Abstract
Non-invasive genomic profiling of tumors may be possible with next-generation sequencing (NGS) of blood-derived circulating tumor DNA (ctDNA), but proof of concept in a large cohort of patients with diverse cancers has yet to be reported. Here we report the results of an analysis of plasma-derived ctDNA from 670 patients with diverse cancers.
The tumors represented in the patient cohort were mainly gastrointestinal (31.8%), brain (22.7%) or lung (20.7%). ctDNA obtained from most patients (N = 423 (63%)) displayed at least 1 alteration. The most frequent alterations seen, as characterized mutations or variations of unknown significance, occurred in TP53 (32.5% of patients), EGFR (13%), KRAS (12.5%) and PIK3CA (9.1%); for characterized alterations, 30.7% (TP53), 7.6% (EGFR), 12.2% (KRAS), and 7.7% (PIK3CA). We found that 32% of brain tumors had at least 1 ctDNA alteration. Head and neck tumors were independently associated with a higher number of alterations in a multivariable analysis (P=0.019). Notably, 320/670 (48%) of patients displayed potentially actionable alterations with 241 patients possible candidates for on-label or off-label treatment with an FDA-approved drug. Several illustrations of the clinical utility of the information obtained for improving treatment of specific patients is provided. Our findings demonstrate the feasibility and impact of genomic profiling of tumors by ctDNA next-generation sequencing, greatly encouraging broader investigations of the application of this technology for precision medicine in cancer management.
Keywords: Cancer, liquid biopsy, ctDNA, actionable alteration, personalized therapy
Introduction
The landscape of the cancer genome features frequently altered genes that are potential clinical targets. Technology to detect circulating tumor DNA (ctDNA) derived from blood (liquid biopsy) is a promising tool to interrogate the molecular characteristics of tumors. The discipline of noninvasive disease monitoring has advanced greatly since cell-free DNA (cfDNA) was first reported in body fluids. In contrast to liquid biopsies, the invasive nature of tissue biopsies poses a risk to patients and can be costly. Tumor sampling for some cancer types is difficult and, as a result, inadequate amount or quality of tissues is often an issue. Indeed, in metastatic non-small cell lung cancers (NSCLC), almost 30% of patients do not have easily accessible tissue (1). In addition, tissue preservation methods such as formalin fixation can cause cytosine to thymine transitions by cytosine deamination, theoretically leading to false-positive results during sequencing (2). Finally, due to tumor heterogeneity, biopsies suffer from their limitation of reflecting only the small piece of tissue sampled (3,4).
The tumor ecosystem is dynamic and is able to change its dominant mutation pattern or acquire new alterations in response to selective pressure from previous therapies. Tumor evolution is of specific concern when stratifying subjects to a specific targeted therapy based on a mutation profile from an old report. For example, approximately 50% of patients with NSCLC become resistant to initial tyrosine kinase inhibitor therapy through an EGFR T790M mutation (5).
Tumor cells often shed their DNA content upon cell death. The result is that there is often ctDNA detectable in the bloodstream. Interestingly, it has been observed that patients with more advanced disease have higher concentrations of ctDNA, and that ctDNA percentage correlates with survival (6). While detection of ctDNA is still challenging since only a very small fraction of total cell-free DNA is derived from tumors (7), next-generation sequencing (NGS) has made analysis more feasible. Indeed, it is now becoming possible to map the genomic makeup of a cancer and identify potential treatment options with just a simple blood test (8,9). ctDNA assessment often (though not always) shows high overall agreement with tissue genomic tests (10) and may be an attractive technology to serially monitor cancer progression (8,9,11,12).
To facilitate studies that explore the utility of ctDNA analysis, a comprehensive understanding of the findings in diverse cancer types is essential.
Herein, we report the results of digital NGS ctDNA analysis in 670 patients. Most patients demonstrated genomic alterations in their ctDNA. We also describe three illustrative patients (gastric and lung cancer) who had EGFR ctDNA alterations and achieved partial responses after EGFR-targeted therapy, and a third patient (colon cancer) in whom EGFR and KRAS resistant alterations in ctDNA emerged with acquired resistance after initial response on cetuximab.
Materials and Methods
Patients
We retrospectively reviewed the liquid biopsy results of 670 consecutive, de-identified patients with diverse cancers who were seen at the University of California San Diego (UCSD), Moores Cancer Center. Pathology was reviewed at UCSD. Blood samples were collected between June 4th, 2014 and December 17th, 2015. This study was performed in compliance with UCSD IRB exempt approval for study of pre-existing de-identified data and the Declaration of Helsinki. [Patient case studies gave consent (NCT02478931)]. Analysis of usage patterns of liquid biopsies at our institution indicates that the majority of patients who have had these tests performed have advanced or metastatic disease.
Sequencing
Digital Sequencing was performed by Guardant Health, Inc. (Guardant360, www.guardanthealth.com/guardant360/), a Clinical Laboratory Improvement Amendment (CLIA)-certified and College of American Pathologists (CAP)-accredited clinical laboratory. For 150 patients, a 54 gene panel was used. This test identifies potential tumor-related alterations within 54 cancer-related genes (Supplemental Table 1) including amplifications in ERBB2, EGFR, and MET through analysis of cell-free DNA extracted from plasma (from two 10 ml blood tubes; indels and fusions were not detected as part of this panel)(13). For 400 patients, a 68-gene version of the original panel (expanded to all four major alteration types) was used, and for 120 patients, the most recent 70 gene panel version (further expanded to amplifications in 18 genes and fusions in 6 genes) was applied (Supplemental Tables 2 and 3). Only non-synonymous alterations were analyzed in our study (variants of unknown significance (VUSs) did not include synonymous alterations). Alterations were dichotomized into “characterized” or “VUS” at the variant level.
The ctDNA assay used in this study has a high clinical sensitivity (detects 85%+ of the single nucleotide variants detected in tissue in advanced cancer patients) and analytic specificity (>99.9999%). A high degree of specificity is critical to eliminate the false positives (noise) that otherwise accompany sequencing DNA at very low concentrations over long targeted regions. All cell-free DNA is sequenced, including the germline cell-free DNA that is derived from leukocyte lysis and the somatic ctDNA. Only non-germline somatic alterations were reported and analyzed in this study (Guardant360 is currently not validated to report germline alterations). Single nucleotide variants are quantitated as mutant allele fraction (MAF), which is the number of ctDNA fragments, divided by the number of wild type DNA fragments that overlap the same mutated nucleotide base position. Gene amplifications are reported as absolute gene copy number in plasma. In each sequencing run, a normal control sample is included (Guardant360 digital sequencing panel, Guardant Health Inc, data on file).
Potential actionability
Actionability implies that the protein product of a genomic abnormality can potentially be impacted by a specific targeted drug (14). A potentially actionable alteration was defined as a characterized alteration that was either the direct target (such as an EGFR inhibitor targeting an EGFR mutation), or a pathway component (such as an mTOR inhibitor for a PIK3CA mutation (since mTOR is downstream of PIK3CA)) that could be targeted by at least one approved (by the Food and Drug Administration–FDA) or investigational drug in a clinical trial. Potential actionability was crosschecked by two investigators; including the senior investigator (RK).
Data extraction and analysis
Demographic information such as gender, age, primary tumor site, as well as the dates of sample collection, test results, list of actionable alterations data (the number of alterations with an approved drug available in the disease (on-label use), the number of alterations with an approved drug in another disease (off-label use), and the number of alterations with experimental drug(s) available (clinical trials) were extracted from the reports and analyzed. When appropriate, median and 95% confidence intervals (95% CI) or range were reported. The sample size was determined by the available patients with genetic testing information at the time of analysis, and most of the statistical analysis (performed by MS; program: SPSS version 24.0) was descriptive in nature.
Results
Patient characteristics
Our population included 670 patients with diverse cancers who had a biopsy-free digital NGS ctDNA test performed on their plasma. Patient’s median age was 62 years old (Range, 5–92). Women and men were well balanced, with about 50% in each group. The most common tumor sites were gastrointestinal (31.8%), followed by brain (22.7%) and lung (20.7%) (Table 1).
Table 1.
Population characteristics
| Characteristics | Total patients, N = 670 |
|---|---|
| Gender | |
| Women | 336 (50.1%) |
| Men | 334 (49.9%) |
| Age (median, range) | 62.0 years (5–92) |
| Turn over timea (median, 95%CI; range) | 15 days (15–16 ; 7–35) |
| Tumor origin | |
| Gastrointestinal | 213 (31.8%) |
| Brain | 152 (22.7%) |
| Lung | 139 (20.7%) |
| Breast | 55 (8.2%) |
| Head and neck | 25 (3.7%) |
| Genitourinary | 19 (2.8%) |
| Carcinoma of unknown primary | 13 (1.94%) |
| Skin/Melanoma | 10 (1.5%) |
| Gynecologic | 7 (1.0%) |
| Erdheim-Chester disease | 6 (0.9%) |
| Mesothelioma | 5 (0.74%) |
| Otherb | 26 (3.88%) |
| Number of patients with ≥ 1 alteration (includes characterized alterations and VUSs) | 423 (63.1%) |
| Median number of alterations (range) (includes characterized alterations and VUSs) | 1 (0–26) |
| Number of patients with ≥ 1 characterized alteration | 322 (48.1%) |
| Median number of characterized alterations (range) in patients with alterations | 1 (0–13) |
Time from blood collection to results.
Other included: Lymphoma, n=4; Neuroendocrine carcinoma, n=4; Sarcoma, n=3; Castleman disease, n=2; Thymoma, n=2; Chordoma, n=1; Desmoid tumor, n=1; Nerve sheath tumor, n=1; Myoepithelial carcinoma, n=1; Thymic carcinoma, n=1, Prolactinoma, n=1; and Unknown diagnosis, n=5.
The majority of lung cancers were adenocarcinomas, with n=118/139 = 84.9% (The 21 remaining cases were squamous cell carcinoma, n=12; small cell carcinoma, n=7; adenosquamous, n=1; carcinoid tumor, n=1)).
Characterized alterations were defined at the gene variant level. VUS: variant of unknown significance.
Circulating tumor DNA (ctDNA) sequencing results
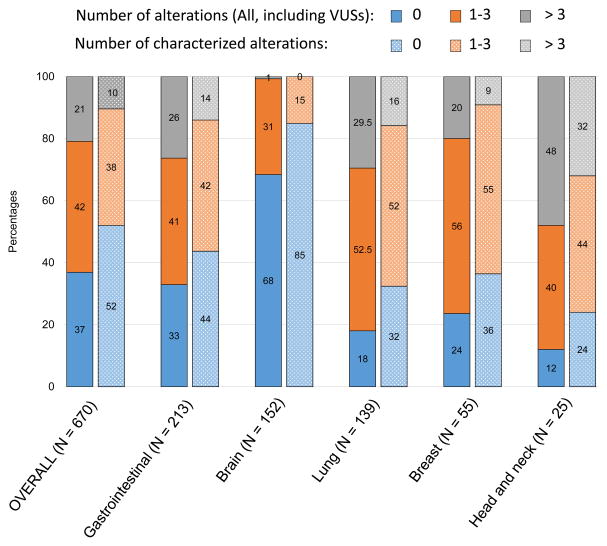
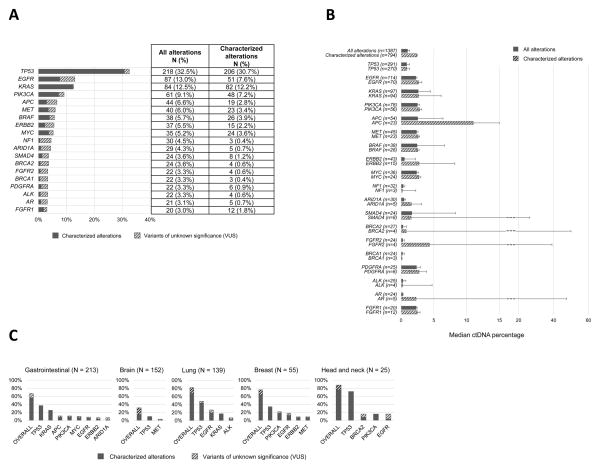
The median testing turnover time, defined as the median time from sample collection until results, was 15 days (95%CI, 15–16 days; range 7–35 days). In the overall population (670 tested patients), 63.1% of patients had at least one alteration detected in their plasma (Table 1 and Figure 1). The most frequent alterations identified were TP53 (32.5%), followed by EGFR (13.0%), KRAS (12.5%), PIK3CA (9.1%) and APC (6.6%) (Figure 2A), and the median percentage of ctDNA detected for all the alterations (N=1,387) in our population was 0.98% (Figure 2B). Of interest, 32% of brain tumors (N = 49/152) had at least one alteration detected in their plasma. Head and neck cancers comprised the tumor type with the highest percentage of patients with alterations detected (88%), as well as the highest frequency of patients with more than three alterations detected (48%) (Figure 1 and 2C). This was confirmed by a multivariable analysis (linear regression model) looking at the correlation between patient’s clinical characteristics and the number of detected alterations; head and neck tumors were an independent predictor of a higher number of alterations (P=0.019, median of 3 alterations (95%CI 1–6)), while brain tumors correlated with less alterations (P=0.017, median of 0 alterations identified) (Table 2). The examination of characterized alterations (excluding VUSs) also concluded that head and neck tumors were associated with a statistically higher number of characterized alterations (P=0.008, median 2 (95%CI 1–4) in a multivariable analysis) (Table 2 and Figure 1). In addition, the presence of either TP53, EGFR, KRAS, and/or PIK3CA alteration(s), as well as the detection of at least one alteration with a ctDNA percentage ≥10% were also statistically independent predictors of an increased number of alterations detected in patient’s plasma (all P<0.001 in the multivariable analyses when both “all alterations” or “characterized alterations only” considered, Table 2).
Figure 1. Percentage of patients with a given number of alterations.
Bar graph representing the percentage of patients carrying the specified number of alterations, for the most common tumor types (at least 25 patients). For each tumor type, the left bar represents the percentages for all non-synonymous alterations, including variant of unknown significance (VUSs); and the right bar represents the percentages for the characterized alterations only (defined at the gene variant level).
Figure 2. Frequency and quantitation of alterations.
A, Frequency of alterations in the overall population (N = 670). Only genes with alterations in ≥ 20 patients are displayed; all alterations were non-synonymous. Alterations of unknown significance (variant of unknown significance; VUSs) versus characterized were considered at the variant level. Some patients had multiple alterations in the same gene. Alterations were also identified in the following genes (albeit in less than 3% of patients): ABL1, AKT1, ARAF, ATM, CCND1, CCND2, CCNE1, CDK4, CDK6, CDKN2A, CDKN2B, CSF1R, CTNNB1, ERBB4, ESR1, EZH2, FBXW7, FGFR3, GATA3, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KIT, MAP2K1, MAP2K2, MLH1, NFE2L2, NOTCH1, NRAS, NTRK1, PROC, PTEN, RAF1, RB1, RET, RHOA, RIT1, ROS1, SMO, SRC, STK11, TERT, TSC1, and VHL.
B, Median ctDNA percentages (95%CI) in patients who had an alteration in the designated gene. Note the break in the scale after 15% of ctDNA to accommodate some of the large 95%CI.
C, Percent of patients that had the designated alteration by tumor type. Bar graph representing the most frequent alterations per tumor type. Percentages given are percent of patients with that tumor type who harbored that alteration. Tumor types with at least 25 patients are represented. Most frequent genes per tumor types are represented (exception for lung tumors: we also included ALK because of its potential driver role in the disease, despite the fact that it was only found in 10 patients). All alterations were non-synonymous. The distinction between characterized alterations and variants of unknown significance (VUS) was done at the variant level.
Table 2.
Correlations between patient characteristics and the number of alterations*
| All alterations considered (including variants of unknown significance)
| |||||
|---|---|---|---|---|---|
| Univariable | Multivariable** | ||||
|
| |||||
| Variables | N of alterations median, (95% CI) | P-value | B coef. | t-statisticb | P-value |
|
| |||||
| Overall (N = 670) | 1 (1–1) | --- | --- | --- | --- |
|
| |||||
| Gender | 0.318 | --- | --- | --- | |
| Women (N = 336) | 1 (1–1) | ||||
| Men (N = 334) | 1 (1–1) | ||||
|
| |||||
| Age at diagnosisa | <0.001 | 0.034 | 1.193 | 0.233 | |
| ≤ 62 years old (N = 349) | 1 (0–1) | ||||
| > 62 years old (N = 321) | 2 (1–2) | ||||
|
| |||||
| Tumor type | |||||
| Gastrointestinal vs not (N = 213) | 2 (1–2) vs 1 (1–1) | 0.011 | −0.030 | −0.815 | 0.415 |
| Brain vs not (N = 152) | 0 (0–0) vs 2 (1–2) | <0.001 | −0.084 | −2.387 | 0.017 |
| Lung vs not (N = 139) | 2 (2–3) vs 1 (1–1) | <0.001 | −0.012 | −0.326 | 0.745 |
| Breast vs not (N = 55) | 1 (1–2) vs 1 (1–1) | 0.147 | --- | --- | --- |
| Head and neck vs not (N = 25) | 3 (1–6) vs 1 (1–1) | 0.001 | 0.069 | 2.359 | 0.019 |
|
| |||||
| Alterations detected | |||||
| TP53 vs not (N = 218) | 3 (3–4) vs 0 (0–1) | <0.001 | 0.268 | 8.868 | <0.001 |
| EGFR vs not (N = 87) | 4 (3–5) vs 1 (1–1) | <0.001 | 0.221 | 7.746 | <0.001 |
| KRAS vs not (N = 84) | 4 (3–5) vs 1 (1–1) | <0.001 | 0.166 | 5.494 | <0.001 |
| PIK3CA vs not (N = 61) | 5 (4–6) vs 1 (1–1) | <0.001 | 0.249 | 8.682 | <0.001 |
|
| |||||
| ≥ 1 gene altered withc | <0.001 | 0.216 | 7.035 | <0.001 | |
| ctDNA ≥ 10% (N = 101) | 5 (4–6) | ||||
| ctDNA < 10% (N = 569) | 1 (1–1) | ||||
|
| |||||
|
Characterized alterations considered (excluding variants of unknown significance)
| |||||
| Overall (N = 670) | 0 (0–1) | --- | --- | --- | --- |
|
| |||||
| Gender | 0.619 | --- | --- | --- | |
| Women (N = 336) | 0.5 (0–1) | ||||
| Men (N = 334) | 0 (0–1) | ||||
|
| |||||
| Age at diagnosisa | 0.001 | 0.015 | 0.673 | 0.501 | |
| ≤ 62 years old (N = 349) | 0 (0–0) | ||||
| > 62 years old (N = 321) | 1 (0.5–1) | ||||
|
| |||||
| Tumor type | |||||
| Gastrointestinal vs not (N = 213) | 1 (0–1) vs 0 (0–0) | 0.001 | −0.004 | −0.119 | 0.905 |
| Brain vs not (N = 152) | 0 (0–0) vs 1 (1–1) | <0.001 | −0.037 | −1.177 | 0.240 |
| Lung vs not (N = 139) | 1 (1–2) vs 0 (0–0) | <0.001 | 0.018 | 0.582 | 0.561 |
| Breast vs not (N = 55) | 1 (1–1) vs 0 (0–1) | 0.048 | −0.003 | −0.111 | 0.911 |
| Head and neck vs not (N = 25) | 2 (1–4) vs 0 (0–1) | <0.001 | 0.064 | 2.668 | 0.008 |
|
| |||||
| Characterized alterations detected | |||||
| TP53 vs not (N = 206) | 2 (2–2) vs 0 (0–0) | <0.001 | 0.358 | 15.163 | <0.001 |
| EGFR vs not (N = 51) | 3 (2–4) vs 0 (0–0) | <0.001 | 0.279 | 12.267 | <0.001 |
| KRAS vs not (N = 82) | 3 (2–3) vs 0 (0–0) | <0.001 | 0.241 | 10.127 | <0.001 |
| PIK3CA vs not (N = 48) | 4 (3–5) vs 0 (0–0) | <0.001 | 0.299 | 13.350 | <0.001 |
|
| |||||
| ≥ 1 gene altered withc | <0.001 | 0.183 | 7.473 | <0.001 | |
| ctDNA ≥ 10% (N = 92) | 3 (3–4) | ||||
| ctDNA < 10% (N = 578) | 0 (0–0) | ||||
Only tumor types with > 25 patients were included; only the most prevalent genes were included (genes altered in > 50 patients were included for the upper table and > 40 patients for the lower table).
Variables with P-values ≤ 0.05 in the univariable analysis were included in the multivariable model.
P-values were calculated using the Mann-Whitney tests for univariable analyses and a linear regression model was computed for the multivariable analysis.
Cut-off of 62 years old corresponds to the median (Table 1).
The t-statistic is the ratio of the B coefficient and the Standard Error; the higher the value, the greater is the importance of the variable in the model.
95% CI, 95% Confidence Interval.
The cut-off of 10% corresponds to ten times the median of percentage of ctDNA detected in all the alterations.
Actionability of the detected alterations
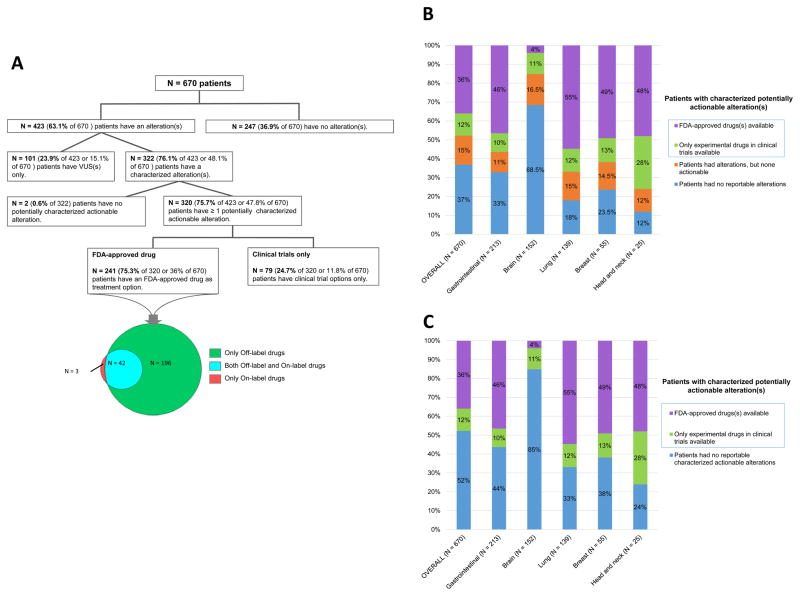
Of the total 670 analyzed patients, 320 had characterized, potentially actionable alterations (48% of total, 76% of the patients with alterations detected), Suppl Table 4. Indeed, amongst these 320 patients, 241 patients (75% of 320 or 36% of total patients) had at least one potentially matched FDA-approved drug as a treatment option (N=45 on-label and N=196 off-label only), Figure 3A. Of note, the majority of patients with gastrointestinal, lung, breast, and head and neck cancers had hypothetically actionable alterations (56%, 67%, 62%, and 76%, respectively), Figures 3B and 3C.
Figure 3. Actionability analysis.
A, Actionability diagram in the overall population. Diagram representing the potential actionability of ctDNA alterations in the overall population. In total, 320 patients (47.8% of 670) had at least one theoretically actionable alteration.
B, Potential actionability of alterations by tumor types. Only tumor types with at least 25 patients were represented. * Most of the time, patients were designated as having alterations, but none potentially actionable, because the alterations represented variants of unknown significance (overall 101 of 103 patients): only 2 patients had non-actionable characterized alterations.
C, Potential actionability of characterized alterations by tumor types. Only tumor types with at least 25 patients were represented. * Included patients with no alterations at all (N=247), patients with variants of unknown significance (VUS) only (N=101), or patients with non-actionable characterized alterations (N=2).
On the other hand, 350 patients (52% of total) had no actionable alterations. This was because no alterations were detected by the ctDNA test (247 patients, 37% of total) or because the alterations detected were variants of unknown significance (101 patients, 24%), Figure 3A.
Case Study 1: A patient with gastric cancer and EGFR amplification on ctDNA treated with anti-EGFR therapy
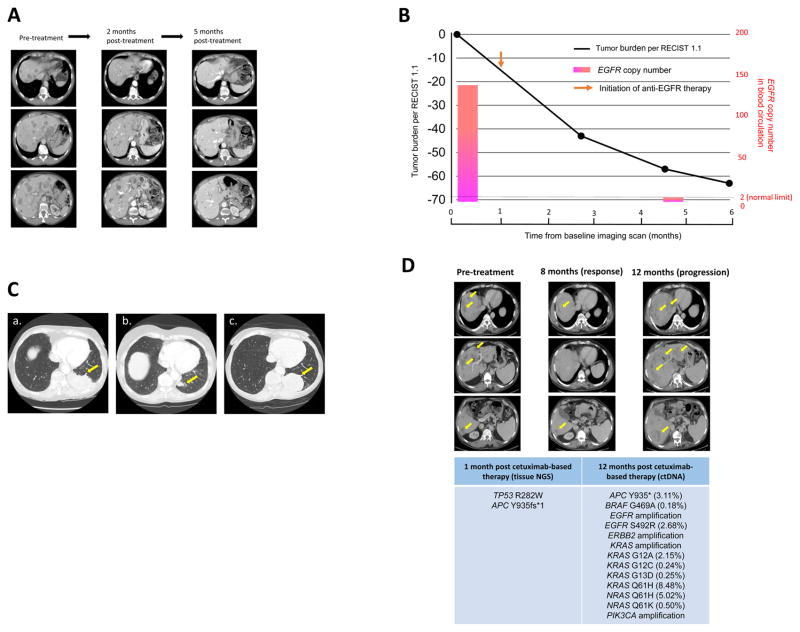
A 68-year-old woman presented to the clinic with advanced gastric cancer and liver metastasis. Tissue NGS (315 genes) revealed multiple amplifications including in EGFR, CCND3, MAP2K1, VEGFA, AURKA, ARFRP1, GNAS, PIK3CG and ZNF217 genes, as well as a TP53 G245S mutation. ctDNA analysis also revealed EGFR amplification. Since EGFR amplification was identified in both tissue and ctDNA profiling, the patient was started on an anti-EGFR-based regimen, which included both erlotinib and cetuximab (15). One week after the initiation of anti-EGFR therapy, the patient noticed significant improvement in her abdominal fullness and pain. Re-staging imaging approximately two months after the initiation of therapy showed reduction of multiple liver metastases (overall 40% decrease by RECIST 1.1. Response is ongoing at the most recent restaging (60% reduction at 5 months) (Figure 4A). Along with the reduction of tumor burden, EGFR copy number in blood circulation rapidly declined (originally 143.9, down to reference level 2.0) (Figure 4B). Carcinoembryonic antigen, which was originally 101 ng/ml also decreased to undetectable levels.
Figure 4. Patient cases.
A, Case Study 1: Serial imaging while receiving anti-EGFR therapy. Tissue NGS also showed EGFR amplification. Patient received erlotinib and cetuximab demonstrated notable improvement in abdominal fullness and pain after one week. Erlotinib dose was 150 mg PO daily and cetuximab dose was 250 mg/m2 intravenously (weekly).
Left, CAT scan of the abdomen before treatment. Middle, CAT scans about two months after treatment shows about 40% reduction in size of liver tumors. Right, CAT scans about five months after treatment showed further reduction of liver tumors (~60 % reduction).
B, Case Study 1: Change in tumor burden and EGFR copy number in blood circulation while receiving anti-EGFR therapy. Patient’s tumor burden assessed by RECIST 1.1 overall showed 60% tumor reduction five months after the initiation of anti-EGFR therapy. Also observed was a decrease in EGFR copy number to reference level (EGFR copy number: pre-treatment: 143.9, post-treatment: 2.0 [germline copy number of EGFR = 2.0]). Copy number was calculated based on the number of EGFR copy molecules in circulation by previously described methods (13).
C, Case Study 2: Serial CT scan images of primary EGFR-positive lung mass of a patient with adenocarcinoma of the lung and EGFR E746_A750 del alteration in ctDNA treated with anti-EGFR therapy (erlotinib).
Tissue NGS later also showed the EGFR alteration. Patient received erlotinib and demonstrated partial response.
a, CAT scan of the chest before treatment.
b, CAT scan after treatment shows significant reduction in tumor size.
c, Eight months after treatment started, patient shows progression. There is no EGFR T790M in the ctDNA or tissue and the patient therefore receives chemotherapy.
D, Case Study 3: Patient with K-RAS wild-type colon cancer treated with cetuximab-based therapy. ctDNA analysis at progression with multiple resistance alterations.
Patient with KRAS wild-type colon cancer was initially responding to the cetuximab-based therapy. Images show, left: pre-treatment, middle: partial response, and right: at progression.
At the time of progression, ctDNA was obtained and showed multiple alterations including EGFR/ERBB2/KRAS/PIK3CA amplifications, EGFR S492R, BRAF G469A, KRAS G12A/G12C/G13D/Q61H and NRAS Q61H alterations that could explain the reason for progression.
Case Study 2: A patient with adenocarcinoma of the lung and an EGFR ctDNA mutation treated with erlotinib
A 77-year-old woman was diagnosed with adenocarcimoma of the lung and multiple brain and bone metastases. ctDNA revealed an EGFR del 19 (E746_A750 del) mutation. Molecular tissue NGS analysis later confirmed EGFR E746_A750del, and also showed EGFR amplification, AURKA amplification, GNAS amplification, TP53 splice site 375+1G >T, and ZNF217 amplification. The patient was started on erlotinib and was expected to begin brain radiation. A week into the erlotinib treatment, brain imaging performed as a part of radiation treatment planning showed near complete resolution of all brain metastases, and radiation treatment plans were aborted. Follow-up chest computerized tomography showed partial response (Figure 4C). Eight months later, imaging indicated progression. Repeat ctDNA analysis demonstrated the original EGFR E746_A750 del mutation, EGFR amplification and no EGFR T790M secondary resistance mutation. Molecular analysis from a post-progression biopsy revealed, EGFR E746_A750del and TP53 splice site 375+1G >T. Because of the lack of EGFR T790M resistance mutation in either ctDNA or tissue, chemotherapy was initiated.
Case Study 3: Patient with K-RAS wild-type colon cancer treated with cetuximab-based therapy. ctDNA analysis at progression with multiple resistance alterations
A 55-year-old man with metastatic colon cancer was initiated on third-line therapy with cetuximab (anti-EGFR antibody) based therapy, along with 5-fluorouracil and irinotecan, after tumor progression on an oxaliplatin, irinotecan and bevacizumab based regimen. Biopsy of the liver mass performed one month after the initiation of cetuximab-based therapy for tissue NGS revealed TP53 R282W and APC Y935fs*1 alterations. The patient initially attained a partial response to the cetuximab-based regimen; however, the tumor gradually progressed after a year of therapy (Figure 4D). ctDNA analysis at the time of progression revealed multiple new alterations including EGFR/ERBB2/KRAS/PIK3CA amplifications, EGFR S492R, BRAF G469A, KRAS G12A/G12C/G13D/Q61H and NRAS Q61H alterations (Figure 4D).
Discussion
In this large study, we describe the molecular alterations identified in 670 patients suffering from various cancers. Targeted NGS that analyzed ctDNA from plasma was utilized. In the overall population, 63% of patients had at least one molecular alteration (in 48% of patients, it was a characterized alteration and, in the others, only a VUS). Consistent with our previous study (8), the most frequent alterations were in the TP53 tumor suppressor (32.5% of our patients), of which nearly all are characterized (30.7%). This frequency data is similar to that found with the use of tissue biopsy (16). The frequency of MET alterations in our population was 6% compared with previous studies where the rate of MET alterations varied from 6% to 32%, depending on whether ctDNA or tissue was analyzed, the tumor type, and the number of patients evaluated (17–20). Other common alterations were in EGFR (13.0%), KRAS (12.5%), PIK3CA (9.1%) and APC (6.6%) genes (Figure 2A). These frequencies were in the range of frequencies described in other studies (8,9,21).
Nearly 48% of all patients had a potentially actionable alteration. Among the 423 patients with alterations detected, 75.7% had a potentially actionable alteration. The rates of potentially actionable alterations in tissue, as reported in other studies, have varied widely (from about 20% to 93%), probably depending on the panel used, the definition of actionability (14) and the time period of the study (21–24). Of special interest was a patient with advanced gastric cancer who showed amplifications in EGFR in her ctDNA, a result that was confirmed by tissue NGS, and attained a rapid partial remission after two months of therapy with an anti-EGFR regimen combining an EGFR antibody and small molecule inhibitor. EGFR amplification is uncommon in gastric cancer (being seen in about 2–3% of patients). Preclinical studies have shown that a subset of gastric cancers and EGFR amplification respond to anti-EGFR agents (25). Further, combining anti-EGFR antibodies such as cetuximab with small molecule inhibitors such as erlotinib may augment clinical responsiveness (15).
In this study, 37% of patients did not have any detectable alteration. This was most prominent for brain tumors, where 68% of patients (N = 152 total tested) had no discernible alteration in their ctDNA. Bettegowda et al (6) determined that ctDNA can be detected over 75% of 640 patients with various cancer types, but in less than 50% of primary brain, renal, prostate, or thyroid cancers, suggesting that blood-brain barrier could prevent ctDNA from entering the circulation. On the other hand, it is interesting that 31% of our patients with brain tumors did have a discernible aberration on ctDNA testing If brain tumors are eliminated from the denominator, 72% of patients (374/518) had at least one discernible ctDNA alteration.
Two hundred forty-one patients (36%) had characterized abnormalities that could conceivably be addressed by at least one FDA-approved drug; an additional 79 patients (11.8%) had at least one alteration potentially druggable via in clinical trial. Overall, 76% of patients with alteration(s) had an aberration potentially actionable either by an experimental agent in clinical trials or by an approved drug. Potential actionability with FDA-approved drugs for characterized alterations was common in lung (55% of patients), breast (49%), head and neck (48%), and gastrointestinal cancers (46%).
For lung cancers, EGFR alterations were frequent and actionable. An illustrative patient is shown (Figure 4C). This patient had lung and brain metastases, and a first-generation kinase sensitive EGFR alteration in the ctDNA (also confirmed by tissue NGS). The patient showed an excellent response to erlotinib in both the brain and the lung, and, as a result, the planned radiation therapy for the brain was aborted. Because ctDNA requires only a small tube of blood, the time to attaining results is generally shorter than for tissue NGS, and the results may therefore give an early readout for therapy that is important for individuals with rapidly progressing disease or with metastases in crucial areas such as the brain.
Other potential clinical implications of ctDNA relate to understanding mechanisms of resistance to targeted agents. We describe an illustrative patient with colon cancer whose tissue NGS initially showed wild-type KRAS and was responding to cetuximab-based therapy. At the time of progression, multiple emerging alterations in ERBB2, BRAF, KRAS and NRAS were revealed; these alterations may lead to and predict cetuximab resistance (Figure 4D) (26,27).
For breast cancers, many of the actionable alterations were in the PTEN/PI3K/mTOR axis. Several mTOR inhibitors are being investigated or are approved, including everolimus, which is on-label for breast cancer. For head and neck cancers, characterized PIK3CA anomalies were also relatively frequent (about 16% of patients) (28). Notably, most of the alterations in BRCA2 and EGFR in head and neck cancer were of unknown significance (and hence not considered potentially actionable).
Although this study included significantly higher numbers of patients than many previous reports, it has several limitations. Most importantly, the database was de-identified and therefore had minimal clinical annotation. As such, staging as well as treatment outcomes and comparison with tissue biopsy results were not feasible. However, our previous work has shown that about 85% of our patients sent for ctDNA testing have metastatic/recurrent disease (9). Furthermore, our prior study in lung cancer demonstrated that approximately 28 percent of patients received therapy matching ≥ 1 ctDNA alteration(s) (29). This rate may be higher in lung cancer than in other tumors because of the availability of matched therapies. In addition, a previous study testing for ctDNA with the current technology demonstrated that important alterations were matched in blood and tissue in 87% of individuals (30).
In conclusion, we have evaluated 670 patients for ctDNA anomalies and demonstrate that a significant subset has detectable alterations. Perhaps surprisingly, about one-third of patients with brain tumors also have discernible altered ctDNA. Many of the abnormalities are at least theoretically druggable. This is exemplified by our patient with gastric cancer and an EGFR amplification, an alteration that occurs in less than three percent of patients with this type of tumor, who achieved a 60% reduction in the size of liver metastases after about five months of anti-EGFR treatment (case 1, Figure 4A and 4B). Furthermore, repeat testing showed normalization in amplitude of EGFR copy number in blood circulation consistent with tumor response. Of interest in this regard, liquid biopsies are non-invasive, and require only a small amount of blood for testing. In contrast, a tissue biopsy can be painful, and may have complications. Even though it has been already published that EGFR ctDNA tests may be useful to select treatment (31,32), our patient with lung cancer and brain metastases (Case 2, Figure 4C) exemplifies well how ctDNA was used prospectively to select therapy and how the patient responded when a rapid readout for genomic tests was needed in a setting of urgent treatment decisions. Serial liquid biopsies may also lend themselves to following therapy response and predicting resistance, and ctDNA testing may provide a technology for evaluating actionable alterations in difficult-to-biopsy tumors.
Supplementary Material
Acknowledgments
Funded in part by the Joan and Irwin Jacobs fund and by National Cancer Institute grant P30 CA016672 (R Kurzrock)
Footnotes
Conflict of Interest Disclosures: Dr. Kurzrock has research funding from Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, and Guardant Health, as well as consultant fees from X-Biotech and Actuate Therapeutics and has an ownership interest in Curematch, Inc. Dr. Bazhenova receives stocks or ownership from Epic Sciences, has a consulting or advisory role for Heat Biologics, Astra Zeneca, Pfizer, and Genoptix, participated in a speaker’s bureau for Genentech, Astra Zeneca and Novartis. Kimberly Banks, Richard Lanman, and AmirAli Talasaz are employees and own stocks at Guardant Health. Richard Lanman and AmirAli Talasaz also have a leadership position at Guardant Health and AmirAli Talasaz owns a patent or other intellectual property at Guardant Health. The other authors have nothing to disclose.
References
- 1.Kim ES, Hirsh V, Mok T, Socinski MA, Gervais R, Wu YL, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372:1809–18. doi: 10.1016/S0140-6736(08)61758-4. [DOI] [PubMed] [Google Scholar]
- 2.Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC clinical pathology. 2004;4:1. doi: 10.1186/1472-6890-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–64. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inukai M, Toyooka S, Ito S, Asano H, Ichihara S, Soh J, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer research. 2006;66:7854–8. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 6.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science translational medicine. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. 2016;7:9707–17. doi: 10.18632/oncotarget.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Use of Liquid Biopsies in Clinical Oncology: Pilot Experience in 168 Patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-16-0318. [DOI] [PubMed] [Google Scholar]
- 10.Janku F, Huang HJ, Claes B, Falchook GS, Fu S, Hong D, et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Molecular cancer therapeutics. 2016;15:1397–404. doi: 10.1158/1535-7163.MCT-15-0712. [DOI] [PubMed] [Google Scholar]
- 11.Hyman DM, Diamond EL, Vibat CR, Hassaine L, Poole JC, Patel M, et al. Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer discovery. 2015;5:64–71. doi: 10.1158/2159-8290.CD-14-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janku F, Angenendt P, Tsimberidou AM, Fu S, Naing A, Falchook GS, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. 2015;6:12809–21. doi: 10.18632/oncotarget.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PloS one. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turski ML, Vidwans SJ, Janku F, Garrido-Laguna I, Munoz J, Schwab R, et al. Genomically Driven Tumors and Actionability across Histologies: BRAF-Mutant Cancers as a Paradigm. Molecular cancer therapeutics. 2016;15:533–47. doi: 10.1158/1535-7163.MCT-15-0643. [DOI] [PubMed] [Google Scholar]
- 15.Wheler JJ, Tsimberidou AM, Falchook GS, Zinner RG, Hong DS, Fok JY, et al. Combining erlotinib and cetuximab is associated with activity in patients with non-small cell lung cancer (including squamous cell carcinomas) and wild-type EGFR or resistant mutations. Molecular cancer therapeutics. 2013;12:2167–75. doi: 10.1158/1535-7163.MCT-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Said R, Hong DS, Warneke CL, Lee JJ, Wheler JJ, Janku F, et al. P53 mutations in advanced cancers: clinical characteristics, outcomes, and correlation between progression-free survival and bevacizumab-containing therapy. Oncotarget. 2013;4:705–14. doi: 10.18632/oncotarget.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HJ, Mok TS, Chen ZH, Guo AL, Zhang XC, Su J, et al. Clinicopathologic and molecular features of epidermal growth factor receptor T790M mutation and c-MET amplification in tyrosine kinase inhibitor-resistant Chinese non-small cell lung cancer. Pathology oncology research : POR. 2009;15:651–8. doi: 10.1007/s12253-009-9167-8. [DOI] [PubMed] [Google Scholar]
- 20.de Melo Gagliato D, Jardim DL, Falchook G, Tang C, Zinner R, Wheler JJ, et al. Analysis of MET genetic aberrations in patients with breast cancer at MD Anderson Phase I unit. Clinical breast cancer. 2014;14:468–74. doi: 10.1016/j.clbc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwaederle M, Daniels GA, Piccioni DE, Fanta PT, Schwab RB, Shimabukuro KA, et al. On the Road to Precision Cancer Medicine: Analysis of Genomic Biomarker Actionability in 439 Patients. Molecular cancer therapeutics. 2015;14:1488–94. doi: 10.1158/1535-7163.MCT-14-1061. [DOI] [PubMed] [Google Scholar]
- 22.Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, et al. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer research. 2016;76:3690–701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DB, Dahlman KH, Knol J, Gilbert J, Puzanov I, Means-Powell J, et al. Enabling a genetically informed approach to cancer medicine: a retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. The oncologist. 2014;19:616–22. doi: 10.1634/theoncologist.2014-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasan N, Yelensky R, Wang K, Moulder S, Dzimitrowicz H, Avritscher R, et al. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. The oncologist. 2014;19:453–8. doi: 10.1634/theoncologist.2013-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Yang J, Cai J, Song X, Deng J, Huang X, et al. A subset of gastric cancers with EGFR amplification and overexpression respond to cetuximab therapy. Scientific reports. 2013;3:2992. doi: 10.1038/srep02992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature medicine. 2012;18:221–3. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 28.Psyrri A, Seiwert TY, Jimeno A. Molecular pathways in head and neck cancer: EGFR, PI3K, and more. American Society of Clinical Oncology educational book/ASCO American Society of Clinical Oncology Meeting; 2013; pp. 246–55. [DOI] [PubMed] [Google Scholar]
- 29.Schwaederle M, Patel SP, Husain H, Ikeda M, Lanman R, Banks KC, et al. Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017 doi: 10.1158/1078-0432.CCR-16-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zill OA, Mortimer S, Banks KC, Nagy RJ, Chudova D, Jackson C, et al. Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016:34. [Google Scholar]
- 31.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nature communications. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu YL, Sequist LV, Hu CP, Feng J, Lu S, Huang Y, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. British journal of cancer. 2017;116:175–85. doi: 10.1038/bjc.2016.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.