Abstract
The oncogenic activation of the ETS related gene (ERG) due to gene fusions is present in over half of prostate cancer (CaP) in Western countries. Due to its high incidence and oncogenic role, ERG and components of ERG network have emerged as potential drug targets for CaP. Utilizing gene expression datasets, from matched normal and prostate tumor epithelial cells, an association of NOTCH transcription factors with ERG expression status was identified; confirming that NOTCH factors are direct transcriptional targets of ERG. Inhibition of ERG in TMPRSS2-ERG positive VCaP cells led to decreased levels of NOTCH-1 and -2 proteins and downstream transcriptional targets and partially recapitulated the phenotypes associated with ERG inhibition. Regulation of NOTCH-1 and -2 genes by ERG were also noted with ectopic ERG expression in LNCaP (ERG-negative CaP) and RWPE-1 (benign prostate derived immortalized) cells. Furthermore, inhibition of NOTCH by the small molecule gamma-Secretase inhibitor 1, GSI-1, conferred an increased sensitivity to androgen receptor (AR) inhibitors (Bicalutamide, Enzalutamide,) or the androgen biosynthesis inhibitor (Abiraterone) in VCaP cells. Combined treatment with Bicalutamide and GSI-1 showed strongest inhibition of AR, ERG, NOTCH1, NOTCH2, and PSA protein levels along with decreased cell growth, cell survival and enhanced apoptosis. Intriguingly, this effect was not observed in ERG-negative prostate cancer cells or immortalized benign/normal prostate epithelial cells. These data underscore the synergy of AR and NOTCH inhibitors in reducing the growth of ERG-positive CaP cells.
Implications
Combinational targeting of NOTCH and AR signaling has therapeutic potential in advanced ERG-driven prostate cancers.
Keywords: ERG, Oncogene, NOTCH, small molecule, inhibitor, prostate cancer
Introduction
Radical prostatectomy or radiation therapies are effective for treatment of localized organ-confined prostate cancer (CaP), inhibition of the androgen receptor (AR) and the androgen biosynthesis remains the major therapeutic strategy for the treatment of metastatic disease (1–3). Androgen deprivation therapy (ADT) is effective initially, but the inevitable transition from ADT responsive to castration resistant prostate cancer (CRCP) remains the most significant challenge. Second generation androgen axis inhibitors such as Abiraterone and Enzalutamide have significantly improved survival of patients with CRPC (4). However, the benefit is short-lived, and resistance to these drugs and treatment side effects usually develops (4,5). There is an urgent need to continue to develop effective and novel strategies to inhibit AR as well as other CaP drivers which contribute to CaP progression (6,7).
Frequent activation of the ETS Related Gene (ERG) represents one of the most validated oncogenic alterations in CaP (8–11). The androgen dependent expression of ERG oncogene in more than half of all CaPs in Western countries plays a major role in the AR oncogenic network. Accumulating evidence has established that ERG is a key regulator that controls the activity of various signaling pathways recognized as key oncogenic drivers in various malignancies including CaP (12–14). Given the high incidence and mounting evidence supporting its oncogenic role, ERG and components of ERG network have emerged as promising drug targets for CaP therapies (15–18). However, the prevailing notion is that oncogenic nuclear transcription factors such as ERG are challenging therapeutic targets. This scenario prompted us to consider an alternative strategy based on the understanding of the interface between ERG and other functionally relevant pathways. Our initial evaluations of transcriptomes from ERG positive and ERG negative CaP suggested up regulation of NOTCH factors by ERG. Mechanistic investigations in TMPRSS2-ERG positive VCaP cells defined NOTCH 1 and 2 receptors as direct transcriptional targets of the ERG.
The NOTCH signaling pathway controls cell-fate decisions during development including differentiation, proliferation, stem cell maintenance and self-renewal of various cell types (19). The NOTCH signaling pathway has complex functions ranging from being tumor suppressor or as an oncogene in a specific cellular context as well as in a signal strength dependent manner (20). NOTCH pathway has been reported to be involved in drug-resistance (21). Pharmacological inhibition of NOTCH signaling has been shown to increase drug sensitivity to conventional therapies of various types of tumors (22–31). Recently Cui et al have shown that inhibition of NOTCH signaling by Gamma-Secretase Inhibitor (GSI-1) enhances the antitumor effects of Docetaxel in CaP (31).
Although NOTCH 1 mutations are associated with ERG overexpression in animal models and human T cell acute lymphoblastic leukemia, direct regulation of NOTCH by ERG noted in this study has not been described before (32–34). This report further describes synergistic effects of NOTCH and androgen axis inhibitors on ERG positive CaP. The report also revealed unexpected observations of synergy between NOTCH and AR signaling inhibitors currently in clinical use.
Materials and Methods
Reagents
Gamma-Secretase inhibitor I (cat.# 565750) was purchased from Calbiochem/EMD Millipore (Billerica, MA). ERG monoclonal antibody developed by our laboratory (ERG-MAb, 9FY) was obtained from Biocare Medical (Concord, CA). Antibodies against AR (cat.# sc-816), GAPDH (cat.# sc-25778), and α-Tubulin (cat.# sc-5286) were purchased from Santa Cruz (Santa Cruz, CA). Anti-PSA (cat.#A056201-2) antibody was obtained from DAKO cytomation (Carpinteria, CA). Antibodies against NOTCH1 (cat.#3268), NOTCH2 (cat.#4530), EMT antibody sampler kit(cat.# 9782) and apoptosis antibody sampler kit (cat.# 9915) were purchased from Cell Signaling Technologies (Danvers, MA). Sheep anti-mouse IgG-HRP (cat #NXA931) and donkey-anti rabbit IgG-HRP (cat #NA934) were from GE Healthcare (Fairfield, CT). Bicalutamide (cat #S1190), Enzalutamide (cat # S1250) and Abiraterone (cat # S1123) were purchased from Selleckchem (Houston, TX).
Cell Lines
VCaP, LNCaP, PC-3, DU145, and RWPE-1 cells were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA) and were grown as recommended by the supplier. LAPC-4 and BPH-1 were generous gifts from Dr. Charles Sawyers (then at University of California at Los Angeles) and Dr. Simon Hayward (Vanderbilt University Medical Center), respectively. Cell lines obtained from ATCC have been authenticated and tested for Mycoplasma contamination by the vendor using Short Tandem Repeat (STR) Profiling kit (cat.# 135-XV) and Universal Mycoplasma Detection Kit (cat.#30-1012K). Each cell line was passaged for fewer than six months after resuscitation. Reference data were not available for authentication of LAPC-4 and BPH-1 cell lines, hence, these cell lines were not authenticated.
Construction of lentiviral ERG expression vectors
LVX tet-on advanced vector system that includes the pLVX-tet-on plasmid with the tet inducible transactivator element (rtTR-advanced) and pLVX-Tight-puro plasmid harboring the tetracycline response element (TRE) upstream of a minimal CMV-multiple cloning site (MCS) cassette were obtained from Clontech (Clontech, San Diego, CA). TMPRSS2-ERG3 cDNA(35) was inserted into the pLVX-Tight-puro MCS, the two plasmids were packaged, high titer lentiviral particles containing the plasmids were generated and transfected into the HeLa cells. The cells were selected using G418 (800 μg/ml) and puromycin (2 μg/ml) for the retention of pLVX-tet-on and pLVX-Tight-puro plasmids(36). Stable transfectants of LNCaP (lentiviral TMPRSS2-ERG: LNCaP-LTE3) and RWPE1 (lentiviral TMPRSS2-ERG: RWPE1-LTE3) cells were maintained in RPMI 1640 medium with 10% Tet-system approved FBS, and KSFM medium, respectively.
Inhibition of target genes by small interfering RNA
Two or more different siRNA were used for each gene transcript knockdown. ERG specific siRNA sequences and conditions were previously reported by us (37). NOTCH1 (NOTCH1-si1: cat.# J-007771-10, and NOTCH1 si-2: cat.# J-007771-12), NOTCH2 (NOTCH2 si-1: cat.#J-012235-05, and NOTCH2 si-2: cat.# J-012235-06) and non-targeting (NT) siRNA duplexes (cat.# D-001206-13-20). All siRNAs were purchased from Thermo Scientific/Dharmacon (Lafayette, CO). Cells were cultured in their respective medium supplemented with 10% of fetal bovine serum for 48 hours followed by transfection with optimized dosage (50 nM) of target siRNA or NT siRNA, using Lipofectamine 2000 (Life Technologies, Carlsbad, CA). Cells were harvested at desired time points post treatment and processed for Western blot analysis.
Immunoblot Assays
Cells were lysed in Mammalian Protein Extraction Reagent (M-PER) (Thermo, Rockford, IL) containing a protease inhibitor cocktail and phosphatase inhibitor cocktails I & III (Sigma, St Louis, MO). Cell lysates equivalent to 50 μg of total protein were separated on 4%–12% bis-tris gel (Life Technologies, Carlsbad, CA) and transferred to PVDF membrane (Life Technologies, Carlsbad, CA). Membranes were incubated with the primary antibodies at 4°C for 12 hours, followed by three 5-minute washes with wash buffer (1xPBST or 1xTBST) before treatment with secondary antibodies at room temperature for one hour. Finally, membranes were washed three times with wash buffer and developed with ECL Western blot detection reagent (GE Healthcare, Fairfield, CT).
Quantitative Reverse Transcription- Polymerase Chain Reaction (QRT-PCR)
Total RNA prepared from ERG siRNA or NT siRNA treated cells was reverse transcribed into cDNA by OmniScript® RT Kit (QIAGEN, Germantown, MD). Primers used are shown in Supplementary Table 1. Primers and probes were designed using the Primer3 online software (Martinsried, Germany)
Cell Proliferation Assay
Cells were grown in their respective media containing 10% FBS for 48 hours and then treated with indicated concentrations of drugs, either alone or in combination at the indicated time points. Cells were harvested by trypsinization at the indicated time points post-treatment and viable cells were counted by hemocytometer by using trypan blue exclusion method (Cat. # 15250-061, Life Technologies, Carlsbad, CA).
Clonogenic cell survival assay
VCaP cells (50K cell/10cm dish) were plated using the recommended medium for 48 hours. Cells were then treated with the indicated concentrations of AR and NOTCH inhibitors as a single agent or in combination for 48 hours with replenishment of inhibitors every 24 hours. After 48 hours the medium was removed, washed with 1xPBS and replaced with regular growth medium. Cells were then allowed to recover for 14 days and evaluated for their survivability. The colonies which consisted of at least 50 cells were selected for assessment. Colonies were fixed with 4% Paraformaldehyde (PF), stained with crystal violet (0.5% w/v), and counted under inverted microscope.
Chromatin Immuno-precipitation (ChIP) assay
VCaP cells transfected with 50 nM of ERG siRNA or 50 nM of NT siRNA were processed for ChIP assay as previously described (37). Amplification reactions were carried out on T-Gradient Thermoblock (Biometra, Göttingen, Germany) by using 95°C, 5 s; ,95°C, 15 s; 54°C 30 s; 72°C 60 s program setting. For detecting genomic input DNA and specific ChIP products 35 and 40 PCR amplification cycles were used, respectively. ETS binding sites within the target regions were identified (Supplementary Table 2) by matrix match analysis using the MatInspector software (Genomatix GmbH, Munich, Germany). Fold enrichment values were calculated by first normalizing the average fold changes of NT or ERGsi to the average of corresponding input values of target sequence amplicons (NOTCH1/V$ETS#1, NOTCH1/V$ETS#2, NOTCH2/V$ETS#3, and C-MYC (37). Then the ratio of normalized fold changes between NT and ERGsi were calculated.
Correlation of ERG and NOTCH expression in human prostate cancer
Gene expression dataset from matched normal epithelial cells and prostate tumor epithelial cells with known ERG gene expression status previously developed in our laboratory (NCBI, GSE32448) were used to identify ERG associated relevant oncogenes that could be utilized as surrogates of ERG targeted therapy in CaP (9,38,39). The transcriptomes were derived from tumor cells with well to moderately differentiated (n=20) and poorly differentiated morphology (n=20) representing two groups of prostate tumor types.
Treatment of cells with NOTCH and AR inhibitors
To evaluate the combined effect of the GSI-1 and the AR inhibitors, cells were grown in recommended media containing 10% FBS for 48 hours. After 48 hours at confluency of 50% cells were then treated with the indicated concentrations of the drugs (0, 1, 5, or 10μM), either alone or in combination at the indicated time points. Initially other γ-secretase inhibitors including DAPT, MK-0752, Semagacestat, and R04929097 (all from Selleckchem, Houston, TX) were also evaluated for the synergistic effect with the AR inhibitors but only GSI-1 show significant synergy with AR inhibitors and selected for further evaluation.
Statistical analysis
Two-tailed Student’s t-test was used to compare between specific groups within a data set. P<0.05 was considered statistically significant difference. Data are presented as the mean ± standard error of the mean.
Results
NOTCH2 expression in prostate cancer correlates with TMPRSS2-ERG status
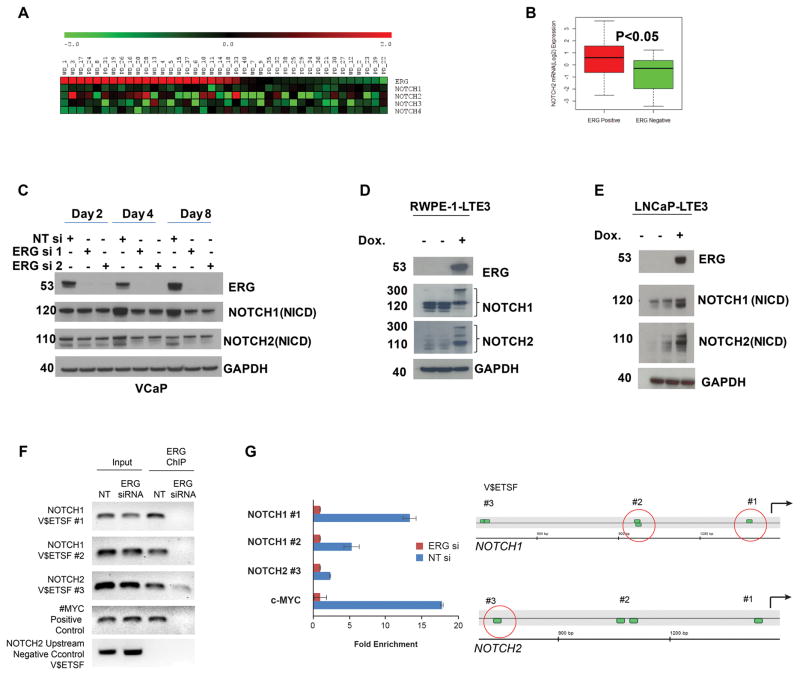
To identify functionally and therapeutically relevant targets of ERG oncogene for CaP, an association study was performed in a gene expression dataset from laser capture microdissected matched prostate tumor and normal epithelial cells. We evaluated the correlation between tumor/benign normalized NOTCH1, NOTCH2 and ERG expression status in the gene expression dataset analysis of 20 well to moderately differentiated (WD) and 20 poorly-differentiated (PD) CaP tumors (37–39). The results revealed that the expression of NOTCH2 correlated with the expression of ERG expression while the expression of NOTCH1, NOTCH3 and NOTCH4 showed no significant correlation with ERG expression (Fig. 1A). Fifty seven percent of ERG positive samples expressed NOTCH2 (15/26) versus 35% of the ERG negative samples (5/16) (Fig. 1B).
Figure 1.
NOTCH1 and NOTCH2 are transcriptionally regulated by ERG. A, Association of NOTCH factors with ERG expression status. Association of ERG and NOTCH expression were evaluated in a cohort of well (WD) and poorly differentiated (PD) CaP samples. B, Among the four NOTCH transcription factors, NOTCH2 showed significant association with ERG status. C, Decreases protein levels NOTCH1 and 2 (NICD: NOTCH intracellular domain) in response to knockdown of ERG in VCaP cells. D&E, Heterologous expression of TMPRSS2:ERG3 (ERG) under the control of stable integrant doxycycline (Dox) inducible promoter resulted in increased protein levels of NOTCH1 and 2 in LNCaP (LNCaP-LTE3) cell line or in benign prostate derived immortalized prostate epithelial cells (RWPE-1). F, ERG is recruited to predicted ERG binding sites of NOTCH1 (NOTCH1_V$ETSF#1, NOTCH1_V$ETSF#2) and NOTCH2 (NOTCH2_V$ETSF#3) gene promoter upstream sequences which was diminished in response to ERG siRNA treatment. The recruitment of ERG to the C-MYC promoter (37) was used as a positive and a distal upstream sequence within 15 kb of the NOTCH2 transcription initiation site lacking ETS motifs was as negative control. Input DNA was used as technical control. In quantitative ChIP PCR the fold enrichment was calculated as the ratio of input normalized fold changes between NTsi (2^ dCT) and ERGsi (2^ dCT) values (48). G, Position of predicted ERG binding sites (V$ETSF) relative to the 5′ end of evaluated NOTCH1 and NOTCH2 gene promoter upstream sequences. Red circles indicate position of detected ERG recruitment.
NOTCH transcription factors are direct targets of ERG
To further understand the regulation of NOTCH transcription factors by ERG, we performed western blot analyses on ERG siRNA treated VCaP cells. Both NOTCH1 and NOTCH2 protein expression levels were reduced in response to inhibition of ERG by the siRNA (Fig. 1C). This effect was even more pronounced in sustained inhibition of ERG expression over eight days. The functional connection between ERG and NOTCH1 and NOTCH2 was further examined in stable transfectants of LNCaP or RWPE-1 cells harboring doxycycline inducible TMPRSS2-ERG3 lentiviral expression vector (LNCaP-LTE3 and RWPE1-LTE3). NOTCH1 and NOTCH2 protein levels were increased in response to doxycycline induced ERG expression (Fig. 1D&E). To gain further insights into regulation of NOTCH transcription factors by ERG, we performed Chromatin Immuno-precipitation (ChIP) analysis examining the promoter upstream regions of the NOTCH1 and 2 genes. Using MatInspector software (Genomatix GmbH, Munich, Germany), a matrix match survey of −1,500bp promoter upstream sequences of the NOTCH1 and NOTCH2 genes identified ETS-matrix matches which are potential binding sites for ERG. ChIP assay employing VCaP cells confirmed the specific recruitment of ERG oncoprotein to multiple distinct clusters of ETS sites upstream of the NOTCH1 and NOTCH2 promoters (see Supplementary Table 2 for locations of the ETS binding sites). The recruitment of ERG to these sites was significantly reduced in response to ERG siRNA- knock-down (Fig. F and G). The recruitment of ERG suggests involvement of ERG oncoprotein on the regulation of NOTCH1 and NOTCH2 in prostate tumor cells.
ERG modulates EMT through NOTCH signaling
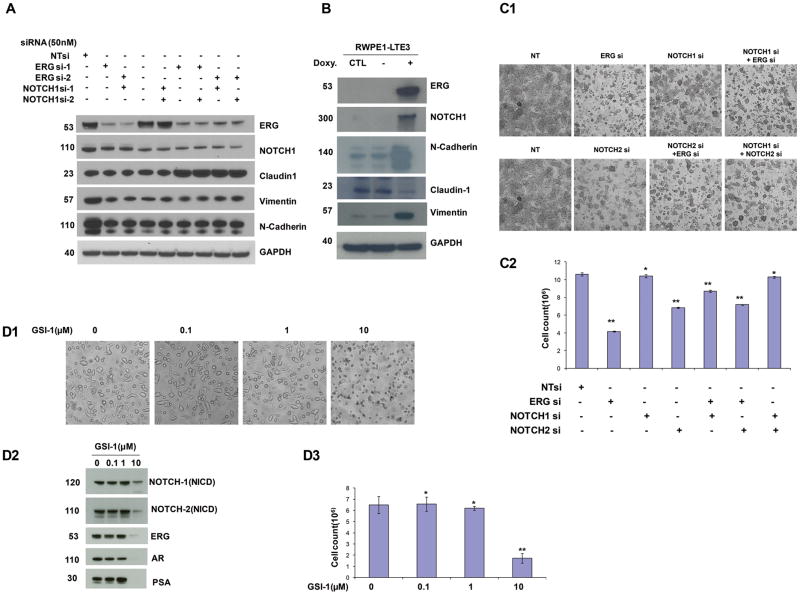
To determine the role of NOTCH transcription factors in ERG positive CaP cell lines, inhibition of ERG, NOTCH1, and NOTCH2 alone, or in combination was examined on cell growth and cell morphology. We have reported previously that suppressing ERG expression induced the differentiation markers of prostate epithelial cells (37). Inhibition of ERG and NOTCH1, either alone or in combination, resulted in the inhibition of the mesenchymal markers N-Cadherin and Vimentin. Simultaneous knockdown of ERG and NOTCH1 resulted in a marked increase of the epithelial marker Claudin-1 (Fig. 2A). Overexpression of ERG protein in the immortalized benign prostate epithelial cells, RWPE-1, resulted in the concomitant overexpression of mesenchymal markers N-Cadherin and Vimentin, and inhibition of the epithelial marker, Claudin-1 (Fig. 2B).
Figure 2.
ERG affects EMT in prostate cancer through modulation of NOTCH transcription factors. A, Inhibition of ERG in the TMPRSS2:ERG positive VCaP cells resulted in the inhibition of NOTCH1 and the mesenchymal markers (N-Cadherin and Vimentin) while it induced the epithelial marker (Claudin-1). B, Overexpression of TMPRSS2:ERG3 in normal immortalized prostate epithelial cells resulted in an increased expression of NOTCH1, N-Cadherin, and Vimentin, while inhibiting Claudin-1 protein. C1&C2, The knock-down of NOTCH1 and NOTCH2 recapitulates the effect of ERG knock-down on prostate epithelial differentiation. Experiments were performed in triplicates and results are shown as mean ±SD of three independent experiments. Student’s t test was used to compare treated cells with control (0μM), * p> 0.05, ** p< 0.05. D1–D3, Treatment of the NOTCH inhibitor GSI-1 significantly reduced expression of AR, ERG and PSA as well as NOTCH1 and NOTCH2 proteins and inhibited cell growth at higher concentrations (10μM). Experiments were performed in triplicates and results are shown as mean ±SD, * p> 0.05, ** p< 0.05, (t test, n=3).
Inhibition of ERG and NOTCH abrogate tumor cell growth
To assess the effect of ERG and NOTCH1 and NOTCH2 on cell proliferation, we treated VCaP cells with ERG, NOTCH1, NOTCH2 specific siRNAs or NT siRNA, either alone or in combination as shown in Fig. 2C1&C2. Knock-down of NOTCH1 or NOTCH2 alone in VCaP cells by siRNA did not show significant cell growth inhibition (Fig. 2C1&C2). However, inhibition of NOTCH1 or NOTCH2 resulted in a characteristic change in cell morphology similar to the morphology observed in response to ERG inhibition resembling reversal of the mesenchymal and non-malignant epithelial shape in VCaP cells (Fig. 2C1). These findings are consistent with reports on the modulation of EMT by NOTCH transcription factors in several cancer types.
Inhibition of NOTCH selectively enhances the effect of AR inhibitors in ERG positive prostate cancer cells
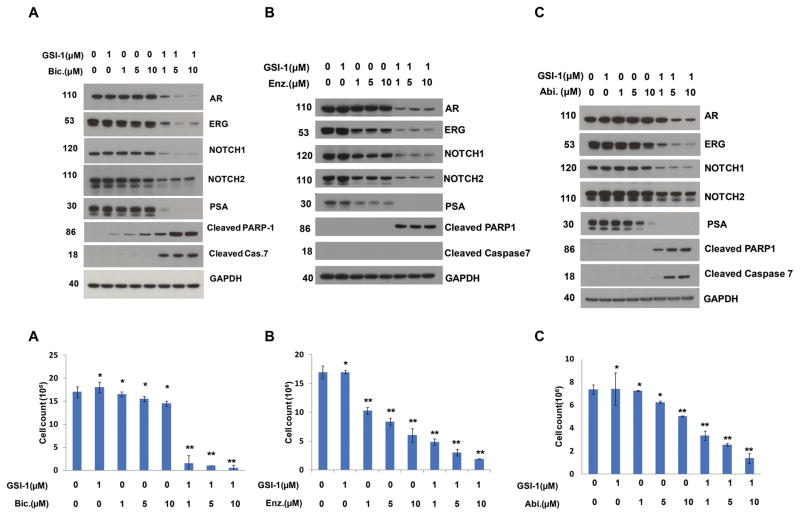
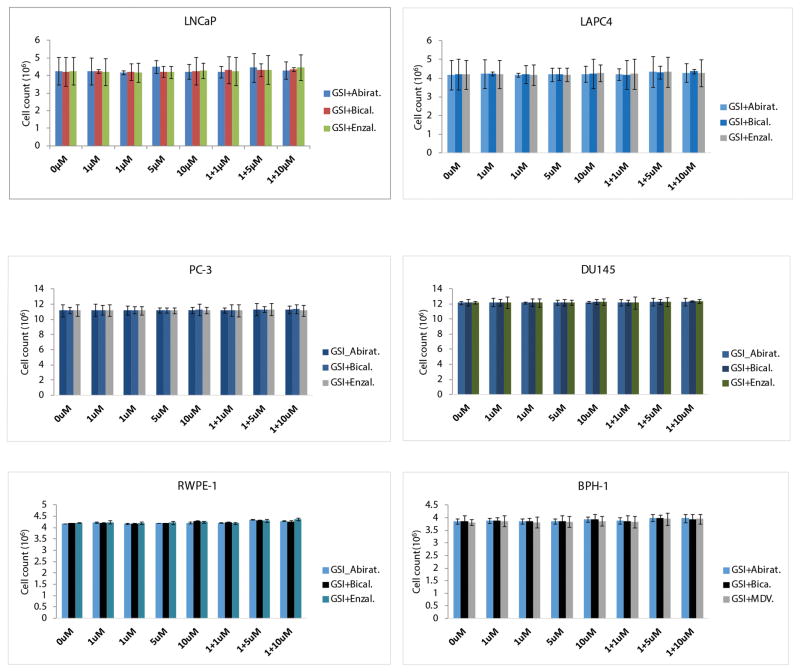
The NOTCH inhibitor, γ-Secretase inhibitors-1 (GSI-1) exhibited inhibition of NOTCH1 and NOTCH2 protein levels and VCaP prostate cancer cell growth in a dose dependent manner (Fig. 2D1–D3). An unexpected observation of this study was the inhibition of AR and ERG by the GSI-1, suggesting for other, as yet unknown, effects of this inhibitor (Fig. 2D2). This prompted us to assess the inhibition of ERG positive prostate cancer cell growth by combining AR axis inhibitors used for the ADT (Abiraterone, or Bicalutamide, or Enzalutamide) with GSI-1. Combination of low dose of GSI-1 with each of the AR axis inhibitors resulted in enhanced inhibition of the ERG and also AR, PSA, NOTCH1 and NOTCH2 protein levels. In addition to these observations, significant reduction of cell growth and increased apoptosis was noted as shown by the presence of cleaved PARP1 and cleaved caspase 7 (Fig. 3A–C). However, combination of GSI-1 and Enzalutamide did not induce cleaved caspase 7, indicating a different mechanism of apoptosis induction by this treatment. Next, a panel of ERG negative prostate or prostate cancer cell lines that included transformed or benign prostate epithelium derived cells were tested with a combination of GSI-1 and AR axis inhibitors (Abiraterone, Bicalutamide, or Enzalutamide). In contrast to the observations response in ERG positive VCaP cells, no appreciable inhibition of cell growth was seen in these cells using the same drug combinations (Fig. 4). These results suggested selectivity of NOTCH and AR inhibitor combinations for ERG positive prostate cancer cells.
Figure 3.
Increased sensitivity of ERG positive tumor cells to AR inhibitors in combination with NOTCH inhibitor. A, B & C (upper panels), Treatment of VCaP cells with NOTCH inhibitor, GSI-1, in combination with either Abiraterone (Abi), Bicalutamide (Bic), or Enzalutamide (Enz) augmented their effects on the inhibition of AR, ERG, PSA, NOTCH1 and NOTCH2 expression and induced cleavage of PARP1 and Caspase 7. A, B & C (lower panels), GSI-1 increases sensitivity of ERG positive CaP cells to AR inhibitors. Concomitant treatment of GSI-1 with Abiraterone, Bicalutamide, or Enzalutamide significantly enhanced the inhibition of cell growth in VCaP cells. Each experiment were performed three times and results are shown as mean ±SD, * p> 0.05, ** p< 0.05.
Figure 4.
GSI-1 does not augment the inhibition of cell growth by AR inhibitors in benign or transformed prostate epithelial cells, or in ERG negative prostate cancer cell lines, irrespective of their AR expression status. Simultaneous treatment of GSI-1 with AR inhibitors (either Abiraterone, Bicalutamide, or Enzalutamide) did not show any synergistic effect in benign prostate epithelial cells, RWPE-1, transformed prostate epithelial cells, BPH-1-1, and a panel of ERG negative prostate cancer cell lines, regardless whether they are AR positive, LNCAP, LAPC-4, or AR negative, DU145 and PC-3.
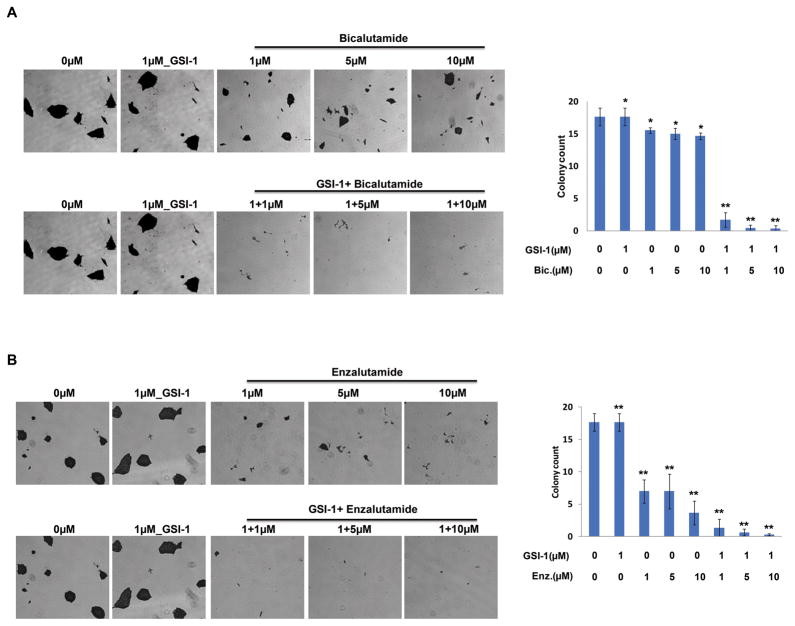
Combination of AR and NOTCH inhibitors delay survival colony formation of VCaP cells
To further evaluate the ability of VCaP cell survival after treatment with combination of AR and NOTCH inhibitors, survival colony formation assay was employed for VCaP cells treated with the AR and NOTCH inhibitors in combination or as a single agent for 48 hours. Combination of low concentrations of GSI-1 and Bicalutamide or Enzalutamide resulted in significant reduction in cell growth and cell survival when compared to mock treated control cells (Fig. 5A&B).
Figure 5.
Combination of GSI-1 and AR inhibitors delays colony survival formation. A&B, Treatment of VCaP cells with NOTCH inhibitor, GSI-1, in combination with either Bicalutamide or Enzalutamide significantly inhibited survival colony formation capability of VCaP cells. Colonies with more than 50 cells were counted (right bar diagrams). Results are shown as mean ± SD of triplicate experiments, * p> 0.05, ** p< 0.05.
Discussion
The dysregulation and high frequency of ERG expression and function in CaP and other malignancies have demonstrated ERG protein and its functional net-work into promising candidates for ERG targeted therapeutic intervention. As a transcription factor, ERG biochemical and biological functions are diverse including maintenance of cellular differentiation and stem cell phenotypes. Within the context of cancer, ERG oncogenic activation influences cancer biology; e.g, activation of cell invasion and other pro-cancer signaling pathways and inhibits cell differentiation (11).
Positive correlation between ERG and NOTCH2 expression was found assessing laser capture microdissected human prostate cancer tumor/normal matched transcriptome datasets. Although, correlation with NOTCH1, 3 and 4 was not apparent, we have evaluated all known NOTCH transcription factors in ERG positive (TMPRSS2-ERG harboring) VCaP cells. Evaluation by ERG knockdown and overexpression approaches suggests that NOTCH1 and NOTCH2 are positively regulated by ERG. In contrast, NOTCH3 and NOTCH4 were non-responsive to ERG overexpression or knockdown.
The NOTCH signaling pathway is critical in controlling cell-fate decisions during development (19,40). The ligands driven NOTCH signaling is activated when the NOTCH receptors of a signal-sending cell physically interacts with a signal-receiving cell through receptor-ligand interaction. Ligand binding triggers a succession of proteolytic events, whereby NOTCH receptor is cleaved twice, first by an extracellular matrix metalloprotease (tumor necrosis factor-α-converting enzyme, TACE) and then by the transmembrane protease γ-Secretase complex. Thus, γ-secretase inhibitor (GSI-1) has been studied for NOTCH targeted treatment of human malignancies(41). NOTCH signaling also plays crucial role in the development of both normal prostate gland and prostate cancer (42,43). Using Notch1 knock-out mouse model it was shown that Notch1- expressing cells are indispensable for prostate branching morphogenesis, growth, differentiation and re-growth, suggesting its role in defining progenitor cells in the prostate (43).
In an earlier study we showed that ERG negatively regulates the expression of a number of prostate differentiation genes and abrogates the prostate epithelial differentiation program (37). In the current study, we further extend these observations by showing that expression of ERG is directly correlated to the expression of NOTCH1 and NOTCH2 factors. Our analysis of the promoter upstream regions of NOTCH1 and NOTCH2 genes showed the specific recruitment of ERG protein to binding sites for ERG suggesting the likely basis for activation by ERG. The knock-down of either NOTCH1 or NOTCH2 or both in TMPRSS2-ERG positive CaP cells recapitulated the phenotypes associated with ERG knock-down. Moreover, we demonstrate that ERG regulates EMT in part through NOTCH signaling pathway and increased NOTCH2 is associated with ERG expression in a cohort of prostate cancer patients.
Our investigations revealed an unexpected finding showing that GSI-1 inhibited ERG, AR and AR targets only in ERG positive CaP Cells. This prompted us to assess potential synergistic effect of a combination of NOTCH and AR inhibitors. When VCaP cells are treated with low doses of the GSI-1 in combination with AR inhibitors, we observed the enhanced inhibition of ERG, AR, PSA, NOTCH1 and NOTCH2 proteins, the induction of apoptosis and significant cell growth inhibition (Fig. 3). These effects were not observed in a panel of similarly treated ERG negative prostate cancer or benign or normal transformed prostate epithelial cells regardless of their AR status (Fig. 4). Among the evaluated AR inhibitors, Bicalutamide showed most robust synergistic effect with GSI-1. These findings imply that pharmacological inhibition of both NOTCH and AR could be used effectively to treat ERG positive CaP patients.
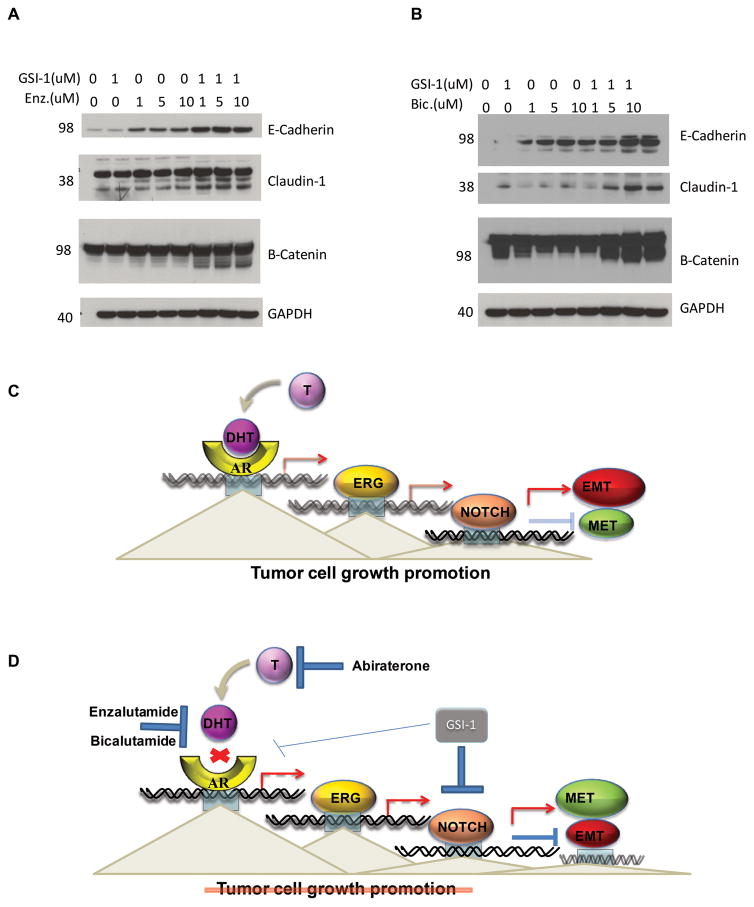
In summary, the results presented in this study show that there is a “functional cross talk” between AR, ERG, NOTCH signaling pathways. This finding further enabled us to uncover the synergistic effect of a combination of NOTCH and AR inhibitors on ERG positive prostate cancer cells. The mechanism responsible for the observed synergy deserves more investigation and is the subject of follow up study. However, we propose that the observed synergy is due to induction of Mesenchymal-Epithelial Transition (MET) and inhibition of Epithelial-Mesenchymal Transition (EMT) markers, which in turn sensitizes cells to the drugs (Fig. 6), as has been reported before, activation of NOTCH signaling is essential for the maintenance of EMT and cancer stem cells(44–47). We propose that the therapeutic approach involving a combination of NOTCH and AR inhibitors will be of potential utility in the treatment of advanced prostate cancer with ERG defects.
Figure 6.
Synergistic inhibition of NOTCH and ERG signaling reverses EMT. A&B, The concurrent inhibition of AR pathway and NOTCH signaling by GSI-1 and either Bicalutamide (Bic), or Enzalutamide (Enz) induces epithelial markers Claudin-1, E-Cadherin and β-Catenin, indicating activation of MET and inhibition of EMT. This might be responsible for the observed enhanced synergy between AR inhibitors and GSI-1. C&D, The proposed mechanism is that inhibition of AR’s tumor promoting function in combination with the pro-tumorigenic functions of NOTCH is more effective therapeutic approach than any of the compounds alone.
Supplementary Material
Acknowledgments
We are grateful to all members of the CPDR, USU, HJF, and especially Mr. Stephen Doyle for the art work; Ms. Chantal Falade for administrative support. This research was supported in-part by the CPDR-USU program HU0001-10-2-0002 to D.G.M. and the National Cancer Institute R01 DK065977 grant to S.S. The views expressed in this manuscript are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the U.S. Government.
Financial support: This work was supported in part by CPDR-USU program HU0001–10–2-0002 to D.G.M., and NIH Grants RO1 DK065977 to S.S.
Abbreviations
- AR
Androgen Receptor
- ERG
EST Related Gene
- PSA
Prostate Specific Antigen
- GSI-1
Gamma-Secretase Inhibitor 1
- CaP
Prostate Cancer
- ADT
Androgen Ablation Therapy
- CRPC
Castration Resistant Prostate Cancer
- EMT
Epithelial-Mesenchymal Transition
- MET
Mesenchymal-Epithelial Transition
- FBS
Fetal Bovine Serum
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest
Authors’ Contributions:
Conception and design: Ahmed A. Mohamed, Albert Dobi and Shiv Srivastava
Development of methodology: Ahmed A. Mohamed, Albert Dobi and Shiv Srivastava
Acquisition of data: Ahmed A. Mohamed, Shyh-Han Tan, Charles P. Xavier, Shilpa Katta, Wei Huang, Lakshmi Ravindranath, Muhammad Jamal, Hua Li, Meera Srivastava, Eri Srivatsan and Gyorgy Petrovics
Analysis and interpretation of data: Ahmed A. Mohamed, Shiv Srivastava, Albert Dobi, Meera Srivastava, Taduru Sreenath, Gyorgy Petrovics, David G. McLeod
Writing, review, and revision of the manuscript: Ahmed A. Mohamed, Shyh-Han Tan, Alagarsami Srinivasan, Albert Dobi, Shiv Srivastava
Administrative, technical or material support: Ahmed A. Mohamed, Shyh-Han Tan, Charles P. Xavier, Shilpa Katta, Wei Huang, Lakshmi Ravindranath
Study supervision: Shiv Srivastava, Albert Dobi
References
- 1.Rodrigues DN, Boysen G, Sumanasuriya S, Seed G, Marzo AM, de Bono J. The molecular underpinnings of prostate cancer: impacts on management and pathology practice. J Pathol. 2016 doi: 10.1002/path.4826.. [DOI] [PubMed] [Google Scholar]
- 2.Yap TA, Smith AD, Ferraldeschi R, Al-Lazikani B, Workman P, de Bono JS. Drug discovery in advanced prostate cancer: translating biology into therapy. Nat Rev Drug Discov. 2016;15(10):699–718. doi: 10.1038/nrd.2016.120. [DOI] [PubMed] [Google Scholar]
- 3.Sharp A, Welti J, Blagg J, de Bono JS. Targeting Androgen Receptor Aberrations in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2016;22(17):4280–2. doi: 10.1158/1078-0432.CCR-16-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francini E, Taplin ME. Prostate cancer: Developing novel approaches to castration-sensitive disease. Cancer. 2016 doi: 10.1002/cncr.30329.. [DOI] [PubMed] [Google Scholar]
- 5.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–11. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri CE, Chinnaiyan AM, Lerner SP, Swanton C, Rubin MA. The Emergence of Precision Urologic Oncology: A Collaborative Review on Biomarker-driven Therapeutics. Eur Urol. 2016 doi: 10.1016/j.eururo.2016.08.024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roychowdhury S, Chinnaiyan AM. Translating genomics for precision cancer medicine. Annual review of genomics and human genetics. 2014;15:395–415. doi: 10.1146/annurev-genom-090413-025552. [DOI] [PubMed] [Google Scholar]
- 8.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 9.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24(23):3847–52. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 10.Barbieri CE, Rubin MA. Genomic rearrangements in prostate cancer. Curr Opin Urol. 2015;25(1):71–6. doi: 10.1097/MOU.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sreenath TL, Dobi A, Petrovics G, Srivastava S. Oncogenic activation of ERG: A predominant mechanism in prostate cancer. J Carcinog. 2011;10:37. doi: 10.4103/1477-3163.91122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamo P, Ladomery MR. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2016;35(4):403–14. doi: 10.1038/onc.2015.109. [DOI] [PubMed] [Google Scholar]
- 13.Abou-Ouf H, Zhao L, Bismar TA. ERG expression in prostate cancer: biological relevance and clinical implication. J Cancer Res Clin Oncol. 2016;142(8):1781–93. doi: 10.1007/s00432-015-2096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Thoms JA, Tursky ML, Knezevic K, Beck D, Chandrakanthan V, et al. MAPK/ERK2 phosphorylates ERG at serine 283 in leukemic cells and promotes stem cell signatures and cell proliferation. Leukemia. 2016;30(7):1552–61. doi: 10.1038/leu.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahim S, Minas T, Hong SH, Justvig S, Celik H, Kont YS, et al. A small molecule inhibitor of ETV1, YK-4-279, prevents prostate cancer growth and metastasis in a mouse xenograft model. PLoS One. 2014;9(12):e114260. doi: 10.1371/journal.pone.0114260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer cell. 2011;19(5):664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Kollipara RK, Humphries CG, Ma SH, Hutchinson R, Li R, et al. The ubiquitin ligase TRIM25 targets ERG for degradation in prostate cancer. Oncotarget. 2016;7(40):64921–31. doi: 10.18632/oncotarget.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Qiao Y, Asangani IA, Ateeq B, Poliakov A, Cieslik M, et al. Development of Peptidomimetic Inhibitors of the ERG Gene Fusion Product in Prostate Cancer. Cancer cell. 2017;31(4):532–48. e7. doi: 10.1016/j.ccell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaFoya B, Munroe JA, Mia MM, Detweiler MA, Crow JJ, Wood T, et al. Notch: A multi-functional integrating system of microenvironmental signals. Developmental biology. 2016;418(2):227–41. doi: 10.1016/j.ydbio.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13(9):654–66. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12(8):445–64. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majidinia M, Alizadeh E, Yousefi B, Akbarzadeh M, Zarghami N. Downregulation of Notch Signaling Pathway as an Effective Chemosensitizer for Cancer Treatment. Drug Res (Stuttg) 2016;66(11):571–9. doi: 10.1055/s-0042-111821. [DOI] [PubMed] [Google Scholar]
- 23.Yahyanejad S, Theys J, Vooijs M. Targeting Notch to overcome radiation resistance. Oncotarget. 2016;7(7):7610–28. doi: 10.18632/oncotarget.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su F, Zhu S, Ruan J, Muftuoglu Y, Zhang L, Yuan Q. Combination therapy of RY10-4 with the gamma-secretase inhibitor DAPT shows promise in treating HER2-amplified breast cancer. Oncotarget. 2016;7(4):4142–54. doi: 10.18632/oncotarget.6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HW, Kim SJ, Choi IJ, Song J, Chun KH. Targeting Notch signaling by gamma-secretase inhibitor I enhances the cytotoxic effect of 5-FU in gastric cancer. Clin Exp Metastasis. 2015;32(6):593–603. doi: 10.1007/s10585-015-9730-5. [DOI] [PubMed] [Google Scholar]
- 26.Vinson KE, George DC, Fender AW, Bertrand FE, Sigounas G. The Notch pathway in colorectal cancer. Int J Cancer. 2016;138(8):1835–42. doi: 10.1002/ijc.29800. [DOI] [PubMed] [Google Scholar]
- 27.Ikezawa Y, Sakakibara-Konishi J, Mizugaki H, Oizumi S, Nishimura M. Inhibition of Notch and HIF enhances the antitumor effect of radiation in Notch expressing lung cancer. Int J Clin Oncol. 2016 doi: 10.1007/s10147-016-1031-8.. [DOI] [PubMed] [Google Scholar]
- 28.Sakakibara-Konishi J, Ikezawa Y, Oizumi S, Kikuchi J, Kikuchi E, Mizugaki H, et al. Combined antitumor effect of gamma-secretase inhibitor and ABT-737 in Notch-expressing non-small cell lung cancer. Int J Clin Oncol. 2016 doi: 10.1007/s10147-016-1060-3.. [DOI] [PubMed] [Google Scholar]
- 29.Yu P, Petrus MN, Ju W, Zhang M, Conlon KC, Nakagawa M, et al. Augmented efficacy with the combination of blockade of the Notch-1 pathway, bortezomib and romidepsin in a murine MT-1 adult T-cell leukemia model. Leukemia. 2015;29(3):556–66. doi: 10.1038/leu.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Armstrong AJ. Docetaxel Resistance in Prostate Cancer: Taking It Up a Notch. Clin Cancer Res. 2015;21(20):4505–7. doi: 10.1158/1078-0432.CCR-15-1613. [DOI] [PubMed] [Google Scholar]
- 31.Cui D, Dai J, Keller JM, Mizokami A, Xia S, Keller ET. Notch Pathway Inhibition Using PF-03084014, a gamma-Secretase Inhibitor (GSI), Enhances the Antitumor Effect of Docetaxel in Prostate Cancer. Clin Cancer Res. 2015;21(20):4619–29. doi: 10.1158/1078-0432.CCR-15-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldus CD, Thibaut J, Goekbuget N, Stroux A, Schlee C, Mossner M, et al. Prognostic implications of NOTCH1 and FBXW7 mutations in adult acute T-lymphoblastic leukemia. Haematologica. 2009;94(10):1383–90. doi: 10.3324/haematol.2008.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben Abdelali R, Asnafi V, Leguay T, Boissel N, Buzyn A, Chevallier P, et al. Pediatric-inspired intensified therapy of adult T-ALL reveals the favorable outcome of NOTCH1/FBXW7 mutations, but not of low ERG/BAALC expression: a GRAALL study. Blood. 2011;118(19):5099–107. doi: 10.1182/blood-2011-02-334219. [DOI] [PubMed] [Google Scholar]
- 34.Stankiewicz MJ, Crispino JD. AKT collaborates with ERG and Gata1s to dysregulate megakaryopoiesis and promote AMKL. Leukemia. 2013;27(6):1339–47. doi: 10.1038/leu.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y, Dobi A, Sreenath T, Cook C, Tadase AY, Ravindranath L, et al. Delineation of TMPRSS2-ERG splice variants in prostate cancer. Clin Cancer Res. 2008;14(15):4719–25. doi: 10.1158/1078-0432.CCR-08-0531. [DOI] [PubMed] [Google Scholar]
- 36.Soh H, Venkatesan N, Veena MS, Ravichandran S, Zinabadi A, Basak SK, et al. Cystatin E/M Suppresses Tumor Cell Growth through Cytoplasmic Retention of NF-kappaB. Molecular and cellular biology. 2016;36(12):1776–92. doi: 10.1128/MCB.00878-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27(40):5348–53. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubovenko A, Serebryiskaya T, Nikolsky Y, Nikolskaya T, Perlina A, JeBailey L, et al. Reconstitution of the ERG Gene Expression Network Reveals New Biomarkers and Therapeutic Targets in ERG Positive Prostate Tumors. J Cancer. 2015;6(6):490–501. doi: 10.7150/jca.8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobi A, Furusato B, Shaheduzzaman S, Chen Y, Vahey M, Nydam T, et al. ERG Expression Levels in Prostate Tumors Reflect Functional Status ofthe Androgen Receptor (AR) as a Consequence of Fusion of ERG with ARRegulated Gene Promoters. The Open Cancer Journal. 2010;3:101–8. [Google Scholar]
- 40.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast cancer research : BCR. 2004;6(6):R605–15. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126(Pt 10):2135–40. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng G, Ma L, Meng Q, Ju X, Jiang K, Jiang P, et al. Notch signaling in the prostate: critical roles during development and in the hallmarks of prostate cancer biology. J Cancer Res Clin Oncol. 2016;142(3):531–47. doi: 10.1007/s00432-015-1946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang XD, Leow CC, Zha J, Tang Z, Modrusan Z, Radtke F, et al. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Developmental biology. 2006;290(1):66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Upadhyay P, Nair S, Kaur E, Aich J, Dani P, Sethunath V, et al. Notch pathway activation is essential for maintenance of stem-like cells in early tongue cancer. Oncotarget. 2016;7(31):50437–49. doi: 10.18632/oncotarget.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Jain S, Azad AK, Xu X, Yu HC, Xu Z, et al. Notch and TGFbeta form a positive regulatory loop and regulate EMT in epithelial ovarian cancer cells. Cellular signalling. 2016;28(8):838–49. doi: 10.1016/j.cellsig.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 46.Shao S, Zhao X, Zhang X, Luo M, Zuo X, Huang S, et al. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Molecular cancer. 2015;14:28. doi: 10.1186/s12943-015-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Zhao X, Shao S, Zuo X, Ning Q, Luo M, et al. Notch1 induces epithelial-mesenchymal transition and the cancer stem cell phenotype in breast cancer cells and STAT3 plays a key role. International journal of oncology. 2015;46(3):1141–8. doi: 10.3892/ijo.2014.2809. [DOI] [PubMed] [Google Scholar]
- 48.McMullin RP, Dobi A, Mutton LN, Orosz A, Maheshwari S, Shashikant CS, et al. A FOXA1-binding enhancer regulates Hoxb13 expression in the prostate gland. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):98–103. doi: 10.1073/pnas.0902001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.