been conducted in patients with advanced solid tumors unresponsive to standard therapies (45) and showed promising results. It is possible that splicing factor inhibitors may also be useful in patients which harbor RBM10 mutations.
In conclusion we report the mutational profile of the largest series of fatal NAT that has not been reported before. We have identified TERT promoter mutations in a very high percentage of tumors indicating its importance in thyroid cancer virulence. We report a high incidence of chromosome 1q gain which highlights its importance in tumor aggressiveness. We have identified 2 novel fusion genes DLG5-RET and OSBPL1A-BRAF. Lastly, we report on many novel genes not previously reported in differentiated thyroid cancer including MED12 and RBM10 suggesting a role for these novel genes in tumor virulence. This new data will clarify the genetic basis of the most virulent forms of thyroid cancer and will therefore help focus future therapeutic directions.
Supplementary Material
Statement of translational relevance.
We report the mutational profile of the largest series of fatal non anaplastic thyroid cancer that has not been reported before. We have identified TERT promoter mutations in a very high percentage of tumors indicating its importance in thyroid cancer virulence. We report a high incidence of chromosome 1q gain which highlights its importance in tumor aggressiveness. We have identified 2 novel fusion genes DLG5-RET and OSBPL1A-BRAF. Lastly, we report on many novel genes not previously reported in differentiated thyroid cancer including MED12 and RBM10 suggesting a role for these novel genes in tumor virulence. These genes are targetable mutations and therefore have translational importance in thyroid cancer management. The study also has diagnostic importance as these genetic alterations may predict for poor outcome in patients presenting with thyroid cancer allowing the early identification of patients who may benefit from more aggressive therapy.
Acknowledgments
Financial support: This work was supported in part by NIH grant P30-CA008748 and the LesLois Family Foundation
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest none
References
- 1.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahimpasic T, Ghossein R, Carlson DL, Nixon I, Palmer FL, Shaha AR, Patel SG, Tuttle RM, Shah JP, Ganly I. Outcomes in patients with poorly differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2014;99:1245–1252. doi: 10.1210/jc.2013-3842. [DOI] [PubMed] [Google Scholar]
- 4.Sanders EM, Jr, LiVolsi VA, Brierley J, Shin J, Randolph GW. An evidence-based review of poorly differentiated thyroid cancer. World journal of surgery. 2007;31:934–945. doi: 10.1007/s00268-007-9033-3. [DOI] [PubMed] [Google Scholar]
- 5.Viola D, Valerio L, Molinaro E, Agate L, Bottici V, Biagini A, Lorusso L, Cappagli V, Pieruzzi L, Giani C, Sabini E, Passannati P, Puleo L, Matrone A, Pontillo-Contillo B, Battaglia V, Mazzeo S, Vitti P, Elisei R. Treatment of advanced thyroid cancer with targeted therapies: ten years of experience. Endocr Relat Cancer. 2016;23:R185–R205. doi: 10.1530/ERC-15-0555. [DOI] [PubMed] [Google Scholar]
- 6.Hiltzik D, Carlson DL, Tuttle RM, Chuai S, Ishill N, Shaha A, Shah JP, Singh B, Ghossein RA. Poorly differentiated thyroid carcinomas defined on the basis of mitosis and necrosis: a clinicopathologic study of 58 patients. Cancer. 2006;106:1286–1295. doi: 10.1002/cncr.21739. [DOI] [PubMed] [Google Scholar]
- 7.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyman DM, Solit DB, Arcila ME, Cheng DT, Sabbatini P, Baselga J, Berger MF, Ladanyi M. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug discovery today. 2015;20:1422–1428. doi: 10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic acids research. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, Lohmann DR, Zeschnigk M. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nature genetics. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer cell. 2014;26:309–317. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen MF, Johansen J, Bjornevoll I, Sylvander AE, Steinsbekk KS, Saetrom P, Sandvik AK, Drablos F, Sjursen W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Familial cancer. 2015;14:437–448. doi: 10.1007/s10689-015-9803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoude LG, Heitzer E, Johansson P, Gartside M, Wadt K, Pritchard AL, Palmer JM, Symmons J, Gerdes AM, Montgomery GW, Martin NG, Tomlinson I, Kearsey S, Hayward NK. POLE mutations in families predisposed to cutaneous melanoma. Familial cancer. 2015;14:621–628. doi: 10.1007/s10689-015-9826-8. [DOI] [PubMed] [Google Scholar]
- 14.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science (New York, NY) 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 15.Landa I, Ganly I, Chan TA, Mitsutake N, Matsuse M, Ibrahimpasic T, Ghossein RA, Fagin JA. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98:E1562–E1566. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, Sun H, El-Naggar AK, Xing M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20:603–610. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song YS, Lim JA, Choi H, Won JK, Moon JH, Cho SW, Lee KE, Park YJ, Yi KH, Park do J, Seo JS. Prognostic effects of TERT promoter mutations are enhanced by coexistence with BRAF or RAS mutations and strengthen the risk prediction by the ATA or TNM staging system in differentiated thyroid cancer patients. Cancer. 2016;122:1370–1379. doi: 10.1002/cncr.29934. [DOI] [PubMed] [Google Scholar]
- 18.EIF1AX eukaryotic translation initiation factor 1A, X-linked [Homo sapiens (human)] - Gene - NCBI. https://www.ncbi.nlm.nih.gov/pubmed/
- 19.Friedrichs F, Stoll M. Role of discs large homolog 5. World journal of gastroenterology. 2006;12:3651–3656. doi: 10.3748/wjg.v12.i23.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Li J, Ren Y, Liu P. DLG5 in cell polarity maintenance and cancer development. International journal of biological sciences. 2014;10:543–549. doi: 10.7150/ijbs.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorsen K, Schepeler T, Oster B, Rasmussen MH, Vang S, Wang K, Hansen KQ, Lamy P, Pedersen JS, Eller A, Mansilla F, Laurila K, Wiuf C, Laurberg S, Dyrskjot L, Orntoft TF, Andersen CL. Tumor-specific usage of alternative transcription start sites in colorectal cancer identified by genome-wide exon array analysis. BMC genomics. 2011;12:505. doi: 10.1186/1471-2164-12-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, Gandhi M, Carty SE, Hodak SP, Luo J, Dacic S, Yu YP, Nikiforova MN, Ferris RL, Altschuler DL, Nikiforov YE. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4233–4238. doi: 10.1073/pnas.1321937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusano H, Togashi Y, Akiba J, Moriya F, Baba K, Matsuzaki N, Yuba Y, Shiraishi Y, Kanamaru H, Kuroda N, Sakata S, Takeuchi K, Yano H. Two Cases of Renal Cell Carcinoma Harboring a Novel STRN-ALK Fusion Gene. Am J Surg Pathol. 2016;40:761–769. doi: 10.1097/PAS.0000000000000610. [DOI] [PubMed] [Google Scholar]
- 24.Yakirevich E, Resnick MB, Mangray S, Wheeler M, Jackson CL, Lombardo KA, Lee J, Kim KM, Gill AJ, Wang K, Gowen K, Sun J, Miller VA, Stephens PJ, Ali SM, Ross JS, Safran H. Oncogenic ALK Fusion in Rare and Aggressive Subtype of Colorectal Adenocarcinoma as a Potential Therapeutic Target. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:3831–3840. doi: 10.1158/1078-0432.CCR-15-3000. [DOI] [PubMed] [Google Scholar]
- 25.Kjellman P, Lagercrantz S, Hoog A, Wallin G, Larsson C, Zedenius J. Gain of 1q and loss of 9q21.3-q32 are associated with a less favorable prognosis in papillary thyroid carcinoma. Genes, chromosomes & cancer. 2001;32:43–49. doi: 10.1002/gcc.1165. [DOI] [PubMed] [Google Scholar]
- 26.Wreesmann VB, Sieczka EM, Socci ND, Hezel M, Belbin TJ, Childs G, Patel SG, Patel KN, Tallini G, Prystowsky M, Shaha AR, Kraus D, Shah JP, Rao PH, Ghossein R, Singh B. Genome-wide profiling of papillary thyroid cancer identifies MUC1 as an independent prognostic marker. Cancer Res. 2004;64:3780–3789. doi: 10.1158/0008-5472.CAN-03-1460. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Rendueles ME, Ricarte-Filho JC, Untch BR, Landa I, Knauf JA, Voza F, Smith VE, Ganly I, Taylor BS, Persaud Y, Oler G, Fang Y, Jhanwar SC, Viale A, Heguy A, Huberman KH, Giancotti F, Ghossein R, Fagin JA. NF2 Loss Promotes Oncogenic RAS-Induced Thyroid Cancers via YAP-Dependent Transactivation of RAS Proteins and Sensitizes Them to MEK Inhibition. Cancer discovery. 2015;5:1178–1193. doi: 10.1158/2159-8290.CD-15-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Molecular and cellular biology. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turunen M, Spaeth JM, Keskitalo S, Park MJ, Kivioja T, Clark AD, Makinen N, Gao F, Palin K, Nurkkala H, Vaharautio A, Aavikko M, Kampjarvi K, Vahteristo P, Kim CA, Aaltonen LA, Varjosalo M, Taipale J, Boyer TG. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell reports. 2014;7:654–660. doi: 10.1016/j.celrep.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Critical reviews in biochemistry and molecular biology. 2015;50:393–426. doi: 10.3109/10409238.2015.1064854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banyai G, Lopez MD, Szilagyi Z, Gustafsson CM. Mediator can regulate mitotic entry and direct periodic transcription in fission yeast. Molecular and cellular biology. 2014;34:4008–4018. doi: 10.1128/MCB.00819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science (New York, NY) 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, Aavikko M, Katainen R, Virolainen E, Bohling T, Koski TA, Launonen V, Sjoberg J, Taipale J, Vahteristo P, Aaltonen LA. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science (New York, NY) 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 36.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Molecular cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J Biol Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 38.Bejar R. Splicing Factor Mutations in Cancer. Advances in experimental medicine and biology. 2016;907:215–228. doi: 10.1007/978-3-319-29073-7_9. [DOI] [PubMed] [Google Scholar]
- 39.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA biology. 2016;13:466–472. doi: 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Gogol-Doring A, Hu H, Frohler S, Ma Y, Jens M, Maaskola J, Murakawa Y, Quedenau C, Landthaler M, Kalscheuer V, Wieczorek D, Wang Y, Hu Y, Chen W. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO molecular medicine. 2013;5:1431–1442. doi: 10.1002/emmm.201302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Molecular cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Zanetti M. A second chance for telomerase reverse transcriptase in anticancer immunotherapy. Nature reviews Clinical oncology. 2016 doi: 10.1038/nrclinonc.2016.67. [DOI] [PubMed] [Google Scholar]
- 44.Malik S, Roeder RG. Epigenetics? Mediator does that too! Molecular cell. 2008;31:305–306. doi: 10.1016/j.molcel.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Eskens FA, Ramos FJ, Burger H, O’Brien JP, Piera A, de Jonge MJ, Mizui Y, Wiemer EA, Carreras MJ, Baselga J, Tabernero J. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.