Abstract
Background
Chronic alcohol abuse, a major risk factor for such diseases as hepatitis and cirrhosis, impairs hepatic alcohol dehydrogenase (ADH, key ethanol metabolizing enzyme). Therefore, differentially altered hepatic and plasma proteomes were identified in chronic ethanol feeding model of hepatic ADH-deficient (ADH−) deer mice to understand the metabolic basis of alcoholic liver disease (ALD).
Methods
ADH− deer mice were fed 3.5g% ethanol via Lieber-DeCarli liquid diet daily for three months, and histology of the liver assessed. Liver and plasma proteins were separated by 2-dimensional gel electrophoresis. The proteins differentially expressed were identified by MALDI-TOF/TOF mass spectrometry (MS).
Results
Histology of the liver showed panlobular steatosis and infiltration of T-lymphocytes. Using the criteria of ≥1.5 for fold change (p value ≤0.05) with expectation value (E ≤10−3) and protein score (≥64), eighteen proteins in the livers and five in the plasma of ethanol-fed mice were differentially expressed and identified. Prolyl 4-hydroxylase, cytochrome b-5, endo A cytokeratin, ATP synthase, heat shock 70kD proteins, enoyl CoA hydratase, stress-70 protein, peroxiredoxin 1 and ornithine carbamoyl-transferase were up regulated in the livers. However, carbonic anhydrase 3, mitochondrial ATP synthase, aldolase 2, actin γ, laminin receptor and carbamoyl-phosphate synthase were down regulated. Contrary to the increased expression of creatine kinase m-type, a decreased expression of serine protease inhibitor A3A precursor, sulfated glycoprotein-2 (clusterin) and apolipoprotein E isoforms were found in the plasma of ethanol group.
Conclusions
Chronic ethanol feeding in ADH− deer mice causes steatosis and infiltration of T-lymphocytes in the livers along with increased expression of proteins involved in ER stress, fibrosis, fatty acid β oxidation and biogenesis, and decreased expression of proteins involved in ATP synthesis, carbohydrate metabolism, in cell regulation and architecture. Reduced expression of various carrier proteins as found in the plasma of ethanol group has a biomarker potential.
Keywords: Ethanol, Liver, Plasma, Proteomics, Deer mice
Introduction
Alcoholic liver disease (ALD) is a serious global health problem causing significant morbidity and mortality among chronic alcohol abusers (NIAAA, 2000). Liver is a primary target organ for the metabolism and toxicity of ingested alcohol (ethanol). While metabolic basis of ALD is not well defined due to a lack of suitable animal model, the disease develops after a long history of chronic alcohol abuse initiating from the lipid dysregulation and resulting into steatohepatitis followed by fibrosis and cirrhosis. Chronic alcohol abuse is also known to impair hepatic alcohol dehydrogenase (ADH), a major enzyme involved in the oxidative metabolism of alcohol (Sharakawi, 1984; Panés et al., 1993; Kaphalia et al., 1996). Published work from our laboratory have shown that chronic ethanol feeding via Lieber–DeCarli liquid diet for 60 days increases hepatic lipids and formation of fatty acid ethyl esters (nonoxidative metabolites of ethanol), and causes notable hepatic injury in ADH− deer mice as compared to normal hepatic ADH (ADH+) deer mice (Bhopale et al., 2006; Fernando et al., 2012).
Alcohol affects turnover of a large number of both resident and secreted liver proteins and inhibits their synthesis (Karinch et al., 2008). Identification of differentially expressed proteins using proteomic analysis is considered to be suitable markers to identify such diseases as Alzheimer (Galasko, 2005), lung carcinogenesis (Gomperts et al., 2011) leukemia (Kanaujiya et al., 2011), asthma and chronic obstructive pulmonary disease (Verrills et al., 2011). Attempts also have been made to identify biomarkers of ALD (Newton et al., 2009; Witzmann and Strother, 2004). An ideal model to explore markers of ALD should mimic the disease and differentially express a plasma proteome indicative of the biochemical and molecular changes occurring at an early stage of ALD (Torrente et al., 2012). Such studies are critical for developing preventive strategies before the diseases progress to a clinically overt stage. ADH− deer mouse model mimics a metabolic condition of hepatic ADH deficiency commonly detected in chronic alcoholic individuals and in experimental animals after chronic ethanol feeding. Therefore, we determined hepatic and plasma proteome differentially expressed in hepatic ADH− deer mouse model after long term ethanol feeding to identify potential biomarker(s) and toxic pathway(s) involved in an early stage ALD.
Materials and Methods
Animal experimental studies
One year old male hepatic ADH deficient (ADH−) deer mice (Peromyscus maniculatus, a genetic variant lacking ADH 1 in the liver) were obtained from Peromyscus Stock Center, University of South Carolina, SC. Mice were housed in UTMB’s animal research center facility maintained at 25±1°C and under a 12 hour light/dark cycle. Animals were randomly divided into two groups, experimental and control. In brief, mice were caged individually and fed with a nutritionally adequate Lieber-DeCarli liquid diet (Dyets, Inc., Bethlem, PA; cat# 710260; Lieber and DeCarli, 1989). Experimental group was fed with 3.5g% ethanol via Lieber-DeCarli liquid diet (Bhopale et al., 2006; Kaphalia et al., 2010). Ethanol concentration was gradually increased from1g% to 3.5g% in a two week period, and thereafter maintained at 3.5g% ethanol for an additional three months. Control group was pair-fed with isocaloric liquid diet (Dyets, cat# 40285) where ethanol calories were replaced with maltose-dextrin. Both groups were euthanized using pentobarbital sodium (Nembutal sodium; 100mg/kg administered intraperitoneally). Blood was collected by cardiac puncture and 50 μl transferred to a gas-chromatography (GC) vial. Blood alcohol concentration (BAC) was analyzed by head space GC using isopropanol as an internal standard as described earlier (Kaphalia et al., 2014). Remaining blood was centrifuged at 1,000 g for 5 min, plasma separated and stored in −80°C. Liver tissue was harvested and processed for histology and proteomic studies as detailed earlier (Bhopale et al., 2006, 2011; Fernando et al., 2013). Formalin fixed thin liver sections from ethanol-fed and control mice were subjected to hematoxylin and eosin (H & E) staining. In addition, immunohistochemistry was performed using rabbit polyclonal antibodies to CD3 antigen (Dako, Santa Clara, CA; cat# A0452 1:100 dilution) and biotinylated goat anti rabbit secondary antibodies (H&L; Vector Shield, Burlingame, CA; Cat # BA-1000; 1:200 dilution) to examine infiltration of T-lymphocytes (Fernando et al., 2013).
Proteomic studies
Preparation of hepatic tissue samples
Ethanol-fed and control mice (four in each group) were used in this study. Frozen liver tissue (75mg) was homogenized in 500μl DeStreak rehydration buffer (GE Healthcare, Life sciences, Pittsburgh, PA) containing protease inhibitor cocktail (Sigma-Aldrich, Milwaukee, WI). The homogenate was treated with 150 U/ml of benzonase (E1014, Sigma) for 30 min at room temperature to remove nucleic acids and centrifuged at 13,000 g for 15 min. The supernatant was collected and protein concentration was measured using RC DC protein assay kit (Bio-Rad, cat# 500-0122). Triplicate sets from each liver sample were prepared for the proteomic analysis.
Preparation of plasma samples
Since a large amount of albumin and IgG present in the plasma interferes with separation and identification of low abundant proteins (Seferovic et al., 2008), albumin and IgG were removed by ProteoExtract™ Albumin/IgG removal kit (cat# 122642, Calbiochem, San Diego, CA) (Bhopale et al., 2011). The proteins were precipitated using ProteoExtract TM protein precipitation kit (cat# 539180, Calbiochem) and measured using RC DC protein assay kit as described earlier.
2-Dimentional gel electrophoresis of liver or plasma proteins
Established methodologies were used for 2-dimentional gel electrophoresis (2-DE) and isoelectric focusing (IEF), fluorescent staining, imaging, gel analysis, matrix assisted laser desorption ionization–time of flight (MALDI-TOF/TOF) mass spectrometry (MS) and protein identification (Forbus et al., 2006; Bhopale et al., 2011; Fernando et al., 2013). In brief, a total of 200 μg protein equivalent from liver homogenate supernatant was loaded on 11-cm long precast immobilized pH gradient (IPG) strips at pH ranges 3–5.4, 5.4–7.0 and 7–10, while the albumin and IgG depleted plasma ran at pH 3–10 range. The strips were rehydrated overnight for IEF and focused, and IEF was performed at 20°C in six steps; 50 V for 11h, 250 V for 1h, 500 V for 1h, 1,000 V for 1h, 8,000 V for 2h, and 8,000 V for 6h for a total of 48,000V hrs. After IEF, the IPG strips were stored at −80°C till the next step. The frozen stored IPG strips, prior to the 2-Dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) procedure, were incubated at 22°C in 4 ml of equilibration buffer (6M urea, 2%SDS, 50mM tris-HCl, pH 8.8, 20% glycerol) containing 10μl of 0.5M tris (2-carboxyethyl) phosphine with shaking for 15 minutes. In the subsequent step, these strips were again incubated in 4 ml of equilibration buffer containing 25 mg/ml of iodoacetamide with shaking for 15 min at 22°C. After completion of equilibration, electrophoresis was performed at 150 V for 2.25h at 4°C with precast 8 to 16% polyacrylamide gels in Tris-glycine buffer (25mM Tris-HCl, 192 mM glycine, 0.1%SDS, pH 8.3).
Plasma sample containing 200 μg protein was applied on IPG strip for IEF followed by 2-DE as described earlier (Bhopale et al., 2011). The strips were incubated in equilibration buffer followed by SDS-PAGE as described for the liver samples.
Gel staining, image analysis, spot excision and in-gel digestion, mass spectrometry and protein identification
All the procedures for gel staining, image analysis, spot excision and in-gel digestion, MALDI TOF/TOF MS and protein identification used in this study have been established in our UTMB’s NHLBI Proteomics Center and used previously by us (Bhopale et al., 2011; Fernando et al., 2013). In brief, 2-DE gels after electrophoresis were fixed in the fixing buffer containing 10% methanol and 7% acetic acid in double distilled water for 2h at room temperature followed by staining with SyproRuby stain (Invitrogen, Carlsbad, CA) over night at room temperature and subsequently de-stained with 10% ethanol for 1h before imaging. The de-stained gels were imaged at 100 μM resolution using ProExpress 2D Proteomic Imaging System (PerkinElmer Life and Analytical Sciences, Waltham, MA) at 480 nm excitation and 620 nm emission filters and adjusting exposure time to achieve 55,000 to 63,000-pixel intensity value for obtaining the most intense protein spots on the gels. Gel images were analyzed using Progenesis SameSpots software version 4.0 (TotalLab, Newcastle Upon Tyne, UK). The program selected one of the Images as the reference gel and all the other gel images were aligned to it based on user-drawn and automatic vectors. SameSpots then performed spot detection based on a selected subset of the aligned gels. Background subtraction and spot volume intensity normalization to reference gel selected by the program were automatically performed. Average protein expression was calculated from the average normalized volumes of the spots. Average values, fold-changes for expression were calculated as assigned ratios of ethanol-fed to control groups. Significance of differential expression was determined by Student t-test (p-value ≤ 0.05) after log 2 transformation and fold change at ≥1.5.
After identification of differentially expressed proteins, gel spots of interest were excised using DigiLab’s (Holliston, MO) ProPic1 robotic instrument by following the manufacturer’s instructions. The excised gel plugs were incubated at 37°C for 24h with trypsin (20 μg/ml in 25 mM ammonium bicarbonate, pH 8.0; Promega Corp., Madison, WI). MALDI-TOF/TOF MS analysis was performed using a Proteomics Analyzer 4700 (Applied Biosystems, Foster City, CA). Protein identification was performed by recording both MS and MS/MS data and searching the NCBI mouse data base of GeneBank with the MASCOT search engine (Matrix Science, London, UK) and probability matches were searched by a high protein score (http://www.matrixscience.com) and a low expectation (E) value (Zhang and Chait, 2000). The lower the E value, higher the probability of identification being valid and for confident identification, E ≤ 10−3 was used as a threshold value. Well-established proteomic criteria (≥1.5 fold change with p value ≤0.05; best protein score of >64, and a MS expectation value of 10−3 or less) were used for a high confidence protein identification.
Western blot analysis
Some of the proteins differentially expressed were also confirmed by Western blot analysis. Frozen liver samples were homogenized in cold RIPA buffer containing protease inhibitors (Boston Bio Product, MA), centrifuged at 15,000 g and the supernatant containing 45 μg protein concentration was subjected to SDS-PAGE (NuPAGE™ Novex™ 4–12% Bis-Tris Protein Gels, 1.5 mm thick, 10 well, Invitrogen) (Kaphalia et al., 2014). The gels were washed with washing buffer and the proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk in phosphate buffered saline containing 5% nonfat dried milk, 0.001% Tween-20 and then incubated overnight at 4°C with primary antibodies. Membrane was incubated either with rabbit polyclonal antibodies against cytochrome b5 (Santa Cruz, Cat # SC33174, 1:500 dilution, MW 15kDa), heat shock protein 70 (EMD Millipore, cat # 386032, 1:1000 dilution, MW 70 kDa) or peroxiredoxin 1 (Abcam, cat # ab59538, 1:1000 dilution, MW: 22kDa), or monoclonal antibodies against enoyl CoA hydratase (Abcam, cat # ab170108, 1:1000 dilution, MW 31kDa). After washing, the blots were again incubated with horseradish peroxidase conjugated secondary antibodies (Cell Signaling, cat # 7075; 1: 1000 dilution), and the target proteins visualized using as Pierce™ ECL Western Blotting Substrate (ThermoFisher Scientific, Rockford, lL). Rabbit monoclonal antibodies against β-actin (Cell Signaling, Cat #8457, 1:1000 dilution, MW 45 kDa) was used as an individual loading control. The protein band intensity was assessed by NIH Image J Software (version 1.50i) and normalized by individual loading control (β-actin) values.
For identification of proteins in the plasma by Western blot analysis, albumin globulin depleted plasma protein (25μg) was subjected to SDS PAGE as described above for the liver samples. Proteins were transferred onto nitrocellulose membrane and incubated with rabbit polyclonal antibodies against apolipoprotein E (Abcam, cat# ab20874, 1:1000 dilution, MW 36kDa,) or mouse monoclonal antibody of creatinine kinase-M (Santa Cruz, cat# 390053,1:500 dilution, MW 43kDa) as described earlier.
Statistical analysis
Statistical significance was determined by using the Student’s t-test and the values are expressed as mean ± SEM (standard error of mean, four animals/group). p-Value ≤0.05 was considered statistically significant.
Results
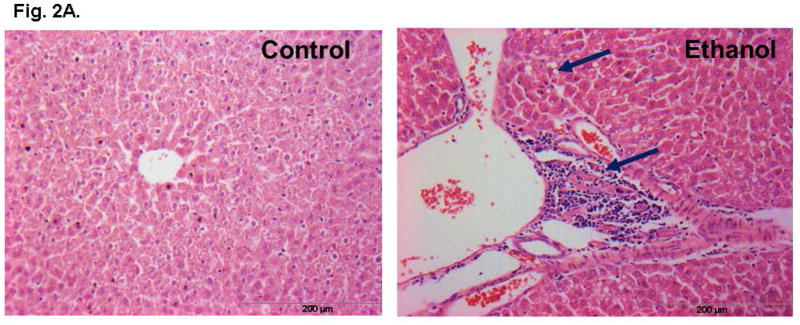
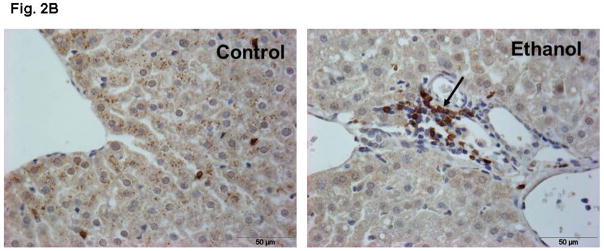
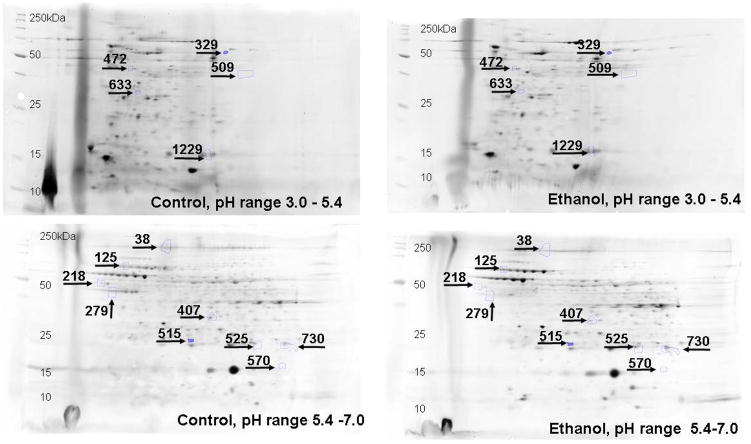
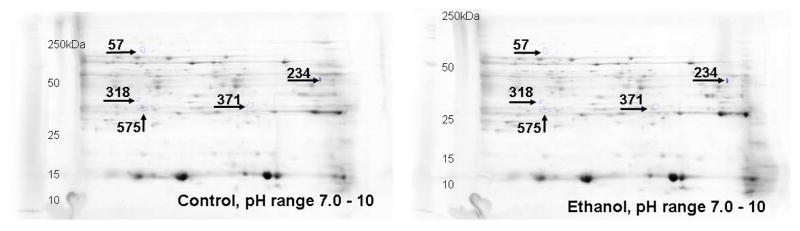
Ethanol fed mice averaged ~200 mg % BAC as compared to ~15 mg% in pair-fed controls (Fig. 1). Remarkable changes including hepatic steatosis comprising macro and micro vesicular lipid vacuoles with panlobular changes along with infiltration of inflammatory/immune cells in some areas of the liver sections (Fig. 2A). These sections also showed infiltration of T-lymphocytes as determined by immunohistochemistry using antibodies against CD3 (Fig. 2B). Representative electrophoretic patterns of differentially expressed proteins for liver tissue fractionated into three pH ranges (3–5.4, 5.4–7.0 and 7–10) are shown in Fig. 3. Similarly, electrophoretic patterns of plasma proteins differentially expressed (pH 3–10) are shown in Fig. 4. A total of 140 proteins in the livers and 58 in the plasma were differentially expressed in ethanol fed group by SameSpots image analysis of 2-DE gels. While a large number of proteins differentially expressed were post translationally modified (PTMs), and few ones were unknown proteins. Using criteria of fold change (≥1.5 with p value ≤0.05), MS expectation value of 10−3 or less and good protein score (>64), a total of 18 and 5 proteins differentially expressed in the livers and plasma, respectively, could be identified (Tables 1 and 2).
Fig. 1.
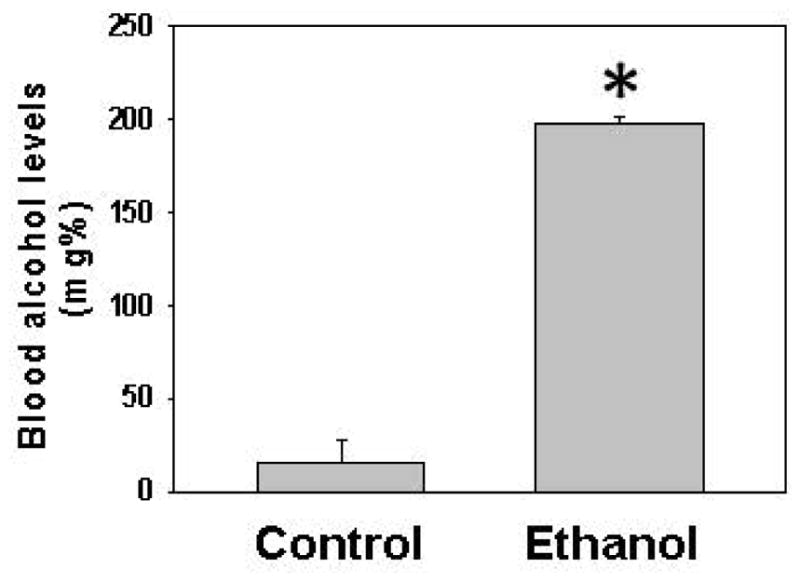
Blood alcohol concentration (BAC) in ADH− deer mice fed ethanol for three months. Values are mean ± standard error of 4 animals/group. * p value ≤ 0.05
Fig. 2.
Representative H & E stained section of liver of ethanol-fed mice showing steatosis and infiltration of inflammatory/immune cells (magnification 20X, A) and CD3 positive T-lymphocytes as shown by arrows (magnification x40, B).
Fig. 3.
Representative 2D gels showing differentially altered proteins (≥1.5 fold, p value ≤ 0.05) identified by MALDI-TOF/TOF at three different pH ranges in the liver tissue homogenate of ADH− deer mice fed ethanol as compared to pair-fed controls. All the gel images have been cropped and contrast/brightness adjusted for better view ability. For this reason, the intensity differences between spots cannot be judged reliably from these pictures. The numbers in the figures are the spot identifiers given by the software (n = 4 animals/group).
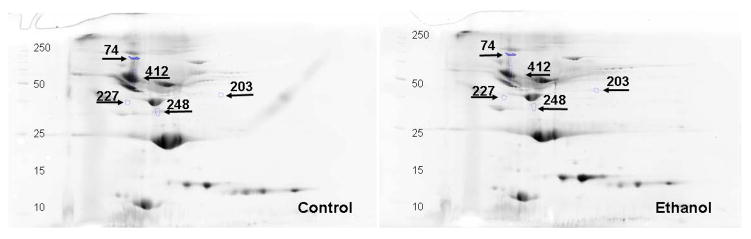
Fig. 4.
Representative 2D gels showing differentially altered proteins (≥1.5 fold, p value ≤0.05) identified by MALDI-TOF/TOF in the albumin and IgG depleted plasma of ADH− deer mice fed ethanol as compared to pair-fed controls. All the gel images have been cropped and contrast/brightness adjusted for better view ability. For this reason, the intensity differences between spots cannot be judged reliably from these pictures. The numbers in the figures are the spot identifiers given by the software (n = 4 animals/group).
Table 1.
Differentially expressed proteins in the livers of ethanol-fed ADH− deer mice (PI-isoelectric point, MW-molecular weight)
| UniProt KB Recommended Protein Name (alternate name) | GenInfo Identifier Number | Gel Spot No | PI | MW (kD) | MS ID Expectation value | Protein Abundance | Protein Function (cellular localization) |
|---|---|---|---|---|---|---|---|
| Fraction 1 (pH 3.0–5.4) | |||||||
| Protein disulfide isomerase (Prolyl 4-hydroxylase, β polypeptide, isoform CRA a) | gi|148702818 | 329 | 4.13 | 54 | 3.15E-04 | 5.03 | Peptide-proline hydroxylation to 4-hydroxy-L-proline, marker of hepatic fibrosis, catalyzes post-translational formation of 4-hydroxyproline in collagens and other protein with collagen sequences in ER lumen |
| Heat shock 70kD protein 5 (glucose-regulated protein), isoform CRA b | gi|148676670 | 472 | 3.48 | 41 | 3.97E-14 | 1.84 | Stress response protein involved in folding and assembly of proteins and monitors protein transport, ATP binding and apoptosis (ER) |
| Keratin type II cytoskeleton 8 | gi|309215 | 509 | 4.22 | 37 | 7.92E-18 | 2.34 | Together with KRT19, helps to link the contractile apparatus to dystrophin at the costameres of striated muscle, structural molecule activity, hepatocyte apoptosis process |
| 4OS ribosomal protein SA (laminin receptor) | gi|293694 | 633 | 3.51 | 30 | 2.50E-23 | −1.62 | Plays a role in cell adhesion to the basement membrane, cell fate determination and tissue morphogenesis (cell membrane, cytoplasm and nucleus). |
| Cytochrome b-5, isoform CRA a | gi|148677405 | 1229 | 3.99 | 15 | 7.92E-17 | 3.42 | Fatty acid metabolic and redox process (cell membrane, mitochondria) |
| Fraction 2 (pH 5.4–7.0) | |||||||
| ATP synthase subunit β, mitochondrial | gi|89574015 | 218 | 5.54 | 49 | 9.98E-29 | −1.93 | Catalyzes the production of ATP from ADP in the presence of proton gradient across the membrane (mitochondria) |
| Actin, gamma, cytoplasmic 1 | gi|123298587 | 279 | 5.62 | 43 | 9.98–10 | −1.64 | ATP binding and structural constituent of cytoskeleton |
| Stress-70 protein, mitochondria (75 kDa glucose-regulated protein; Short=GRP-75) | gi|14917005 | 126 | 5.7 | 71 | 3.15E-22 | 1.78 | Implicated in the control of cell proliferation and cellular aging, and may act as chaperone (mitochondria, nucleus) |
| Carbamoyl-phosphate synthase [ammonia], mitochondrial | gi|124248512 | 38 | 6.01 | 143 | 3.97E-18 | −1.56 | Involved in the urea cycle of ureotelic animals where the enzyme plays an important role in removing excess ammonia from the cell (mitochondria and nucleus) |
| Peroxiredoxin 1 (not listed in UniProt KB) | gi|123230136 | 515 | 6.15 | 24 | 5.00E-05 | 1.66 | Cell proliferation, possesses thioredoxin peroxidase activity and redox process (cytoplasm, mitochondria) |
| mCG124077, isoform CRA_f | gi|148706617 | 407 | 6.29 | 31 | 1.58E-11 | −1.63 | ?? not found |
| ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit, isoform 1, isoform CRA_g | gi|148677503 | 525 | 6.6 | 23 | 3.15E-09 | 1.85 | ATP synthesis, H+ ion transport and respiration |
| Ornithine carbamoyl transferase, mitochondrial | gi|19353187 | 570 | 6.79 | 18 | 3.97E-13 | 1.56 | Catalyzes the second step in urea cycle |
| Glutathione S-transferase | gi|340545553 | 730 | 6.81 | 23 | 6.29E-05 | 1.8 | Catalyzes the conjugation of reduced form of glutathione to xenobiotic substrates in detoxification process (cytoplasm) |
| Fraction 3 (pH 7–10) | |||||||
| Carbonic anhydrase 3 | gi|31982861 | 57 | 7.71 | 70 | 1.99E-09 | −2.13 | Reversible hydration of carbon dioxide, involved in formation of double bonds by removing groups from a substrate other than by hydrolysis or that adds groups to double bonds (cytoplasm) |
| Aldolase 2, B isoform, isoform CRA_e (oxidoreductase) | gi|148670368 | 234 | 9.55 | 39 | 5.00E-10 | −1.67 | Catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another the oxidant, an electron acceptor. This group of enzymes usually utilizes NADP or NAD+ as cofactors (cytoplasm) |
| ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit, isoform 1, isoform CRA_e (not listed in UniProt KB) | gi|148677501 | 318 | 7.68 | 31 | 3.15E-11 | 1.97 | Synthesis of ATP from ADP in the presence of a proton gradient across the membrane, hydrogen ion transport (cell and mitochondrial inner membrane) |
| Enoyl Coenzyme A hydratase, mitochondrial | gi|148685962 | 371 | 8.81 | 28 | 2.51E-12 | 1.82 | Catalyzes the hydration of 2-trans-enoyl-coenzyme A (CoA) intermediates to L-3-hydroxyacyl-CoAs in β-oxidation of fatty acids |
Table 2.
Differentially expressed proteins in plasma of ethanol fed ADH-deer mice
| UniProt KB Recommended Protein Name (alternate name) | GenInfo Identifier Number | Gel Spot No | PI | MW (kD) | MS ID Expectation value | Protein Abundance | Protein Function (cellular localization) |
|---|---|---|---|---|---|---|---|
| Serine protease inhibitor A3A precursor (serpina 3a) | gi|267844860 | 74 | 4.88 | 100 | 5.00E-06 | −5.64 | Inhibitor of serine protease (secreted) |
| Creatine Kinase M-type (creatine kinase M-type) | gi|6671762 | 203 | 7.2 | 42 | 9.98E-10 | 1.92 | Reversibly catalyzes the transfer of phosphate between ATP and various phosphogens (e.g. creatinine phosphate) and energy transduction in tissues (cytoplasm) |
| Clusterin (sulfated glycoprotein-2 isoform 2) | gi|7677495 | 227 | 4.83 | 37 | 2.50E-04 | −1.54 | Functions as extracellular chaperone that prevents aggregation of nonnative proteins and cell death (in all cellular components) |
| Apolipoprotein E (apolipoprotein E, isoform CRA_e) | gi|148691233 | 248 | 5.58 | 34 | 6.29E-08 | −3.35 | Lipid metabolism and transport (secreted) |
| Serine protease inhibitor A3A (Serpin A3A) | gi|68053300 | 412 | 5.02 | 53 | 1.58E-06 | −1.6 | Regulation of proteolysis and inhibits serine-type endopeptidase (secreted, extracellular) |
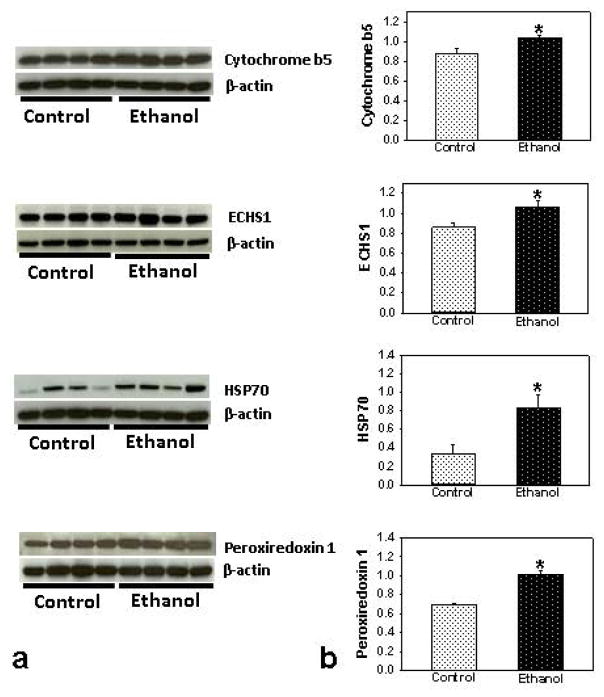
Out of total 18 identified proteins in the livers of ethanol-fed deer mice, 5 proteins were from fraction 1 (pH 3 – 5.4), 9 from fraction 2 (pH 5.4 – 7.0) and 4 from fraction 3 (pH 7 – 10) (Table 1). A total of 11 proteins were up regulated and 7 down regulated. Highly abundant proteins were prolyl 4-hydroxylase (protein disulfide isomerase, ~5 fold), cytochrome b-5 (~3 fold) and ATP synthase, heat shock 70kD protein, enoyl CoA hydratase and peroxiredoxin 1 (~2 fold). In contrast, decreased abundance was found for carbonic anhydrase 3, mitochondrial ATP synthase, subunit β (mitochondrial); laminin receptor and aldolase 2, B isoform. Up-regulated proteins indicate collagen formation, initiation of fibrosis, ER stress and increased fatty acid metabolism, cell proliferation and transferase activities. The down regulated proteins indicate decreased urea removal process, mitochondrial ATP synthesis and its binding, tissue morphogenesis and hydration of carbon dioxide. Western blot analysis of some differential expressed proteins supports proteomic data (Fig. 5).
Fig. 5.
Western blot analyses (a) and relative intensities (b) of some target proteins differentially expressed in the livers of ethanol fed ADH− deer mice using antibodies against cytochrome b5, ECHS1, HSP70 and peroxiredoxin 1. * p value ≤0.05 as compared to the controls (n = 4 animals/group).
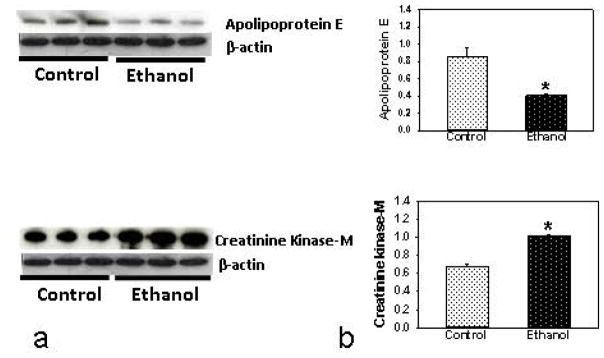
Only 5 out of 58 differentially altered proteins could be identified in the plasma of ethanol group using the criteria applied for the proteins identified in the liver (Table 2 and Fig. 4). Down regulated proteins were identified to be serine protease inhibitor A3A precursor (~6 fold), apolipoprotein E, isoform CRA-e (~3 fold), clusterin or sulfated-2 isoform 2 (~1.5 fold) and serine protease inhibitor A3A (~2 fold) in ethanol group. Only protein up-regulated was identified to be creatine kinase M-type (~2 fold). Western blot analysis supports the proteomic data of apolipoprotein and creatine kinase M-type (Fig. 6).
Fig. 6.
Western blot analysis (a) and relative intensity (b) of differentially expressed proteins in the plasma of ethanol-fed ADH− deer mice using antibodies against creatinine kinase-M or apolipoprotein E. * p value ≤0.05 as compared to the pair-fed controls (n = 3 animals/group).
Interrelationship between differentially altered proteins in the livers with those in the plasma could not be derived, but the ethanol-induced liver injury appears to be remarkable in our animal model. However, the changes in plasma proteome have potential for developing as biomarkers of ethanol induced liver injury.
Discussion
Liver is a major organ system for the toxicity and metabolism of the ingested alcohol (Lieber, 1997). However, hepatic ADH deficiency resulting into greater BAC as found in ethanol group is supported by our previous work (Bhopale et al., 2006; Kaphalia et al., 2010). Hepatic steatosis as found in the ethanol group could be due to increased lipogenesis, diminished transport of lipids from the liver and impaired β oxidation of fatty acids. This in part can be explained by the proteomic findings presented in this study. Infiltration of monocytes/macrophages, natural killer, NKT cells and T cells is central to pathogenic activity following acute and chronic liver injury. Upon activation, several hepatic cell populations, including hepatocytes, Kupffer cells, sinusoidal endothelial cells and hepatic stellate cells can secrete chemotactic cytokines and chemokines, which direct the migration of immune cells. Differentially altered proteome in the liver apparently reflects ethanol-induced injury and infiltration of inflammatory/immune cells including T-lymphocytes in our chronic ethanol feeding model of ADH− deer mice.
The general consensus on chronic alcoholic liver injury is thought to be mainly mediated via formation of acetaldehyde, catalyzed by hepatic ADH and/or Cyp2E1, resulting into oxidative stress (Lieber, 1997). Neither the levels of blood acetaldehyde nor oxidation of proteins as determined by Western blot analysis using antibodies against 4-hydroxynonenal (4-HNE, marker of oxidative stress) were significantly altered between the ethanol-fed mice and pair-fed controls (data not shown). ADH− deer mice cannot efficiently metabolize ethanol oxidatively resulting into an increased body burden of ethanol as found in this study is parallel to our previous reports (Bhopale et al., 2006; Kaphalia et al., 2010). This also contradicts a significant role of microsomal Cyp2E1 in ethanol oxidation to acetaldehyde and associated liver injury in our animal model. Overall, inhibition of hepatic ADH facilitates ethanol metabolism via nonoxidative pathway, which may be associated with observed liver injury in ethanol-fed ADH− deer mice. Further experiments regarding inhibition of hepatic nonoxidative metabolism and resultant liver injury can clarify the role of oxidative vs. nonoxidative ethanol metabolism in ethanol-induced hepatic injury. In this study, we focused on identification of proteins differentially altered in the liver and plasma of ADH− deer mice after chronic ethanol feeding.
Increased expression of prolyl 4-hydroxylase (protein disulfide isomerase), heat shock 70kD proteins, endo A cytokeratin, cytochrome b5 and peroxiredoxin 1 as found in the livers of ethanol fed mice indicate fibrotic response, ER stress, changes in structural proteins and their conformation and contractile apparatus, increased fatty acid metabolic process and peroxidase activity, respectively. Peroxiredoxin 1 is involved in cell proliferation, and possesses thioredoxin peroxidase activity and catalyzes the reduction of hydrogen and alkyl peroxide (Rhee et al., 2005; Wood et al., 2003). Similarly, up regulation of mitochondrial ATP synthase (H+ transporting F1 complex α subunit), ornithine carbamoyl-transferase (OTC ) and enoyl Co enzyme as found in the livers suggest impact of chronic ethanol feeding on ATP synthesis, transferase activity and fatty acid β oxidation, respectively. Glucose regulated protein (GRP)-75, a mitochondrial matrix protein, generally recognized as a member of the heat shock protein 70 class of proteins, is induced under conditions of low glucose and other nutritional and environmental stresses including ethanol-induced pancreatic injury (Kaphalia et al., 2010). GRP-75 is involved in various chaperoning functions in protein translocation, folding and mitochondrial functions, and implicated in the control of cell proliferation and cellular aging. Therefore, an up regulation of GRP-75 and other heat shock proteins is complementary and are closely associated. However, an increased expression of mitochondrial ATP synthase subunit (H+ transporting F1 complex α subunit) expression, a decreased expression of mitochondrial ATP synthase sub unit β (H+ transporting F1 complex β subunit) in ethanol-fed mice appears contradictory, since α (regulatory) and β (catalytic) subunits forms catalytic core of the F1 complex in F1-ATPases. Overall, the findings of this study suggest that chronic ethanol feeding targets a broad range of metabolic and structural proteins in the liver.
40S Ribosome protein SA or Laminin receptor plays an essential role as cell surface receptor and expresses laminin binding activity, which facilitates cell adhesion to the basement membrane and consequent activation of signaling transduction pathways (Rao et al., 1983). This protein may induce CD8 T-suppressor cells secreting IL-10 (Coggin et al., 1999), and its down regulation may result in detachment and migration of cells leading to tissue morphogenesis (Khalfaoui et al., 2013). Similarly, down regulation of carbamoyl phosphate synthase (CPS), which removes excess cellular ammonia, is consistent with our earlier finding in ethanol-fed rats (Fernando et al., 2013). CPS1 has been suggested as a biomarker for an early diagnosis of fatty liver, nonalcoholic steatohepatitis; and seems to be involved in regulations of triacylglycerol, ATP, phospholipid and fatty acid binding (Rodriguez-Suárez et al., 2010; Fernando et al., 2013). Therefore, deficiency of CPS1 could cause an accumulation of ammonia in the blood resulting into neurotoxicity (Ah Mew et al., 2013; Skowrońska and Albercht, 2013). Cytochrome B5 is involved in a number of oxidative reactions and probably also in mitochondrial anabolic metabolism of fats and steroids (Perez-Vázquez et al., 2014). Contrary to an increased expression in ethanol-fed group as found in this study, cytochrome B5 is shown to be down regulated in ethanol-induced fatty liver and hepatocellular carcinoma in rat models (Newton et al., 2009; Liu et al., 2011; Fernando et al., 2013). Currently, we do not have clear understanding about such variations in different animal species, which will need further studies.
Carbonic anhydrase (CA, cytosolic as well as membrane bound protein) maintains acid-base balance and exists in 14 different isoforms. CA III and other isoforms catalyze inter conversion of CO2 and HCO3 to maintain the acid–base balance in tissues, and scavenges oxygen radical (Yamada et al., 2013; Cabiscol and Levine, 1995). CA III has been reported to have an antioxidant activity and prevents H2O2–induced apoptosis, and is down-regulated in Cu-Zn superoxide dismutase-deficient mice (Elchuri et al., 2005; Ishii et al., 2005). Therefore, down regulation of CA III as reported in alcoholic liver injury supports our finding (Yamada et al., 2013). Keratin, type II cytoskeleton 8 (Krt8) expressed in the liver (Zhou et al., 2003) perhaps is involved in apoptosis of hepatocytes and tumor necrosis factor-mediated signaling pathway (Caulin et al., 2000). Therefore, an increased expression of endo A′ cytokeratin as found in this study could be associated with cell death process in ethanol-induced liver injury by chronic alcohol ingestion.
Significantly low expression of actin γ, cytoplasmic 1, an ATP binding protein and structural constituent of cytoskeleton; suggests structural injury (Belyantseva et al., 2009). Ornithine carbamoyl-transferase (OTC), like as glutathione S-transferase, possesses transferase activity and may be involved in urea cycle, which itself is involved in nitrogen metabolism, and plays important role in ammonia homeostasis (Petrak et al., 2007). Over expression of hepatic OTC has been reported to be a response to drugs and also ethanol intake (Murayama et al., 2009). Similarly, glutathione S-transferase (one of the key enzymes involved in phase II metabolism) could be deemed important for the ethanol excretory mechanisms.
The second step of fatty acid β-oxidation, an important cellular process to generate energy in the mitochondrial matrix, is catalyzed by enoyl CoA hydratase. An impaired β oxidation of fatty acids can result in accumulation of lipids in the hepatocytes as reported in ethanol-fed ADH− deer mice (Bhopale et al., 2006; Fernando et al., 2012). However, over expression of enoyl CoA hydratase as found in the livers of ethanol group could be a biological response to compensate an impaired β oxidation of fatty acids.
Plasma proteome regulates physiological status of various organs in the body, and has been applied for biomarker discovery and validation (Jacobs et al., 2005; Hanash et al., 2008). Serine protease inhibitor (inter-alpha-trypsin inhibitor heavy chain H4 produced in the liver) negatively regulates endopeptidase activity and its precursor protein (serine protease inhibitor A3A precursor). However, ethanol fed group showing no significant differential expression of serine protease inhibitor diminishes its utility as a potential biomarker (Freeman et al., 2011). Amongst many other plasma proteins, down regulation of clusterin or sulfated glycoprotein-2 isoform 2 or apolipoprotein E isoform CRA_e in ethanol fed group is also supported by data from nonhuman primate model (Freeman et al., 2011), although precise function of clusterin is not established yet.
Creatine Kinase (CK) M-type, a cytoplasmic isoenzyme, plays an important role in energy transduction in tissues and phosphocreatine biosynthetic and metabolic processes. CK catalyzes the production of ATP from ADP using phosphocreatine, and acts as an energy buffer (Wallimann et al., 1992). Increased expression of CK in the plasma probably indicates high demand of energy in ethanol-fed mice. Clinically, elevated CK level in the blood may be an indication of damage to CK–rich tissues including myocardial damage (Armstrong et al., 2008).
Apolipoproteins, mainly synthesized in the liver, are involved in lipid transport and maintenance of lipoproteins (Tietge et al., 1998; Sherlock, 1995; Getz and Rearon, 2009). Apolipoprotein E, isoform CRA_e has been identified as a potential biomarker in patients with hepatitis, cirrhosis and/or hepatocellular carcinoma induced by hepatitis C virus (Mas et al., 2009; Gőbel et al., 2006). Serum lipoprotein profile of patients with nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis reflected as hypercholesterolemia and dyslipidemia, and has been used as a diagnostic biomarker for identifying and staging of NAFLD (Bell et al., 2010). Therefore, decreased expression of apolipoprotein suggests impaired lipid transport from the liver in our model of chronic alcohol feeding.
Conclusion
Chronic ethanol feeding to ADH− deer mice results in very high levels of blood alcohol and significant liver injury as evident by steatosis and inflammatory response. This is the first proteomic study in deer mouse model showing that long-term ethanol feeding targets lipid and carbohydrate metabolism, ER stress and structural proteins in the livers. However, the proteins differentially expressed in the livers were not associated with those in the plasma. Heat shock/stress proteins and laminin receptor, and the proteins involved in ATP synthesis were found to be major target proteins in the livers of ethanol-fed deer mice. Significantly increased expression of propyl 4-hydroxylase, which catalyzes the formation of hydroxyproline (a marker of hepatic fibrosis), is an important finding suggesting that ADH− deer mouse model could be a suitable model to pursue the mechanistic aspect of ALD. Nevertheless, long term ethanol feeding in ADH− deer mouse model decreases the expression of proteins involved in the transport of lipids, which may be linked to reduced expression of apolipoproteins in the plasma. Therefore, reduced abundance of various carrier proteins as found in the plasma of ethanol group may have potential to be developed as biomarker(s) of ALD.
Acknowledgments
This work is supported by funds from grant AA24699 from National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health. The analysis of FAEEs was conducted in the Exposure Assessment and Biomarker Development laboratory supported by Sealy Center of Environmental Health & Medicine and NIEHS Center grant P30ES06676. Authors also acknowledge the support of Research Histopathology Core Laboratory, Department of Pathology for histology and immunohistochemistry work. The proteomic analysis was performed in the core laboratories of UTMB’s NHLBI-supported by Proteomics Center (N01-HV-00245, A).
Footnotes
Conflict of Interest
The authors have no conflict of interests to declare.
References
- Ah Mew N, Krivitzky L, McCarter R, Batshaw M, Tuchman M. Clinical outcomes of neonatal onset proximal versus distal urea cycle disorders do not differ. J Pediatr. 2013;162:324–329. doi: 10.1016/j.jpeds.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong AW, David EG. Pharmacology of Hemostasis and Thrombosis. In: Golan David E, Tashjian Armen H, Armstrong Ehrin J, Armstrong April W., editors. Principles of pharmacology: the pathophysiologic basis of drug therapy. Philadelphia: Lippincott Williams & Wilkins; 2008. p. 388. OCLC 76262148. [Google Scholar]
- Bell LN, Theodorakis JL, Vuppalanchi R, Saxena R, Bemis KG, Wang M, Chalasani N. Serum proteomics and biomarker discovery across the spectrum of nonalcoholic fatty liver disease. Hepatology. 2010;51:111–120. doi: 10.1002/hep.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva IA, Perrin BJ, Sonnemann KJ, Zhu M, Stepanyan R, Mc Gee J, Frolenkov GI, Walsh EJ, Friderick KH, Friedman TB, Ervasti JM. Gamma-actin is required for cytoskeletal maintenance but not development. Proc Natl Acad Sci USA. 2009;106:9703–9708. doi: 10.1073/pnas.0900221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhopale KK, Wu H, Boor PJ, Popov VL, Ansari GA, Kaphalia BS. Metabolic basis of ethanol-induced hepatic and pancreatic injury in hepatic alcohol dehydrogenase deficient deer mice. Alcohol. 2006;39:179–188. doi: 10.1016/j.alcohol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Bhopale KK, Nauduri D, Soman KV, Sood GK, Okorodudu A, Ansari GAS, Kaphalia BS. Differentially altered plasma proteins in patients diagnosed with alcoholic and nonalcoholic fatty liver disease. Euroasian J Hepato-Gastroenterology. 2011;1:89–99. [Google Scholar]
- Cabiscol E, Levine RL. Carbonic anhydrase III. Oxidative modification in vivo and loss of phosphatase activity during aging. J Biol Chem. 1995;270:14742–14747. doi: 10.1074/jbc.270.24.14742. [DOI] [PubMed] [Google Scholar]
- Caulin C, Ware CF, Magin TM, Oshima RG. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J Cell Biol. 2000;149:17–22. doi: 10.1083/jcb.149.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggin JH, Jr, Barsoum AL, Rohrer JW. 37 kiloDalton oncofetal antigen protein and immature laminin receptor protein are identical, universal T-cell inducing immunogens on primary rodent and human cancers. Anticancer Res. 1999;19:5535–5542. [PubMed] [Google Scholar]
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson RL, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- Fernando H, Bhopale KK, Paul J, Boor PJ, Shakeel Ansari GA, Kaphalia BS. Hepatic lipid profiling of deer mice fed ethanol using 1H and 31P NMR spectroscopy: a dose-dependent subchronic study. Toxicol Appl Pharmacol. 2012;264:361–369. doi: 10.1016/j.taap.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando H, Wiktorowicz JE, Soman KV, Kaphalia BS, Khan MF, Shakeel Ansari GA. Liver proteomics in progressive alcoholic steatosis. Toxicol Appl Pharmacol. 2013;266:470–480. doi: 10.1016/j.taap.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Vanguilder HD, Guidone E, Krystal JH, Grant KA, Vrana KE. Plasma proteomic alterations in non-human primates and humans after chronic alcohol self-administration. Int J Neuropsychopharmacol. 2011;14:899–911. doi: 10.1017/S1461145711000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes J, Spratt H, Wiktorowicz J, Wu Z, Boldogh I, Denner L, Kurosky A, Brasier RC, Luxon B, Brasier AR. Functional analysis of the nuclear proteome of human A549 alveolar epithelial cells by HPLC-high resolution 2-D gel electrophoresis. Proteomics. 2006;6:2656–2672. doi: 10.1002/pmic.200500652. [DOI] [PubMed] [Google Scholar]
- Galasko D. Biomarkers for Alzheimer’s disease-clinical needs and application. J Alzheimers Dis. 2005;8:339–346. doi: 10.3233/jad-2005-8403. [DOI] [PubMed] [Google Scholar]
- Göbel T, Vorderwülbecke S, Hauck K, Fey H, Häussinger D, Erhardt A. New multi protein patterns differentiate liver fibrosis stages and hepatocellular carcinoma in chronic hepatitis C serum samples. World J Gastroenterol. 2006;12:7604–7612. doi: 10.3748/wjg.v12.i47.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz GS, Reardon CA. Apoprotein E as a lipid transport and signaling protein in the blood, liver, and artery wall. J Lipid Res. 2009;50:S156–S161. doi: 10.1194/jlr.R800058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts BN, Spira A, Massion PP, Walser TC, Wistuba II, Minna JD, Dubinett SM. Evolving concepts in lung carcinogenesis. Semin Respir Crit Care Med. 2011;32:32–43. doi: 10.1055/s-0031-1272867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Akazawa D, Aoki Y, Yamada H, Oguri K. Suppression of carbonic anhydrase III mRNA level by an aryl hydrocarbon receptor ligand in primary cultured hepatocytes of rat. Biol Pharm Bull. 2005;28:1087–1090. doi: 10.1248/bpb.28.1087. [DOI] [PubMed] [Google Scholar]
- Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- Kanaujiya JK, Lochab S, Pal P, Christopeit M, Singh SM, Sanyal S, Behre G, Trivedi AK. Proteomic approaches in myeloid leukemia. Electrophoresis. 2011;32:357–367. doi: 10.1002/elps.201000428. [DOI] [PubMed] [Google Scholar]
- Kaphalia BS, Khan MF, Carroll RM, Aronson J, Ansari GAS. Subchronic toxicity of 2-chloroethanol and 2-bromoethanol in rats. Res Commun Pharmacol Toxicol. 1996;1:173–186. [Google Scholar]
- Kaphalia BS, Bhopale KK, Kondragunti S, Wu H, Boor PJ, Ansari GAS. Pancreatic injury in hepatic alcohol dehydrogenase-deficient deer mice after subchronic exposure to ethanol. Toxicol Appl Pharmacol. 2010;246:152–163. doi: 10.1016/j.taap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaphalia L, Boroumand N, Hyunsu J, Kaphalia BS, Calhoun WJ. Ethanol metabolism, oxidative stress, and endoplasmic reticulum stress responses in the lungs of hepatic alcohol dehydrogenase deficient deer mice after chronic ethanol feeding. Toxicol Appl Pharmacol. 2014;277:109–117. doi: 10.1016/j.taap.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karinch AM, Martin JH, Vary TC. Acute and chronic ethanol consumption differentially impact pathways limiting hepatic protein synthesis. Am J Physiol Endocrinol Metab. 2008;295:E3–9. doi: 10.1152/ajpendo.00026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfaoui T, Groulx JF, Sabra G, GuezGuez A, Basora N, Vermette P, Beaulieu JF. Laminin receptor 37/67LR regulates adhesion and proliferation of normal human intestinal epithelial cells. PLoS One. 2013;8:e74337. doi: 10.1371/journal.pone.0074337. eCollection 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber C. Ethanol metabolism, cirrhosis and alcoholism. Clini Chim Acta. 1997;257:59–84. doi: 10.1016/s0009-8981(96)06434-0. [DOI] [PubMed] [Google Scholar]
- Lieber CS, DeCarli LM. Liquid diet technique of ethanol administration: 1989 update. Alcohol Alcohol. 1989;24:197–211. [PubMed] [Google Scholar]
- Liu Z, Ma Y, Yang J, Qin H. Upregulated and downregulated proteins in hepatocellular carcinoma: a systematic review of proteomic profiling studies. OMICS. 2011;15:61–71. doi: 10.1089/omi.2010.0061. [DOI] [PubMed] [Google Scholar]
- Mas VR, Maluf DG, Archer KJ, Yanek K, Bornstein K, Fisher RA. Proteomic analysis of HCV cirrhosis and HCV-induced HCC: identifying biomarkers for monitoring HCV-cirrhotic patients awaiting liver transplantation. Transplantation. 2009;87:143–152. doi: 10.1097/TP.0b013e318191c68d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama H, Ikemoto M, Hamaoki M. Ornithine carbamyltransferase is a sensitive marker for alcohol-induced liver injury. Clin Chim Acta. 2009;401:100–104. doi: 10.1016/j.cca.2008.11.027. [DOI] [PubMed] [Google Scholar]
- Newton BW, Russell WK, Russell DH, Ramaiah SK, Jayaraman A. Liver proteome analysis in a rodent model of alcoholic steatosis. J Proteome Res. 2009;8:1663–1671. doi: 10.1021/pr800905w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIAAA (National Institute on Alcohol Abuse and Alcoholism) 10th Special Report to the U.S Congress on Alcohol and Health. U.S. Department of Health and Human Services, NIH; 2000. Alcohol Health Services Research. [Google Scholar]
- Panés J, Caballería J, Guitart R, Parés A, Soler X, Rodamilans M, Navasa M, Parés X, Bosch J, Rodés J. Determinants of ethanol and acetaldehyde metabolism in chronic alcoholics. Alcohol Clin Exp Res. 1993;17:48–53. doi: 10.1111/j.1530-0277.1993.tb00725.x. [DOI] [PubMed] [Google Scholar]
- Pérez-Vázquez V, Guzmán-Flores JM, Mares-Álvarez D, Hernández-Ortiz M, Macías-Cervantes MH, Ramírez-Emiliano J, Encarnación-Guevara S. Differential proteomic analysis of the pancreas of diabetic db/db mice reveals the proteins involved in the development of complications of diabetes mellitus. Int J Mol Sci. 2014;15:9579–9593. doi: 10.3390/ijms15069579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrak J, Myslivcova D, Man P, Cmejla R, Cmejlova J, Vyoral D, Elleder M, Vulpe CD. Proteomic analysis of hepatic iron overload in mice suggests dysregulation of urea cycle, impairment of fatty acid oxidation, and changes in the methylation cycle. Am J PhysiolGastrointest Liver Physiol. 2007;292:G1490–1498. doi: 10.1152/ajpgi.00455.2006. [DOI] [PubMed] [Google Scholar]
- Rao NC, Barsky SH, Terranova VP, Liotta LA. Isolation of a tumor cell laminin receptor. Biochem Biophys Res Commun. 1983;111:804–808. doi: 10.1016/0006-291x(83)91370-0. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Suárez E, Duce AM, Caballería J, MartínezArrieta F, Fernández E, Gómara C, Alkorta N, Ariz U, Martínez-Chantar ML, Lu SC, Elortza F, Mato JM. Non-alcoholic fatty liver disease proteomics. Proteomics Clin Appl. 2010;4:362–371. doi: 10.1002/prca.200900119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferovic MD, Krughkoc V, Pinto D, et al. Quantitative 2Dgel electrophoresis-based expression proteomics of albumin and IgG immunodepletedplasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;865:147–152. doi: 10.1016/j.jchromb.2008.01.052. [DOI] [PubMed] [Google Scholar]
- Sharakawi M. In vivo inhibition of liver alcohol dehydrogenase by ethanol administration. Life Sci. 1984;35:2353–2357. doi: 10.1016/0024-3205(84)90527-7. [DOI] [PubMed] [Google Scholar]
- Sherlock S. Alcoholic liver disease. Lancet. 1995;345:227–229. doi: 10.1016/s0140-6736(95)90226-0. [DOI] [PubMed] [Google Scholar]
- Skowrońska M, Albrecht J. Oxidative and nitrosative stress in ammonia neurotoxicity. Neurochem Int. 2013;62:731–737. doi: 10.1016/j.neuint.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Tietge UJ, Boker KH, Bahr MJ, Weinberg S, Pichlmayr R, Schmidt HH, Manns MP. Lipid parameters predicting liver function in patients with cirrhosis and after liver transplantation. Hepatogastroenterology. 1998;45:2255–2260. [PubMed] [Google Scholar]
- Torrente PM, Freeman WM, Vrana KE. Protein biomarkers of alcohol abuse. Expert Rev Proteomics. 2012;9:425–436. doi: 10.1586/epr.12.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrills NM, Irwin JA, He XY, Wood LG, Powell H, Simpson JL, McDonald VM, Sim A, Gibson PG. Identification of novel diagnostic biomarkers for asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:1633–1643. doi: 10.1164/rccm.201010-1623OC. [DOI] [PubMed] [Google Scholar]
- Wallimann T, Wyss M, Brdiczka D, Nicolay K, Eppenberger HM. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the ‘phosphocreatine circuit’ for cellular energy homeostasis. Biochem J. 1992;281:21–40. doi: 10.1042/bj2810021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzmann FA, Strother WN. Proteomics and alcoholism. Int Rev Neurobiol. 2004;61:189–214. doi: 10.1016/S0074-7742(04)61008-7. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxin. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Yamada M, Satoh M, Seimiya M, Sogawa K, Itoga S, Tomonaga T, Nomura F. Combined proteomic analysis of liver tissue and serum in chronically alcohol-fed rats. Alcohol Clin Exp Res. 2013;37(Suppl 1):E79–87. doi: 10.1111/j.1530-0277.2012.01883.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chait BT. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Toivola DM, Feng N, Greenberg HB, Franke WW, Omary MB. Keratin 20 helps maintain intermediate filament organization in intestinal epithelia. Mol Biol Cell. 2003;214:2959–2971. doi: 10.1091/mbc.E03-02-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]