Abstract
Purpose
MUC1, an oncogene overexpressed in multiple solid tumors including pancreatic cancer, reduces overall survival and imparts resistance to radiation and chemotherapies. We previously identified that MUC1 facilitates growth promoting metabolic alterations in pancreatic cancer cells. The present study investigates the role of MUC1-mediated metabolism in radiation resistance of pancreatic cancer by utilizing cell lines and in vivo models.
Experimental design
We used MUC1 knockdown and overexpressed cell line models for evaluating the role of MUC1-mediated metabolism in radiation resistance through in vitro cytotoxicity, clonogenicity, DNA damage response and metabolomic evaluations. We also investigated if inhibition of glycolysis could revert MUC1-mediated metabolic alterations and radiation resistance using in vitro and in vivo models.
Results
MUC1 expression diminished radiation-induced cytotoxicity and DNA damage in pancreatic cancer cells by enhancing glycolysis, pentose phosphate pathway, and nucleotide biosynthesis. Such metabolic reprogramming resulted in high nucleotide pools and radiation resistance in in vitro models. Pre-treatment with the glycolysis inhibitor, 3-bromopyruvate (BrPA) abrogated MUC1-mediated radiation resistance both in vitro and in vivo, by reducing glucose flux into nucleotide biosynthetic pathways and enhancing DNA damage, which could again be reversed by pre-treatment with nucleoside pools.
Conclusions
MUC1-mediated nucleotide metabolism plays a key role in facilitating radiation-resistance in pancreatic cancer and targeted effectively through glycolytic inhibition.
Keywords: MUC1, mucin, pancreatic cancer, cancer metabolism, radiation resistance, bromopyruvate, nucleotide metabolism
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the United States (1). It has the lowest survival rate of 6% and is projected to be the second leading cause of cancer-related deaths by 2030 (1,2). Current therapeutic options for advanced pancreatic cancer include chemotherapy and radiation therapy (RT). However, these options provide only marginal increases in the survival rate due to the therapeutic resistance of pancreatic tumors (3). Clinical trials indicate a significant response to radiation in only 20% of primary pancreatic tumors (4,5). While multiple factors cause resistance to RT, the biological mechanisms mediating such innate resistance are currently being explored.
Pancreatic tumors present elevated DNA damage responses (6). Also, pancreatic cancer cell models can be sensitized to radiation through inhibition of the non-homologous end joining mode of DNA repair (7). Efficient DNA damage responses would depend on anabolic alterations that could provide cancer cells with nucleotide pools upon radiation-induced DNA damage (8). Pancreatic tumors are highly glycolytic, as indicated by high [18F]-fluorodeoxyglucose uptake in positron emission tomography (9–11). The success of RT in pancreatic cancer patients inversely correlates to the overall glycolytic index of the tumors (12,13). Although the exact role of glycolysis in imparting radiation resistance remains poorly defined, the increased glycolytic index may provide cells with increased nucleotide pools by routing carbon flux through the pentose phosphate pathway. Factors that could mediate the radiation-induced DNA repair mechanisms through upregulation of nucleotide metabolism are least studied in pancreatic cancer.
Mucin1 (MUC1) is a transmembrane glycoprotein that is overexpressed and aberrantly localized in pancreatic ductal adenocarcinoma (14,15). MUC1 serves as a sensor of extracellular signaling cues and transmits these signals to the nucleus, modulating the transcriptional profile of cancer cells (15). These functions allow MUC1 to facilitate tumor cell growth, invasion, motility, and cell survival under harsh conditions. Furthermore, MUC1 mediates DNA damage response by interacting with ataxia telangiectasia mutated (ATM) in breast cancer cells (16). MUC1 also interacts with c-Abl and prevents its nuclear targeting to block DNA damage-induced apoptosis (17). Besides, its role in mediating DNA damage repair, MUC1 also facilitates metabolic reprogramming in pancreatic cancer cells (18,19). We previously reported that the MUC1 cytoplasmic domain directly interacts with and stabilizes Hypoxia-inducible factor-1 alpha, which can, in turn, alter the metabolic gene expression in pancreatic cancer cells (18,20). We also identified that MUC1-mediated metabolic alterations induce increased flux of carbon through the pentose phosphate pathway, which primarily generates the precursor metabolites for de novo nucleotide synthesis in cancer cells (18,21). Based on our previous studies, we hypothesized that MUC1-mediated metabolic alterations play a role in radiation resistance in pancreatic cancer cells. Our present study demonstrates that radiation resistance in pancreatic cancer is dependent on MUC1-mediated metabolic flux. We utilized mass spectrometry-based metabolomics technology and pharmacological inhibition of MUC1-induced aberrant metabolism to decipher the role of MUC1-mediated metabolic alterations in imparting radiation resistance to pancreatic cancer.
Methods
Cell culture
Pancreatic cancer cell lines S2-013 and Capan2 with MUC1-overexpression (S2-013.MUC1) and MUC1-knockdown (Capan2-sh.MUC1) have been described previously (18). Additional MUC1-knock downs of FG and HPAF2 were developed using established shMUC1 constructs. Osteosarcoma cell line U2OS (U2OS SA-GFP) was provided by Dr. Jeremy Stark (Beckman Research Institute of the City of Hope, Duarte, CA) (22). After transfection of pcDNA3.MUC1F construct (23) into these cells, the transfected cells were cultured in pyruvate-free DMEM, supplemented with 10% fetal bovine serum and 1 mg/ml G418, for 7–14 days. G418- resistant cells were isolated and utilized for the experiments. An empty vector was stably transfected to establish the control cell-lines. The cell lines were validated by STR profiling and tested for mycoplasma every six months.
Irradiation experiments
Cells maintained in DMEM with 10% fetal bovine serum were irradiated by utilizing a linear accelerator available in the Department of Radiation Oncology at UNMC. Briefly, cells were seeded in culture dishes 16 h before irradiation. Irradiation performed by placing culture plates on 10 cm of solid water (phantom material used for radiation beam calibration) by positioning plates in the center of the 40 cm × 40 cm radiation field with dose verifications evaluated using Metal Oxide Semiconductor Field Effect Transistor (MOSFET) detectors which enables the dose verification through the radiation-induced threshold shifts. Irradiation was conducted with 6 MV X-rays at a rate of 2.73 Gy/min from the posterior direction, with the cells on the flask base being 100 cm from the X-ray source.
Cell survival assays
Cells irradiated with fractionated or single-dose radiation were trypsinized and subsequently cultured for cell survival assays. The medium was replaced with fresh DMEM with or without inhibitors before and immediately after irradiation. Cells cultured in the respective media for 72 h were evaluated for cell survival using either MTT or trypan blue exclusion method using BIO-RAD TC20™ automated cell counter.
Clonogenic survival assay
Clonogenic assays were performed to determine cellular response to radiation (24). Experimental (MUC1 knockdown and overexpressed cell line models) and control cells with similar seeding densities were chosen for clonogenic assays and colonies formed at the end of experiments were washed, fixed in methanol and stained with 0.4% crystal violet in 25% methanol. Colonies containing >50 cells for each well were counted (in triplicate). Surviving fraction at each dose was determined by using the formula: [(number of surviving colonies in dose X)/(number of cells seeded for dose X (average colonies arising from the non-irradiated cells (0Gy)/number of non-irradiated cells seeded)] (25).
Western blotting
Cells were washed twice with cold PBS and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer at 4°C for 30 min. Pellets were separated by centrifugation at 13,000 rpm for 10min supernatants were collected for protein estimation. Equal quantities of denatured proteins were separated by electrophoresis using SDS-PAGE gels and transferred to activated, PVDF membranes. Western blotting was performed using primary antibodies against MUC1-CT (Armenian Hamster monoclonal antibody), actin and beta-tubulin (Clones J5 and E7, respectively, from Developmental Studies Hybridoma Bank, Iowa City, IA).
Staining for DNA damage
Cells seeded at 40% density on sterile glass coverslips in 24-well plates were used for evaluating DNA damage response. Fresh media with or without inhibitors was added immediately before and after irradiation. Cells were rinsed with PBS to remove the media at specific time points after treatment and fixed in 4% paraformaldehyde. Cells were permeabilized with 0.2% tween-20 for 5 min at room temperature and washed thrice with PBS, followed by blocking at room temperature with 1% non-fat milk in PBS containing 0.05% Tween. Fixed cells were subsequently subjected to incubation at 4°C overnight with the primary antibody (Phospho-Histone H2A.X (Ser139) (20E3) Rabbit mAb #9718 from Cell Signaling Technology, 1:1000 in blocking solution), followed by three washes with PBS and then incubated with secondary antibody conjugated to Alexa fluor-644 for 30 min, in dark at room temperature. After washing three times with PBS, coverslips were mounted on DAPI-fluoro mount-G. Immunofluorescence imaging capturing was performed at 20X magnification by using a DMI6000 Leica microscope.
In vivo studies
Animal studies were conducted according to an approved protocol by the University of Nebraska Medical Center Institutional Animal Care and Use Committee (IACUC). Female athymic nude mice were injected subcutaneously with one tumor each with 2 million S2-013.Neo or S2-013.MUC1 cells at the left flank. Tumor volume was calculated using the formula: volume = [length *(width^2)]/2. Mice were grouped into 4 treatment groups and treated with saline control, bromopyruvate (BrPA) alone, radiation alone, and BrPA + radiation, once all tumors reached 100 mm3 or greater. BrPA treatment started on day 0 and given i.p. daily at 7.5mg/kg. Radiation treatments were started on day 1 and given at a dose of 4 Gy for 5 consecutive days (20 Gy total). Mice were anesthetized using ketamine/xylazine mix, and irradiated with the body shielded using a lead shield while tumor remained exposed to radiation. Tumors irradiation was performed with a 160 kV RAD Source (Suwanee, GA) RS 2000 X-Ray irradiator at a dose rate of 1.2 Gy/min. Tumor sizes were monitored for every 3 days until 27 days, post day 0.
Liquid chromatography and tandem mass spectrometry analysis
Cells (0.75×107) were cultured for 24 h in normal DMEM, and culture medium was exchanged with fresh media before irradiation. Cells were exposed to IR (4 Gy) followed by rinsing with PBS and frozen on dry ice for metabolomic studies. Polar metabolomics and data analyses were performed as described previously (26–28).
Results
MUC1 expression diminishes the response to radiation therapy in cancer cells
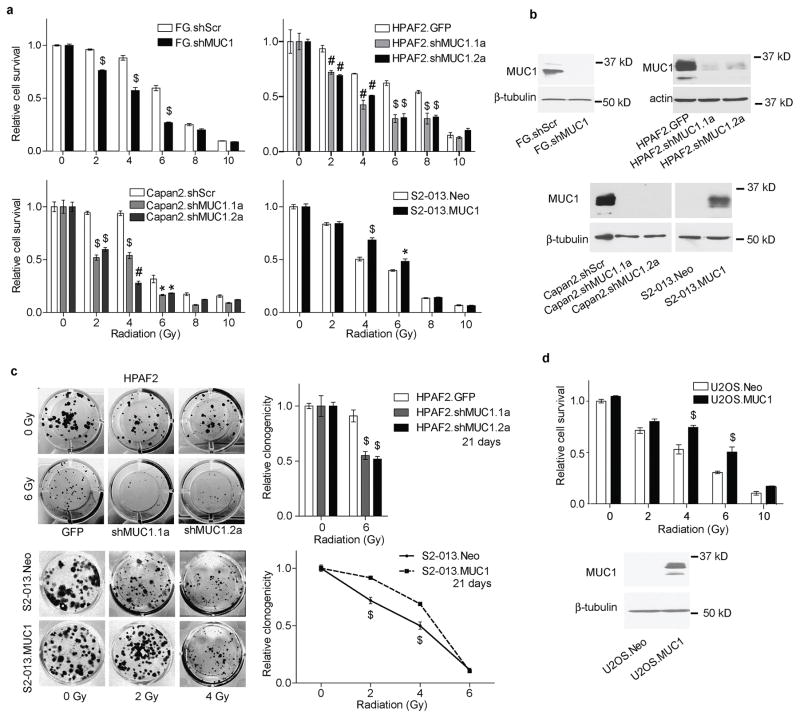
Radiation-induced DNA damage would resultantly inhibit tumor cell proliferation (29). To determine the role of MUC1 in regulating radiation resistance in pancreatic cancer, we evaluated cell survival using MUC1-knock down and overexpressed cell line models. Reducing MUC1 expression using MUC1-knock down resulted in sensitization of FG, HPAF2, and Capan2 pancreatic cancer cell lines to radiation. The survival of FG.shMUC1 cells upon 2–6 Gy radiation exposure was lesser than the control FG.shScr cells (Figure 1a and b). Similarly, HPAF2 and Capan2 cells exposed to radiation (2–8 Gy) showed a significant reduction in survival upon MUC1 knockdown. Reciprocally, we observed increased cell survival in S2-013 cells under conditions of MUC1 expression when irradiated with 2–6 Gy doses of irradiation as shown in Figure 1a and b. MUC1-expressing HPAF2 and S2-013 cells also exhibited higher clonogenic potential at lower doses of irradiation (2–6 Gy), compared to controls (Figure 1c). Furthermore, to evaluate if the effect of MUC1-expression on radiation-resistance is only specific for pancreatic cancer cell lines, we utilized an osteosarcoma cell line model, U2OS, which expresses low levels of MUC1. Exogenous expression of MUC1 in U2OS (U2OS.MUC1) cells enhanced their survival upon irradiation compared to the cells stably expressing vector control (U2OS.Neo) (Figure 1d).
Figure 1. MUC1-expression decreases cancer cell sensitivity to radiation.
(a) Survival of control and experimental (MUC1-knockdown/MUC1-overexpression) pancreatic cancer cells at 72 h post-exposure to indicated radiation doses, relative to untreated cells. (b) Western blot analysis of MUC1 expression levels in pancreatic cancer cell lines used as MUC1-knock down and overexpression models. β-tubulin or actin were utilized as loading controls. (c) Representative images and relative quantification of clonogenicity, 21 days post-exposure to the indicated radiation doses. (d) Effect of MUC1 overexpression in U2OS osteosarcoma-cell survival upon treatment with the indicated doses of radiation and western blot analysis of MUC1 expression in U2OS cells. Values in bar graphs and line plots indicate mean ± SEM of three replicates. ‘*’ p<0.05, ‘#’ p<0.01, and ‘$’ p<0.001 obtained through Two-way ANOVA and Tukey’s multiple comparison tests.
MUC1 expression suppresses radiation-induced DNA damage
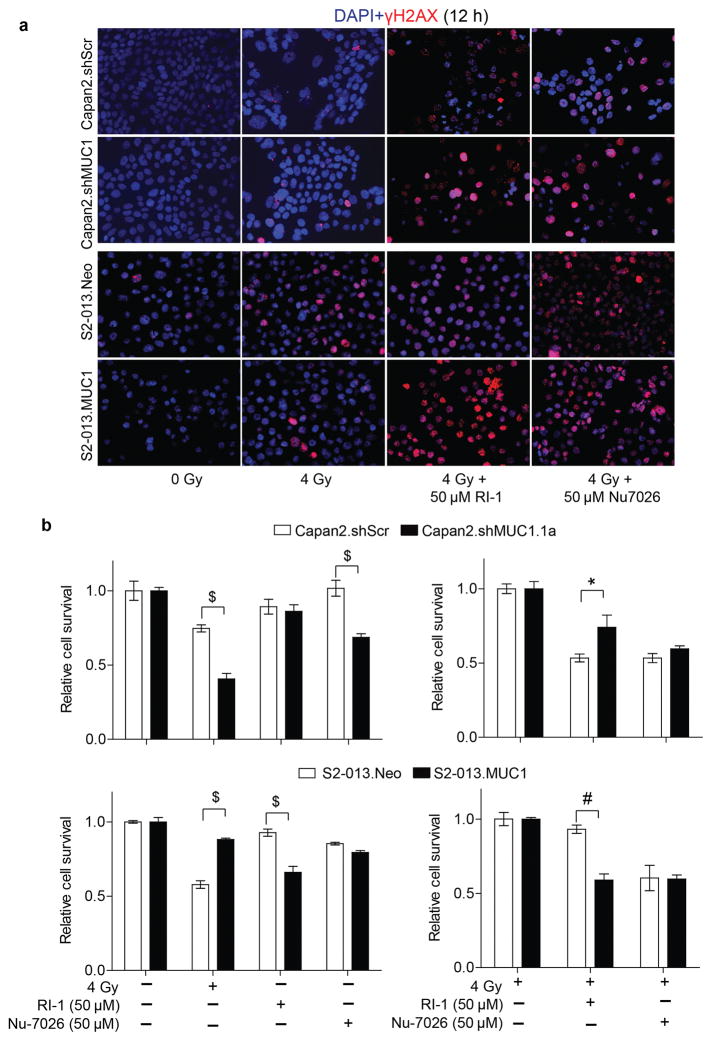
Radiation-induced cellular damage includes the modification of DNA bases resulting in DNA strand breaks (29). Phosphorylation of histone 2A (γH2AX) is an indicator of DNA damage. Also, low levels of γH2AX are indicative of effective DNA damage repair occurring in irradiated cells. In our irradiation study, we observed that MUC1-expressing cells stained less positively for γH2AX twelve hours post-irradiation in comparison to either control or MUC1-knock down cells (Figure 2a). Furthermore, we evaluated the role of non-homologous end joining repair (NHEJ) and homologous repair (HR) in DNA damage repair of MUC1-expressing cells using NHEJ inhibitor Nu-7026 and HR inhibitor RI-1, respectively (30,31). Both the treatments abrogated MUC1-induced differences in γH2AX staining as the level of γH2AX staining increased by combination treatments with DNA damage repair inhibitors and radiation (Figure 2a). Inhibition of DNA damage repair by RI-1 significantly reduced the survival of irradiated MUC1-expressing cells. However, Nu-7026 diminished the survival of irradiated pancreatic cancer cells irrespective of their MUC1 expression status (Figure 2b).
Figure 2. MUC1 expression reduces radiation-induced DNA damage.
(a) Representative immunofluorescence images indicating γH2AX staining (red) as a measure of DNA damage upon irradiation and treatment with NHEJ or RI inhibitors for 12h; DAPI staining (blue) indicates nuclei. (b) Bar charts representing relative cell survival upon irradiation in the presence of solvent control, either the HR or NHEJ repair inhibitors, as measured by MTT assay. Fluorescence images represent representative fields of respective treatments captured at 20X magnification. All the bars represent mean ± SEM of 3 replicates. ‘*’ p<0.05, ‘#’ p<0.01, and ‘$’ p<0.001 obtained through two-way ANOVA and Bonferroni method for multiple comparisons.
MUC1 expression facilitates radiation-resistance by upregulating nucleotide metabolism
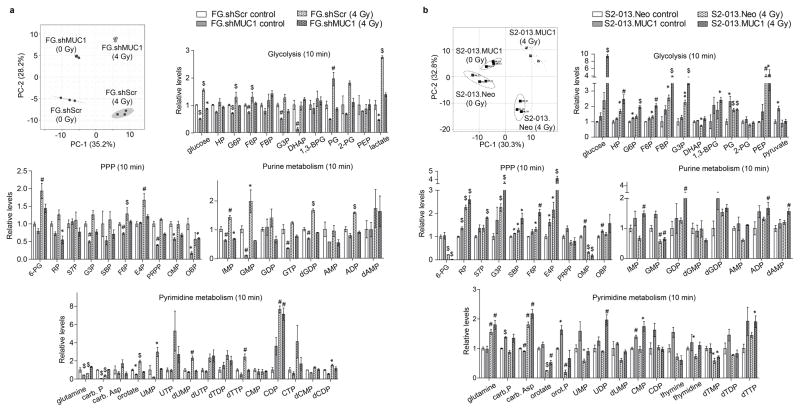
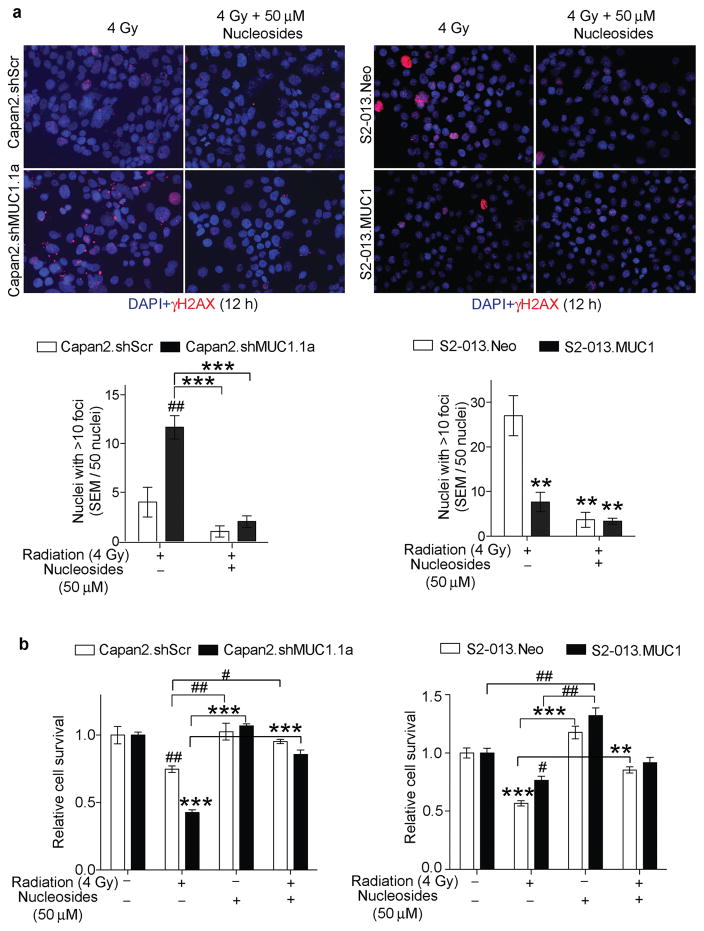
Our previous studies demonstrated that MUC1 expression promotes glucose uptake and glycolytic metabolism in pancreatic cancer cells (18). To determine if, the MUC1 expression also induces differential metabolic changes upon irradiation, we performed LC-MS/MS-based metabolomics analysis of polar metabolite extracts from control and irradiated cell lines. Partial least square discriminant analysis (PLS-DA) of the polar metabolite components indicated an overall metabolic shift in pancreatic cancer cells with MUC1 knockdown or overexpression, under control and irradiated conditions (Figure 3a and b, PLS-DA plots). Further comparison of metabolite levels among the control and irradiated cells indicated higher levels of glycolytic, PPP and nucleotide biosynthetic pathway intermediates in MUC1-expressing cells, which increase within 10 min of irradiation (4 Gy) (Figure 3a and b, bar graphs). Based on the increased nucleotide levels observed in irradiated MUC1-expressing cells, we evaluated if nucleoside supplementation to the MUC1-knock down or S2-013.Neo cells would decrease their responsiveness to radiation, to the level observed in MUC1-expressing cells. As expected, nucleoside supplementation prevented the radiation-induced DNA damage in pancreatic cancer cells irrespective of MUC1 expression (Figure 4a). Nucleoside supplementation along with radiation significantly enhanced survival of the Capan2.shMUC1 and S2-013.Neo cells in comparison to the radiation treatments (Figure 4b). Furthermore, comparison of relative survival between Capan2.shMUC1 cells and combination-treated Capan2.shMUC1 cells and S2-013.Neo cells with combination-treated S2-013.Neo cells showed that nucleosides recovered the survival in combination-treated cells.
Figure 3. MUC1 facilitates radiation-induced carbon flux into glycolysis and de novo nucleotide biosynthesis pathways.
Partial least squares discriminant analysis (PLS-DA) plots in panels a and b represent differential clustering of control and irradiated cells based on their total polar metabolite content. Each filled circle/square indicates an individual sample. The three closely clustered circles or squares represent three biological replicates of an individual group of samples extracted from one cell line. The axes of the PLS-DA plots represent principal component-1 (PC-1) and -2 (PC-2), which indicate the variance among the groups. PLS-DA analyses were performed utilizing Metaboanalyst algorithm with mean intensities and pareto scaling distribution. Bar charts in a and b show fold change in metabolite levels of glycolysis, pentose phosphate pathway (PPP), and purine and pyrimidine metabolic pathways upon irradiation in experimental cells compared to their respective wild-type cells. Bars represent mean ± SEM of three biological replicate values; *, # and $-indicate p<0.05, p<0.01, and p<0.001, respectively, obtained through one-way ANOVA and Benjamini-Hochberg’s procedure to control the false discovery rate.
Figure 4. MUC1 facilitates radiation resistance through nucleoside-mediated DNA damage repair.
(a) Representative immunofluorescence images indicating γH2AX staining (red) as a measure of DNA damage upon irradiation and treatment with nucleosides for 12 h. DAPI staining (blue) indicates nuclei. Fluorescence images represent representative fields of respective treatments captured at 20X magnification. Bar charts indicate quantification of the number of nuclei with >10 foci from a total of 50 nuclei considered for each condition. (b) Bar charts show relative cell survival under radiation and/or nucleoside supplementation. All bars represent mean ± SEM of three biological replicates. */#, **/##, and ***/### indicate p<0.05, p<0.01, and p<0.001, respectively, obtained through one-way ANOVA with Tukey’s multiple comparison test. *, **, and *** indicate statistical significance in comparison to wild type or MUC1-kd cells, whereas, #, ##, and ### indicate statistical significance with respect to the MUC1-expressing cells.
Metabolic inhibition sensitizes MUC1-expressing pancreatic cancer cells to radiation
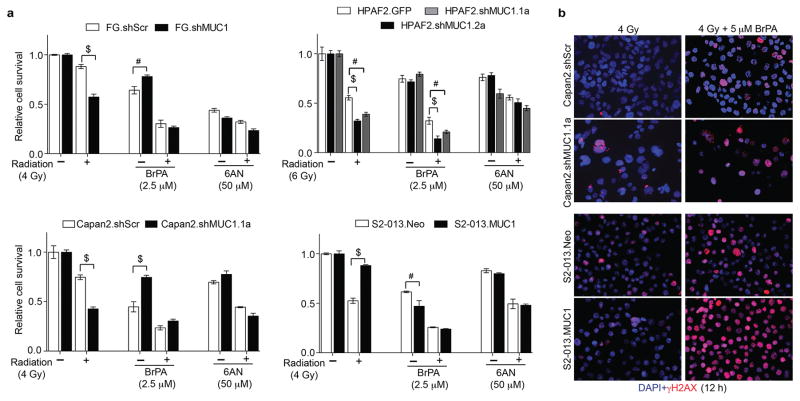
Our findings indicated a significant increase in glycolysis, PPP, and nucleotide metabolism in irradiated MUC1-expressing pancreatic cancer cells. Hence, we next determined if metabolic inhibition of glycolysis with 3-bromopyruvate (BrPA) and PPP with 6-amino nicotinamide (6AN) would sensitize MUC1-expressing cells to radiation. Cell survival analyses showed a reduction in cell survival with BrPA (5 μM) and 6AN (50 μM) treatments in both control and experimental cell lines, similar to the radiation treatment alone. Treatment with BrPA reduced cell survival in four different pancreatic cancer cells, and the combination of BrPA along with radiation was more effective in reducing cell survival irrespective of MUC1 expression (Figure 5a). Though PPP inhibitor, 6AN enhanced the sensitivity of MUC1-expressing cells to radiation; its combination with radiation was less effective in comparison to the combinations of radiation and BrPA (Figure 5a). Glycolytic flux into PPP provides intermediates for de novo nucleotide biosynthesis required for proliferating cancer cells. We observed that the pretreatment with glycolytic inhibitor, BrPA, before irradiation diminished the levels of metabolites in glycolysis and PPP, in FG and S2-013 cell types (Supplementary Figure S1). Relative levels of glycolysis pathway metabolites including G6P, F6P, FBP and G3P decreased significantly in control and experimental cell line models upon treatment with either BrPA alone or in combination with radiation (Supplementary Figure S1). Dual treatment with BrPA and radiation reduced most of the PPP-metabolite levels and de novo nucleotide biosynthesis pathway, as some of the nucleotides were relatively decreased in BrPA and combination-treated MUC1-expressing FG and S2-013 cells (Supplementary Figure S1). Based on our BrPA-treated metabolomic studies, we anticipated that the inhibitory effect of BrPA on MUC1-mediated glycolysis and PPP would suppress MUC1-regulated DNA damage upon irradiation. Therefore, we evaluated the effects of BrPA on DNA damage in combination with radiation. We observed that BrPA-treatment enhanced DNA damage in both control and experimental cell lines (Figure 5b).
Figure 5. BrPA abrogates MUC1-mediated radiation-resistance.
(a) Bar charts represent relative cell survival in wild-type and experimental cells upon irradiation and treatments with either BrPA or 6-AN. (b) Representative immunofluorescence images indicating γH2AX staining (red) as a measure of DNA damage upon irradiation and treatment with solvent control or BrPA treatments of cells for 12 h. DAPI staining (blue) indicates nuclei. Bars represent mean±SEM of three biological replicate values. # and $-indicate p<0.01 and p<0.001, respectively, obtained through two-way ANOVA with Bonferroni posttests for cell survival analysis. Fluorescence images represent representative fields of respective treatments captured at 20X magnification.
Bromopyruvate abrogates MUC1-induced radiation-resistance
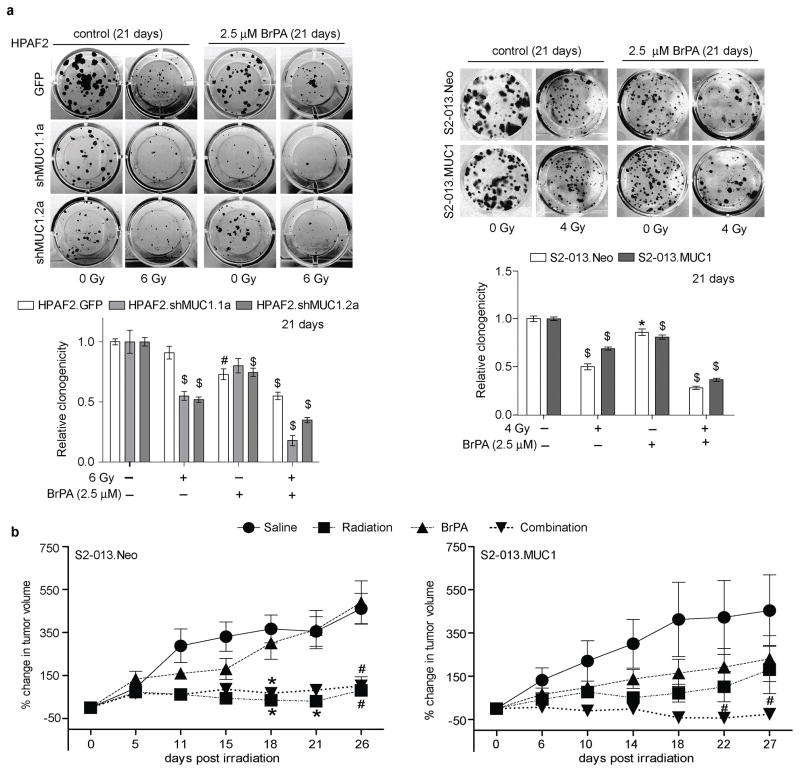
We further evaluated the effects of BrPA and radiation on clonogenicity. The combination of BrPA and radiation significantly reduced clonogenicity of MUC1-expressing cells in both HPAF2 and S2-013 cell line models (Figure 6a). Based on our in vitro findings, we evaluated the effects of BrPA on MUC1-induced radiation resistance using sub-cutaneous xenograft tumor models. Tumor growth studies showed that irradiation reduced tumor growth in S2-013.Neo group tumors in comparison to the saline treated S2-013.Neo group (Figure 6b, left panel). In contrast to the irradiated S2-013.Neo group mice, tumors in mice implanted with S2-013.MUC1 cells did not exhibit significant differences in tumor growth among the saline, radiation, and BrPA treated groups (Figure 6b, right panel). Interestingly, both S2-013.Neo and S2-013.MUC1 groups treated with the combination of radiation and BrPA showed a significant decrease in tumor growth in comparison to their respective saline-treated groups from 18 days of combination treatments. The tumors were validated for MUC1 expression by IHC upon necropsy (Supplementary Figure S2).
Figure 6. BrPA inhibits MUC1-mediated radiation resistance.
(a) Representative images (top panels) of crystal violet stained colonies at 21 days post treatment with radiation and/or BrPA treatment. Bar charts in bottom panels indicate the normalized relative clonogenicity levels in the HPAF2 and S2-013 cell line models. (b) Tumor growth rate plots indicate the percent change in S2-013.Neo and S2-013.MUC1 tumor growth in comparison to the day 0 of the treatment (BrPA treatment initiation was set as the day 0). Each value indicates mean±SEM of either eight or nine biological replicates; *, # and $ -indicate p<0.05, <0.01 and <0.001, respectively, obtained through t-test analysis.
Discussion
Exposure of tumor cells to stress conditions such as radiation therapy can induce oncogene-regulated stress responses, which in turn lead to radiation-resistance in tumor cells (32,33). Oncogene-regulated mechanisms allow tumor cells in overcoming radiation-induced DNA damage, thus imparting radiation resistance to tumor cells (34). However, the exact nature of signaling pathways and mechanisms that confer radiation resistance remains poorly defined. Our present findings demonstrate that MUC1 oncogene abrogates cytotoxic effects of radiation therapy. Cell survival and clonogenicity upon irradiation were enhanced by the MUC1 expression, whereas reduction was observed by the MUC1 knockdown in pancreatic cancer cells (Figure 1). Furthermore, MUC1 expression increased survival of osteosarcoma cells upon irradiation ascertaining the role of MUC1 in mediating radiation-resistance in different cancer cell types, which is in agreement with another study wherein MUC1 knockdown radio-sensitized breast cancer cell lines (16).
Cancer cells overcome DNA damage induced by irradiation through efficient DNA damage repair mechanisms (35). Such repair facilitates radiation resistance as DNA strand breaks induced by radiation are fixed, allowing the cells to survive and proliferate (36). Our present findings indicate that the MUC1-expression enhances DNA damage repair in pancreatic cancer cells as evident from relatively low γH2AX staining within the same interval (12 h) of irradiation in MUC1-expressing cells compared to the control cells (Figure 2). A previous study demonstrated that pancreatic cancer cells innately possess high DNA damage (7). NHEJ is the primary mechanism that facilitates DNA damage repair in pancreatic cancer cells upon irradiation followed by HR mechanism (7). Hence, we investigated the relative contribution of NHEJ and HR in MUC1-mediated DNA damage repair. Sustenance of radiation induced-γH2AX staining with inhibition of DNA-PK (involved in NHEJ repair) or RAD51 (involved in HR repair) in both control and MUC1-expressing cells indicated that both NHEJ and HR play roles in DNA damage response in pancreatic cancer cells independent of MUC1 expression. However, we noticed a significant decrease in the survival of irradiated MUC1-expressing cells exposed to RI-1, in comparison to the MUC1-low cells. Such difference was not observed in the case of Nu-7026 exposure indicating that MUC1-expressing cells are dependent on HR-mediated DNA damage repair; but the NHEJ-mediated repair is independent of MUC1-expression in irradiated pancreatic cancer cells (Figure 2). Thus, we further investigated possible mechanisms leading to the decrease in radiation sensitivity upon MUC1 expression.
Pancreatic cancer cells exhibit strong glycolytic phenotype (11). We previously reported that MUC1 expression alters the metabolism in pancreatic cancer cells (18,20). MUC1-expressing pancreatic cancer cells exhibit enhanced glycolysis, TCA cycle, and PPP under steady-state conditions (18). Such metabolic alterations in tumor cells can facilitate DNA damage repair induced by radiation (29). Metabolomic analysis performed in the present study demonstrates that pancreatic cancer cells alter their metabolism upon irradiation independent of MUC1 expression (Figure 3). Furthermore, our current findings show that radiation-induced metabolic alterations were distinct under conditions of MUC1 expression. These findings ascertain that MUC1 expression coordinates up-regulation of the interconnected pathways and mediates the metabolic response of cancer cells in response to radiation. Our results demonstrate that the levels of nucleotide precursors in PPP (including the ribose-phosphate/ribulose-5-phosphate, RP) and that of nucleotides in both purine (GDP and dAMP) and pyrimidine biosynthesis pathways are enhanced in both control and MUC1-expressing cell lines. However, such changes are more pronounced in MUC1-expressing cells, suggesting that de novo nucleotide biosynthesis pathways are upregulated upon irradiation in a MUC1-dependent manner. We anticipated that the MUC1 mediated radiation resistance could be due to the enhancement of nucleotide metabolism upon irradiation. This could facilitate efficient DNA damage repair by supplementing the nucleotide pools essential for the replacement of damaged nucleotides. As expected, nucleoside supplementation reduced the DNA damage in control cells similar to that of the MUC1-expressing cells (Figure 4a). Also, nucleoside supplementation mostly enhanced the cell survival in both MUC1-knock down/MUC1-low and MUC1-expressing cells upon irradiation, to a similar extent (Figure 4b). These results indicate that the MUC1-mediated resistance to radiation response is at least in part due to the increase in nucleotide metabolism. Irradiation leads to MUC1-mediated increase of glycolytic flux into PPP and downstream nucleotide metabolism pathways which are essential for the DNA damage repair and survival of the irradiated MUC1 expressing cells.
Oncogene-mediated aberrant metabolism provides a survival advantage for tumor cells (37). In addition, it also causes the susceptibility of tumor cells to inhibitors of metabolic pathways (38). Our present findings demonstrate radiosensitization of MUC1-expressing cells with BrPA; a metabolic inhibitor characterized as an efficient chemotherapeutic agent against pancreatic cancer (39,40). Our results also show that BrPA-treatment reduced the metabolite levels in glycolysis, PPP and nucleotide biosynthetic pathways 12h post-treatment in MUC1-expressing cells (Supplementary Figure S1). Furthermore, inhibition of glycolysis resulted in radiosensitization of MUC1-expressing cells by preventing DNA damage repair (Figure 5). Thus, inhibition of nucleotide metabolism by BrPA treatment resultantly inhibited survival and clonogenicity of MUC1-expressing cells by enhancing DNA damage (Figures 5 and 6a).
MUC1 is expressed in most of the pancreatic tumors and promotes pathological progression of the disease (41–45). MUC1 expression also confers drug resistance in pancreatic cancer (46). However, the role of MUC1 in regulating radiation responsiveness in pancreatic cancer is least studied, compared to other cancer models (16,47,48). Our present findings establish the role of MUC1 in imparting poor radiation responsiveness to pancreatic cancer cells. We identified that enhancement of glycolysis and PPP by MUC1 expression diminishes radiation responsiveness in pancreatic cancer cells. Our in vivo studies substantiate the role of MUC1 in imparting radiation resistance to pancreatic cancer. Notably, we also identified that MUC1 expression diminishes radiation responsiveness in pancreatic tumors (Figure 6b). We determined that combination of BrPA along with radiation sensitizes pancreatic tumors to radiation. Thus, our findings establish the efficacy of targeting MUC1-mediated nucleotide metabolism for radio-sensitization of pancreatic tumor cells. A potential molecular mechanism that could facilitate the MUC1-induced metabolic reprogramming could be through HIF-1α. Low dose irradiation enhances HIF-mediated metabolic responses as reported in a recent study (49) and our previous study showed that MUC1-mediated HIF-1α stabilization leads to upregulation of glycolytic and PPP metabolic pathway genes in MUC1-expressing pancreatic cancer cells (18). Our results establish that metabolic responses elicited in MUC1-expressing cells can be effectively targeted using BrPA, which reduces radiation-resistance in pancreatic cancer cells. This modality needs further evaluation in clinical studies wherein glycolysis-dependent pancreatic tumors can be pre-treated with glycolytic inhibitors before irradiation, for enhancing the efficacy of fractionated-radiation therapies.
Supplementary Material
Translational Relevance.
Pancreatic cancer is currently the fourth leading cause of cancer-related deaths in the United States, and is projected to become the second leading cancer killer by 2030. Finding targets to radiosensitize pancreatic tumors will provide improved curative hope for resectable patients and could significantly improve outcome for patients with the locally advanced disease by increasing the percentage of patients down-staged to have a resection. We demonstrate that MUC1-mediated nucleotide metabolism impacts radiation resistance by suppressing radiation-induced DNA damage in pancreatic cancer. Furthermore, we show that inhibiting MUC1-mediated glycolytic flux with 3-bromopyruvate sensitizes pancreatic cancer to radiation. Hence, our studies provide a better understanding of the metabolic mechanisms responsible for reduced clinical radiation response in pancreatic cancer. Since metabolic inhibitors are currently in clinical trials for different conditions, the outcomes of these studies are directly translatable to human pancreatic cancer patients.
Acknowledgments
Financial Support: This work was supported in part by funding from the National Institutes of Health grant (R01 CA163649, R01 CA210439, and R01 CA216853, NCI) to PKS; the Pancreatic Cancer Action Network-AACR Career Development Award (13-20-25-SING) to PKS; the Specialized Programs of Research Excellence (SPORE, 2P50 CA127297, NCI) to PKS. We would also like to acknowledge the Fred & Pamela Buffett Cancer Center Support Grant (P30CA036727, NCI) for supporting shared resources.
Abbreviations
- HP
hexose phosphate
- G6P
glucose 6-phosphate
- F6P
fructose 6-phosphate
- FBP
fructose 1,6-bis-phosphate
- G3P
glyceraldehyde 3-phosphate
- DHAP
dihydroxy acetone phosphate
- 1,3-BPG
1,3-bis-phosphoglycerate
- PG
phosphoglycerate
- 2PG
2-phosphoglycerate
- PEP
phosphoenol pyruvate
- 6PG
6-phosphogluconate
- PRPP
phosphoribosyl pyrophosphate
- E4P
erythrose 4-phosphate
- SBP
sedoheptulose 1,7-bis-phosphate
- S7P
sedoheptuolose 7-phosphate
- RP
ribulose/ribose phosphate
- OMP
octulose monophosphate
- OBP
octulose 1,8 bis-phosphate
- IMP
inosine monophosphate
- GMP
guanosine monophosphate
- GDP
guanosine diphosphate
- dGMP
deoxyguanosine monophosphate
- AMP
adenosine monophosphate
- ADP
adenosine diphosphate
- dAMP
deoxyadenosine monophosphate
- carb.P
carbamoyl phosphate
- carb.Asp
carbamoyl aspartate
- orot.P
orotidylate phosphate
- UMP
uridine monophosphate
- UDP
uridine diphosphate
- dUMP
deoxyuridine monophosphate
- CMP
cytidine monophosphate
- CDP
cytidine diphosphate
- dTMP
deoxythymidine monophosphate
- dTDP
deoxythymidine diphosphate
- dTTP
deoxythymidine triphosphate
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363(9414):1049–57. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 4.Hazard L. The role of radiation therapy in pancreas cancer. Gastrointest Cancer Res. 2009;3(1):20–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Roldan GE, Gunderson LL, Nagorney DM, Martin JK, Ilstrup DM, Holbrook MA, et al. External beam versus intraoperative and external beam irradiation for locally advanced pancreatic cancer. Cancer. 1988;61(6):1110–6. doi: 10.1002/1097-0142(19880315)61:6<1110::aid-cncr2820610610>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Osterman M, Kathawa D, Liu D, Guo H, Zhang C, Li M, et al. Elevated DNA damage response in pancreatic cancer. Histochemistry and cell biology. 2014;142(6):713–20. doi: 10.1007/s00418-014-1245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YH, Wang X, Pan Y, Lee DH, Chowdhury D, Kimmelman AC. Inhibition of non-homologous end joining repair impairs pancreatic cancer growth and enhances radiation response. PloS one. 2012;7(6):e39588. doi: 10.1371/journal.pone.0039588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isebaert SF, Swinnen JV, McBride WH, Begg AC, Haustermans KM. 5-aminoimidazole-4-carboxamide riboside enhances effect of ionizing radiation in PC3 prostate cancer cells. International journal of radiation oncology, biology, physics. 2011;81(5):1515–23. doi: 10.1016/j.ijrobp.2011.06.1964. [DOI] [PubMed] [Google Scholar]
- 9.Inokuma T, Tamaki N, Torizuka T, Magata Y, Fujii M, Yonekura Y, et al. Evaluation of pancreatic tumors with positron emission tomography and F-18 fluorodeoxyglucose: comparison with CT and US. Radiology. 1995;195(2):345–52. doi: 10.1148/radiology.195.2.7724751. [DOI] [PubMed] [Google Scholar]
- 10.Rose DM, Delbeke D, Beauchamp RD, Chapman WC, Sandler MP, Sharp KW, et al. 18Fluorodeoxyglucose-positron emission tomography in the management of patients with suspected pancreatic cancer. Annals of surgery. 1999;229(5):729–37. doi: 10.1097/00000658-199905000-00016. discussion 37–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaika NV, Yu F, Purohit V, Mehla K, Lazenby AJ, DiMaio D, et al. Differential expression of metabolic genes in tumor and stromal components of primary and metastatic loci in pancreatic adenocarcinoma. PLoS One. 2012;7(3):e32996. doi: 10.1371/journal.pone.0032996. PONE-D-11-16061 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dholakia AS, Chaudhry M, Leal JP, Chang DT, Raman SP, Hacker-Prietz A, et al. Baseline metabolic tumor volume and total lesion glycolysis are associated with survival outcomes in patients with locally advanced pancreatic cancer receiving stereotactic body radiation therapy. International journal of radiation oncology, biology, physics. 2014;89(3):539–46. doi: 10.1016/j.ijrobp.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schellenberg D, Quon A, Minn AY, Graves EE, Kunz P, Ford JM, et al. 18Fluorodeoxyglucose PET is prognostic of progression-free and overall survival in locally advanced pancreas cancer treated with stereotactic radiotherapy. International journal of radiation oncology, biology, physics. 2010;77(5):1420–5. doi: 10.1016/j.ijrobp.2009.06.049. [DOI] [PubMed] [Google Scholar]
- 14.Besmer DM, Curry JM, Roy LD, Tinder TL, Sahraei M, Schettini J, et al. Pancreatic ductal adenocarcinoma mice lacking mucin 1 have a profound defect in tumor growth and metastasis. Cancer research. 2011;71(13):4432–42. doi: 10.1158/0008-5472.CAN-10-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16(9):467–76. doi: 10.1016/j.tcb.2006.07.006. S0962-8924(06)00195-4 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Liao X, Beckett M, Li Y, Khanna KK, Wang Z, et al. MUC1-C Oncoprotein Interacts Directly with ATM and Promotes the DNA Damage Response to Ionizing Radiation. Genes Cancer. 2010;1(3):239–50. doi: 10.1177/1947601910368059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raina D, Ahmad R, Kumar S, Ren J, Yoshida K, Kharbanda S, et al. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. The EMBO journal. 2006;25(16):3774–83. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaika NV, Gebregiworgis T, Lewallen ME, Purohit V, Radhakrishnan P, Liu X, et al. MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America; 2012; pp. 13787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehla K, Singh PK. MUC1: A Novel Metabolic Master Regulator. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbcan.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehla K, Singh PK. MUC1: a novel metabolic master regulator. Biochim Biophys Acta. 2014;1845(2):126–35. doi: 10.1016/j.bbcan.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin L, Kosugi M, Kufe D. Inhibition of the MUC1-C oncoprotein induces multiple myeloma cell death by down-regulating TIGAR expression and depleting NADPH. Blood. 2012;119(3):810–6. doi: 10.1182/blood-2011-07-369686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods in molecular biology. 2012;920:379–91. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 23.McDermott KM, Crocker PR, Harris A, Burdick MD, Hinoda Y, Hayashi T, et al. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int J Cancer. 2001;94(6):783–91. doi: 10.1002/ijc.1554. [DOI] [PubMed] [Google Scholar]
- 24.Boothman DA, Greer S, Pardee AB. Potentiation of halogenated pyrimidine radiosensitizers in human carcinoma cells by beta-lapachone (3,4-dihydro-2,2-dimethyl-2H-naphtho[1,2-b]pyran-5,6-dione), a novel DNA repair inhibitor. Cancer research. 1987;47(20):5361–6. [PubMed] [Google Scholar]
- 25.Souchek JJ, Baine MJ, Lin C, Rachagani S, Gupta S, Kaur S, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J Cancer. 2014 doi: 10.1038/bjc.2014.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla SK, Gunda V, Abrego J, Haridas D, Mishra A, Souchek J, et al. MUC16-mediated activation of mTOR and c-Myc reprograms pancreatic cancer metabolism. Oncotarget. 2015;6(22):19118–31. doi: 10.18632/oncotarget.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla SK, Dasgupta A, Mehla K, Gunda V, Vernucci E, Souchek J, et al. Silibinin-mediated metabolic reprogramming attenuates pancreatic cancer-induced cachexia and tumor growth. Oncotarget. 2015;6(38):41146–61. doi: 10.18632/oncotarget.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunda V, Yu F, Singh PK. Validation of Metabolic Alterations in Microscale Cell Culture Lysates Using Hydrophilic Interaction Liquid Chromatography (HILIC)-Tandem Mass Spectrometry-Based Metabolomics. PloS one. 2016;11(4):e0154416. doi: 10.1371/journal.pone.0154416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int J Med Sci. 2012;9(3):193–9. doi: 10.7150/ijms.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budke B, Kalin JH, Pawlowski M, Zelivianskaia AS, Wu M, Kozikowski AP, et al. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. Journal of medicinal chemistry. 2013;56(1):254–63. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willmore E, de Caux S, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, et al. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103(12):4659–65. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- 32.McKenna WG, Weiss MC, Bakanauskas VJ, Sandler H, Kelsten ML, Biaglow J, et al. The role of the H-ras oncogene in radiation resistance and metastasis. International journal of radiation oncology, biology, physics. 1990;18(4):849–59. doi: 10.1016/0360-3016(90)90407-b. [DOI] [PubMed] [Google Scholar]
- 33.Bernhard EJ, Stanbridge EJ, Gupta S, Gupta AK, Soto D, Bakanauskas VJ, et al. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer research. 2000;60(23):6597–600. [PubMed] [Google Scholar]
- 34.Kasid U, Pirollo K, Dritschilo A, Chang E. Oncogenic basis of radiation resistance. Advances in cancer research. 1993;61:195–233. doi: 10.1016/s0065-230x(08)60959-8. [DOI] [PubMed] [Google Scholar]
- 35.Skvortsov S, Debbage P, Lukas P, Skvortsova I. Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Bai J, Guo XG, Bai XP. Epidermal growth factor receptor-related DNA repair and radiation-resistance regulatory mechanisms: a mini-review. Asian Pac J Cancer Prev. 2012;13(10):4879–81. doi: 10.7314/apjcp.2012.13.10.4879. [DOI] [PubMed] [Google Scholar]
- 37.Nagarajan A, Malvi P, Wajapeyee N. Oncogene-directed alterations in cancer cell metabolism. Trends Cancer. 2016;2(7):365–77. doi: 10.1016/j.trecan.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell death & disease. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isayev O, Rausch V, Bauer N, Liu L, Fan P, Zhang Y, et al. Inhibition of glucose turnover by 3-bromopyruvate counteracts pancreatic cancer stem cell features and sensitizes cells to gemcitabine. Oncotarget. 2014;5(13):5177–89. doi: 10.18632/oncotarget.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko YH, Verhoeven HA, Lee MJ, Corbin DJ, Vogl TJ, Pedersen PL. A translational study “case report” on the small molecule “energy blocker” 3-bromopyruvate (3BP) as a potent anticancer agent: from bench side to bedside. J Bioenerg Biomembr. 2012;44(1):163–70. doi: 10.1007/s10863-012-9417-4. [DOI] [PubMed] [Google Scholar]
- 41.Tinder TL, Subramani DB, Basu GD, Bradley JM, Schettini J, Million A, et al. MUC1 enhances tumor progression and contributes toward immunosuppression in a mouse model of spontaneous pancreatic adenocarcinoma. Journal of immunology. 2008;181(5):3116–25. doi: 10.4049/jimmunol.181.5.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30(12):1449–59. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohr AM, Bailey JM, Lewallen ME, Liu X, Radhakrishnan P, Yu F, et al. MUC1 regulates expression of multiple microRNAs involved in pancreatic tumor progression, including the miR-200c/141 cluster. PloS one. 2013;8(10):e73306. doi: 10.1371/journal.pone.0073306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, et al. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283(40):26985–95. doi: 10.1074/jbc.M805036200. M805036200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, et al. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer research. 2007;67(11):5201–10. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- 46.Nath S, Daneshvar K, Roy LD, Grover P, Kidiyoor A, Mosley L, et al. MUC1 induces drug resistance in pancreatic cancer cells via upregulation of multidrug resistance genes. Oncogenesis. 2013;2:e51. doi: 10.1038/oncsis.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu CF, Li Y, Song YJ, Rizvi SM, Raja C, Zhang D, et al. MUC1 expression in primary and metastatic pancreatic cancer cells for in vitro treatment by (213)Bi-C595 radioimmunoconjugate. Br J Cancer. 2004;91(12):2086–93. doi: 10.1038/sj.bjc.6602232. 6602232 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kao CJ, Wurz GT, Monjazeb AM, Vang DP, Cadman TB, Griffey SM, et al. Antitumor effects of cisplatin combined with tecemotide immunotherapy in a human MUC1 transgenic lung cancer mouse model. Cancer Immunol Res. 2014;2(6):581–9. doi: 10.1158/2326-6066.CIR-13-0205. [DOI] [PubMed] [Google Scholar]
- 49.Lall R, Ganapathy S, Yang M, Xiao S, Xu T, Su H, et al. Low-dose radiation exposure induces a HIF-1-mediated adaptive and protective metabolic response. Cell Death Differ. 2014;21(5):836–44. doi: 10.1038/cdd.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.