Abstract
Purpose
The majority of genomic alterations causing intratumoral heterogeneity (ITH) in colorectal cancer (CRC) are thought to arise during early stages of carcinogenesis as a burst but only after truncal mutations in APC have expanded a single founder clone. We have investigated if the initial source of ITH is consequent to multiple independent lineages derived from different crypts harboring distinct truncal APC and driver KRAS mutations, thus challenging the prevailing monoclonal monocryptal model.
Experimental design
High-depth next-generation sequencing and SNP arrays were performed in whole lesion extracts of 37 FAP colorectal adenomas. Also, ultra-sensitive genotyping of hotspot mutations of APC and KRAS was performed using nanofluidic PCRs in matched bulk biopsies (n=59) and crypts (n=591) from 18 adenomas and 7 carcinomas and adjacent normal tissues.
Results
Multiple co-occurring truncal APC and driver KRAS alterations were uncovered in whole lesion extracts from adenomas and subsequently confirmed to belong to multiple clones. Ultra-sensitive genotyping of bulk biopsies and crypts revealed novel undetected APC mutations that were prominent among carcinomas, whereas abundant wild-type APC crypts were detected in adenomas. KRAS mutational heterogeneity within crypts was evident in both adenomas and carcinomas with a higher degree of concordance between biopsy and crypt genotyping in carcinomas. Non-random heterogeneity among crypts was also observed.
Conclusions
The striking degree of non-random intercrypt heterogeneity in truncal and driver gene mutations observed in adenomas and carcinomas is consistent with a polycryptal model derived from multiple independent initiation linages as the source of early ITH in colorectal carcinogenesis.
Keywords: adenoma, colorectal cancer, clonality, intestinal crypt, tumorigenesis
Introduction
Intestinal homeostasis is based on the balance of two cellular processes: 1. the continuous replacement of epithelial cells within crypts, which is maintained by the pool of stem cells located at the base (1); 2. the generation of new crypt units, which is secondary to fission (2, 3). Based on the adenoma-to-carcinoma model of linear evolution it has been assumed that the pool of normal colorectal stem cells in the crypts is likely the initial target of genomic events that initiates intestinal carcinogenesis, thus becoming founder clones (4–6). Moreover, the process of crypt fission, which is infrequently observed in normal colorectal mucosa (7), is definitely increased in colorectal adenomas (8), thus providing a plausible explanation for the geographical expansion of mutant clones harboring genomic events (9, 10). Therefore, the study of the mutational events involving the intestinal crypt will inform the dynamics of the initial steps of colorectal tumorigenesis.
Peter Nowell laid the foundation of the concept of clonal evolution in cancer by applying Darwinian evolution logic to the process of carcinogenesis and progression (11). In principle, owing to the accumulation of somatic alterations, one particular genetic change will provide a selective advantage to a cancer cell (driver event) and will give rise to a dominant population (clone), thus making this alteration truncal (founding alteration present in all cancer cells, which is an APC alteration in the vast majority of CRC). Subsequent mutations will affect discrete groups of cells (subclonal and therefore private mutations) and depending on the timing of the acquisition of the mutation will be present in a small (private and restricted mutations) or large number of cells (private and pervasive such as KRAS mutations). Progression along the normal mucosa-adenoma-carcinoma sequence in colorectal cancer (CRC) is fostered by progressive accumulation of genomic events in well-known driver genes with the truncal events being the acquisition of APC alterations in approximately 85% of cases (12). APC hits targeting the stem cells of the colorectal crypt will induce proliferation of founder clones carrying these alterations in all dysplastic, and later cancerous cells, across all stages of carcinogenesis (truncal and driver events) (5, 6).
The ‘Big Bang’ model, which is based on the concept of punctuated evolution (13, 14), has provided a novel framework to understand the clonal evolution and the timing of the emergence of intratumoral heterogeneity (ITH) in CRC. This model posits that the vast majority of genomic alterations accumulate during early stages of carcinogenesis, while the tumor mass is still less than one million cells, and before progressing into an advanced neoplasm. However, this model assumes that this accumulation could only happen after truncal alterations in genes that are essential for colorectal carcinogenesis, such as APC, have occurred in a founder clone, thus consistent with a monocryptal and monoclonal origin (15). However, the presence of multiple founder clones (lineages) from distinct crypts observed in both animal models and human samples (16–18) challenges the monocryptal monoclonal origin posed by the ‘Big Bang’ model to explain the source of ITH. The observation of multiple initiating clones implies the recruitment of different distinct dysplastic crypts from which one will emerge as dominant, thus driving carcinogenesis and contributing another source of ITH at the premalignant stage that will have important consequences for the design of chemopreventive strategies and to understand resistance to therapies.
KRAS mutations are acquired during subsequent stages of colorectal carcinogenesis and drive further tumor progression. KRAS mutations are present in a restricted, although significant, proportion of dysplastic cells (private and pervasive events), generating the presence of subclones with different mutational profiles that will form mixtures and contribute another level of ITH early in carcinogenesis. However, the interplay between inactivated APC and mutant KRAS is still debated and challenges the sequence of acquisition of these events. In fact, it has been shown that there is a cross-talk between both pathways when mutant KRAS activates the translocation of β-catenin to the nucleus (19), thus inducing cancer stemness (20) and increasing the number of fissions among neighboring colorectal crypts (2, 21) thus proving that KRAS mutations are also essential for the expansion of a founder clone. Therefore, the assessment of the origin of ITH in colorectal premalignancy requires an in-depth analysis of mutations in both APC and KRAS at the crypt level.
We posit that early acquisition of ITH is consequent to a polyclonal origin of premalignant lesions derived from the interaction of multiple independent crypts within adenomas that acquire distinct alterations in APC. Moreover, another level of heterogeneity is added by the acquisition of multiple competing somatic KRAS mutations as an essential driver event. Therefore, we aimed to assess whether the type and degree of mutational heterogeneity in both APC and KRAS in whole lesion extracts, multi-region biopsies and crypts at early and late stage lesions are consistent with our hypothesis that challenges a single cell origin in colorectal carcinogenesis.
Material and Methods
Samples and Patients
Ampliseq sequencing and SNP arrays were performed on 37 colorectal adenomas and matched normal mucosa samples from 14 patients with Familial Adenomatous Polyposis (FAP) collected at MD Anderson Cancer Center (Figure 1). Adenomas and normal mucosa samples were acquired at the time of endoscopic surveillance and were either fully resected or biopsied with snare forceps (Supplementary Methods, Tables S2–3). Fresh colectomy specimens from 23 sporadic colorectal tumors (14 right and 9 left colon) were collected at the time of the surgery in Bellvitge University Hospital. Sampling of the intraluminal surface of carcinomas and adenomas macroscopically separated from the margin of the tumor was performed by an experienced pathologist. Normal distal mucosa was also collected. A total of 6 cases were selected for further analysis by KRAS genotyping using digital PCR. In addition, 17 adenoma samples from the three regions of the colon were obtained from 4 patients diagnosed with hereditary colorectal polyposis (Tables S2–3). Moreover, we obtained one adenoma from a patient with constitutional mismatch repair (MMR) gene deficiency (CMMR-D), and one tumor from an unrelated patient diagnosed with Lynch syndrome. Therefore, a total of 18 adenomas (all coming from patients diagnosed with hereditary cancer syndromes) and 7 carcinomas were analyzed (6 from sporadic cases and one from a Lynch syndrome case). Matched normal mucosa samples were available for all patients (Supplementary Methods and Table S2). All premalignant samples were classical tubular adenomas per pathology assessment. Informed consent was obtained from all patients and the Institutional Review Board approved this study at both institutions.
Figure 1.
Experimental design and molecular analysis performed in bulk biopsies, crypts and whole lesion extracts of adenomas and colorectal carcinomas. Adenoma analyses were performed only in samples obtained from patients diagnosed with hereditary colorectal cancer syndromes. Carcinoma analysis were performed in a total of 6 tumors from sporadic cases and one tumor from a hereditary case. Note that both adenomas and carcinomas were analyzed from different locations of the colon. WES, whole exome sequencing.
Ampliseq sequencing and SNP Array
Ampliseq sequencing was performed with an IT Personal Genome Machine (PGM; Life Technologies, Carlsbad, CA) by the Sequencing and Non-Coding RNA Program at MD Anderson using the Ion PGM 200 Sequencing Kit on an Ion 318 Chip Kit (Life Technologies).. In parallel, SNP array genotyping of the same samples was performed using the HumanOmniExpressExome beadChip array (Illumina, San Diego, CA) (See Supplementary Methods for details).
Colony Genotyping Assay
We performed a colony genotyping assay to validate the presence of the double somatic hits detected in APC by Ampliseq sequencing in adenoma MDAC33_P04 and also to determine whether the two somatic mutations (Somatic #1 and Somatic #2) in one adenoma belong to different clones. The regions containing the two somatic mutations [Somatic #1 (c.4267_4280del) and Somatic #2 (c.4348C>T)] and the germline mutation (c.2894del) were amplified by Long-Range PCR and ligated into pGEM-Teasy Vector System II (Promega, Madison, WI). DNA from more than 100 clones was extracted using QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and genotyped using one Taqman assay per mutation. We performed power calculations to determine the minimum number of assays required to observe Somatic #1 and Somatic #2 hits separately at least in 3 different colonies with 95% power and determined that at least 82 clones had to be analyzed (Supplementary Methods).
Crypt isolation
Fresh samples from normal mucosa, adenomas and tumors (Table S4) were minced into 3 mm pieces, incubated in 30mM EDTA in HBSS for 20 min, and vigorously shaken to obtain a supernatant enriched for crypts. The same day of the sample collection individual crypts were picked up under microscope and were placed into 0.5 mL microfuge tubes. Normal crypts were easily distinguishable from tumor crypts based on their characteristics (normal crypts were small and nonbranched, while tumor crypts were large and displayed sheets of epithelium with thick clusters of different sizes) (22). This method limits the contamination of non-neoplastic cells by an efficient separation of lamina propria mucosa or stroma (23). Isolation of individual crypts was verified by microscopy in a selection of cases to ensure that the molecular analysis was representative of individual crypts and that the methodology for isolation was consistent (Figure 1).
APC Mutation Cluster region and LOH analysis in bulk and crypt samples
A fragment of 1650 base pairs (bp) of the mutational cluster region (MCR) was amplified and then sequenced in biopsy sections. For crypt analysis, a nested PCR approach was performed to increase the amount of DNA template. All detected alterations were validated in at least 3 independent reactions and verified by two independent reviewers. Normal mucosa biopsies and normal crypts from the same cases were also analyzed in parallel to rule out random acquisition of APC mutations or PCR artifacts. In addition, LOH analysis of APC was performed following standard procedures (See Supplementary Methods and Table S9 for primers).
Ultrasensitive detection of KRAS hotspot mutations in bulk and crypt samples
In bulk biopsies, genotyping of KRAS hotspot mutations was performed using allele-specific probes for seven KRAS mutational hotspots (G12A, G12C, G12D, G12R, G12S, G12V and G13D) in the Digital Array platform (Fluidigm Corporation, South San Francisco, CA; Supplementary Methods) with an analytical sensitivity from 0.05% to 0.1%, depending on the mutation analyzed (24). In crypts, the same assays were performed using the 48.48 BioMark Dynamic Array (Fluidigm Corporation) after a pre-amplification step. The analytical sensitivity of the Dynamic Array for the detection of the mutant allele oscillated between 5–12,5% depending upon the probe used (Table S5) in the same range of conventional real-time PCR genotyping (see Supplementary Methods). In the absence of detectable DNA contamination, consistent results were obtained in sixtuplicates allowing for robust observations. Median values of replicates were used to quantify the amount of mutant alleles. The quantitation of the signal was extrapolated from standard curves generated from reconstitution experiments that also allowed the definition of conservative thresholds (See Supplementary Methods for details).
Of note, we carefully ruled out the introduction of systematic DNA contamination as a consequence of performing the pre-amplification of DNA using a nested PCR approach by using multiple independent steps. First, mastermix preparation, DNA pipetting, first PCR amplification, and second PCR amplification were all performed in four separate rooms, where different instrumentation was used. Second, we always kept a unidirectional flow for all the samples. Third, controls for contamination were included in every single PCR reaction. Contamination controls for the first external KRAS PCR reaction were re-amplified together including always specific positive and wild-type controls. Fourth, random sets of amplicons were tested specific KRAS mutation probes using conventional real-time PCR analysis by LightCycler 480 to add an additional contamination control, always ruling out the presence of contamination. Finally, we performed a total of six replicates for each assay using the 48.48 Dynamic Array on the BioMark platform (Fluidigm Corporation) in order to re-assure on the robustness of our results (See Supplementary Figures S11–S15 and Supplementary Methods for technical details).
Metapopulations analysis in crypts
Hierarchical clustering of crypts was performed using percentages of KRAS mutant cells extrapolated from the allele frequencies of the mutations in each crypt (See Supplementary Methods for details).
Results
High-depth next generation sequencing detects multiple somatic events in adenomas from FAP patients
Next-generation sequencing at high depth using Ion Torrent AmpliSeq was performed in a total of 37 adenomas from 14 patients diagnosed with FAP to detect public somatic mutations in APC and KRAS (Figure 1). An advantage of studying adenomas of FAP patients with confirmed germline alterations of APC to detect hints of multiclonality is that one APC event is already given from the germline, so any new additional APC events will be acquired (somatic events under the assumption that adenomas are diploid). Therefore, accumulation of multiple somatic events either by mutation or allelic imbalance (loss of 5q) will be suggestive of polyclonality. The average sequencing depth obtained per sample for APC and KRAS was 3,640x and 6,685x, respectively. We identified a total of 32 somatic events in APC with an average allelic frequency of 21%. The majority were mutations generating a stop codon (29 out of 32); although we detected other events such as a splicing mutation (c.1744-1G>A) and two missense mutations (c.2438A>G; p.Asn813Ser and c.2258A>G; p.His753Arg), which were all predicted to have a functional impact (Table S6). Using SNP arrays, loss of 5q was identified in 4 adenomas with an average allelic frequency of 4% (Table S3). Overall, we were able to detect a second hit in APC in 81% of adenomas (30/37). Six adenomas (16%) presented double somatic events in APC in the form of two different mutations or the combination of one mutation associated with a 5q loss (Table S1). In addition, we detected 15 KRAS somatic mutations in 14 adenomas with an average allelic frequency of 29%.
One adenoma presented two different KRAS mutations: c.35G>A (p.Gly12Asp) and c.40G>A (p.Val14Ile) with allelic frequencies of 5% and 16%, respectively. Interestingly, this adenoma also presented 2 independent somatic events in APC: 5q loss and c.4464_4467delInsGTAAT (p.Leu1489*) (Table S1). We selected the APC alterations detected in one adenoma (MDAC33_P04) to validate the multiclonal origin of these events using colony analysis. Of note, this adenoma was euploid and the region of APC containing the two somatic mutations (c.4348C>T and c.4267_4280del) and the germline mutation (c.2894delA) was amplified and cloned into pGEM-T plasmid. A total of 100 colonies were genotyped and each colony carried a single mutational event: 25% were wild-type, 36% carried the germline mutation (c.2894del), 15% one of the somatic hits c.4348C>T and 24% the other somatic hit c.4267_4280del (Figure 2). These cloning results confirmed that the somatic APC mutations arose from separate and independent clones (polyclonal adenomas).
Figure 2.
A. Schematic representation of the multiple APC mutations identified in the adenoma selected for assessment of polyclonality by a cloning approach. The germline event is depicted in green and the somatic events are depicted in red; B. Frequency of colonies harboring wild-type (wt), germline, and somatic mutated genotypes (mut). One hundred colonies were analyzed using Taqman assay. X-axis represents intensity of FAM fluorescence (wt allele) and Y-axis VIC fluorescence (mut allele). On the scatter plot diagrams, end signals from each sample have been presented as a single dot. Homozygous samples of the wt allele exhibit an increased fluorescence along the X-axis, homozygous samples of the mut allele along the Y-axis, and heterozygous samples should appear within the centre of the plot. Every clone displayed a unique genotype that was consistent with the expected allelic frequencies from next-generation sequencing results.
Paired analysis of multi-region biopsies and crypts of adenomas and carcinomas reveal high degree of heterogeneity
Next-generation sequencing results from whole lesion DNA extracts provided evidence that the mutational heterogeneity observed in adenomas was derived from multiple different clones harbor distinct mutations in truncal driver genes. This led us to hypothesize that ITH emerges early on colorectal carcinogenesis due to the interaction of multiple mutated crypts (independent lineages) that is not captured by bulk biopsies, thus requiring individual crypt analysis. Therefore, we decided to assess the presence of multiple somatic events in paired multi-region bulk biopsies and crypts extracted from the same lesions by applying ultrasensitive genotyping techniques within mutational hotspots of the APC and KRAS genes.
APC gene analysis
Since our goal was to discover the origin of mutational heterogeneity in the continuum of colorectal carcinogenesis we studied a total of 17 adenomas from patients diagnosed with hereditary polyposis syndromes [1 case with FAP, 1 with MAP and 2 with UFP (UFP1 and UFP2), Table S4] and a total of 165 paired colorectal crypts. Multi-region biopsy analysis revealed the presence of somatic APC mutations in almost all of the regions of 2 adenomas from the FAP and UFP2 cases (Figure 3, Figure S3 and Table S7). Crypt analysis of these adenomas revealed mixtures of novel somatic APC alleles, which were not present in the bulk biopsies, and also an abundance of wild-type alleles of the MCR of APC. The percentage of crypts with APC mutations varied across adenomas but overall there was a predominance of wild-type ones (average 20%, range 13–100%, Table S7). In contrast, adenomas from the MAP and UPF1 cases showed a good correlation between bulk biopsy and crypt analyses with all the biopsies and crypts displaying a wild-type status for the MCR region of APC (Figures S1–2 and Table S7).
Figure 3. APC and KRAS genotyping of crypts and bulk biopsies in adenomas from the FAP case.
On the left side is displayed the study of the mutator cluster region (MCR) of APC performed in 3 adenomas (Ad1.1, Ad2.1, and Ad3.1). The upper panel shows the results of the bulk biopsy analysis and the lower panel the results obtained from crypts that correspond to the same lesions. One dominant mutation was observed in the bulk biopsies of two adenomas (Ad1.1. and Ad3.1), while the analysis performed in individual crypts observed an abundance of wt crypts and one novel APC mutation in Ad1.1. In addition, co-existence of the dominant mutation from the biopsy with wt crypts was evident in both lesions Ad1.1 and Ad3.1. On the right side are presented the results of the KRAS genotyping. Each column represents one of the KRAS hotspot mutations that were tested by the digital or the dynamic array, and each row represents the results of bulk biopsies (one biopsy per adenoma, upper panel) or single crypts (10 crypts per adenoma, lower panel). The upper panel shows the digital PCR results from bulk biopsies, which identify a single KRAS mutation at low frequency in each adenoma. The lower panel shows the widespread heterogeneity observed in crypts which were not detected in the biopsies. The color scale depicts the mutational frequency of KRAS events.
Among carcinomas, all of them harbored APC mutations (2 frameshift, 3 missense and 2 silent changes) in all regions analyzed (5 different areas were analyzed in each tumor) (Figures S5–7). In addition, we detected LOH in 5q in one case (Figure S6, SP4), which also harbored a silent somatic change. Strikingly, a high degree of APC heterogeneity was evident at the crypt level. An average of 72% of the carcinoma crypts analyzed were APC mutated in a background of wild-type crypt alleles, despite having selected the intraluminal surface of the tumors distal to adjacent healthy mucosa making it unlikely the sampling of normal crypts (Figures S5–S7). Only in one of the six cases analyzed, we observed that all crypts shared the same APC clonal mutation (SP4). In two cases additional APC mutations not detected in the bulk biopsies were evidenced in the crypt analysis (Table S7), thus showing a very significant degree of APC mutational intracarcinoma heterogeneity (cases SP5 and SP6; Figure 4 and S7). Our APC studies revealed a high degree of heterogeneity in both adenomas and carcinomas, thus pointing towards interactions and potential competition among independent lineages from different crypts.
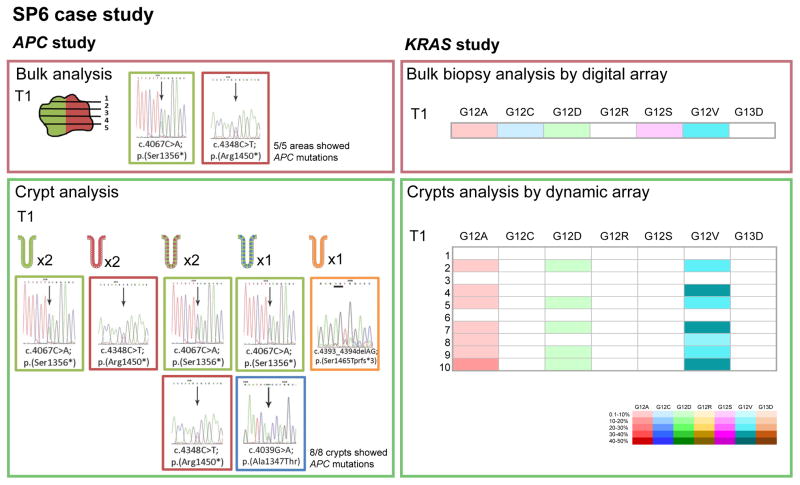
Figure 4. APC and KRAS genotyping of crypts and bulk biopsies from sporadic colorectal carcinoma case #6 (SP6).
The APC analysis in bulk biopsies (5 areas) revealed two APC mutations. The same mutations were observed in crypts (total 8 crypts) with different proportions. In addition, there were two crypts showing novel mutations that were undetected in the bulk biopsies. KRAS genotyping capture the breath of the mutational heterogeneity present in the carcinoma which was similar to the crypt analysis. The coexistence of different crypt populations contributing to the biopsy results were evident from the crypt analysis. Each column represents one of the KRAS hotspot mutations that were tested by the digital or the dynamic array, and each row represents the results of bulk biopsies (one biopsy per tumor, upper panel) or single crypts (10 crypts per tumor, lower panel).
KRAS gene analysis
We then turned our attention to KRAS as a main driver in early carcinogenesis and due to its cross-talk interactions with APC. In general, all bulk biopsies analyzed in adenomas from the MAP, UFP1 and UFP2 cases unveiled multiple co-existing KRAS mutations (mutational load average 11%) while FAP adenomas only harbored single mutations (Figure 3, S1–3 and Table S8). However, when we studied crypts obtained from the same lesions a striking intracrypt mutational heterogeneity was evident in 76% of adenomas (13/17). An average of 44% of the crypts per adenoma displayed multiple KRAS mutations (range 10–100%) and the proportion of mutant alleles was much higher compared to the bulk biopsies (average 26%, versus 7.6%). Moreover, we also observed KRAS mutational heterogeneity in the matched surrounding normal mucosa that may reflect a ‘genetic field effect’, which may be acquired prior to the APC alterations or as a consequence of underlying deficiencies in DNA repair that are the basis of these syndromes (Table S8).
A high great degree of consistency between the results of bulk biopsies and crypts in terms of KRAS mutational diversity and proportion of mutated alleles was observed among carcinomas (29% in biopsies versus 22% in crypts). In this regard, no additional mutations were detected in the crypts analyzed. However, in terms of mutational load, the proportion of KRAS mutant alleles among crypts was variable (Figure 4, Table S8). Intriguingly, we noted in several carcinomas (SP2, SP3 and SP6) the presence of a variable fraction of wild-type KRAS crypts that co-existed with crypts with a high KRAS mutational load (Table S8), depicting another level of genetic heterogeneity.
In an effort to provide a complete picture of the evolutionary dynamics of APC and KRAS mutations in all pathways of carcinogenesis in CRC, two different hereditary cases displaying MMR deficiency were included. We analyzed one adenoma from a patient with CMMR-D and one carcinoma from an unrelated Lynch Syndrome patient (Table S2). The bulk biopsy analysis in these two samples was limited to one biopsy rather than being multiregional as in the rest of hereditary adenomas and sporadic tumors included in this report. The analysis of the MCR of APC in both cases did not reveal any mutation at the biopsy and crypt level (Figure S4 and Table S7). Absence of heterogeneity with a predominance of wild-type KRAS crypts was observed in the adenomas while the carcinoma showed a relatively low level of heterogeneity among the crypts (Figure S4 and Table S8). Therefore, APC and KRAS analysis did not capture mutational heterogeneity in MMR deficient lesions but these results do not rule out that polyclonality that may be observed by alternative studies analyses other drivers more relevant to MMR deficient carcinogenesis. Nonetheless, these results reinforce the robustness of our technical approach as it did not observe any random variation among the MMR deficient samples, thus providing a negative control.
Hierarchical clustering of crypts reveals a striking degree of non-random inter-crypt heterogeneity with several crypt metapopulations
Unsupervised clustering of all crypts based on KRAS mutations for each patient revealed a non-random pattern (Figure 5 and Figures S8–11). A discrete and similar number of metapopulations was evident in both adenomas (mean 3.80 metapopulations per case) and carcinomas (mean 2.84 metapopulations per case). This fact reflects that a stable number of founder clones emerge early in carcinogenesis and remains stable during progression of carcinogenesis.
Figure 5. Hierarchical clustering analysis of crypts based on KRAS genotyping results from FAP adenomas (A) and one sporadic carcinoma (B).
Trees have been generated from the crypt genotyping results presented in Figures 3 (FAP) and 4 (SP6). This analysis was performed to capture the presence of heterogeneity reflected by the co-existence of different metapopulations. The color scale reflects the percentage of cells harboring the mutation. The color bar indicates the number of groups detected in the clustering analysis.
Crypts from MAP, UFP1 and UFP2 cases clustered around 9, 2 and 4 metapopulations, respectively, compared to FAP, which showed 2 main ones (Figure 5 and Figure S8). The difference observed between the number of clones in MAP and FAP may be related to the mechanism of genetic instability (driven by deficiency in base excision repair versus chromosomal instability). Particularly striking was the fact that crypts from adenomas that were physically located at different parts of the colon clustered together, once again pointing towards the existence of a ‘genetic field’ effect.
Discussion
The approach presented in this manuscript combines for the first time high-depth next-generation sequencing of whole lesion extracts, and analyses of matched bulk biopsies and crypts from adenomas and carcinomas using highly sensitive genotyping techniques to assess simultaneously the mutational status of APC and KRAS. This approximation provides a deeper insight into the timing and dynamics of the somatic acquisition of ancestral mutations in colorectal carcinogenesis. Our data is consistent with the co-existence of multiple truncal APC mutations harbored by different crypts that will lead to the establishment of a dominant clone through complex interactions either though recruitment or cooperation, which is consistent with previous murine and human observations (16–18, 25). In fact, our ultra-sensitive approach has uncovered the highest degree of ITH ever reported as a result of the presence of multiple independent initiating events, which allowed us to make the following observations: (i) co-occurring somatic truncal APC hits detected in pre-malignant samples belong to distinct clones; (ii) wild-type APC and KRAS alleles are predominantly observed in crypts of premalignant lesions; (iii) the initial source of ITH is derived from competing truncal APC and driver KRAS mutations that are not captured by bulk biopsies. These mutations belong to independent lineages and are present in a low proportion of cells within the crypt; (iv) the identification of a putative ‘genetic field effect’ in premalignant adenomas of hereditary cases that is evident among lesions from the same patient and are even physically distant within the colon.
We have uncovered the presence of multiple co-occurring somatic APC mutations using whole lesion extracts from colorectal adenomas of FAP patients, which is consistent with previous observations. However, using cloning approaches we have confirmed for the first time that they belong to independent clones (25–27). Furthermore, the fact that is observed only in pre-malignant samples, which have not acquired yet chromosomal instability and are still diploid, confirms that the theory of the presence of ‘three-hits’ inactivating APC is still plausible in carcinomas that may have acquired multiple copies of APC (28, 29).
We have detected the presence of mutational heterogeneity in APC at the crypt level among early stage adenomas with only a minority of the crypts harboring the clonal APC mutation identified in their paired bulk biopsies. In addition, the majority of crypts from adenomas were APC wild-type. Both results are in line with previous observations made in more advanced lesions such as adenoma-in-carcinoma samples, where the degree of heterogeneity was higher, and only a minority of crypts were wild-type (18). This is not unexpected and reflects further progression into the clonal evolution presented in more advanced stages of carcinoma progression. The fact that we have consistently identified wild-type APC crypts in the intraluminal surface of carcinomas and adenomas distal to normal mucosa is compatible with the coexistence of normal crypts inside of the dysplastic cancerous mass that, based on the work of Thliveris et al, could represent wild-type crypts recruited by the mutant APC founder clone to foster neoplastic growth through transformation later on (16). Alternatively, we cannot rule out that these normal crypts were entrapped by the hyperproliferating clone as our method of analysis is based on mechanical separation of crypts. Therefore, this lack of spatial resolution also accounts for our inability to determine if the expansion of a mutant APC founder clone follows a ‘top-down’ versus a ‘bottom-up’ model of expansion in the colonic crypt (30). Finally, Thirlwell et al identified bona fide wild-type APC crypts in FAP adenomas that were attributed to biased tissue sampling capturing normal areas that were intermixed with the carcinoma (18).
Our KRAS genotyping analysis using digital PCR has proved both robust and sensitive, thus allowing the detection of a striking degree of heterogeneity at the biopsy and crypt levels within adenomas and carcinomas, which is in line previous results that used less sensitive techniques with lower resolution (31–36). Clustering analysis has revealed that the observed KRAS mutational pattern in crypts was not random and depicted the presence of a limited number of crypt metapopulations in adenomas and carcinomas. It was notable that crypts obtained from distinct adenomas arising from hereditary patients displayed a similar mutational pattern, irrespective of the location of the lesion consistent with a ‘genetic field effect’. There are several factors that could explain this observation: (i) the asymmetric expansion associated to crypt fission or other mechanisms of crypt interactions yet to be determined (2, 8, 16, 17, 21, 34, 37); (ii) the underlying mechanism of genetic instability present in hereditary cases such as chromosomal instability in FAP, base excision repair deficiency in the case of MAP, and MMR deficiency in Lynch syndrome and CMMRD (38), the latter being notorious for an absence of KRAS heterogeneity. However, we cannot rule out that at least part of these KRAS mutations emerge prior to the acquisition of APC mutations and the establishment of a founder clone. In fact, KRAS mutations have been detected in normally appearing tissues and in aberrant crypt foci that may not progress into adenomas (39). Lastly, this diversity of KRAS mutations acquired early in carcinogenesis has multiple ramifications for the design of chemoprevention strategies and to explain the emergence of resistance to therapies.
Our study has several limitations. First, the mechanical isolation of crypts is subject to sampling bias and selection of specific cell populations. However, we believe that we have minimized this problem by involving dedicated expert gastrointestinal pathologists to collect samples following established procedures that are unlikely to be contaminated by normal crypts. Second, performing genomic analysis in relatively small groups of cells is challenging due to the limited amounts of DNA rendered. We have evaluated different approaches (whole genome amplification versus nested PCR) to amplify the nucleic acid material prior to our ultrasensitive genotyping and demonstrated that our approach using nested PCR is capable to render robust analytical results. Furthermore, all of our tests have been performed in sextuplicate and included multiple internal controls, thus minimizing uncertainty. Therefore, we are confident that this rigorous and reproducible approach minimizes concerns for contamination. Third, in our crypts analysis we have only assessed the MCR of APC and therefore the possibility that we have missed mutations located outside of this region reflecting additional heterogeneity is obvious. In the future, single cell sequencing analysis will be the most appropriate tool to interrogate the dynamics and clonal heterogeneity of crypts in premalignancy (40), thus helping to clarify the level of mutational diversity within the crypt and establishing a cellular hierarchy based on mutational events. Alternatively, the use of next-generation sequencing including multiple molecular barcodes will help to discriminate between the high background error rate generated by conventional NGS and emerging clones. These approaches will have the potential to shed light in the ‘top-down’ versus ‘bottom-up’ debate of expansion of founder clones among adjacent crypts (30).
Taken together, the body of evidence presented here demonstrates that the presence of ITH in colorectal premalignancy is abundant and secondary to the presence of multiple independent lineages derived from crypt progenitors, which is highlighted by the mutational heterogeneity detected in APC and KRAS. This observation contrasts with the majority of tumor evolution models that are based on the expansion of a single dominant clone that harbors a truncal mutation and several subsequent driver alterations (mainly in APC but also KRAS, TP53 and PIK3CA). These models ignore the degree of ITH that has been already acquired at the premalignant stage with multiple founder clones awaiting for one to get selected over the others, thus making the ITH acquired at the carcinoma level just the ‘tip of the iceberg’. Certainly, our observations are complementary to the ‘Big Bang’ model and precede it (15). We speculate that polyclonality in premalignancy depends on crypt interactions by mechanisms involving crypt fission that leads to the recruitment of different independent lineages, which may not contribute further to progression but rather be engulfed in the dominant clone (Figure 6). This is in line with observations made in skin exposed to UV light (41). In addition, this model of crypt dynamics supports the continuous emergence of KRAS mutant clones from a ‘genetic field effect’ that are occasionally selected and serves as an engine for tumor progression in the context of APC/KRAS mutant tumors. Finally, the conceptual challenges posed by the identification of wild-type APC crypts in carcinomas cannot be ignored and warrants further investigations.
Figure 6. Model of polyclonal polycryptal evolution in colorectal carcinogenesis.
The graph represents a population of colonic crypts (population level, top view) that progress through the acquisition of a series of somatic mutations starting by multiple truncal APC and then followed by multiple KRAS that can be acquired at different time points (1 through 4). Normal colonic crypts acquire a truncal mutation in APC followed by a driver event in KRAS at different time points (blue and green clones). The sequential acquisition of an APC and KRAS event generates an intrinsic growth advantage and the expansion of a clone that will eventually dominate the neoplastic mass, entrapping the other clones that may continue contributing to the growth of the mass as well. Some crypts acquire APC mutations without subsequent hits and their proliferation is outpaced by more dominant and faster growing clones (yellow clone). Note that normal crypts may acquire KRAS driver mutations without deriving a growth advantage and that are eventually engulfed by the major dominant clone (orange clone). These KRAS mutations could be secondary to DNA repair deficiencies that are intrinsic to the pathogenesis of hereditary syndromes (such as deficiencies in the Base Excision Repair system). Finally, inside of each crypt KRAS mutational diversity can arise generating inter-crypt mutational heterogeneity as depicted in the inset (crypt level, side view).
Supplementary Material
Statement of Translational Relevance.
Colorectal carcinogenesis models assume that neoplasms develop from a single colorectal crypt that has acquired truncal APC alterations, which expand the founder clone, and subsequently driver KRAS mutations. We performed ultra-sensitive genotyping of matched bulk biopsies and individual crypts of colorectal adenomas and carcinomas, and observed the presence of novel APC mutations, abundant wild-type APC crypts co-existing with mutant crypts, and mutational heterogeneity in KRAS within crypts, all of them not observed in the analysis of biopsies. We conclude that the non-random heterogeneity among crypts support a polycryptal model as the origin of multiclonality in colorectal carcinogenesis. The fact that intratumor heterogeneity arises at premalignant stages from the convergence and admixtures of multiple different crypts harboring different APC and KRAS mutations informs our knowledge on tumor evolution of colorectal cancers, which is critical for designing new chemopreventive measures, predict resistance to therapies, and design early detection strategies addressing this diversity.
Acknowledgments
Funding support: This work was supported by grants R03CA176788 (US National Institutes of Health/National Cancer Institute), The University of Texas MD Anderson Cancer Center Institutional Research Grant (IRG) Program and a gift from the Feinberg Family to E.V.; U01GM92666 and R01HG005859 (US National Institutes of Health) to P.S.; the Janice Davis Gordon Memorial Postdoctoral Fellowship in Colorectal Cancer Prevention (Division of Cancer Prevention/The University of Texas MD Anderson Cancer Center) to E.B.; Cancer Prevention Educational Award (R25T CA057730, U.S. National Institutes of Health/National Cancer Institute) to K.C.; Spanish Ministry of Economy and Competitiveness (SAF2012-33636 GC), the Carlos III Health Institute (PI13/00285 CL), RTICC (RD12/0036/0031 and RD12/0036/0008), the Scientific Foundation Asociación Española Contra el Cáncer, the Government of Catalonia (2009SGR290 and 2014 SGR 338) and the Scientific Foundation Asociación Española Contra el Cáncer to G.C.; P30 CA016672 (US National Institutes of Health/National Cancer Institute) to the University of Texas Anderson Cancer Center Core Support Grant.
We thank the patients and their families for their participation. We thank the staff of the ncRNA and Sequencing Core at MD Anderson Cancer Center for the assistance with IonTorrent sequencing. We thank the Clinical Cancer Prevention Research Core for their assistance obtaining the samples for this study.
Abbreviations
- CRC
colorectal cancer
- FAP
Familial Adenomatous Polyposis
- UFP
Unexplained Familial Polyposis
- MAP
MUTYH-associated Polyposis
- bp
base pair
- MCR
mutator cluster region
- LOH
loss of heterozygosity
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
- wt
wild-type
- Ad
adenoma
- MMR
mismatch repair
- CMMR-D
constitutional mismatch repair deficiency
- WES
whole exome sequencing
Footnotes
Presented in part at the AACR Special Conference: Colorectal Cancer: From Initiation to Outcomes, Tampa, FL, September 17–20, 2016.
Conflict of Interest: The authors declare no conflicts of interest.
Contributions:
M.G., E.B., K.C., G.C. and E.V. designed the study and experiments.
E.V., P.M.L., S.G., M.P., Y.N.Y. and G.C. identified, consented and recruited study subjects and provided clinical information.
M.G., E.B., M.W.T., X.S., M.J.P. processed case samples and performed pathology assessments.
M.G., E.B., D.A., A.D.A. performed molecular experiments.
G.E.D. and E.E. performed SNP array experiments.
K.C., F.A.S.L., J.F., and P.S. analyzed SNP array and sequencing data.
K.C., P.S., A.L-D., and V.M. performed statistical analysis.
M.G., E.B., K.C., V.M., E.T.H., C.L., P.S., G.C., N.N., and E.V. analyzed and discussed the data.
M.G., E.B., K.C., G.C. and E.V. wrote the manuscript.
All authors: edited and approved the content of this manuscript.
References
- 1.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 2.Wasan HS, Park HS, Liu KC, Mandir NK, Winnett A, Sasieni P, et al. APC in the regulation of intestinal crypt fission. J Pathol. 1998;185:246–55. doi: 10.1002/(SICI)1096-9896(199807)185:3<246::AID-PATH90>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415–24. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 4.Huels DJ, Sansom OJ. Stem vs non-stem cell origin of colorectal cancer. Br J Cancer. 2015;113:1–5. doi: 10.1038/bjc.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 6.Powell AE, Vlacich G, Zhao ZY, McKinley ET, Washington MK, Manning HC, et al. Inducible loss of one Apc allele in Lrig1-expressing progenitor cells results in multiple distal colonic tumors with features of familial adenomatous polyposis. Am J Physiol Gastrointest Liver Physiol. 2014;307:G16–23. doi: 10.1152/ajpgi.00358.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totafurno J, Bjerknes M, Cheng H. The crypt cycle. Crypt and villus production in the adult intestinal epithelium. Biophysical journal. 1987;52:279–94. doi: 10.1016/S0006-3495(87)83215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong WM, Mandir N, Goodlad RA, Wong BC, Garcia SB, Lam SK, et al. Histogenesis of human colorectal adenomas and hyperplastic polyps: the role of cell proliferation and crypt fission. Gut. 2002;50:212–7. doi: 10.1136/gut.50.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez-Gonzalez L, Deheragoda M, Elia G, Leedham SJ, Shankar A, Imber C, et al. Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J Pathol. 2009;217:489–96. doi: 10.1002/path.2502. [DOI] [PubMed] [Google Scholar]
- 10.Preston SL, Wong WM, Chan AO, Poulsom R, Jeffery R, Goodlad RA, et al. Bottom-up histogenesis of colorectal adenomas: origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res. 2003;63:3819–25. [PubMed] [Google Scholar]
- 11.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 12.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 13.Davis A, Gao R, Navin N. Tumor Evolution: Linear, Branching, Neutral or Punctuated? Biochim Biophys Acta. 2017 doi: 10.1016/j.bbcan.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, Sun R, Curtis C. A population genetics perspective on the determinants of intra-tumor heterogeneity. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbcan.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–16. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thliveris AT, Schwefel B, Clipson L, Plesh L, Zahm CD, Leystra AA, et al. Transformation of epithelial cells through recruitment leads to polyclonal intestinal tumors. Proc Natl Acad Sci U S A. 2013;110:11523–8. doi: 10.1073/pnas.1303064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thliveris AT, Halberg RB, Clipson L, Dove WF, Sullivan R, Washington MK, et al. Polyclonality of familial murine adenomas: analyses of mouse chimeras with low tumor multiplicity suggest short-range interactions. Proc Natl Acad Sci U S A. 2005;102:6960–5. doi: 10.1073/pnas.0502662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thirlwell C, Will OC, Domingo E, Graham TA, McDonald SA, Oukrif D, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–54. 54 e1–7. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–34. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon BS, Jeong WJ, Park J, Kim TI, Min do S, Choi KY. Role of oncogenic K-Ras in cancer stem cell activation by aberrant Wnt/beta-catenin signaling. J Natl Cancer Inst. 2014;106:djt373. doi: 10.1093/jnci/djt373. [DOI] [PubMed] [Google Scholar]
- 21.Snippert HJ, Schepers AG, van Es JH, Simons BD, Clevers H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO reports. 2014;15:62–9. doi: 10.1002/embr.201337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habano W, Sugai T, Nakamura S, Yoshida T. A novel method for gene analysis of colorectal carcinomas using a crypt isolation technique. Lab Invest. 1996;74:933–40. [PubMed] [Google Scholar]
- 23.Cheng H, Bjerknes M, Amar J. Methods for the determination of epithelial cell kinetic parameters of human colonic epithelium isolated from surgical and biopsy specimens. Gastroenterology. 1984;86:78–85. [PubMed] [Google Scholar]
- 24.Azuara D, Ginesta MM, Gausachs M, Rodriguez-Moranta F, Fabregat J, Busquets J, et al. Nanofluidic digital PCR for KRAS mutation detection and quantification in gastrointestinal cancer. Clin Chem. 2012;58:1332–41. doi: 10.1373/clinchem.2012.186577. [DOI] [PubMed] [Google Scholar]
- 25.Thliveris AT, Clipson L, White A, Waggoner J, Plesh L, Skinner BL, et al. Clonal structure of carcinogen-induced intestinal tumors in mice. Cancer Prev Res (Phila) 2011;4:916–23. doi: 10.1158/1940-6207.CAPR-11-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, et al. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: a new facet to Knudson’s ‘two-hit’ hypothesis. Nat Med. 1999;5:1071–5. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 27.Lamlum H, Papadopoulou A, Ilyas M, Rowan A, Gillet C, Hanby A, et al. APC mutations are sufficient for the growth of early colorectal adenomas. Proc Natl Acad Sci U S A. 2000;97:2225–8. doi: 10.1073/pnas.040564697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segditsas S, Rowan AJ, Howarth K, Jones A, Leedham S, Wright NA, et al. APC and the three-hit hypothesis. Oncogene. 2009;28:146–55. doi: 10.1038/onc.2008.361. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree M, Sieber OM, Lipton L, Hodgson SV, Lamlum H, Thomas HJ, et al. Refining the relation between ‘first hits’ and ‘second hits’ at the APC locus: the ‘loose fit’ model and evidence for differences in somatic mutation spectra among patients. Oncogene. 2003;22:4257–65. doi: 10.1038/sj.onc.1206471. [DOI] [PubMed] [Google Scholar]
- 30.Leedham SJ, Wright NA. Expansion of a mutated clone: from stem cell to tumour. J Clin Pathol. 2008;61:164–71. doi: 10.1136/jcp.2006.044610. [DOI] [PubMed] [Google Scholar]
- 31.Zhu D, Keohavong P, Finkelstein SD, Swalsky P, Bakker A, Weissfeld J, et al. K-ras gene mutations in normal colorectal tissues from K-ras mutation-positive colorectal cancer patients. Cancer Res. 1997;57:2485–92. [PubMed] [Google Scholar]
- 32.Sakurazawa N, Tanaka N, Onda M, Esumi H. Instability of X chromosome methylation in aberrant crypt foci of the human colon. Cancer Res. 2000;60:3165–9. [PubMed] [Google Scholar]
- 33.Leedham SJ, Graham TA, Oukrif D, McDonald SA, Rodriguez-Justo M, Harrison RF, et al. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology. 2009;136:542–50. e6. doi: 10.1053/j.gastro.2008.10.086. [DOI] [PubMed] [Google Scholar]
- 34.Humphries A, Cereser B, Gay LJ, Miller DS, Das B, Gutteridge A, et al. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proc Natl Acad Sci U S A. 2013;110:E2490–9. doi: 10.1073/pnas.1220353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giaretti W, Rapallo A, Sciutto A, Macciocu B, Geido E, Hermsen MA, et al. Intratumor heterogeneity of k-ras and p53 mutations among human colorectal adenomas containing early cancer. Analytical cellular pathology : the journal of the European Society for Analytical Cellular Pathology. 2000;21:49–57. doi: 10.1155/2000/747524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giaretti W, Monaco R, Pujic N, Rapallo A, Nigro S, Geido E. Intratumor heterogeneity of K-ras2 mutations in colorectal adenocarcinomas: association with degree of DNA aneuploidy. Am J Pathol. 1996;149:237–45. [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KM, Shibata D. Methylation reveals a niche: stem cell succession in human colon crypts. Oncogene. 2002;21:5441–9. doi: 10.1038/sj.onc.1205604. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pretlow TP, Pretlow TG. Mutant KRAS in aberrant crypt foci (ACF): initiation of colorectal cancer? Biochim Biophys Acta. 2005;1756:83–96. doi: 10.1016/j.bbcan.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–6. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.