Abstract
Background
Chronic ethanol consumption is a major cause of liver disease worldwide. Oxidative stress is a known consequence of ethanol metabolism and is thought to contribute significantly to alcoholic liver disease (ALD). Therefore, elucidating pathways leading to sustained oxidative stress and downstream redox imbalances may reveal how ethanol consumption leads to ALD. Recent studies suggest that ethanol metabolism impacts mitochondrial antioxidant processes through a number of proteomic alterations, including hyperacetylation of key antioxidant proteins.
Methods
In order to elucidate mechanisms of ethanol-induced hepatic oxidative stress, we investigate a role for protein hyperacetylation in modulating mitochondrial superoxide dismutase (SOD2) structure and function in a 6-week Lieber-DeCarli murine model of ethanol consumption. Our experimental approach includes immunoblotting, immunohistochemistry, activity assays, mass spectrometry, and in silico modeling.
Results
We found that ethanol metabolism significantly increased the acetylation of SOD2 at two functionally relevant lysine sites, K68 and K122, resulting in a 40% decrease in enzyme activity while overall SOD2 abundance was unchanged. In vitro studies also reveal which lysine residues are more susceptible to acetylation. Immunohistochemical analysis demonstrates that SOD2 hyperacetylation occurs near zone 3 within the liver, which is the main ethanol-metabolizing region of the liver.
Conclusion
Overall, the findings presented in this study support a role for ethanol-induced lysine acetylation as an adverse post-translational modification within the mitochondria that directly impacts SOD2 charge-state and activity. Lastly, the data presented here indicate that protein hyperacetylation may be a major factor contributing to an imbalance in hepatic redox homeostasis due to chronic ethanol metabolism.
Keywords: Superoxide Dismutase, Alcoholic Liver Disease, Lysine Acetylation, Sirtuin, Oxidative Stress
Introduction
Ethanol consumption is currently the fourth highest preventable cause of death in the United States and is a severe public health concern (Mokdad et al., 2004, CDC, 2013). Furthermore, ethanol is a significant hepatotoxicant and oxidative stress plays a central role in the pathogenesis of alcoholic liver disease (ALD) (Cederbaum et al., 2009). Oxidative stress is defined as an imbalance between pro-oxidants and anti-oxidants, resulting in dysregulated redox signaling and control (Jones, 2006). This persistent oxidative imbalance can alter redox sensitive pathways and can result in damage to cellular macromolecules, including lipids, proteins and DNA, when reactive oxygen species are in excess (Fritz and Petersen, 2011, Lobo et al., 2010). Elucidating cellular mechanisms of altered hepatic redox homeostasis due to ethanol metabolism is essential to understanding the initiation and progression of ALD.
In addition to oxidative stress, chronic ethanol metabolism significantly perturbs mitochondrial metabolic pathways and induces protein hyperacetylation (Fritz et al., 2012, Picklo, 2008, Shukla and Aroor, 2006, Shepard et al., 2010). Mitochondrial lysine acetylation is regarded as a footprint of metabolic status as demonstrated in a model of caloric restriction, arising in part, as a consequence of altered acetyl-CoA flux through glycolysis and lipid metabolism (Hebert et al., 2013, Pougovkina et al., 2014, Shi and Tu, 2015). The acetylation of lysine residues has also recently been described as an important post-translational modification that can modulate protein activity, protein-protein interactions, protein turnover, and cell cycle progression (Friedmann and Marmorstein, 2013). Lastly, in a number of instances, lysine acetylation has been shown to suppress the enzymatic activity of critical mitochondrial proteins, including those relevant to antioxidant defense, thus supporting our hypothesis that ethanol-induced mitochondrial protein hyperacetylation may inhibit critical antioxidant processes (Baeza et al., 2016).
A number of original reports characterized ethanol-induced changes in protein acetylation (Picklo, 2008, Shukla and Aroor, 2006, Shepard et al., 2010). Recently, our laboratory demonstrated that mitochondrial superoxide dismutase (SOD2) is an acetylation target that is hyperacetylated as a result of ethanol consumption (Fritz et al., 2012). Additionally, several studies have shown that SOD2 enzymatic activity is negatively altered through acetylation of lysine residues. For instance, Zhu et al. demonstrated that acetylation of SOD2 K122 decreased the enzymatic activity via masking the positively charged amino acids of the active site (Zhu et al., 2012). Lu et al. also showed that acetylation of SOD2 K68 reduced its activity via modulating structure and charge state (Lu et al., 2015). SOD is responsible for detoxifying superoxide radicals by catalyzing the conversion of the superoxide anion into hydrogen peroxide and molecular oxygen. Hydrogen peroxide is then further converted into H2O and molecular oxygen via both enzymatic and non-enzymatic processes (Lobo et al., 2010). In mammals, SOD is present as three isoforms that differentially localize within the cell: SOD1 in the cytoplasm, SOD2 in the mitochondria, and SOD3 in the extracellular matrix (Miao and St Clair, 2009). SOD2 is the major mitochondrial antioxidant enzyme that prevents oxidative damage resulting from the generation and accumulation of superoxide radicals (Flynn and Melov, 2013). Mice with inactivated SOD2 die prematurely and exhibit severe mitochondrial defects, demonstrating this enzyme’s critical importance in sustaining life (Melov et al., 2001). Interestingly, overexpression of SOD2 prevented liver damage due to ethanol metabolism by neutralizing certain toxicological markers such as GSH oxidation, ALT elevation, and the formation of free radical adducts in bile (Wheeler et al., 2001). Furthermore, SOD2 overexpression also ameliorated alcohol-induced pathologies like steatosis, necrosis, apoptosis, and inflammation (Wheeler et al., 2001). Another study demonstrated that chronic alcohol feeding slightly reduced hepatic SOD2 activity; however, the exact mechanism was not revealed (Roede et al., 2009).
Ethanol metabolism increased protein acetylation within the mitochondria, which may represent a critical mechanism underlying an altered mitochondrial state (Fritz et al., 2013). The present study examines the effect of ethanol-induced protein hyperacetylation on SOD2, a protein with 16 lysine residues prone to acetylation. Importantly, it has been reported that SOD2 K68 and K122 are key regulators of enzymatic activity due to alterations in protein charge-state dynamics (Ozden et al., 2011, Zou et al., 2016, Lu et al., 2015, Zhu et al., 2012). Since oxidative stress is a central mechanism of ethanol-induced liver injury, and we previously observed SOD2 hyperacetylation upon ethanol administration, we investigated the hypothesis that chronic ethanol consumption increases SOD2 acetylation, leading to alterations in SOD2 protein structure and activity, potentially contributing to overall mitochondrial dysregulation and the progression of ALD (Fritz et al., 2012).
Materials and Methods
Animal studies
Animal studies were approved by the Institutional Animal Care and Use Committee of the University of Colorado. Male, C57BL/6J mice (8–12 week old) were purchased from Jackson Laboratories (Bar Harbor, ME) for chronic ethanol feeding as previously described (Fritz et al., 2013). Briefly, 12 mice were fed a modified Lieber-DeCarli liquid diet (Bio-Serv, Frenchtown, NJ) for 6 weeks, where ethanol was ramped from 1% to 6% (v/v) over the 6-week period. After six weeks, mice were anesthetized via an intraperitoneal injection of pentobarbital (65 mg/kg). Tissues were either collected and flash frozen for biochemical analysis, or formalin fixed for histopathology assessment. An aliquot of liver tissue was extracted, weighed, and subcellular fractionation was performed as previously described (Harris et al., 2015). Oxidized and reduced glutathione were measured via HPLC and fluorescence detection as previously detailed (Harris et al., 2015).
Immunohistochemistry
Freshly excised liver tissues were fixed in 10% formalin (Sigma, Saint Louis, MO) overnight. The tissue was then embedded in paraffin, cut and mounted on slides by the University of Colorado Anschutz Medical Campus Histology Core. Standard hematoxylin and eosin (H&E) staining was performed. Immunohistochemistry was performed using the ABC citrate antigen retrieval system (Vector labs, Burlingame, CA) and by incubating liver tissue sections with primary antibodies raised against CYP2E1 (AB1252; Millipore, Darmstadt, Germany), SOD2 (ab13534; Abcam, Cambridge, MA, USA), and SOD2 acetyl-K68 (ab137037; Abcam, Cambridge, MA, USA) overnight at 4 °C via a protocol described by Harris et. al. (Harris et al., 2015). Histologic images were captured on an Olympus BX51 microscope equipped with a 4 megapixel Macrofire digital camera using the PictureFrame Application 2.3 (Optronics). All images were cropped and assembled using Photoshop CS2 (Adobe Systems, Inc.). Images were then imported into Slidebook (3I, Denver CO) for quantification, which was performed on 5 images per animal, 3–4 animals per group.
Western blotting
Protein samples (10–60 µg) from ethanol-fed and pair-fed controls were separated using 12% SDS-PAGE at 150 V for 1 hour, then transferred to activated Hybond-PVDF membrane (GE Healthcare, Buckinghamshire, UK). Membranes were blocked using 5% (w/v) non-fat dry milk in Tris-Buffered Saline-Tween 20 TBS/0.1 % (v/v) Tween (TBS-T) for 1 hour at room temperature. Membranes were incubated with primary antibodies against SOD2 (ab13534), SOD2 acetyl K68 (ab137037), or SOD2 acetyl K122 (ab214675) (Abcam, Cambridge, MA) overnight at 4°C, then washed three times with TBS-T, and incubated with horseradish peroxidase conjugated secondary antibody at room temperature for one hour. Membranes were washed again with TBS-T three times and then Clarity Western ECL Substrate (BioRad) was applied before imaging via Chemidoc® MP (Bio-Rad, Hercules, CA). 2,2,2-trichloroethanol (Sigma, Saint Louis, MO) stain was used to visualize overall protein load.
Two-dimensional SDS-PAGE was performed using 200 µg of liver whole cell extract and was separated on IPG strips (pH 3–11) (Bio-Rad), then resolved using 10% SDS-PAGE. The proteins were then transferred onto PVDF membrane and probed with anti-SOD2 (ab13534) and SOD2 acetyl-K68 (ab137037) antibody (Abcam, Cambridge, MA) using standard Western blot analysis as described above.
Superoxide dismutase activity assay
In vitro SOD2 activity was performed using 1 µg of murine recombinant SOD2 (Enzo Farmingdale, NY). SOD2 was incubated with and without increasing concentrations of acetic anhydride for one hour at room temperature to induce protein acetylation and then the activity assay was performed according to the manufacturer’s protocol (Cayman Ann Arbor, MI).
In vivo SOD2 activity was measured in mitochondria enriched fractions from control and ethanol-fed mice. Protein concentration was measured using the BCA assay (Thermo scientific, Rockford IL). Samples were diluted to (10 µg/µl) and 20 mM of KCN (Sigma, Saint Louis, MO) was added to each sample then incubated for 1 hour to eliminate trace activity from SOD1 and SOD3. SOD2 activity was measured using the method previously described by Misra, et al. (Misra and Fridovich, 1972).
In vitro acetylation
In order to determine preferential sites of acetylation, in vitro acetylation of SOD2 was carried out using 1 µg of recombinant murine SOD2 (Enzo Farmingdale, NY). SOD2 was subjected to non-enzymatic acetylation via acetic anhydride (Sigma, Saint Louis, MO) using three different concentrations of acetic anhydride (50 µM, 500 µM, 5000 µM) (Harris et al., 2017). SOD2 was incubated with acetic anhydride for one hour at room temperature. The samples were then subjected to reducing SDS-PAGE gel electrophoresis using a 12% gel. Once completed in duplicate, one of the gels was used for Western blotting with an anti-acetyl antibody (Abcam, Cambridge, MA, USA) and the other one was stained with Coomassie blue where protein bands were visualized and excised from the gel. Isolated protein was then digested with trypsin as previously described (Fritz et al., 2011). The resulting peptides were desalted using ZipTip C18 tips (EMD Millipore, Darmstadt, Germany) and analyzed by nHPLC coupled to a nano-ESI source on an Impact HD Q-TOF tandem mass spectrometer (Bruker, Billerica, MA). Data analysis was performed using ProteinScape® and DataAnalysis software (Bruker, Billerica, MA).
Computational modeling
All computational analyses were performed using Accelrys Discovery Studio 4.0 (Biovia, San Diego, CA). The crystal structure coordinates for SOD2 were downloaded from the Protein Data Bank (www.pdb.org; PDB IDs:1MSD) (Wagner et al., 1993). Figures were rendered using Lightwave 2015.3 (NewTek, Salford, UK).
Statistical analysis
Statistical analyses including unpaired T-test and graphs were calculated using Prism 6 (Graphpad, La Jolla, CA). Graphs represent the average of three independent experiments with error bars indicating the SEM. Results were considered significant if p<0.05.
Results
Chronic ethanol feeding increases pathological markers of liver damage
In order to assess the impact of chronic alcohol consumption on relevant pathological markers, we examined liver triglycerides, alanine aminotransferase (ALT), body weight, and liver weight to body weight ratio. As a result of chronic ethanol treatment liver triglyceride levels were significantly increased from 0.025 to 0.058 mmol/L/mg and ALT was increased with ethanol treatment from 25.67 to 141.2 IU/L (Table 1). These results demonstrated that the initiation of alcoholic liver disease was robust in this model.
Table 1.
Biochemical markers of alcohol induced liver injury
| Parameter | Control | Ethanol |
|---|---|---|
| ALT (IU/L) | 25.67 ± 12.27 | 141.2 ± 37.31* |
| Liver triglycerides (mmol/L/mg) | 0.025 ± 0.012 | 0.058 ± 0.005* |
| Body weight changes (g) | 5.13 ± 0.72 | 2.25 ± 0.703* |
| Liver weight / body weight (g) | 0.034 ± 0.001 | 0.040± 0.001* |
Mean ± SEM,
P<0.05, n=4
Chronic ethanol metabolism induces steatosis, CYP2E1 expression, and SOD2 acetyl-K68 in hepatic zone 3
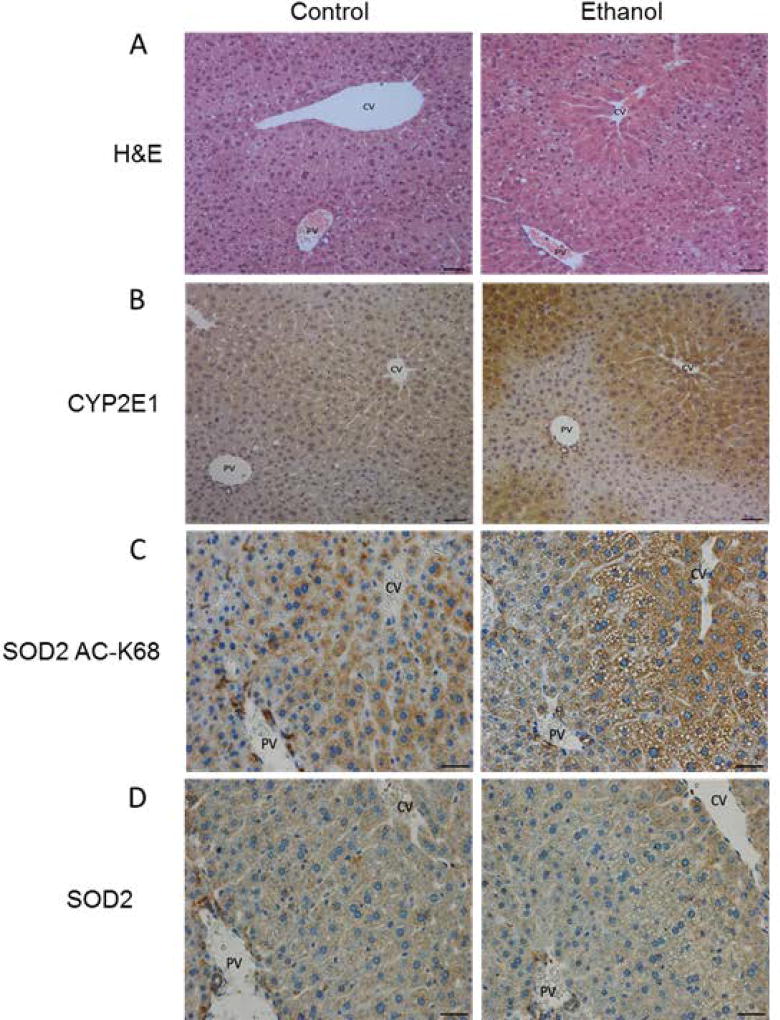
Structurally, the liver is divided into three zones: zone 1 is referred to as the periportal zone and surrounds the portal vein, whereas zone 3 is adjacent to the central vein. Zone 2 occupies the transitional region between zones 1 and 3. Chronic ethanol consumption is well known to induce hepatic steatosis and significantly induces the expression of cytochrome P450 2E1 (CYP2E1) in zone 3. Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) were utilized to assess these markers in this model. As shown in Figure 1, this chronic ethanol model resulted in steatosis (Figure 1A) and induced an increase in zone 3 CYP2E1 expression (Figure 1B) as a result of sustained ethanol metabolism. While mitochondrial protein acetylation has been shown to occur independent of Cyp2E1 activation, the zonal distribution of hyperacetylation suggests that ethanol metabolism and resulting hypoxia likely play a role in this marker as an ethanol-induced occurrence (Picklo, 2008).
Figure 1.
Chronic ethanol metabolism in a 6-week Lieber-DeCarli model induces steatosis and CYP2E1 expression in liver tissue of alcohol fed mice, as demonstrated by (A) Hematoxylin and eosin staining and (B) CYP2E1 immunohistochemistry. Immunohistochemistry analysis of liver tissue from control- and ethanol-fed mice demonstrates an increase in (C) acetylation of SOD2 K68 and (D) the panlobular distribution of SOD2. CV= central vein, PV=portal vein Scale bar size = 200 micron in (A,B). and 40 micron in (C,D). Quantification of CYP2E1, SOD2-acK68 and SOD2 expression are shown in Supplemental Figure 1.
Utilizing IHC, we found the majority of SOD2 hyperacetylation to occur in zone 3 of the hepatic lobule (Figure 1C), and was moderately increased due to ethanol consumption. Global SOD2 analysis by IHC demonstrated that SOD2 expression and distribution remained unaltered between control and ethanol groups (Figure 1D). SOD2 and SOD2 ac-K68 IHC images were quantified and demonstrate that SOD2 abundance was unchanged between control and ethanol hepatic tissue. Furthermore, IHC quantification revealed that SOD2 ac-K68 was increased in the ethanol group, however the increase did not reach significance (Supplemental Figure S1). These findings show that SOD2 is hyperacetylated at the regulatory site of lysine 68 predominantly in zone 3, where the majority of hepatic ethanol metabolism occurs (Cederbaum, 2012). This regional impairment of SOD2 activity suggested that there could be a strong link between ethanol metabolism, protein hyperacetylation, and the pathogenesis of ALD.
Ethanol consumption results in depletion of reduced glutathione
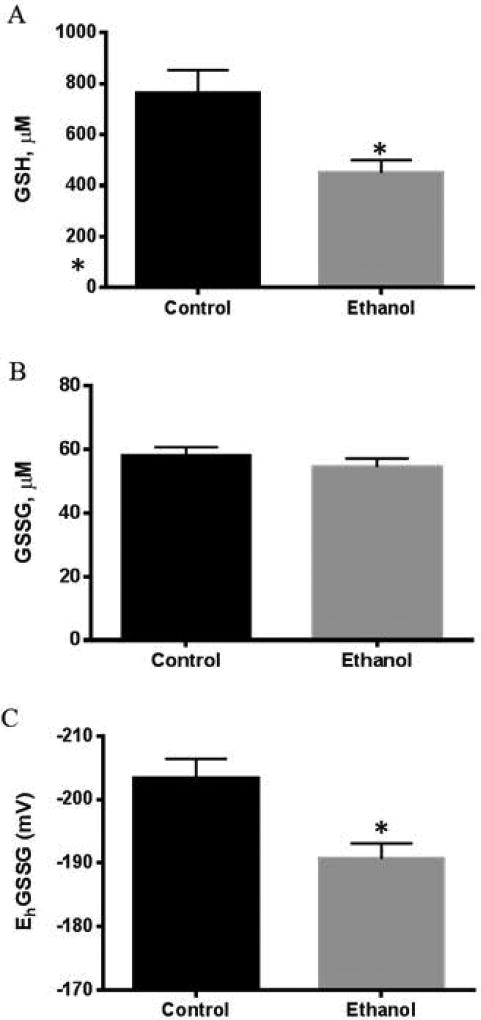
Since oxidative stress plays a major role in ALD, we studied the potential impact of ethanol consumption on GSH/GSSG to determine the role of SOD2 acetylation on liver glutathione redox status. Our results demonstrated that chronic ethanol consumption decreased hepatic GSH levels (Figure 2A) without any alteration in GSSG levels (Figure 2B), leading to a significant oxidation of the hepatic glutathione redox potential (EhGSSG) (Figure 2C). These results show that chronic ethanol consumption induced sustained hepatic oxidative stress, as determined by the depletion of GSH and oxidized redox potential (EhGSSG).
Figure 2.
Chronic ethanol metabolism in a 6-week Lieber-DeCarli model significantly impacts hepatic glutathione homeostasis as determined by (A) a decrease in GSH concentration in ethanol fed group compared to controls (P< 0.05), (B) unchanged GSSG levels between groups, and (C) a significantly decreased redox potential in ethanol-fed animals (P< 0.05). (Mean ± SEM, n=4).
Chronic ethanol consumption increases SOD2 acetylation and inhibits SOD2 activity
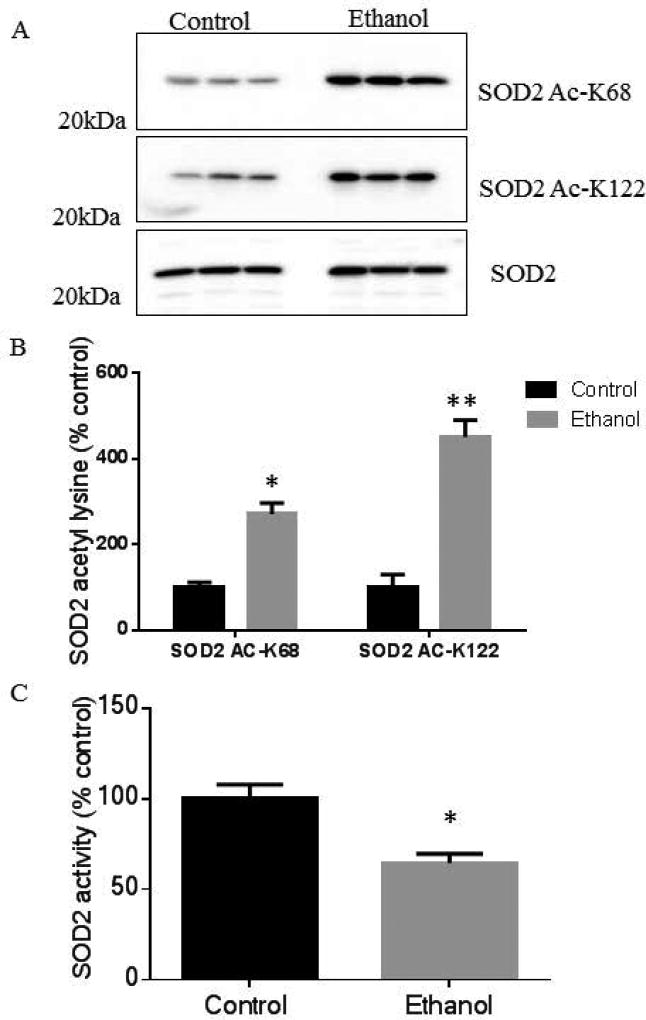
To elucidate the role of ethanol-induced acetylation on the regulatory sites, K68 and K122, Western blot analyses of liver mitochondrial fractions using anti-SOD2 acetyl-K68 and anti-SOD2 acetyl-K122 antibodies revealed that chronic ethanol consumption increased the acetylation of both residues (Figure 3A). Furthermore, total SOD2 protein abundance was unchanged. Immunoblots were quantified and normalized to total SOD2. These results demonstrate that chronic ethanol consumption significantly increased the acetylation of SOD2 K68 by 3-fold and increased the acetylation of SOD2 K122 by 5-fold (Figure 3B) compared to pair-fed controls. To investigate the functional impact of lysine hyperacetylation on SOD2 function, an activity assay was performed on liver mitochondrial extract from control and ethanol-fed mice. Compared to the pair-fed control group, chronic ethanol consumption significantly decreased SOD2 activity by 40%, implicating the observed increase in SOD2 lysine acetylation as a potential inhibitory mechanism (Figure 3C). A critical aspect of our SOD2 activity assay was that mitochondrial fractions were enriched from whole liver tissue. Accounting for our IHC analysis, inhibition of SOD2 would be much higher around zone 3 where SOD2 hyperacetylation is focused. Additionally, since SOD2 is a reported substrate for the mitochondrial deacetylase SIRT3, we examined the effect of recombinant SIRT3 on mitochondrial lysates from both control and ethanol-fed hepatic murine mitochondrial fractions (Chen et al., 2011). Supplemental Figure S2 demonstrates that SIRT3 can remove the ethanol-induced acetylation of SOD2 K68.
Figure 3.
Acetylation of SOD2 is increased in hepatic tissue of ethanol-fed mice in this 6-week chronic ethanol model, as shown via (A) Western blot on liver mitochondrial lysates obtained from 3 controls and 3 ethanol fed mice using specific antibodies against SOD2, SOD2 acetyl-K68, and SOD2 acetyl-K122 and (B) quantification of SOD2 acetyl-K68 (P<0.05) and SOD2 acetyl-K122 (P<0.01). (C) SOD2 activity was significantly decreased in hepatic mitochondrial fractions of ethanol-fed mice when compared to control animals (P<0.05). (Mean ± SEM, n=3).
Ethanol consumption alters the isoelectric point profile of SOD2
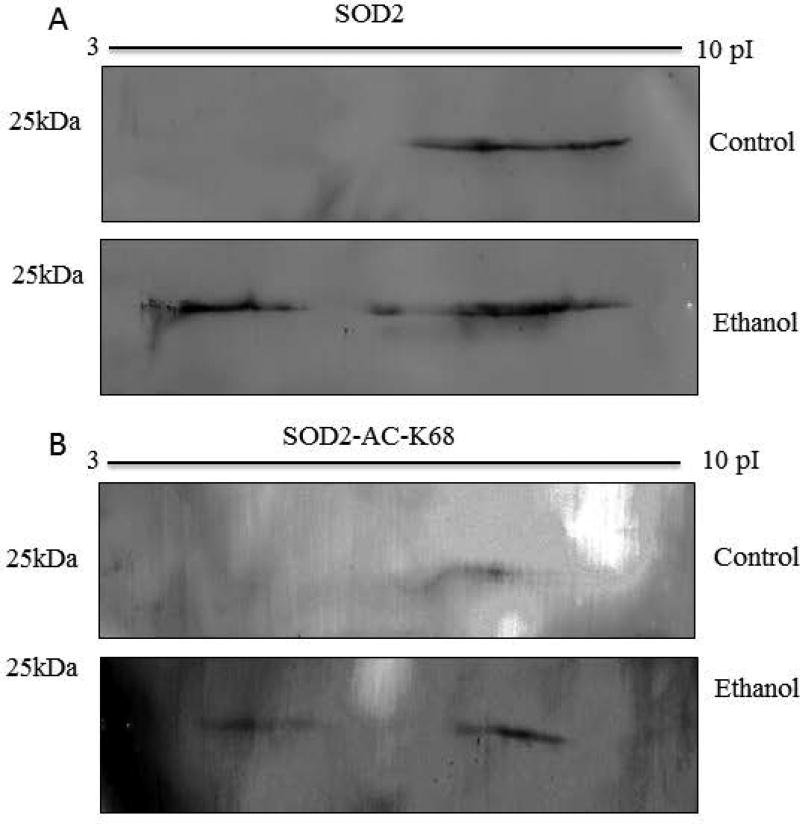
The positive charge-state of certain lysine residues of SOD2 is crucial for enzymatic activity (Zhu et al., 2012). Since acetylation abrogates this charge, acetylated SOD2 would be predicted to both possess decreased enzymatic activity and a change in the isoelectric point of the protein. In order to assess this, liver mitochondrial extracts from both control- and ethanol-fed mice were separated using 2D electrophoresis and probed with anti-SOD2 and anti-SOD2-K68 antibodies. Figure 4 shows that chronic ethanol feeding altered the isoelectric point of SOD2 to a more acidic state (less positively charged), likely due in part to protein acetylation which would change positively charged lysine residues to a neutral amino acid. These results suggested that chronic ethanol metabolism inhibited the activity of SOD2, in part, by masking many positively charged lysine residues. Previous reports demonstrate that SOD2 is modified on multiple lysine residues by numerous acyl species, including acetylation and succinylation (Weinert et al., 2013). Acetylation would neutralize the positive charge on lysine while succinylation would switch the charge of that residue from positive to negative, further perturbing the isoelectric point of the SOD2. If this were to occur across many lysine residues on SOD2, in conjunction with other modifications like succinylation and phosphorylation, it would likely result in the broadly smeared band observed in these samples. While these Westerns were probed with a specific acetyl-K68-SOD2 antibody, many other PTMs likely impact the isoelectric point of SOD2. The overall result of myriad acylation events and other PTMs on SOD2 remains a key point of interest for further investigation.
Figure 4.
Two-dimensional SDS-PAGE is utilized to study the impact of ethanol metabolism on the isoelectric point of SOD2. Liver mitochondrial extracts from both control and ethanol-fed mice were separated using 2D electrophoresis and probed with (A) anti-SOD2 and (B) anti-SOD2 ac-K68 antibodies results here reveal that chronic ethanol consumption induces a major shift in the isoelectric point of SOD2 in murine hepatic mitochondria due in large part to protein acetylation and other PTMs.
In vitro acetylation and computational modeling of recombinant SOD2
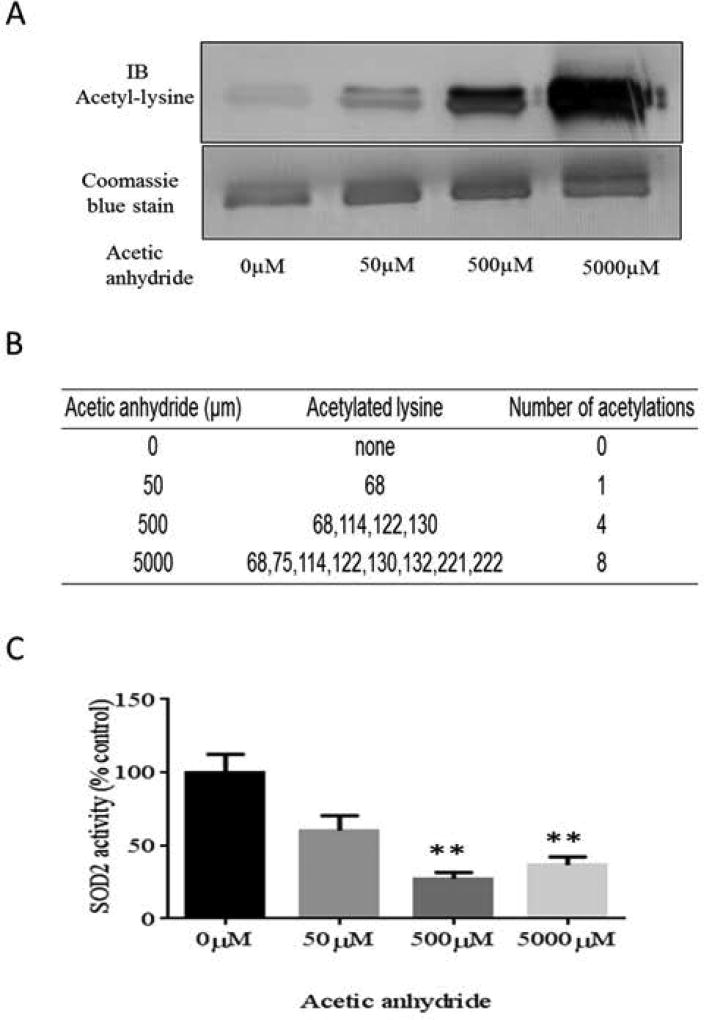
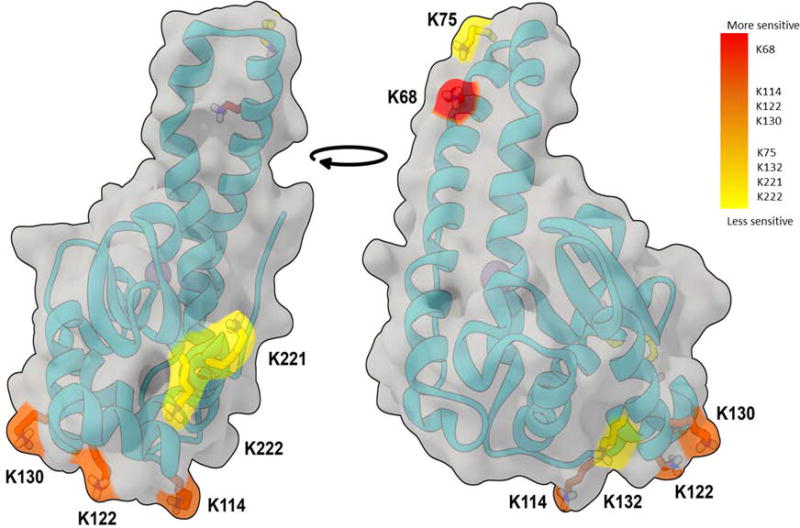
To study the mechanisms of SOD2 acetylation, recombinant murine SOD2 was incubated with increasing concentrations (0–5 mM) of acetic anhydride. Immunoblotting demonstrated that the resulting acetylation of recombinant SOD2 occurred in a concentration-dependent manner (Figure 5A). LC-MS/MS analyses of the acetylated recombinant protein (Figure 5B) also confirmed that acetic anhydride-mediated acetylation of SOD2 was concentration dependent. The LC-MS/MS results also indicated that K68 is the lysine residue most susceptible to acetylation, suggesting it was a mitochondrial site of allosteric regulation and is susceptible to altered acetylation status in response to mitochondrial metabolic alterations. As anticipated, acetic anhydride-mediated acetylation inhibited SOD2 activity by 40%, 73%, and 63% at 50µM, 500µM, and 5 mM acetic anhydride, respectively (Figure 5C). The locations of each of the major targets of acetylation were highlighted in the SOD2 crystal structure and are colored by relative sensitivity to modification from high (red) to low (yellow) (Figure 6). These results illustrate that K68 is highly sensitive to lysine acetylation, suggesting that K68 is a major allosteric site that regulates SOD2 activity among the 8 sites we identified.
Figure 5.
In vitro acetylation of rSOD2 by acetic anhydride to determine susceptibility of each lysine residue toward acetylation and enzyme inhibition. (A) Western blotting using anti-acetyl-lysine antibody demonstrates an increase in SOD2 acetylation due to non-enzymatic acetylation and Coomassie blue staining shows protein abundance. (B) Acetylated rSOD2 was examined via LC-MS/MS analysis to identify acetylated lysine residues at each concentration of acetic anhydride. (C) SOD2 activity was inhibited as a result of lysine acetylation (P< 0.01). (Mean ± SEM, n=3).
Figure 6.
Computational modeling of SOD2 tertiary structure illustrates the experimentally observed sensitivity of lysine residues to in vitro acetylation and supports our finding that one of the regulatory amino acids, K68, was found to have the highest degree of sensitivity to non-enzymatic acetylation.
In vivo LC-MS/MS quantification
To examine the effect of ethanol-induced hepatic mitochondrial protein acetylation on liver SOD2, a quantitative acetylomics approach was employed to determine the relative increase in ethanol-induced acetylation of SOD2 K68. Chronic ethanol consumption resulted in a 5-fold increase in hepatic SOD2 K68 acetylation when compared to controls, which confirmed the results of our Western blot analyses. A representative fragmentation spectra identifying acetyl-K68 of SOD2 demonstrated b- and y-ion coverage of this site of acetylation (Supplemental Figure 3).
Discussion
Oxidative stress and mitochondrial dysfunction are proposed to play a major role in the pathogenesis of ALD (Wu and Cederbaum, 2009). Supporting this overall model, our data demonstrated that as a result of chronic ethanol metabolism hepatic GSH was decreased and EhGSSG was significantly oxidized. SOD2 is a key antioxidant defense enzyme that protects mitochondria from superoxide radicals and the negative consequences of unchecked free radical species generation (Miao and St Clair, 2009). Importantly, SOD2 can be regulated by numerous transcriptional, translational, and posttranslational mechanisms, including protein acetylation (Miao and St Clair, 2009, Huang et al., 1997, Chen et al., 2011, Gao et al., 2016, Qiu et al., 2010). Specifically, acetylation of K68 and K122, among other sites, can decrease SOD2 activity and remains an interesting target for numerous disease pathologies (Ozden et al., 2011, Zhu et al., 2012, Zou et al., 2016, Lu et al., 2015).
Our laboratory has recently reported that chronic ethanol consumption induced global SOD2 acetylation (Fritz et al., 2012). Here, we demonstrated that chronic ethanol consumption altered both SOD2 charge-state and activity, in part, through acetylation of K68 and K122. These findings provide new mechanistic insight regarding ethanol-induced hepatic oxidative stress. While ethanol metabolism results in the hyperacetylation of mitochondrial proteins, the exact biochemical mechanism underlying this process remains unknown (Fritz et al., 2012). Two likely candidate pathways include: 1) acetylation derived directly from the breakdown of ethanol via acetaldehyde and acetate; and 2) a secondary effect of altered lipid metabolism and glycolysis impacting acetyl-CoA flux. Importantly, recently published in vivo analyses of mitochondrial sirtuin expression and activity suggest that impaired deacetylation activity is an unlikely candidate pathway (Fritz et al., 2013). Sirtuins (SIRT) are nutrient sensing, NAD+-dependent deacetylases and SIRT3 is the primary mitochondrial deacetylase (Qiu et al., 2010, Chen et al., 2011, Gao et al., 2016). Although sirtuin activity is a critical factor in a number of pathologies, a direct role for mitochondrial deacetylase activity in ethanol metabolism remains an intriguing area of investigation (Gertz and Steegborn, 2016). The data presented here further demonstrate that hyperacetylation of SOD2 inhibits its activity, suggesting a key mechanism contributing to oxidative stress and altered mitochondrial metabolism, all of which are key characteristics of ALD etiology (Vasquez-Vivar et al., 2000, Armstrong et al., 2004, Gardner, 1997, Pias et al., 2003, Murphy et al., 2003). In 2001, it was reported that SOD2 activity did not change as a result of intragastric alcohol administration in rats. (Wheeler et al., 2001). However, another study conducted by Ge et al. reported that ethanol metabolism significantly decreased SOD2 activity (Ge et al., 2013) and Roede et al. demonstrated similar findings, in agreement with our findings (Roede et al., 2009). In aggregate, it is apparent that examining SOD2 activity in varying models of alcohol is complicated and further studies are needed to reveal the underlying mechanisms of how ethanol metabolism and resulting epiproteomic alterations regulate SOD2 activity.
Elucidating a role for both global and site-specific mitochondrial protein acetylation remains a key focus towards understanding mitochondrial fidelity during oxidative and metabolic stress due to ethanol metabolism. Deciphering the intricate relationship of lysine acetylation and protein function on a molecular scale remains a complex challenge. Indeed, the charge state of SOD2 is crucial for enzyme activity (Zhu et al., 2012). Under physiological conditions, the active site of SOD2 exhibits an overall positive charge. This localized positive charge is hypothesized to attract and guide the superoxide anion towards the active site of SOD2. Acetylation of K68 and other lysine residues eliminates this localized positive charge, leading to a reduced affinity of the superoxide anion at the active site (Zhu et al., 2012, Lu et al., 2015). Importantly, our data demonstrate that chronic ethanol metabolism significantly alters the isoelectric point of hepatic SOD2; however, further studies are needed in order to explore the impact of lysine acetylation, succinylation, and other PTMs on SOD2 charge-state dynamics and quaternary structure. In addition to SOD2, previous results demonstrate that numerous other antioxidant enzymes are affected by protein hyperacetylation induced by ethanol metabolism (Harris et al., 2015), such as thioredoxin reductase, glutathione S-transferase, glutathione peroxidase, and glutathione reductase (Fritz et al., 2013). Further studies are needed to examine the mechanisms by which ethanol-induced protein hyperacetylation impacts these other antioxidant-related enzymes. While antioxidant therapies have yet to demonstrate effectiveness in ALD, a targeted intervention of antioxidant protein activity such as SOD2 activation may provide a more positive outcome.
In conclusion, the results of this study demonstrate that chronic ethanol consumption impacts SOD2 structure and activity via hyperacetylation. Mass spectrometry, Western blot, and IHC data demonstrate that acetylation of SOD2 was increased as a result of chronic ethanol consumption and that this posttranslational modification was enriched in hepatic zones known to be relevant to ALD. Examination of SOD2 activity showed that chronic ethanol consumption and protein hyperacetylation leads to a significant decrease in hepatic SOD2 activity by 40%. Indeed, chronic ethanol metabolism alters the overall charge state of SOD2, as demonstrated by 2D SDS-PAGE. In aggregate, these findings further support the ability of lysine acetylation to alter protein charge-state dynamics and enzymatic function. Additionally, our in vitro analyses demonstrate that K68 appears to be the lysine residue most susceptible to non-enzymatic acetylation and further supports the hypothesis that ethanol-induced protein acetylation plays a role in regulating antioxidant protein function and mitochondrial antioxidant capacity.
Supplementary Material
Acknowledgments
The authors acknowledge the support of Dr. Li-Ping Liang and Dr. Manisha Patel for assistance with SOD activity assays. Molecular modeling was performed in collaboration with the University of Colorado Computational Chemistry and Biology Core Facility, which is funded in part by NIH/NCATS Colorado CTSI Grant Number UL1TR001082. The authors wish to thank Joe Gomez and Cole Michel in the Skaggs School of Pharmacy and Pharmaceutical Sciences Proteomics Mass Spectrometry Core for assistance with our HPLC-MS/MS analysis and E. Erin Smith of the University of Colorado Denver Cancer Center Research Histology Core (UCDCCRHC) for assistance in preparing histology slides. The UCDCCRHC is supported in part by NIH/NCRR Colorado CTSI Grant Number UL1 RR025780 and the University of Colorado Cancer Center Grant P30 CA046934. This research was funded using support from NIH/NIAAA AA022146 (KSF, MDH), and King Saud University, Riyadh Saudi Arabia (MAA).
Footnotes
Conflict of Interest
The authors have no conflict of interest to disclose.
References
- Armstrong JS, Whiteman M, Yang H, Jones DP. The redox regulation of intermediary metabolism by a superoxide-aconitase rheostat. Bioessays. 2004;26:894–900. doi: 10.1002/bies.20071. [DOI] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. Mechanisms and Dynamics of Protein Acetylation in Mitochondria. Trends Biochem Sci. 2016;41:231–44. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Average for United States 2006–2010 Alcohol-Attributable Deaths Due to Excessive Alcohol Use. Centers for Disease Control and Prevention (CDC); 2013. Alcohol and Public Health: Alcohol-Related Disease Impact (ARDI) [Google Scholar]
- Cederbaum AI. Alcohol metabolism. Clin Liver Dis. 2012;16:667–85. doi: 10.1016/j.cld.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum AI, Lu Y, Wu D. Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol. 2009;83:519–48. doi: 10.1007/s00204-009-0432-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–41. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Melov S. SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic Biol Med. 2013;62:4–12. doi: 10.1016/j.freeradbiomed.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann DR, Marmorstein R. Structure and mechanism of non-histone protein acetyltransferase enzymes. FEBS J. 2013;280:5570–81. doi: 10.1111/febs.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J Proteome Res. 2012;11:1633–43. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem Res Toxicol. 2011;24:651–62. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz KS, Green MF, Petersen DR, Hirschey MD. Ethanol metabolism modifies hepatic protein acylation in mice. PLoS One. 2013;8:e75868. doi: 10.1371/journal.pone.0075868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz KS, Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol. 2011;24:1411–9. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zheng Z, Gu Q, Chen X, Liu X, Xu X. Deacetylation of MnSOD by PARP-regulated SIRT3 protects retinal capillary endothelial cells from hyperglycemia-induced damage. Biochem Biophys Res Commun. 2016;472:425–31. doi: 10.1016/j.bbrc.2015.12.037. [DOI] [PubMed] [Google Scholar]
- Gardner PR. Superoxide-driven aconitase FE-S center cycling. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- Ge N, Liang H, Liu Y, Ma AG, Han L. Protective effect of Aplysin on hepatic injury in ethanol-treated rats. Food Chem Toxicol. 2013;62:361–72. doi: 10.1016/j.fct.2013.08.071. [DOI] [PubMed] [Google Scholar]
- Gertz M, Steegborn C. Using mitochondrial sirtuins as drug targets: disease implications and available compounds. Cell Mol Life Sci. 2016;73:2871–96. doi: 10.1007/s00018-016-2180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PS, Gomez JD, Backos DS, Fritz KS. Characterizing Sirtuin 3 Deacetylase Affinity for Aldehyde Dehydrogenase 2. Chem Res Toxicol. 2017;30:785–793. doi: 10.1021/acs.chemrestox.6b00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PS, Roy SR, Coughlan C, Orlicky DJ, Liang Y, Shearn CT, Roede JR, Fritz KS. Chronic ethanol consumption induces mitochondrial protein acetylation and oxidative stress in the kidney. Redox Biol. 2015;6:33–40. doi: 10.1016/j.redox.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–99. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Peng J, Oberley LW, Domann FE. Transcriptional inhibition of manganese superoxide dismutase (SOD2) gene expression by DNA methylation of the 5' CpG island. Free Radic Biol Med. 1997;23:314–20. doi: 10.1016/s0891-5849(97)00095-6. [DOI] [PubMed] [Google Scholar]
- Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–79. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev. 2010;4:118–26. doi: 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Cheng K, Zhang B, Xu H, Cao Y, Guo F, Feng X, Xia Q. Novel mechanisms for superoxide-scavenging activity of human manganese superoxide dismutase determined by the K68 key acetylation site. Free Radic Biol Med. 2015;85:114–26. doi: 10.1016/j.freeradbiomed.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Melov S, Doctrow SR, Schneider JA, Haberson J, Patel M, Coskun PE, Huffman K, Wallace DC, Malfroy B. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J Neurosci. 2001;21:8348–53. doi: 10.1523/JNEUROSCI.21-21-08348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, St Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med. 2009;47:344–56. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra HP, Fridovich I. The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J Biol Chem. 1972;247:3170–5. [PubMed] [Google Scholar]
- Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–45. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- Murphy MP, Echtay KS, Blaikie FH, Asin-Cayuela J, Cocheme HM, Green K, Buckingham JA, Taylor ER, Hurrell F, Hughes G, Miwa S, Cooper CE, Svistunenko DA, Smith RA, Brand MD. Superoxide activates uncoupling proteins by generating carbon-centered radicals and initiating lipid peroxidation: studies using a mitochondria-targeted spin trap derived from alpha-phenyl-N-tert-butylnitrone. J Biol Chem. 2003;278:48534–45. doi: 10.1074/jbc.M308529200. [DOI] [PubMed] [Google Scholar]
- Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 2011;3:102–7. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pias EK, Ekshyyan OY, Rhoads CA, Fuseler J, Harrison L, Aw TY. Differential effects of superoxide dismutase isoform expression on hydroperoxide-induced apoptosis in PC-12 cells. J Biol Chem. 2003;278:13294–301. doi: 10.1074/jbc.M208670200. [DOI] [PubMed] [Google Scholar]
- Picklo MJ. Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376:615–9. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Pougovkina O, Te Brinke H, Ofman R, Van Cruchten AG, Kulik W, Wanders RJ, Houten SM, De Boer VC. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum Mol Genet. 2014;23:3513–22. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–7. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Roede JR, Orlicky DJ, Fisher AB, Petersen DR. Overexpression of peroxiredoxin 6 does not prevent ethanol-mediated oxidative stress and may play a role in hepatic lipid accumulation. J Pharmacol Exp Ther. 2009;330:79–88. doi: 10.1124/jpet.109.152983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard BD, Tuma DJ, Tuma PL. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol Clin Exp Res. 2010;34:280–91. doi: 10.1111/j.1530-0277.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125–31. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Aroor AR. Epigenetic effects of ethanol on liver and gastrointestinal injury. World J Gastroenterol. 2006;12:5265–71. doi: 10.3748/wjg.v12.i33.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Vivar J, Kalyanaraman B, Kennedy MC. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. J Biol Chem. 2000;275:14064–9. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]
- Wagner UG, Pattridge KA, Ludwig ML, Stallings WC, Werber MM, Oefner C, Frolow F, Sussman JL. Comparison of the crystal structures of genetically engineered human manganese superoxide dismutase and manganese superoxide dismutase from Thermus thermophilus: differences in dimer-dimer interaction. Protein Sci. 1993;2:814–25. doi: 10.1002/pro.5560020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–51. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Wheeler MD, Nakagami M, Bradford BU, Uesugi T, Mason RP, Connor HD, Dikalova A, Kadiiska M, Thurman RG. Overexpression of manganese superoxide dismutase prevents alcohol-induced liver injury in the rat. J Biol Chem. 2001;276:36664–72. doi: 10.1074/jbc.M105352200. [DOI] [PubMed] [Google Scholar]
- Wu D, Cederbaum AI. Oxidative stress and alcoholic liver disease. Semin Liver Dis. 2009;29:141–54. doi: 10.1055/s-0029-1214370. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Park SH, Ozden O, Kim HS, Jiang H, Vassilopoulos A, Spitz DR, Gius D. Exploring the electrostatic repulsion model in the role of Sirt3 in directing MnSOD acetylation status and enzymatic activity. Free Radic Biol Med. 2012;53:828–33. doi: 10.1016/j.freeradbiomed.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Santa-Maria CA, O'brien J, Gius D, Zhu Y. Manganese Superoxide Dismutase Acetylation and Dysregulation, Due to Loss of SIRT3 Activity, Promote a Luminal B-Like Breast Carcinogenic-Permissive Phenotype. Antioxid Redox Signal. 2016 doi: 10.1089/ars.2016.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.