Abstract
Ferredoxin-NADP+ reductase (FNR) is an FAD-containing enzyme best known for catalyzing the transfer of electrons from ferredoxin (Fd) to NADP+ to make NADPH during photosynthesis. It is also the prototype for a broad enzyme superfamily, including the NADPH oxidases (NOXs) that all catalyze similar FAD-enabled electron transfers between NAD(P)H and one-electron carriers. Here we define further mechanistic details of the NAD(P)H ⇌ FAD hydride-transfer step of the reaction based on spectroscopic studies and high resolution (~1.5 Å) crystallographic views of the nicotinamide-flavin interaction in crystals of corn root FNR Tyr316Ser and Tyr316Ala variants soaked with either nicotinamide, NADP+, or NADPH. The spectra obtained from FNR crystal complexes match those seen in solution and the complexes reveal active site packing interactions and patterns of covalent distortion of the FAD that imply significant active site compression that would favor catalysis. Furthermore, anisotropic B-factors show that the mobility of the C4 atom of the nicotinamide in the FNR:NADP+ complex has a directionality matching that expected for boat-like excursions of the nicotinamide ring thought to enhance hydride transfer. Arguments are made for the relevance of this binding mode to catalysis, and specific consideration is given to how the results extrapolate to provide insight to structure-function relations for the membrane-bound NOX enzymes for which little structural information has been available.
Keywords: flavoenzyme, hydride transfer, enzyme mechanism, protein crystallography, NADPH oxidase
Graphical Abstract
Ferredoxin-NADP+ reductases (FNRs) catalyze hydride transfers between NADP(H) and ferredoxin. Combined spectroscopic studies and high resolution crystal structures of corn root FNR with a C-terminal aromatic placeholder residue mutated implicate active site compression – as evidenced by mobility changes, flavin distortion, and short interaction distances – as a key factor promoting hydride transfer in FNRs and the broader FNR superfamily.

INTRODUCTION
Plant ferredoxin NADP+ oxidoreductases (FNR) are flavoenzymes that catalyze the reversible electron transfer between NADP(H) and the iron-sulfur protein ferredoxin (Fd). In photosynthesis, plastidic FNRs serve to catalyze electron transfer from Fd to NADP+ via the FAD prosthetic group, and in non-photosynthetic tissues distinct FNRs use NADPH as an electron source for the reverse reaction [1–3]. Plant-type FNRs – as seen in three-dimensional structures of FNRs from cyanobacteria [4], spinach (leaf [5]), pea (leaf [6]), paprika (leaf [7]), and corn (leaf [8] and root [9]) – are highly similar and contain two distinct domains: one for binding FAD and one for binding NADP+ [10]. Importantly, this two-domain FNR-fold is also the prototype for an enzyme superfamily that uses a bound FAD or FMN to transfer redox equivalents between the hydride carrying NAD(P)+ cofactors and diverse one-electron carriers [10–12]. Included among the FNR superfamily are the NADPH oxidases (NOXs), a biomedically important group of membrane bound enzymes that produce superoxide or hydrogen peroxide as part of many signaling pathways as well as during the oxidative burst of macrophages that is a key part of our immune defenses (recently reviewed in [13–16]).
Structures of plant type FNRs and other superfamily members, such as nitric oxide synthase [17], cytochrome P450 reductase [18, 19], and phthalate dioxygenase reductase [20] showed that NADP(H) appears to bind non-productively to the wild-type enzymes because an aromatic side chain (a C-terminal Tyr residue in the case of plant type FNRs) sits in the site the nicotinamide ring must occupy for hydride transfer (Figure 1A). Deng et al. [6] resolved this mystery by showing for pea leaf FNR at 1.8 Å resolution that a mutant missing the aromatic placeholder residue (Tyr308Ser or Y308S) could bind NADP(H) tightly and in an apparently productive manner with the nicotinamide ring C4 atom adjacent to the FAD N5 atom and with a geometry reasonable for hydride transfer. The Y308S mutant bound its cofactor so tightly that NADP+ co-purified with the enzyme. This mutant also showed an unexpected 500-fold change in cofactor specificity as the stronger binding of NAD(H) increased its steady-state turnover [21], but the stronger binding of NADP(H), decreased its steady state turnover, not because of impaired hydride transfer but because the NADP binding was so tight that koff became rate-limiting [4, 21].
Figure 1. Aromatic placeholder and nicotinamide binding in three FNR superfamily members.
(A) Stacking of aromatic placeholder side chain onto the flavin in wild-type forms of spinach FNR (PDB 1FNC; Tyr314 in forest green, FAD in orange), cytochrome P450 reductase (PDB 1AMO; Trp677 in green, FAD in dandelion), and NO synthase (PDB 1F20; Phe1395 in pale green, FAD in sand). (B) Mode of nicotinamide binding in the NADP+ complexes of Y303S Anabaena FNR (PDB 2BSA; NADP+ in blue), W677X cytochrome P450 reductase (PDB 1JA0; NADP+ in cerulean), and Y303S pea FNR (1QFY; NADP+ in seafoam); FAD colors as in panel A.
With the support of spectroscopic studies, it was concluded that the wild-type enzyme binds NADP(H) in a bipartite fashion in which the 2′-P-AMP binds strongly to anchor the cofactor to the enzyme and the thermodynamics of nicotinamide displacing the aromatic placeholder is such that the nicotinamide ring remains largely in a solvent exposed conformation (“nicotinamide-out”) and is in rapid equilibrium with a smaller population of molecules in which the aromatic placeholder swings out and the nicotinamide ring takes its place (“nicotinamide-in” conformation) to allow for hydride transfer. The Tyr to Ser mutant changes the binding thermodynamics so that the nicotinamide is ~100% “in”. Since that report, studies of aromatic blocker variants of other superfamily members have shown a consistent geometry for the “nicotinamide-in” nicotinamide-flavin interaction [4, 19] (Figure 1B) as well as changes from 50 to 1000-fold in NADP+ vs NAD+ specificity [4, 22–25].
Although there is considerable structural information about FNRs and FNR-like modules in related enzymes, the mechanism of hydride transfer is still not fully understood. For instance, the previous NADP(H) complexes of the pea FNR mutants gave indications that the C4 in the nicotinamide ring of NADP+ was mobile in a way that could favor hydride transfer [6], but higher resolution data are needed to better define the details of this mobility. Also, it has been proposed that the geometry of the complex seen in the structures that have the aromatic placeholder mutated are artifacts that are not relevant to catalysis [26–28]. Here, we obtain further insight into the hydride-transfer step in FNR-like enzymes by combining spectroscopic studies and a series of higher resolution structures of FNR crystals soaked with nicotinamide, NADP+, and NADPH. To accomplish this, we chose corn root FNR, for which crystals of the wild-type enzyme diffract to near 1 Å resolution [29]. We also argue for the general relevance of these insights for broader members of the FNR superfamily, including the NOX enzymes, which is further supported by the very recent structures of the core catalytic subunit domains of NOX5, published while this work was under review [30].
RESULTS AND DISCUSSION
Strategy
An inspection of the molecular packing in the well-diffracting crystals of corn root FNR that we reported earlier [9, 29] revealed that the NADP(H) binding site was not involved in crystal packing interactions. We hypothesized that we could obtain high resolution structures of NADP(H) complexes of this enzyme by making mutants of Tyr316, the aromatic blocking residue. Not knowing which mutant would be more informative, we mutated Tyr316 to Ala (Y316A), Ser (Y316S), and Phe (Y316F). Crystal structures of NADP+ soaks of Y316F FNR showed no nicotinamide binding, so this variant was not subjected to further study. For Y316S and Y316A, however, we present extensive characterization of the in-solution spectroscopic and catalytic properties, as well as crystal structures and in some cases the in-crystal spectroscopic properties of their complexes with NADP(H).
Solution Properties of Mutants
Wild-type and mutant enzyme forms were produced in Escherichia coli and purified essentially as reported elsewhere [9]. The two protein variants were expressed at slightly lower levels than that the wild-type enzyme, and their purification required additional hydrophobic interaction chromatography on butyl Sepharose. This resulted in a substantial drop in overall yields, which in the case of Y316A was low enough to preclude its full functional characterization. As shown in Figure 2A, the two replacements induced very similar perturbations in the visible absorption spectrum of the flavoprotein, strongly reminiscent of those observed in the equivalent Tyr to Ser replacement in both pea and Anabaena FNRs [9, 31]. Phenol – a mimic of the Tyr side-chain – was found to induce a spectral change in Y316S that qualitatively matched the difference between the absorption spectra of the wild-type and the variant protein (Figure 2B), implying that it can stack against the isoalloxazine ring in the active-site pocket. However, the affinity of Y316S FNR for phenol was too low to allow the accurate determination of the Kd of the complex.
Figure 2. Effect of the replacements of the Tyr316 residue on the visible absorption spectrum of FNR and rescue of the wild-type spectral features by phenol binding to the Y316S variant.
All spectra were determined in 50 mM Tris-HCl, pH 7.4 at 25 °C. (A) Extinction coefficients in the visible region of Y316S (solid line) and Y316A (dotted line) FNRs in comparison to that of wild-type enzyme (thin line). (B) Perturbation of the visible absorption spectrum of the Y316S variant (ca. 9 μM) induced by the presence of 150 mM phenol. The difference spectrum was rescaled (solid line) to match that expected for a theoretical enzyme concentration of 1 mM in order to be compared to the difference between the extinction coefficients of wild-type and Y316S FNR forms (dashed lines).
Titrations with NADP(H)
Anaerobic titration of oxidized Y316S FNR with NADP+ induced perturbations in the visible absorption spectrum of the FAD prosthetic group (Figure 3) very similar to those observed for the Y308S mutant of pea FNR [6]. The Kd of the complex was estimated to be ≤0.02 μM, well below that of the wild-type complex (0.3 μM) [9]. Whereas no interaction between NAD+ and wild-type FNR was detectable spectrophotometrically, NAD+ induced in Y316S FNR a spectral shift virtually identical to that produced by NADP+, displaying a Kd of 145 ± 5 μM (Figure 3).
Figure 3. Interaction of Y316S FNR with nicotinamide-containing ligands.
(A) Progress of the titrations of 10 μM Y316S FNR with NADP+ (hollow circles), NAD+ (filled circles), nicotinamide (hollow squares), and methyl-nicotinamide (filled squared). All titrations were performed in Tris-HCl, pH 7.7, at 15 °C. Data points and fitting curves were corrected to account for protein dilution. The logarithmic concentration scale was chosen to allow comparison of ligands displaying huge differences in affinity. (B) Computed difference spectra in the visible region of protein-ligand complexes. The difference between extinction coefficients of complexed and uncomplexed protein forms is shown for NADP+ (solid line), NAD+ (dashed line), nicotinamide (dash-dotted line), and methyl-nicotinamide (dotted line) are shown. (C) Overlay of Y316Anic (slate) and Y316Snic (hot pink) with nicotinamide bound, and as a reference, bound NADP+ (ghosted grey). Hydrogen bonds to bound nicotinamide are shown (slate and hot pink dashed lines in the respective structures).
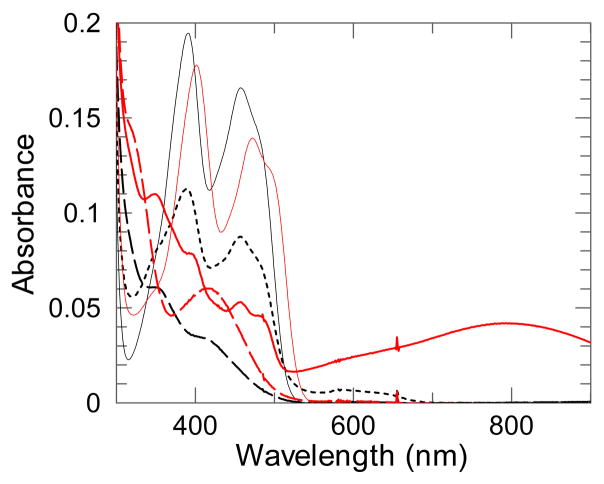
Next, using an EDTA-deazariboflavin system, stepwise anaerobic photoreductions of the FAD group of wild-type and mutant FNR forms were done in the absence and in the presence of roughly equimolar amounts NADP+ or NAD+ (Figure 4). These showed that the Y316S replacement decreased the amount of FAD semiquinone accumulated during the process and greatly increased the intensity of the broad long wavelength (~800 nm) absorption band attributable to charge-transfer electronic transitions of the interacting oxidized nicotinamide and 2-electron reduced flavin [9]. Upon complete reduction of the system, the charge-transfer band disappeared implying that the limited amount of NADP+ present became reduced and no charge transfer interaction occurs between the reduced flavin and NADPH. Furthermore, the presence of NADP+ favored the protonation of the dihydroquinone form of the prosthetic group of Y316S FNR, as indicated by the shift from 360 to 420 nm of the local maximum of the spectra of the fully reduced species (Figure 4). The results with NAD+ (not shown) are qualitatively similar to those obtained with NADP+, although a significantly less intense charge-transfer band was observed, consistent with the lower affinity of the enzyme for NAD+ as compared to NADP+.
Figure 4. Effect of the presence of NADP+ on photoreduction of Y316S FNR.
Significant spectra recorded during the stepwise photoreduction of the FAD prosthetic group in the absence (black lines) and in the presence of a slight excess of NADP+ (red lines). About 16 μM protein solutions in 10 mM HEPES, pH 7.0, were photoreduced at 15 °C in the presence of an EDTA-deazariboflavin system. Spectra corresponding to the maximal accumulation of semiquinone (dotted line) and charge-transfer complex (solid line) species are shown, and compared to those of the oxidized (thin lines) and fully reduced (dashed lines) respective enzyme mixtures.
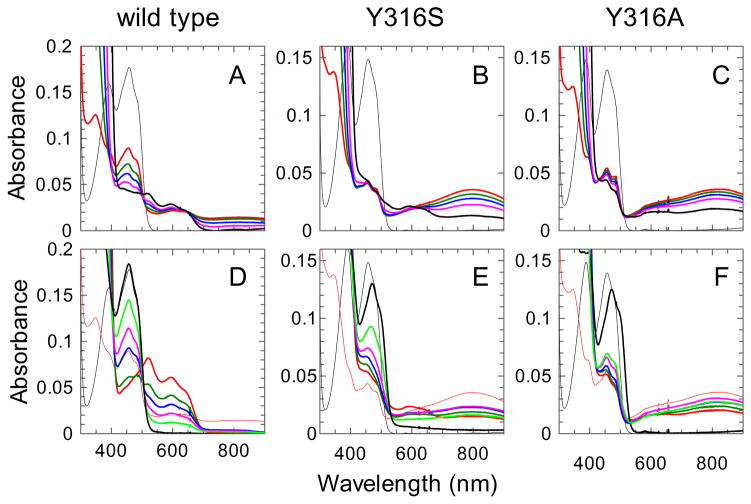
Anaerobic titrations of wild-type FNR and its variants with NADPH were performed to further analyze the interactions between FAD and NADP(H). In such titrations, at one equivalent of NADPH added, for the “nicotinamide-in” species an equilibrium should exist between two charge transfer complexes depending on whether the nicotinamide or the flavin is in the reduced state. Charge-transfer complex 1 (CTC-1) has the nicotinamide ring of NADPH interacting with oxidized flavin (i.e. FNRox:NADPH) and an absorption band with a maximum near 600 nm, and charge transfer complex-2 (CTC-2) has the nicotinamide ring of NADP+ interacting with reduced flavin (i.e. FNRred:NADP+) and a broad absorption band with a maximum at ca. 800 nm [32, 33]. The spectra at one equivalent of NADPH show that for wild-type the CTC-1 absorption band dominates (Figure 5A), whereas for both mutants the CTC-2 band dominates (Figure 5B,C), apparently accounting for ~90% of the population, based on the level of reduced flavin reduction as indicated by A460. For the two mutants, the CTC-2 band is maximal at roughly one equivalent NADPH and decreases to about half of that height as the concentration of NADPH is increased to an ~135-fold excess (Figure 5B,C). The further additions of NADPH presumably result in the progressive decrease in the amount of CTC-2 due to the partial displacement of NADP+ by NADPH. For the wild-type enzyme, the amount of CTC-1 similarly decreases as NADPH concentration increases, but it is progressively replaced by a species attributable to a complex between NADPH and FNR carrying FAD semiquinone. We do not understand how the semiquinone is formed under these conditions, but wonder if it may reflect some flavin disproportionation between the CTC-2 and CTC-1 complexes or is the result of a small amount of residual oxygen or other one-electron acceptor in the titrating NADPH solution. Prolonged incubation of NO synthase with NADPH was also seen to promote semiquinone buildup [34].
Figure 5. Spectra of wild-type FNR and its variants recorded during anaerobic titrations with NADPH and during NADPH-O2 turnover.
(Upper panels) titrations of wild-type (A), Y316S (B) and Y316A (C) FNRs showing the spectra of the free oxidized enzymes (thin black traces) and those recorded after the addition of ca. 1.2, 2.5, 5, 12, and 120-fold excess of NADPH (red, green, blue, magenta, and black bold traces, respectively). (Lower panels) Spectra recorded at different reaction times after air was admitted into the cuvettes containing the previously anaerobic solutions of the FNR forms in the presence of 120-fold excess NADPH. The spectra of the free oxidized FNR forms (thin black traces) and those of the respective CT complexes recorded in the presence of 1.2-fold NADPH excess (thin red traces) are reported for comparison. (D) Spectra recorded after 15 s, 25 min, 35 min, 40 min, 45 min, and 60 min (red, green, blue, magenta, lime, and black bold traces, respectively) turnover catalyzed by wild-type FNR. (E) Spectra recorded after 2.5 min, 6 min, 11 min, 19 min, 46 min, and 100 min (red, green, blue, magenta, lime, and black bold traces, respectively) turnover catalyzed by Y316S FNR. (F) Spectra recorded after 1 min, 4 min, 53 min, 2.5 h, 3.5 h, and 17 h (red, green, blue, magenta, lime, and black bold traces, respectively) turnover catalyzed by Y316A FNR.
Spectral properties during turnover as an NADPH oxidase
To monitor the spectral changes that occur during steady-state NADPH oxidase activity of FNR, air was admitted into the anaerobic cell after the titration was complete and NADPH was >100-fold in excess. In each case, the enzyme underwent turnover until the NADPH was all converted to NADP+. For wild-type FNR (Figure 5D), as has been seen before [9], there was substantial stabilization of the blue semiquinone form of FAD in the early parts of the reaction and CTC bands were hardly detectable. For Y316S FNR (Figure 5E), less FAD semiquinione built up and the reaction had two distinct phases. During the first phase, the CTC-2 concentration progressively increased until at ~15 min it reached ~65% of the maximal value observed during the anaerobic titration (thin red line), then it decreased to near zero. The final spectrum (at ~1.7 h) had the 456 nm band shifted as is characteristic of an oxidized enzyme in complex with NADP+ (Figure 5E). The behavior of Y316A FNR (Figure 5F) was qualitatively similar to that of the Y316S form, but with the reaction slowed by about 10-fold as the steady-state CTC-2 formation was reached after about 2.5 h (vs ~15 min) and the reaction was complete in 17 rather than 1.7 h. The Y316A variant also had still less stabilization of the FAD semiquinione, and its maximal CTC-2 accumulation during turnover was ~85% of the highest amount observed in the anaerobic titration (thin red line). The slower reaction and higher CTC-2 buildup are consistent with the koff for NADP+ being slower for the Y316A mutant. Also, the destabilization of the semiquinone seen in these variants mirrors what was seen for the corresponding mutants of Anabaena FNR [31] and NO synthase [22, 34].
Ferricyanide reductase activity
For the wild-type enzyme, ferricyanide is the most effective electron acceptor from reduced FNR, with kcat = 520 s−1 [9]. The Y316S and Y316A replacements impaired the steady-state diaphorase activity of FNR to a similar extent, and kcat and KmNADPH values determined for the Y316S variant were ca. 170 and 9-fold lower than those of the wild-type enzyme (Table 1) [9], respectively. Such large catalytic impairment parallels that reported for the Tyr to Ser variants of pea and Anabaena FNRs [21, 31], with the slower turnover shown to be limited by the rate of NADP+ (i.e. product) release. Consistent with this, we found that nicotinamide partially rescued the diaphorase activity of Y316S FNR in a concentration-dependent fashion, increasing its kcat to 280 s−1 at 800 mM nicotinamide (Table 1). As has been noted previously [2, 6], by competing with the corresponding moiety of NADP+ for stacking onto the flavin, nicotinamide will favor product dissociation and speed enzyme turnover if the koff of NADP+ is rate limiting.
Table 1.
Effect of the Y316S replacement on the kinetic parameters of the NADPH—K3Fe(CN)6 reductase reaction catalyzed by FNR.a
| FNR form | kcat s−1 | KmNADPH μM | kcat/KmNADPH s−1 μM−1 |
|---|---|---|---|
| Wild typeb | 520 ± 10 | 12 ± 1 | 43 ± 4 |
| Y316S | 3 ± 0.3 | 1.4 ± 0.3 | 2.1 ± 0.5 |
| Y316S, 0.8 M nicotinamide | 280 ± 18 | 45 ± 10 | 6 ± 1 |
Initial rate data were measured as moles of ferricyanide reduced per second.
Data on wild type FNR, taken from [9], are reported here for comparison.
Interactions with small-molecule ligands
The findings that added nicotinamide increased the catalytic activity of Y316S FNR, prompted us to quantify the affinities of nicotinamide and N-methyl-nicotinamide. As shown in Figure 3A, both were found able to interact with Y316S FNR, although only the former had an affinity high enough for its Kd value to be determined (3 ± 0.1 mM). The difference spectrum of the Y316S-nicotinamide complex was blue-shifted by ca. 7 nm with respect to that of the NAD(P)+ complexes, but the spectral change induced by N-methyl-nicotinamide was almost identical to that induced by NAD(P)+ (Figure 3B). This indicates that for Y316S FNR, N-methyl-nicotinamide but not nicotinamide is an excellent mimic of the redox-active moiety of the dinucleotides.
Reanalysis of Stopped-Flow Results Reported for Anabaena FNR
As noted in the introduction, in earlier work Lans et. al. [26] called into question the relevance of the NADP(H) complexes that form in FNR when the aromatic placeholder is mutated. The origin of this view came from stopped-flow results for the Tyr303Ser (Y303S) mutant of Anabaena variabilis FNR. Per their interpretation, these results showed that Y303S promoted the hydride transfer from NADPH to FNRox “slightly slower than the wild-type enzyme” but that “the reverse process is undetectable” (e.g. hydride transfer from FNRred to NADP+ does not occur). This interpretation was based on the observation that when NADP+ was mixed with Y303S FNRred the CTC-2 (~800 nm) charge transfer band that formed upon mixing did not substantially decrease over time (see Figure 3A,B of Lans et al. [26]). While the conclusion that no hydride transfer occurs is consistent with the unchanging CTC-2 band, there could be other explanations. We were skeptical of the inference both based on the thermodynamic principle that if the Y303S mutant catalyzed hydride transfer from NADPH to FNRox it must equally well catalyze the reverse hydride transfer from FNRred to NADP+, and based on our photoreduction studies (Figure 4 above) implying that Y316S-bound NADP+ is converted to NADPH. To clarify this question, we reanalyzed the Lans et al. [26] stopped-flow data for NADPH mixed with Y303S FNRox and for NADP+ mixed with Y303S FNRred.
Importantly, in the original work, when NADP+ is mixed with Y303S FNRred, even though the 800 nm band changes little over time, the ~460 nm peak systematically increases indicating that oxidation of FAD is occurring. Lans et al. attributed this to a “side effect” rather than enzymatic activity; however, when comparing these spectra with those for NADPH mixed with Y303S FNRox, it is striking that both reactions appear to reach the same endpoint, as would be expected if Y303S FNR were reaching the same equilibrium state independent of the direction of approach. Given this observation, we reanalyzed the reported stopped-flow results for both reactions using the changes in A460 as an indicator of FAD oxidation/reduction. This reanalysis yielded excellent fits of kobs of 164 ± 16 s−1 for NADP+ mixed with Y303S FNRred and 146 ± 21 s−1 for NADPH mixed with Y303S FNRox (Figure 6). These two kobs values should both equal the sum of the elementary forward and back reaction rate constants (kobs = k1+k−1) and are equal within the error of the analysis, leading us to conclude that the forward and back reactions both function as would be expected. We suggest the best number to use for kobs for the Anabaena Y303S FNR is 190 ± 15 s−1 which is the value originally reported by Lans et al. [26] for the forward reaction using the much more extensive original dataset.
Figure 6. Reanalysis of stopped-flow results reported for Anabaena FNR.
Data extrapolated from [26] were fit to a single-exponential decay. (Upper panels) Time-course of the approach to equilibrium upon mixing (A) wild-type FNRred and NADP+, and (B) wild-type FNRox and NADPH. Curve fitting yielded the kobs values of 235 ± 47 s−1 and 230 ± 40 s−1, respectively, equivalent to each other within the error of the analysis. Lans et al. [26] reported the rate constants for these reactions to be 285 s−1 and 270 s−1, respectively. (Lower panels) Time-course of the approach to equilibrium upon mixing (C) Y303S FNRred and NADP+, and (D) Y303S FNRox and NADPH. Curve fitting yielded the kobs values of 165 ± 16 s−1 and 145 ± 20 s−1, respectively, equivalent to each other within the error of the analysis. The raw extrapolated data are available at figshare.
This revised interpretation is consistent with all the data and with basic thermodynamic principles, and implies that the nicotinamide-flavin interaction consistently formed in the aromatic placeholder mutants is in fact productive and relevant for understanding catalysis.
Crystal structures of Y316S and Y316A NADP(H) complexes
Co-crystallization of both FNR variants with nicotinamide yielded crystals that grew readily and could be soaked with NADP+ or NADPH to obtain those complexes. Interestingly, the variants crystallized in two space groups, both the P3221 form seen for wild-type protein [9] and a new P3121 crystal form that had remarkably similar unit cell dimensions and related crystal packing. Here, we report a set of eight refined crystal structures of complexes that represent the Y316S and Y316A variants in complex with three ligands (NADP+, NADPH, and nicotinamide) with most structures refined at between 1.35 Å and 1.6 Å resolution (Table 2) and having very well defined active site electron density (Figure 7). As the corresponding complexes of Y316S and Y316A largely show equivalent features, we focus mainly on the highest resolution set of structures that provide unique information.
Table 2.
Data and refinement statistics for FNR variant structuresa
| Ligand soaked | wt | Y316S | Y316A | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Nicotinamide | Nicotinamide | NADP+ | NADP+ | NADPH | Nicotinamide | NADP+ | NADPH | |
| Space group | P3221 | P3221 | P3121 | P3221 | P3121 | P3221 | P3221 | P3221 | P3221 |
| Data Statistics | |||||||||
| Wavelength (Å) | 1.00 | 1.00 | 1.54 | 1.00 | 1.54 | 1.00 | 1.54 | 1.00 | 1.00 |
| Unit cell a, c axes (Å) | 59.1, 186.7 | 58.9, 184.8 | 58.9, 184.4 | 58.8, 187.5 | 58.9, 185.0 | 58.9, 187.3 | 59.4, 187.5 | 58.7, 186.7 | 59.2, 188.0 |
| Resolution (Å) | 50–1.05 (1.07–1.05) | 51–1.35 (1.42–1.35) | 12 – 1.90 (1.97–1.90) | 51–1.45 (1.53–1.45) | 40–1.80 (1.90–1.80) | 51–1.45 (1.53–1.45) | 26–1.95 (2.06–1.95) | 49–1.50 (1.55–1.50) | 49–1.60 (1.66–1.60) |
| Unique reflections | 177114 (8768) | 75008 (9295) | 29752 (2603) | 65831 (8739) | 35032(5039) | 66408 (8907) | 24769 (1325) | 44421 (1371) | 50821 (4336) |
| Multiplicity | 5.2 (2.9) | 5.8(2.6) | 8.2 (3.8) | 9.6(2.5) | 7.3(3.9) | 8.4(2.5) | 9.1(2.3) | 1.7 (1.1) | 4.0 (3.2) |
| Average I/σ | 57.0(3.4) | 6.2 (1.7) | 19.4 (6.9) | 14.7(3.0) | 6.5(2.2) | 12.7(2.7) | 34.3(9.6) | 10.4 (0.82) | 24.5 (6.0) |
| Rmeas (%) | 9.3 (50.6) b | 30.2 (46.9) | 4.0 (15.3) b | 9.5(29.0) | 33.5(53.2) | 13.0(39.5) | 4.4(6.4) | 5.4 (71.1) b | 2.8 (16.5) b |
| Completeness (%) | 99.8 (99.5) | 91.0 (78.4) | 97.6 (86.6) | 97.9 (91.7) | 99.9 (99.9) | 98.1 (92.1) | 85.8 (32.5) | 72.9 (23.0) | 98.6 (86.4) |
| Refinement Statistics | |||||||||
| Refinement method | Aniso | Aniso | TLS | Aniso | TLS | Aniso | TLS | TLS | TLS |
| Amino acid residues | 302 | 309 | 309 | 309 | 309 | 309 | 309 | 309 | 309 |
| Solvent atoms | 646 | 484 | 413 | 491 | 297 | 500 | 556 | 451 | 530 |
| Non-H atoms | 3252 | 3117 | 3035 | 3160 | 2923 | 3217 | 3165 | 2999 | 3155 |
| RMS bonds (Å) | 0.017 | 0.009 | 0.009 | 0.008 | 0.010 | 0.008 | 0.010 | 0.009 | 0.009 |
| RMS angles (˚) | 2.3 | 1.0 | 1.1 | 1.2 | 1.3 | 1.2 | 1.1 | 1.2 | 1.2 |
| <Bprotein> (Å2) | 21.7 | 14.6 | 28.9 | 20.9 | 34.5 | 21.3 | 17.8 | 26.7 | 24.0 |
| <BFAD> (Å2) | 15.1 | 10.4 | 18.3 | 16.6 | 22.0 | 16.7 | 13.1 | 21.1 | 18.5 |
| <BNADP(H)> (Å2) | – | 13.8 | 22.7 | 24.1 | 28.3 | 25.5 | 23.7 | 27.0 | 24.7 |
| Rwork (%) | 12.5 | 13.5 | 13.1 | 10.8 | 17.2 | 11.0 | 13.0 | 14.9 | 13.1 |
| Rfree(%) | 15.5 | 17.3 | 17.3 | 14.4 | 21.1 | 14.7 | 18.0 | 18.5 | 15.6 |
| PDB code | 3LO8 | 5VW4 | 5VW9 | 5VW3 | 5VW8 | 5VW2 | 5VW5 | 5VW6 | 5VW7 |
Numbers in parentheses are in the highest resolution shell
Rmerg reported in place of Rmeas
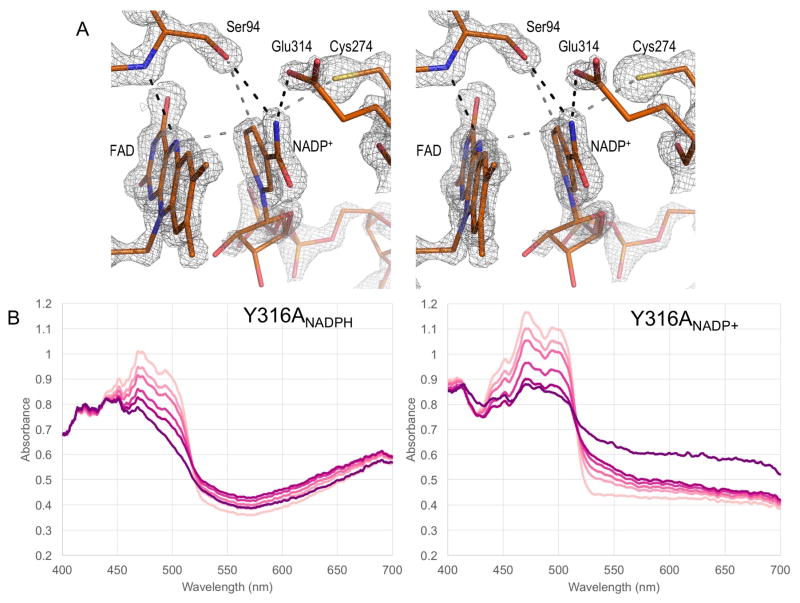
Figure 7. Active site environment of FNR.
(A) A stereo view of the active site environment of FNR represented by the structure of Y316SNADP+ with the 2Fo-Fc map shown at 3.0*ρrms. Key residues within the active site, Ser94, Cys274, and Glu314, which primarily interact with NADP(H), NADP(H), and FAD are shown along with hydrogen bonds (black dashed lines) and interaction distances (gray dashed lines). (B) Single crystal visible absorption spectra of Y316A FNR soaks with NADPH (right-hand panel) and NADP+ (left-hand panel) and their change during data collection. Spectra collected corresponding to oscillation images 0, 10, 20, 50, 100 and 360 go from light to dark shades of pink. These spectra qualitatively match those seen in solution in that the CTC-2 absorption band is seen for the crystals soaked in NADPH but not in those soaked in NADP+. The decrease in A460 over time indicates reduction of the flavin in the X-ray beam during data collection. Over time the CTC-2 band does not disappear in the NADPH soaked crystal and does not appear in the NADP+ soaked crystal so, we infer that at the cryo-temperatures of data collection, the flavin reduction is not leading to the same structural changes it would lead to in solution.
To check whether the complexes in the crystal represent those that form in solution, we obtained single-crystal absorption spectra [35] during data collection for a pair of Y316A crystals soaked with NADP+ and NADPH. These spectra qualitatively match those obtained in solution and indicate that even though the X-ray beam does cause reduction of the FAD, this does not undermine the relevance of the structures (Figure 7). Also, since Y316S and Y316A both predominantly form CTC-2 after an NADPH soak (e.g. Figure 5B,C), we have modeled the NADPH soak structures as an NADP+:FADH2 complex rather than NADPH:FAD.
In the following sections, we first provide a basic description of the complexes and then focus on three detailed aspects of complexes that provide evidence of compression in the active site for enhancing catalysis. In our structural comparisons, the unliganded wild-type structure we use is a 1.05 Å resolution structure refined much earlier (PDB entry 3LO8; released in 2010) and used in tests of new refinement strategies [36]. As the statistics for this structure have not yet been described in the literature, they are included in Table 2.
Overview of the NADP(H) complexes
Globally, all Y316S and Y316A complexes, independent of space group, are very similar to the wild-type structure with rms Cα deviations between 0.1 Å and 0.4 Å. The broad features of NADP(H) binding match those that have been elaborated elsewhere [4, 6, 10, 12], interacting mostly with the NADP-binding domain. Briefly, the 2′-phosphate anchor hydrogen bonds with the side chains of Ser237, Arg238, Lys247, and Tyr249, and the adenine is sandwiched between Tyr249 and Leu276. The 5′-phoshoryl group is largely exposed and the nicotinamide-side phosphoryl group hydrogen bonds with the Thr175 N and the Arg114 guanidinium group. The nicotinamide is surrounded by a triad of key side-chains, with Ser94 and Glu314 hydrogen bonding with the nicotinamide carboxamide nitrogen and Cys274 and Ser94 close to the C4 atom (Figure 7A). In the higher resolution structures of Y316S, an alternate conformation for the nicotinamide ribose is also observed. In both Y316SNADP+ and Y316SNADPH soaks in both space groups, the nicotinamide ribose is modeled as 60% 2′-endo and 40% 3′-endo. In each Y316A structure, the nicotinamide ribose adopts a single conformation, but in Y316ANADP+ it is 2′-endo and in Y316ANADPH it is 3′-endo. The reason for these conformational differences is not readily apparent but their presence suggests that they are the result of real but subtle differences in nicotinamide binding.
The complexes with free nicotinamide
Surprisingly, the binding mode seen in nicotinamide soaks differ for Y316A and Y316S (Figure 3C). In Y316Anic, the nicotinamide orientation and hydrogen bonding roughly match how the NADP(H) nicotinamide binds. However, in Y316Snic, the nicotinamide binds in a flipped orientation that is stabilized by a hydrogen bond between Ser316-Oγ and nicotinamide-N1 as well as hydrogen bonds of the nicotinamide hydroxyamide with Ser94-Oγ and Thr 175-Oγ and O atoms (Figure 3C). Since this binding mode involves Ser316 as a hydrogen-bond partner, we conclude it is an artifact of the Y316S mutation. Also, the existence of the nicotinamide N1 to Ser316-Oγ hydrogen bond in this complex means that neither NADP(H) nor even N1-methyl-nicotinamide could adopt it. This provides a satisfying explanation for why nicotinamide binding to Y316S yields different spectral changes than NAD(P)+, whereas methyl-nicotinamide binds less tightly, but with spectral changes that match those of NAD(P)+ (Figure 3B). It also serves as a reminder of the need to be cautious in inferring the binding mode of a molecule of interest based on that of an analog.
The nicotinamide-flavin interactions in the NADP+/NADPH complexes
While the overall structures of the FNR variants are virtually superimposable, subtle differences exist in the details of nicotinamide binding seen in the NADP+ and NADPH soaks, and having the structures for the two mutants allows us to gain confidence about which variations are reliably due to the difference between the active site redox state rather than the specific mutation or to experimental uncertainty. In terms of the active site, the difference between the structures is a single H-atom: the NADP+ soaks with oxidized flavin and nicotinamide have one H-atom on the nicotinamide C4 and no H-atom on the flavin N5, and the NADPH soaks to produce CTC-2 have one H-atom on the nicotinamide C4 as well as one H-atom on the flavin N5. (The CTC-1 complex would have the same number of H-atoms but distributed with two on the nicotinamide C4-atom and none on the flavin N5-atom.) As described in the following paragraphs, the high resolution structures provide evidence that the single additional H-atom increases active site crowding and a directional compression that we propose is a key promotor of catalysis.
A first line of evidence is provided by the temperature factors of the C4-atom of NADP+ compared to NADPH. In our earlier work on pea FNR [6], a higher conformational freedom of the NADP+ C4-atom was indicated by weaker electron density (and higher B-factors) for this atom in NADP+ compared to NADPH; but at 1.8 Å resolution, this was not conclusive. Our higher resolution analyses here similarly show the additional mobility of the C4 atom of NADP+ but also provide refined anisotropic B-factors that give a more complete description of dynamics than do isotropic B-factors. The anisotropic B-factors very clearly show that the increased movement for the C4 atom of NADP+ is perpendicular to the plane of the ring and towards N5 of the FAD (Figure 8A). This type of motion exactly corresponds to motions that would allow the nicotinamide ring to forms a boat-like conformation that calculations show will promote efficient hydride transfer [37].
Figure 8. Active site compression in corn root FNR as seen by mobility, covalent distortion, and interaction distances.
(A) Mobility of atoms in the active site of Y316S FNR. Atoms in the active sites of Y316SNADP+ (orange) (i) and overlaid with Y316SNADPH (semi-transparent green) (ii) shown with ellipsoids, denoting the use of anisotropic B-factors. The nitrogen and oxygen atoms are shown in blue and red, respectively, in the complexes. The mobility of the C4 of NADP+ is in the direction of the N5 of the FAD, forming a boat-like conformation which is favored for hydride transfer. (B) Active site compression in corn root FNR as seen by covalent distortion. The angle of distortion of the FAD moiety is shown in: (i) FAD in wild-type corn root FNR (mauve), (ii) FAD in Y316SNADP+ (orange) and the corresponding angles from Y316SNADP+ (P3121, olive) and Y316ANADP+ (salmon), (iii) FAD in Y316SNADPH (green) and the corresponding angle from Y316ANADPH (violet). (C) Active site interactions in FNR. Structural overlay of Y316SNADP+ (orange) and Y316SNADPH (green) with relevant average distances shown in corresponding colors. The average distances from Y316SNADP+ (P3121, olive), Y316ANADP+ (salmon), and Y316ANADPH (violet) are also shown. The distances are averages from ten independent refinements of each complex (see Materials and Methods), and all standard deviations were < 0.02 Å. The C4H and N5H distance in Y316SNADPH is 3.0 Å (not labeled). If modeled as FADH2 instead of FAD, the theoretical distance between C4H and N5H in Y316SNADP+ would be 2.5 Å. Nitrogen, oxygen, and sulfur atoms colored blue, red, and yellow, respectively.
A second observation of crowding derives from a visible distortion of the FAD isoalloxazine. In wild-type FNR, the isoalloxazine group appears to be pushed slightly by the Tyr316 side chain so that it is not coplanar with the N10-C1 ribose bond, but is about 3.5° non-planar (Figure 8B). In all FNR variant structures soaked with NADP+, this non-planarity increases to ~10°, and in the structures soaked with NADPH it increases further to ~14° (Figure 8B). These deviations from planarity imply that the nicotinamide displacing the Tyr316 side chain leads to a more tightly packed active site and that the additional H-atom in the active site present in the NADPH soaks increases crowding even more, with the isoallozaxine bending to relieve apparent pressure. It should be noted that the highly crowded complex seen in the NADPH soak is the one that is present during normal catalysis.
A third observation that completes the view of the tightly packed active site of FNR are the interaction distances of the nicotinamide C4 and H4 atoms that are involved in hydride transfer (Figure 8C). In all structures, these atoms are sandwiched between the flavin N5 atom in front and the Cys274-Sγ sulfhydryl behind, where it can act as a backstop. In comparing the Y316SNADP+ complex to the Y316SNADPH complex which has an additional H-atom on the flavin N5-atom, the C4…N5 distance increases by ~0.3 Å while the C4H… Sγ distance decreases by ~0.1 Å (Figure 8C). Taking into account the Y316A complexes (Figure 8C), the consistent increase of the C4…N5 distance in the NADPH complex leads us to conclude that Cys274 acts as a firm backstop while the isolloxazine bends away from the nicotinamide by an additional ~4° in order to increase the distance between the flavin and nicotinamide and relieve the pressure due to the presence of the extra H-atom.
These observations lead us to conclude that the active site is experiencing compression, distorting the FAD and pushing the substrates together to enhance catalysis by aligning the substrates in an optimal position and, as has been noted by others, potentially promoting a quantum tunneling mechanism of hydride transfer [26].
Insights into factors promoting hydride transfer in FNRs
Taken along with our reanalysis of previous stopped-flow kinetics work that called the relevance of aromatic placeholder FNR mutants into question, these in-solution spectroscopy, single-crystal spectroscopy, and high-resolution structures provide insights into hydride transfer in FNRs using variants which have near wild-type hydride-transfer kinetics, but importantly, allow us to capture the productive binding mode of NADP(H) in the active site. Such productive complexes have not been possible to capture in any wild-type member of the FNR superfamily with an aromatic placeholder residue (e.g. [5, 17–20]) or that instead have a C-terminal peptide extension as an alternate way to block nicotinamide binding [3, 38]. They have also been challenging to obtain for superfamily enzymes that have a shifted domain-domain interaction [39, 40] instead of an aromatic blocking residue, but for one of these enzymes, cytochrome b5 reductase, anaerobic co-crystallization has yielded a productive, wild type complex with NADH in which the nicotinamide-flavin interaction geometry is much like that seen here [41].
The high-resolution structures reported here illustrate the role of specific anisotropic motions as well as active site compression as catalytic strategies that promote hydride transfer. Key evidences are the anisotropic motion of the C4 atom of NADP+, that indicates boat-like perturbations in conformation are a preferred mode of vibrational freedom, as well as a strong decrease in the amplitude of that motion in the NADPH complex. Additionally, there is an incrementally increasing deviation from planarity at the flavin N10 atom in wild type, Y316SNADP+, and Y316SNADPH indicating increased crowding and pressure associated with oxidized nicotinamide binding and even more so with reduced nicotinamide binding that is being relieved to some extent by bending of the flavin. Interestingly, a very similar flavin non-planarity was seen in the cytochrome b5 reductase-NADH complex (see Figure 3a of [41]). Finally, comparison of Y316SNADP+ and Y316SNADPH reveal close sub van der Waals contact distances consistent with a tightly packed active site. The central role of Cys274 in these interactions provides a rationale for its conservation across the whole superfamily. While steric compression has occasionally been noted as a key factor promoting catalysis for other enzymes [42–45], there are a limited number of studies which provide direct structural evidence for this [46–50], as this typically requires atomic or near-atomic resolution crystal structures. Also, that the Y316S and Y316A variants of corn root FNR, like similar mutants of other FNR superfamily members [22, 31, 34], show much less stabilization of the FAD seminquinone, is consistent with an earlier proposal [31] that the aromatic side chain is not solely a passive placeholder for the nicotinamide group, but, through its stacking interaction with the flavin, is an active agent that stabilizes the semiquinone form of the flavin to enhance the one-electron transfers required for these dehydrogenase-electron transferases.
Extrapolation of the results to the FNR superfamily members such as NOX enzymes
Clarifying the relevance of the NADP+ binding mode seen in the Tyr316 mutants is not just of interest for understanding catalysis for the whole superfamily, but can guide further studies of these enzymes as well as the generation of superfamily member variants that can be valuable tools. For instance, for the NOX enzymes that play crucial roles in the production of superoxide and hydrogen peroxide for diverse biological processes, the FNR-like module is at the C-terminus of a membrane-bound flavocytochrome catalytic subunit that has been relatively difficult to study [14]. Although a NOX structure has long eluded structural biologists, a crystal structure of the FNR-like module of NOX5 from Cylindrospermum stagnale (referred to as the NADPH-dehydrogenase domain) was solved while this work was under review [30]. Sequence alignments of FNR with human NOXs aided by the C. stagnale NOX5 structure (Figure 9A) show that the NOX isozymes conserve many NADP+ binding residues including the nicotinamide-interacting residues equivalent to corn root FNR Ser94, Cys274, Glu314 and Tyr316 (Phe693 in C. stagnale NOX5).
Figure 9. Sequence alignment of corn root FNR with NADPH oxidases.
(A) Sequence alignment of segments of corn root FNR with human NOX1, NOX2, NOX3, NOX4, and NOX5 and C. stagnale NOX5. Select FNR functional residues and similar aligned residues (in bold) have their function denoted as: aromatic placeholder (a), other catalytic center (c), 2′-phosphate (p) or other NADP interactions (n). Residue numbers are given for the structurally known corn root FNR and C. stagnale NOX5. (B) Overlay of the C. stagnale NOX5 dehydrogenase domain (PDB 5O0X; pink with bound FAD and the natural C-terminal Phe693 and C-terminal extenstion Trp side chains in purple) and the Y316SNADP+ structure (orange with bound FAD and NADPH in yellow). The anticipated fifth β-strand of the NADP+-binding domain of NOX5 which is perturbed and allows the Trp residue to stack against the isoalloxazine ring is highlighted (purple trace) and an arrow indicates the direction we expect it to move in the full-length NOX5 context so that it will align with the rest of the β-sheet as does the fifth β-strand in FNR. The deposited structure of the NADPH binding domain of NOX2 gp91(phox), determined through structural genomics efforts (PDB 3A1F) but not yet described in the literature, also aligns well in a structural overlay.
The NOX5 FNR-like module itself was unstable and crystals were only obtained for a construct with a “hyperstabilizing” C-terminal extension that included a Trp two residues after Phe693. Notably, in this structure, the expected natural C-terminal aromatic placeholder (Phe693) does not stack against the isoalloxazine, and, because the anticipated fifth β-strand of the NADP+-binding domain is perturbed and moved away from the protein core, the Trp residue is able to stack against the isoalloxazine ring instead (Figure 9B). However, if the NOX5 chain followed the path of the β-strand seen in other FNR superfamily members, Phe693 could stack against the isoalloxazine and we propose that this is what occurs in the native enzyme. Magnani et al [30] noted that the isolated dehydrogrenase domain is deregulated and predicted that the normal role of the strictly conserved C-terminal aromatic residue would “emerge only in the context of a full-length protein.” We agree and further suggest that the instability of the isolated dehydrogenase domain and the loose association of the C-terminal segment (including Phe693) is related to the missing cytochrome transmembrane domain and associated lipid bilayer that interact with this surface of the dehydrogenase domain in a full-length complex (see Fig 5A of [30]).
Consistent with the weak association observed for Phe693 with the protein core is in the isolated dehydrogenase domain, a Phe693Ser mutant had higher than wild-type activity and the stabilizing C-terminal extension mutant (adding the Trp695) dropped activity by 5-fold [30]. Nevertheless, we predict that in the context of a full-length membrane-bound NOX, Phe693 will act as an aromatic placeholder residue and that Ser or Ala mutants will bind both NADPH and NADH more tightly. Such mutants could provide a useful handle for purification (through the tight binding of NADP+) or be a useful tool for probing/controlling the physiological roles of NOXs. A solely NADH responsive version could potentially be made through additional mutations such as the equivalent of FNR Ser237 to Asp (Figure 9), as this position has been shown to discriminate against binding the 2′-phosphoryl group in multiple superfamily members [51–54]. Interestingly, a full-length human NOX2 C-terminal Phe mutant (F570A) was characterized in 1998 and found to retain ~50% of the wild-type activity [55] rather than dropping 300–800 fold in activity as had been seen for the equivalent pea FNR mutant, and so it was not studied further. However, this study predated the knowledge that the equivalent FNR mutant lost activity due to the slow dissociation of NADP+ and also gained activity with NADH [9, 21], so those qualities of the variant were never characterized. In retrospect, given that the NOX2 Kd for NADP+ is ~40 μM (over 10-fold higher than is typical for FNRs) and the turnover number is slower, it would not be surprising if the expected enhanced binding of NADP+ in the F570A variant would not be enough to make its dissociation highly rate limiting. Indeed, for NO synthase the equivalent mutation only showed about a 3-fold decrease in steady-state turnover, yet still showed a 50-fold change in specificity [22, 34].
MATERIALS AND METHODS
Production of Recombinant Corn Root FNR and its Variants
Plasmids for the bacterial expression of the Y316A and Y316S proteins were generated from the pETrFNR2 [9]using the QuikChange II Site-Directed Mutagenesis kit (Agilent) and two appropriate oligonucleotide couples following the manufacturer directions. The wild-type and variant proteins were produced in E. coli HMS174(DE3) by induction with 0.1 mM IPTG for 4 h at 30 °C, and purified through a procedure similar to that previously reported for the wild-type protein [9] using an ÄKTA FPLC (GE Healthcare) apparatus. Briefly, the crude cell lysate was brought to 40% NH4SO4 and centrifuged. The supernatant was applied to a Sepharose 4B column (GE Healthcare), eluted with appropriate buffer and precipitated with 75% (NH4)2SO4. In the case of the protein variants, the pellet was resuspended in 40% (NH4)2SO4 and chromatographed on a butyl Sepharose column (GE Healthcare) with a descending salt concentration gradient. The omission of this step prevented the adsorption of the FNR forms on the next cation exchange resin. After desalting, the sample was loaded on an SP-Sepharose HP column (GE Healthcare) and eluted through a NaCl concentration gradient. Notably, the visible spectra of both mutant forms, but not the wild-type enzyme, underwent a blue shift during the ion exchange step, suggesting release of some unidentified bound ligand at that stage. The resulting proteins were homogenous as judged by SDS-PAGE. The FNR forms were concentrated to ~25 mg/mL and stored at −20 °C in 10 mM HEPES, pH 7.0.
Spectral Analyses and Ligand Binding
All spectrophotometric measurements and steady-state enzyme kinetics were performed using an 8453 diode-array spectrophotometer (Agilent). Titration with NADP+, NAD+, phenol, nicotinamide and N-methyl-nicotinamide of the enzyme forms in their oxidized state were performed in 10 mM Tris-HCl, pH 7.7, at 15 °C.
Photoreductions, Anaerobic Titrations and Activity Assays
Flavin photoreduction experiments were carried out both in the absence and in the presence of NADP+ or NAD+ in anaerobic cuvettes in 10 mM HEPES-NaOH, pH 7.0, at 15 °C, according to the procedure described elsewhere [9].
Anaerobic titrations with NADPH of 14–18 μM wild-type, Y316A or Y316S FNR variants were performed on 1.2 mL samples in 50 mM HEPES-NaOH buffer, pH 7.0, at 15 °C, in a sealed cuvette. After recording the spectrum of the oxidized enzyme, five successive additions of anaerobic NADPH were made leading to ligand concentrations of 29 μM, 39 μM, 76 μM, 182 μM, and 1.9 mM for wild-type titrations, 23 μM, 45 μM, 90 μM, 210 μM, and 2.2 mM for FNR Y316A titrations, and 24 μM, 48 μM, 95 μM, 225 μM, and 2.3 mM for FNR Y316S titrations, respectively. In each case, the volume after the final addition of NADPH was 1.4 mL. For each FNR form, the cuvette was then opened to air, and the spectrum of the mixture (kept at 15 °C) was monitored at over time until all the NADPH was oxidized. For technical reasons, the spectra for wild-type FNR during O2 turnover was produced from a separate sample also prepared with 2.4 mM NADPH.
NADPH—K3Fe(CN)6 reductase activity assays were performed in 100 mM Tris-HCl, pH 8, at 25 °C, at different concentrations of both substrates, as reported elsewhere [56]. As needed, nicotinamide was included in the assays at concentrations ranging from 50 to 800 mM.
Reanalysis of Stopped-Flow Results Reported for Anabaena FNR
Spectra (Figure 3 in [26]) were enlarged by ~345% and printed. The height of each peak at ~460 nm corresponding to the flavin oxidation state was measured in cm from the baseline of the spectra (−0.025 AU). The peak height at time 0 for FNRred with NADP+ (corresponding to fully reduced flavin) was taken as a baseline and used to normalize the measurements by subtracting this height from the other peak heights of the corresponding variant. Whereas the final spectra (0.2547 s) for wild-type FNR for both FNRred with NADP+ and FNRox with NADPH matched in both peak height and shape, the final spectra (0.2547 s) for Y303S FNR varied slightly in peak height but both correspond to achieved equilibrium so were normalized to each other by subtracting the difference between the final peak heights from the measured peak heights for Y303S FNRred with NADP+.
Taking the peak height at time 0 for FNRox with NADPH to correspond to fully oxidized flavin, the percentage of oxidized and reduced flavin and corresponding concentration of product (using the provided concentrations of 25 μM FNR and 125 μM NADP(H)) were calculated. A plot of concentration of product (μM) versus time (s) were fitted using GraFit 5 (Erithacus Software Limited) with a single-exponential decay equation to estimate the apparent rate constants of the hydride-transfer reactions.
Crystallization and Structure Determinations
Initial co-crystallization trials of the corn root FNR variants with NADP+ used similar conditions to those used for the wild-type enzyme [9] but yielded only ill-formed small crystals that grew very slowly. Solving their structure revealed that they contained a bound nicotinamide moiety rather than NADP+, so co-crystallization with nicotinamide was tried, and crystals grew readily. The best crystallizations were at room temperature in hanging drops formed by mixing equal volumes of the protein stock (Y316F: 20.2 mg/mL; Y316A: 7.3 mg/mL; Y316S: 10.8 mg/mL; all in 50 mM Tris-HCl, pH 7.4) with a reservoir solution containing 22–24% PEG 8000, 0.1 M sodium cacodylate (pH 6–7), 0.18–0.22 M magnesium acetate, and 100 mM nicotinamide. Typical crystals were ~0.3 mm on each side and grew within 1 week. Both FNR variants formed many prism-shaped crystals belonging to the space group P3221 (same as wild type; [9]) and fewer hexagon-shaped crystals belonging to space group P3121. Soaks of the nicotinamide-bound crystals were done aerobically at room temperature with either 10 mM NADP+ or 10 mM NADPH for 1 h to obtain desired complexes before being flash frozen.
Data collections
For data collection, crystals were pulled through oil and flash-frozen in liquid nitrogen. In-house data were collected at 140 K on a Rigaku RU300 Cu-Kα rotating anode X-ray source running at 50 kV and 100 mA equipped with a Raxis IV image plate detector. Synchrotron data were collected at beamline 5.0.1 or 5.0.3 at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) at 100 K. Oscillation images were collected with Δφ = 1° and were processed using Denzo and Scalepack [57] or iMosflm [58] and SCALA [59]. Unit cell parameters and data reduction statistics for each FNR variant are given in Table 2. In addition, data sets with Δφ = 0.5° and simultaneous single crystal visible absorption spectra were collected by Dr. Allen Orville at a National Synchotron Light Source beamline as described in [35].
Structure solutions and refinements
All P3221 FNR variant crystals were isomorphous with the published wild-type corn root FNR structure (PDB code 3LO8 [36]), and this was used as the starting model for the first refinement, with additional refinements built on partially refined models of the most similar structure. The P3121 crystal form was trivial to solve by molecular replacement and these structures were then similarly refined. The same 10% of data were set aside for cross-validation [60] for each structure as had been set aside for the ~1 Å resolution refinement of the wild-type structure. Refinements were carried out by a number of researchers over many years using various versions of REFMAC [61] with Coot [62] used for manual fitting and Molprobity [63] used to help identify model problems. During iterative manual rebuilding, water molecules were added in Coot using standard criteria (>1 ρrms intensity in the 2Fo−Fc map, >2.4 Å distance from nearest contact, no B-factors >80 Å2). For making a consistent set of structures for publication, a set of final refinements of each structure were done by the lead author using Phenix [64]. During this stage, we decided to model all NADPH soaks as having an oxidized nicotinamide (i.e. NADP+) and a reduced flavin since the spectra show that CTC-2 is present at higher amounts than is CTC-1. For structures refined with anisotropic B-factors (the P3221 Y316S:NADPH, Y316S:NADP+, and Y316S:nicotinamide structures), no anisotropic restraints were used for the ligand in the final round of refinement to ensure the anisotropic B-factors of the nicotinamide atoms were based as much as possible on the diffraction data. Refinement statistics for all models are shown in Tables 2 and S1. Structural overlays were performed using the protein structure visualization software PyMOL [65].
Also, in order to obtain a reliable representation of active site distances for Y316SNADPH, Y316SNADP+ (P3221 and P3121), Y316ANADPH, and Y316ANADP+ structures, ten different starting models for each were generated using the “shake” algorithm of Phenix with the setting “modify.sites.shake = 0.5”. This level of coordination disruption resulted in starting R/Rfree values of ~40%. Each of these models was re-refined and the distances between Cys274 Sγ and NADP+ C4H and between NADP+ C4 and FAD N5 were measured. The average distances and standard deviations for each set were calculated and reported.
Accession Numbers
Coordinates and structure factors for the wild-type corn root FNR, Y316S:nicotinamide (P3221), Y316S:nicotinamide (P3121), Y316S:NADP+ (P3221), Y316S:NADP+ (P3121), Y316S:NADPH (P3221), Y316A:nicotinamide (P3221), Y316A:NADP+ (P3221), Y316A:NADPH (P3221), Y316F (P3221), and Y316F:NADP+ (P3121) models have been deposited in the Protein Data Bank with accession numbers 3LO8, 5VW4, 5VW9, 5VW3, 5VW8, 5VW2, 5VW5, 5VW6, 5VW7, 5VWA, and 5VWB, respectively.
Figure S1: Measurements used for the reanalysis of Anabaena FNR Y303S and wild type kinetics.
Acknowledgments
This work was supported in part by National Science Foundation grant MCB-9982727 and National Institutes of Health grant R01-GM119227. The authors would also like to thank Allen Orville for collecting absorption spectra from single crystals of FNR variants and Peter Zwart for collecting and processing some of the data sets reported here. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Abbreviations
- FNR
ferredoxin:NADP+ reductase
- NOX
NADPH oxidase
- Fd
ferredoxin
- CTC-1
charge-transfer complex 1
- CTC-2
charge-transfer complex 2
Footnotes
Database: Structural data are available in the PDB database under the accession numbers 3LO8 (wild type), 5VW4 (Y316S:nicotinamide (P3221)), 5VW9 (Y316S:nicotinamide (P3221)), 5VW3 (Y316S:NADP+ (P3221)), 5VW8 (Y316S:NADP+ (P3121)), 5VW2 (Y316S:NADPH (P3221)), 5VW5 (Y316A:nicotinamide (P3221)), 5VW6 (Y316A:NADP+ (P3221)), 5VW7 (Y316A:NADPH (P3221)), 5VWA (Y316F (P3221)), and 5VWB (Y316F:NADP+ (P3121)).
Author Contributions. KMK analyzed data and wrote the paper; RAC planned and performed experiments and analyzed data; VP, ARH, and RF planned and performed experiments; GZ planned experiments; AA planned and performed experiments, analyzed data, and wrote the paper; PAK planned experiments, analyzed data and wrote the paper.
Enzyme Commission number: ferredoxin:NADP+ reductase – EC 1.18.1.2
Data Accessibility: Research data pertaining to this article are located at figshare.com: [URL to be added]
References
- 1.Aliverti A, Pandini V, Pennati A, de Rosa M, Zanetti G. Structural and functional diversity of ferredoxin-NADP(+) reductases. Arch Biochem Biophys. 2008;474:283–91. doi: 10.1016/j.abb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Paladini DH, Musumeci MA, Carrillo N, Ceccarelli EA. Induced fit and equilibrium dynamics for high catalytic efficiency in ferredoxin-NADP(H) reductases. Biochemistry. 2009;48:5760–8. doi: 10.1021/bi9004232. [DOI] [PubMed] [Google Scholar]
- 3.Bortolotti A, Perez-Dorado I, Goni G, Medina M, Hermoso JA, Carrillo N, Cortez N. Coenzyme binding and hydride transfer in Rhodobacter capsulatus ferredoxin/flavodoxin NADP(H) oxidoreductase. Biochim Biophys Acta. 2009;1794:199–210. doi: 10.1016/j.bbapap.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Tejero J, Perez-Dorado I, Maya C, Martinez-Julvez M, Sanz-Aparicio J, Gomez-Moreno C, Hermoso JA, Medina M. C-terminal tyrosine of ferredoxin-NADP+ reductase in hydride transfer processes with NAD(P)+/H. Biochemistry. 2005;44:13477–90. doi: 10.1021/bi051278c. [DOI] [PubMed] [Google Scholar]
- 5.Bruns CM, Karplus PA. Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2′-phospho-5′-AMP bound states. J Mol Biol. 1995;247:125–45. doi: 10.1006/jmbi.1994.0127. [DOI] [PubMed] [Google Scholar]
- 6.Deng Z, Aliverti A, Zanetti G, Arakaki AK, Ottado J, Orellano EG, Calcaterra NB, Ceccarelli EA, Carrillo N, Karplus PA. A productive NADP+ binding mode of ferredoxin-NADP + reductase revealed by protein engineering and crystallographic studies. Nat Struct Biol. 1999;6:847–53. doi: 10.1038/12307. [DOI] [PubMed] [Google Scholar]
- 7.Dorowski A, Hofmann A, Steegborn C, Boicu M, Huber R. Crystal structure of paprika ferredoxin-NADP+ reductase. Implications for the electron transfer pathway. J Biol Chem. 2001;276:9253–63. doi: 10.1074/jbc.M004576200. [DOI] [PubMed] [Google Scholar]
- 8.Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimata-Ariga Y, Hase T. Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP(+) reductase. Nat Struct Biol. 2001;8:117–21. doi: 10.1038/84097. [DOI] [PubMed] [Google Scholar]
- 9.Aliverti A, Faber R, Finnerty CM, Ferioli C, Pandini V, Negri A, Karplus PA, Zanetti G. Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues. Biochemistry. 2001;40:14501–8. doi: 10.1021/bi011224c. [DOI] [PubMed] [Google Scholar]
- 10.Karplus PA, Daniels MJ, Herriott JR. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991;251:60–6. [PubMed] [Google Scholar]
- 11.Correll CC, Ludwig ML, Bruns CM, Karplus PA. Structural prototypes for an extended family of flavoprotein reductases: comparison of phthalate dioxygenase reductase with ferredoxin reductase and ferredoxin. Protein Sci. 1993;2:2112–33. doi: 10.1002/pro.5560021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karplus PA, Bruns CM. Structure-function relations for ferredoxin reductase. J Bioenerg Biomembr. 1994;26:89–99. doi: 10.1007/BF00763221. [DOI] [PubMed] [Google Scholar]
- 13.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 14.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275:3249–77. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 15.Panday A, Sahoo MK, Osorio D, Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 2015;12:5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brewer TF, Garcia FJ, Onak CS, Carroll KS, Chang CJ. Chemical approaches to discovery and study of sources and targets of hydrogen peroxide redox signaling through NADPH oxidase proteins. Annu Rev Biochem. 2015;84:765–90. doi: 10.1146/annurev-biochem-060614-034018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004;279:37918–27. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94:8411–6. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard PA, Shen AL, Paschke R, Kasper CB, Kim JJ. NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer. J Biol Chem. 2001;276:29163–70. doi: 10.1074/jbc.M101731200. [DOI] [PubMed] [Google Scholar]
- 20.Correll CC, Batie CJ, Ballou DP, Ludwig ML. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–10. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 21.Piubelli L, Aliverti A, Arakaki AK, Carrillo N, Ceccarelli EA, Karplus PA, Zanetti G. Competition between C-terminal tyrosine and nicotinamide modulates pyridine nucleotide affinity and specificity in plant ferredoxin-NADP(+) reductase. J Biol Chem. 2000;275:10472–6. doi: 10.1074/jbc.275.14.10472. [DOI] [PubMed] [Google Scholar]
- 22.Adak S, Sharma M, Meade AL, Stuehr DJ. A conserved flavin-shielding residue regulates NO synthase electron transfer and nicotinamide coenzyme specificity. Proc Natl Acad Sci U S A. 2002;99:13516–21. doi: 10.1073/pnas.192283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dohr O, Paine MJ, Friedberg T, Roberts GC, Wolf CR. Engineering of a functional human NADH-dependent cytochrome P450 system. Proc Natl Acad Sci U S A. 2001;98:81–6. doi: 10.1073/pnas.98.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neeli R, Roitel O, Scrutton NS, Munro AW. Switching pyridine nucleotide specificity in P450 BM3: mechanistic analysis of the W1046H and W1046A enzymes. J Biol Chem. 2005;280:17634–44. doi: 10.1074/jbc.M413826200. [DOI] [PubMed] [Google Scholar]
- 25.Meints CE, Gustafsson FS, Scrutton NS, Wolthers KR. Tryptophan 697 modulates hydride and interflavin electron transfer in human methionine synthase reductase. Biochemistry. 2011;50:11131–42. doi: 10.1021/bi2012228. [DOI] [PubMed] [Google Scholar]
- 26.Lans I, Peregrina JR, Medina M, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM. Mechanism of the hydride transfer between Anabaena Tyr303Ser FNR(rd)/FNR(ox) and NADP+/H. A combined pre-steady-state kinetic/ensemble-averaged transition-state theory with multidimensional tunneling study. J Phys Chem B. 2010;114:3368–79. doi: 10.1021/jp912034m. [DOI] [PubMed] [Google Scholar]
- 27.Peregrina JR, Lans I, Medina M. The transient catalytically competent coenzyme allocation into the active site of Anabaena ferredoxin NADP+-reductase. Eur Biophys J. 2012;41:117–28. doi: 10.1007/s00249-011-0704-5. [DOI] [PubMed] [Google Scholar]
- 28.Mulo P, Medina M. Interaction and electron transfer between ferredoxin-NADP+ oxidoreductase and its partners: structural, functional, and physiological implications. Photosynth Res. 2017 doi: 10.1007/s11120-017-0372-0. [DOI] [PubMed] [Google Scholar]
- 29.Aliverti AFC, Spinola M, Raimondi D, Zanetti G, Finnerty CM, Faber R, Karplus PA. Structural and functional properties of corn root ferredoxin-NADP+-reductase. Paper presented at the Flavins and Flavoproteins; Konstanz, Germany. 1999. [Google Scholar]
- 30.Magnani F, Nenci S, Millana Fananas E, Ceccon M, Romero E, Fraaije MW, Mattevi A. Crystal structures and atomic model of NADPH oxidase. Proc Natl Acad Sci U S A. 2017;114:6764–6769. doi: 10.1073/pnas.1702293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogues I, Tejero J, Hurley JK, Paladini D, Frago S, Tollin G, Mayhew SG, Gomez-Moreno C, Ceccarelli EA, Carrillo N, Medina M. Role of the C-terminal tyrosine of ferredoxin-nicotinamide adenine dinucleotide phosphate reductase in the electron transfer processes with its protein partners ferredoxin and flavodoxin. Biochemistry. 2004;43:6127–37. doi: 10.1021/bi049858h. [DOI] [PubMed] [Google Scholar]
- 32.Batie CJ, Kamin H. Association of ferredoxin-NADP+ reductase with NADP(H) specificity and oxidation-reduction properties. J Biol Chem. 1986;261:11214–23. [PubMed] [Google Scholar]
- 33.Batie CJ, Kamin H. Electron transfer by ferredoxin:NADP+ reductase. Rapid-reaction evidence for participation of a ternary complex. J Biol Chem. 1984;259:11976–85. [PubMed] [Google Scholar]
- 34.Dunford AJ, Marshall KR, Munro AW, Scrutton NS. Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in the region of the FAD shielding residue Phe1395. Eur J Biochem. 2004;271:2548–60. doi: 10.1111/j.1432-1033.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 35.Orville AM, Buono R, Cowan M, Heroux A, Shea-McCarthy G, Schneider DK, Skinner JM, Skinner MJ, Stoner-Ma D, Sweet RM. Correlated single-crystal electronic absorption spectroscopy and X-ray crystallography at NSLS beamline X26-C. J Synchrotron Radiat. 2011;18:358–66. doi: 10.1107/S0909049511006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tronrud DE, Berkholz DS, Karplus PA. Using a conformation-dependent stereochemical library improves crystallographic refinement of proteins. Acta Crystallogr D Biol Crystallogr. 2010;66:834–42. doi: 10.1107/S0907444910019207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young L, Post CB. Catalysis by entropic guidance from enzymes. Biochemistry. 1996;35:15129–33. doi: 10.1021/bi961875m. [DOI] [PubMed] [Google Scholar]
- 38.Prasad GS, Kresge N, Muhlberg AB, Shaw A, Jung YS, Burgess BK, Stout CD. The crystal structure of NADPH : ferredoxin reductase from Azotobacter vinelandii. Protein Science. 1998;7:2541–2549. doi: 10.1002/pro.5560071207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu G, Lindqvist Y, Schneider G, Dwivedi U, Campbell W. Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain. J Mol Biol. 1995;248:931–48. doi: 10.1006/jmbi.1995.0273. [DOI] [PubMed] [Google Scholar]
- 40.Bewley MC, Marohnic CC, Barber MJ. The structure and biochemistry of NADH-dependent cytochrome b5 reductase are now consistent. Biochemistry. 2001;40:13574–82. doi: 10.1021/bi0106336. [DOI] [PubMed] [Google Scholar]
- 41.Yamada M, Tamada T, Takeda K, Matsumoto F, Ohno H, Kosugi M, Takaba K, Shoyama Y, Kimura S, Kuroki R, Miki K. Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer. J Mol Biol. 2013;425:4295–306. doi: 10.1016/j.jmb.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Bruice TC, Lightstone FC. Ground state and transition state contributions to the rates of intramolecular and enzymatic reactions. Accounts Chem Res. 1999;32:127–136. [Google Scholar]
- 43.Bruice TC, Pandit UK. Intramolecular Models Depicting the Kinetic Importance of “Fit” in Enzymatic Catalysis. Proc Natl Acad Sci U S A. 1960;46:402–4. doi: 10.1073/pnas.46.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajagopalan PT, Benkovic SJ. Preorganization and protein dynamics in enzyme catalysis. Chem Rec. 2002;2:24–36. doi: 10.1002/tcr.10009. [DOI] [PubMed] [Google Scholar]
- 45.Almarsson O, Bruice TC. Evaluation of the Factors Influencing Reactivity and Stereospecificity in Nad(P)H Dependent Dehydrogenase Enzymes. Journal of the American Chemical Society. 1993;115:2125–2138. [Google Scholar]
- 46.Berkholz DS, Faber HR, Savvides SN, Karplus PA. Catalytic cycle of human glutathione reductase near 1 A resolution. J Mol Biol. 2008;382:371–84. doi: 10.1016/j.jmb.2008.06.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quaytman SL, Schwartz SD. Reaction coordinate of an enzymatic reaction revealed by transition path sampling. Proc Natl Acad Sci U S A. 2007;104:12253–8. doi: 10.1073/pnas.0704304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Klinman JP. Enzymatic methyl transfer: role of an active site residue in generating active site compaction that correlates with catalytic efficiency. J Am Chem Soc. 2011;133:17134–7. doi: 10.1021/ja207467d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hay S, Pudney CR, McGrory TA, Pang J, Sutcliffe MJ, Scrutton NS. Barrier compression enhances an enzymatic hydrogen-transfer reaction. Angew Chem Int Ed Engl. 2009;48:1452–4. doi: 10.1002/anie.200805502. [DOI] [PubMed] [Google Scholar]
- 50.Tejero I, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM, York DM. Enzyme dynamics and tunneling enhanced by compression in the hydrogen abstraction catalyzed by soybean lipoxygenase-1. J Phys Chem B. 2006;110:24708–19. doi: 10.1021/jp066263i. [DOI] [PubMed] [Google Scholar]
- 51.Medina M, Luquita A, Tejero J, Hermoso J, Mayoral T, Sanz-Aparicio J, Grever K, Gomez-Moreno C. Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis. J Biol Chem. 2001;276:11902–12. doi: 10.1074/jbc.M009287200. [DOI] [PubMed] [Google Scholar]
- 52.Shiraishi N, Croy C, Kaur J, Campbell WH. Engineering of pyridine nucleotide specificity of nitrate reductase: mutagenesis of recombinant cytochrome b reductase fragment of Neurospora crassa NADPH:Nitrate reductase. Arch Biochem Biophys. 1998;358:104–15. doi: 10.1006/abbi.1998.0827. [DOI] [PubMed] [Google Scholar]
- 53.Elmore CL, Porter TD. Modification of the nucleotide cofactor-binding site of cytochrome P-450 reductase to enhance turnover with NADH in Vivo. J Biol Chem. 2002;277:48960–4. doi: 10.1074/jbc.M210173200. [DOI] [PubMed] [Google Scholar]
- 54.Marohnic CC, Bewley MC, Barber MJ. Engineering and characterization of a NADPH-utilizing cytochrome b5 reductase. Biochemistry. 2003;42:11170–82. doi: 10.1021/bi034819b. [DOI] [PubMed] [Google Scholar]
- 55.Zhen L, Yu L, Dinauer MC. Probing the role of the carboxyl terminus of the gp91phox subunit of neutrophil flavocytochrome b558 using site-directed mutagenesis. J Biol Chem. 1998;273:6575–81. doi: 10.1074/jbc.273.11.6575. [DOI] [PubMed] [Google Scholar]
- 56.Aliverti A, Piubelli L, Zanetti G, Lubberstedt T, Herrmann RG, Curti B. The role of cysteine residues of spinach ferredoxin-NADP+ reductase As assessed by site-directed mutagenesis. Biochemistry. 1993;32:6374–80. doi: 10.1021/bi00076a010. [DOI] [PubMed] [Google Scholar]
- 57.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 58.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992:26. [Google Scholar]
- 59.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 60.Brunger AT. Free R value: cross-validation in crystallography. Methods Enzymol. 1997;277:366–96. doi: 10.1016/s0076-6879(97)77021-6. [DOI] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The PyMOL Molecular Graphics System, Version 1.3. Schrödinger, LLC; [Google Scholar]