Abstract
Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen that infects immunocompromised and cystic fibrosis patients. Treatment is difficult due to antibiotic resistance, and new antimicrobials are needed to treat infections. The alternative sigma factor 54 (σ54, RpoN), regulates many virulence-associated genes. Thus, we evaluated inhibition of virulence in P. aeruginosa by a designed peptide (RpoN molecular roadblock, RpoN*) which binds specifically to RpoN consensus promoters. We expected that RpoN* binding to its consensus promoter sites would repress gene expression and thus virulence by blocking RpoN and/or other transcription factors. RpoN* reduced transcription of approximately 700 genes as determined by microarray analysis, including genes related to virulence. RpoN* expression significantly reduced motility, protease secretion, pyocyanin and pyoverdine production, rhamnolipid production, and biofilm formation. Given the effectiveness of RpoN* in vitro, we explored its effects in a Caenorhabditis elegans–P. aeruginosa infection model. Expression of RpoN* protected C. elegans in a paralytic killing assay, whereas worms succumbed to paralysis and death in its absence. In a slow killing assay, which mimics establishment and proliferation of an infection, C. elegans survival was prolonged when RpoN* was expressed. Thus, blocking RpoN consensus promoter sites is an effective strategy for abrogation of P. aeruginosa virulence.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that causes disease in soft tissue, burns, and in immunocompromised individuals, with wound infection rates from 17% to 59% in hospitals worldwide1–3. Cystic fibrosis patients are particularly susceptible to P. aeruginosa infections, with almost 80% of adults infected4. P. aeruginosa is naturally resistant to many antibiotics, including certain penicillins and cephalosporins5, easily acquires resistance through mutations or acquisition of genes and has been identified on a recently released WHO “Global Priority List” as a critical pathogen and top priority for research and development of new antibiotics6. According to the CDC, increased antibiotic resistance in P. aeruginosa limits effective treatments in hospital-acquired infections, highlighting the need for novel antimicrobials. Preventing the expression or activity of virulence factors has emerged as a promising approach to identify and develop novel agents that would impair the ability of P. aeruginosa to cause disease7–11.
P. aeruginosa controls gene expression through 24 sigma factors, each with a defined regulon12. Multiple sigma factors are involved in controlling global regulators which, in turn, regulate expression of virulence factors13. The alternative sigma factor, σ54 or RpoN, was initially discovered as part of the nitrogen utilization pathway, but is now associated with regulation of many virulence factors12, including motility14,15, quorum sensing16,17, mucoidy18, and biofilms19. In fact, motility, mucoidy, and quorum sensing are under dual regulation with RpoN and another transcriptional regulator18,20,21. RpoN is also associated with P. aeruginosa virulence in nematodes and mice22. Furthermore, some genes necessary for host infection have been identified as part of the RpoN regulon, including vfr, kinB, and rhlR 23,24. While not all virulence factors are universally required to infect potential hosts, some are common for infections in both humans and Caenorhabditis elegans 23.
RpoN has a highly conserved Region III that recognizes a unique −24/−12 promoter with the consensus sequence TGGC-N9-GC12,25,26. Notably, the −24 element, which is centered on the conserved ‘GG’ dinucleotides, is specifically recognized by a motif known as the ‘RpoN box’ which is located in the C-terminal portion of RpoN27. A study by Doucleff, et al. demonstrated that a peptide comprised of the C-terminal 60-amino acids of the Aquifex aeolicus RpoN protein specifically binds the -24 element with high affinity (Kd~109 nM)27. This interaction is considered a main driving force for promoter recognition by RpoN. Additionally, unlike other sigma factors, RpoN can bind its DNA sequence without RNA polymerase (RNAP), although binding is 10-fold less efficient than the RpoN:RNAP complex28.
Here, we describe an engineered peptide, RpoN molecular roadblock or RpoN*, that antagonizes gene transcription by binding to RpoN specific promoters to reduce expression of RpoN-related P. aeruginosa virulence factors. RpoN* is identical to the 60 aa C-terminal DNA binding domain of A. aeolicus, except for a methionine to initiate translation27. The molecular roadblock was first evaluated for its effects on gene transcription in P. aeruginosa. The phenotype of RpoN* expression was then evaluated in vitro and in vivo using a P. aeruginosa–C. elegans infection model. We report that the roadblock reduced transcription of many virulence-related genes, which was verified in relevant in vitro assays. Furthermore, the roadblock improved survival of C. elegans exposed to P. aeruginosa. These results demonstrate that effective binding of an engineered peptide to RpoN consensus promoters is a novel and effective method for reducing P. aeruginosa virulence.
Results
Engineering the molecular roadblock RpoN*
We expected that a peptide consisting of only the C-terminal domain of RpoN would be sufficient to antagonize genome-wide transcription from RpoN consensus promoters. Therefore, a gene encoding a peptide resembling the last 60 amino acids of RpoN from the thermophile A. aeolicus was synthesized and codon-optimized for expression in E. coli (Fig. 1). This gene, rpoN*, was cloned into a broad-host range plasmid under an inducible trc-promoter for controlled expression (see Supplementary Table S1 for a list of plasmids and oligonucleotides). The sequence of RpoN* has previously been shown to specifically bind −24 RpoN consensus promoter sites27. When used with transcriptomic profiling and predictive modeling, RpoN* was expected to help identify sites of RpoN regulation, including points of regulation not previously detected, such as genes with multiple promoters and/or under negative control by RpoN.
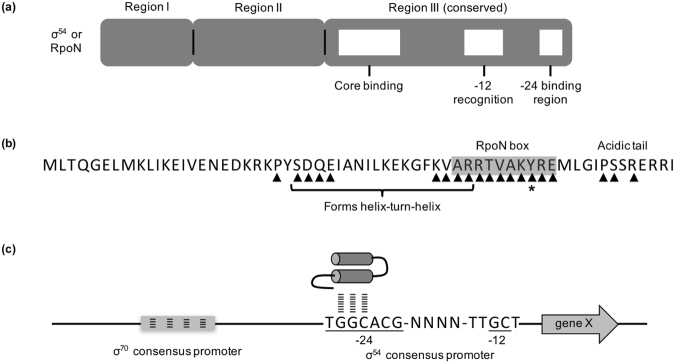
Figure 1.
Schematic of σ54 (RpoN) and the molecular roadblock, RpoN*. (a) σ54 (RpoN) is composed of three regions. Region III is highly conserved, and is necessary for recognizing and binding −24/−12 promoter elements. (b) Amino acid sequence of RpoN*. Peptide sequence includes AA376-400 of RpoN Region III from A. aeolicus and includes amino acids (▴) that specifically bind to the −24 promoter DNA. Point mutation (Y48A, asterisk) attenuates binding and transcriptional activity of RpoN*. (c) Schematic of the interaction between RpoN* and the −24 element of the σ54 consensus promoter.
RpoN* affects gene transcription
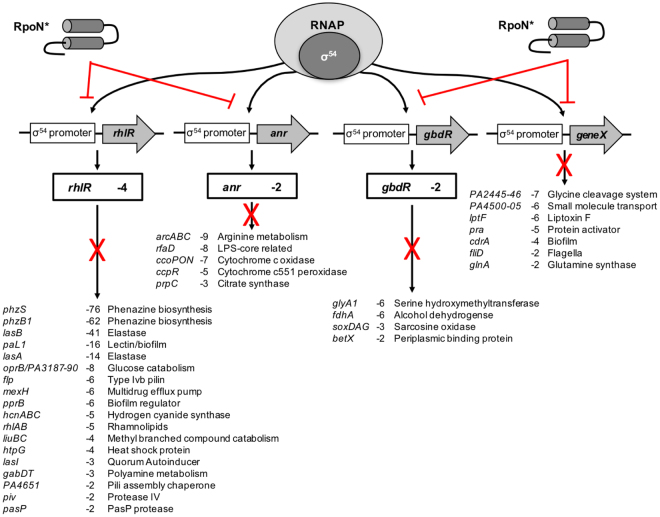
To determine the effects RpoN* has on gene transcription, microarray analysis was performed. P. aeruginosa PAO1 and an isogenic ΔrpoN strain were transformed with an empty vector or vector expressing RpoN*. The bacteria were grown to mid-exponential phase in rich media with IPTG to induce RpoN* expression, and then RNA was purified and analyzed using commercially available genome arrays (Agilent). Overall, approximately 700 genes were differentially transcribed at least 2-fold in the presence of RpoN* (Supplementary Table S2; full data available at GEO accession: GSE35632). Most were downregulated (>400), whereas the majority of upregulated genes involved lipopolysaccharide (LPS) and ribosomal biosynthesis. A selection of genes affected by the roadblock are displayed in Fig. 2. These genes encode proteins involved in metabolism, virulence, stress/survival, or cell signaling. Some genes affected by the RpoN* molecular roadblock have been shown to directly bind RpoN24. A few of these genes encode transcriptional regulators with their own regulons, including those of the las and rhl quorum signaling systems29–31, and the transcription factors anr 32 and gbdR 33. Many genes in each of these signaling regulons were also affected by the roadblock, as would be expected (Fig. 2). The expression of the native rpoN gene was also reduced by approximately one-third, which is expected as rpoN has a consensus promoter for its gene product24.
Figure 2.
Summary of genes affected by expression of RpoN* in P. aeruginosa PAO1. Expression of RpoN* interferes with the ability of RNA polymerase and native RpoN to bind to sig54 promoters altering the transcriptional profile of P. aeruginosa PAO1. Summaries of areas of gene expression that are specifically altered by RpoN* are shown and include the genes encoding the transcriptional regulators RhlR, Anr, GbdR, and their respective regulons. These results are consistent with RpoN binding sites identified by ChIP-seq.24. Expression of RpoN* was shown to alter expression of a number of other genes including hypothetical (~250) and ribosomal (~70) genes. A full list of these genes is available in Supplementary Table S2.
Deletion of native RpoN altered transcription of over 1,800 genes by 1.5-fold or more, with most of these genes downregulated (>950) (Supplementary Table S3, first column) (GEO Accession: GSE35632). Prior studies suggest that multiple sigma factors can bind promoter sites and control transcription of a single gene in P. aeruginosa 12,16,24. For instance, deletion of RpoN did not affect the levels of rhlR transcription because RpoS, a σ70-like factor, can also bind a −35/−10 consensus promoter at the same gene and regulate transcription24. A large overlap was observed between gene transcription downregulated by RpoN* in PAO1 and loss of RpoN in the ΔrpoN mutant (Supplementary Fig. S1). However, some genes were exclusively regulated by the roadblock, suggesting RpoN* reduced transcription of genes under dual regulation. When the microarray analysis of the ΔrpoN mutant harboring the empty vector was compared to ΔrpoN expressing RpoN*, more than 300 genes were differentially transcribed 1.5-fold or more (Supplementary Table S3, second column). Again, RpoN* altered transcription of global regulators, including rhlR, vfr and anr, subsequently affecting genes in their respective regulons demonstrating the specificity of this peptide to bind to RpoN promoter sequences (Supplementary Table S3, second column). Additional comparisons and overlap of genes identified for the different conditions in the microarray is shown in Supplementary Fig. S1.
Based on the microarray analysis, we propose a model for the RpoN* mechanism of action (Fig. 3). The expected mechanism of action of the cis-acting molecular roadblock is that it binds at the −24 site of RpoN consensus promoters, blocking RpoN from binding, thus reducing gene expression at these sites, as well as blocking downstream signaling effects (Fig. 3a). However, in the absence of native RpoN, gene transcription was also reduced. Thus, we also propose that the molecular roadblock can bind −24 consensus promoter sites, blocking transcription by other sigma factors with −35/−10 consensus promoter sites at the same gene (Fig. 3b). Furthermore, unlike full-length RpoN, RpoN* cannot interact with native RNAP, so we expect expression of RpoN* to minimally impact the transcriptional machinery of P. aeruginosa.
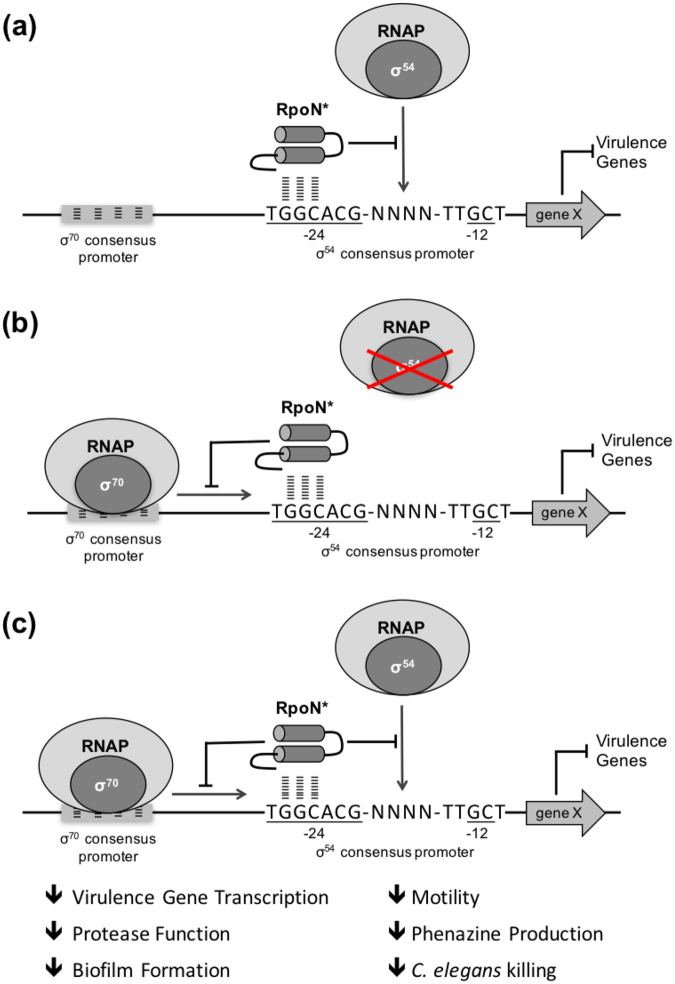
Figure 3.
Model of predicted RpoN* mechanism of action. (a) RpoN*, a cis-acting peptide, binds -24 promoter sites, blocking gene transcription by RpoN. (b) Additional sigma factors can compensate for absence of native RpoN, resulting in transcription of associated genes. In the absence of native RpoN, the RpoN* blocks transcription by other sigma factors with a promoter binding site at the same gene. (c) RpoN* blocks transcription by obstructing RpoN:RNAP binding, as well as halting transcription of other sigma factors binding at the same gene.
RpoN* reduces virulence phenotypes
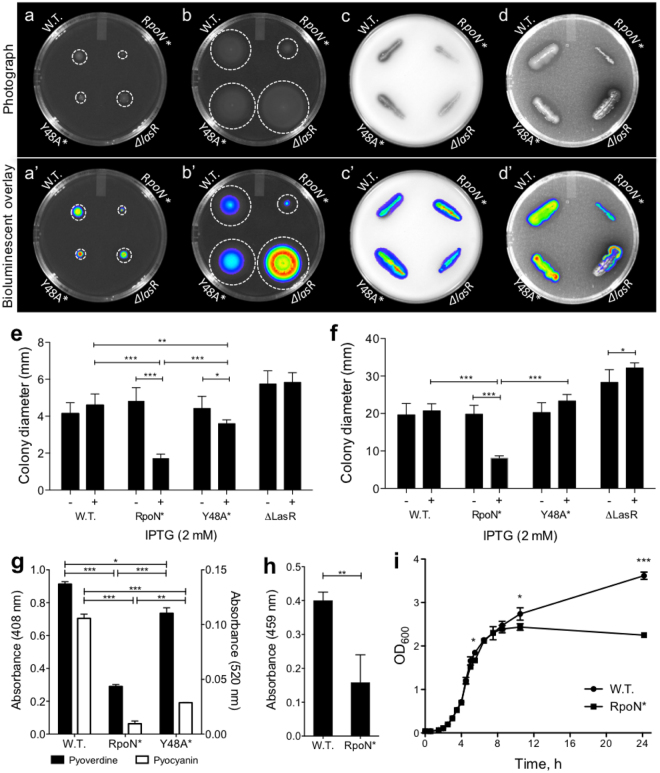
Genes associated with motility, proteases, pyoverdine biosynthesis, phenazine production, and rhamnolipid production were found to have lower transcript levels in the presence of RpoN*. We validated these findings by evaluating P. aeruginosa harboring the empty vector, RpoN*, or the variant Y48A in RpoN*, with an expectation that if RpoN* reduced expression of specific genes, decreases in the associated phenotypes would be observed. The tyrosine residue of the RpoN Box is crucial for DNA binding34, therefore it was expected that the Y48A variant would be less effective than RpoN* at antagonizing RpoN-related functions. Two wild type P. aeruginosa strains were tested: PAO1 and PA19660, a bioluminescent strain to better visualize the bacteria in the motility and protease assays. The negative control for virulence was P. aeruginosa ΔlasR:gent R that was not transformed with plasmids. As expected, P. aeruginosa harboring the empty vector plasmid was motile and exhibited protease activity (Fig. 4). P. aeruginosa ΔlasR significantly increased motility but lacked protease and elastase activity (Fig. 4). Introduction of RpoN* significantly decreased swimming and twitching motility (Students t-test, p ≤ 0.0001) (Fig. 4). Additionally, expression of RpoN* reduced protease activity, and significantly decreased extracellular levels of the siderophore pyoverdine, the phenazine pyocyanin (Students t-test, p ≤ 0.0001), as well as elastase (Students t-test, p ≤ 0.01). RpoN* expression also reduced rhamnolipid production (Supplementary Fig. S2). Finally, induction of RpoN* in a liquid culture modestly reduced growth rate, with significant differences between growth for only a few time points (Student’s t-test, at 5.5 h and 10.5 h p ≤ 0.05, at 24 h p ≤ 0.0001). In comparison, Y48A RpoN* significantly reduced P. aeruginosa twitching motility (Student’s t-test, p ≤ 0.05), pyocyanin production (Students t-test, p ≤ 0.001), and pyoverdine production (Students t-test, p ≤ 0.05), but did not affect swimming motility, or protease activity. The intermediate effects of Y48A RpoN* were expected due to its reduced binding affinity for the −24 element of the RpoN promoter. The RpoN* molecular roadblock was also effective at reducing virulence-associated phenotypes in the P. aeruginosa ΔrpoN mutant (Supplementary Fig. S3, see Supplementary Note for detailed explanation). These results support the findings from our microarray study that demonstrated that RpoN* could effectively reduce transcription of genes involved in the production of various virulence factors in P. aeruginosa.
Figure 4.
RpoN* expression reduced virulence phenotypes. (a–d) Photograph (top) plus bioluminescent overlay (bottom) for phenotype assays: (a,a’) twitching, or pili, motility conducted on semi-hard agar; (b,b’) swimming, or flagellar, motility assays conducted on soft agar; (c,c’) protease assays conducted on milk agar; (d,d’) elastase assays conducted on LB agar with elastin. Strains used: P. aeruginosa PA19660 (a-f) and PAO1 (g-i) wild-type (empty vector), RpoN*, Y48A* point mutant, and P. aeruginosa PAO1 ΔlasR. All assays were conducted at 37 °C for 24–48 h, with 30 mg/L gentamicin and 2 mM IPTG. Colony diameter of P. aeruginosa strains in twitching (e) and swimming (f) motility assays, with (+) and without (−) 2 mM IPTG. Pyocyanin and pyoverdine (g) production assays conducted in LB or King’s B broth, respectively, with 30 mg/L gentamicin and 1 mM IPTG. Elastase (h) production assay in peptone-tryptic soy broth with 1 mM IPTG. Growth kinetics (i) with RpoN* expression induced with 1 mM IPTG at 0.5 OD600. Statistics used were Student’s t-test (***p ≤ 0.0001; **p ≤ 0.01; *p ≤ 0.05). Bars indicate mean colony diameter of replicates; error bars represent one standard deviation of the mean. n = 4 to 7 replicates per assay.
RpoN* decreases biofilm formation in vitro
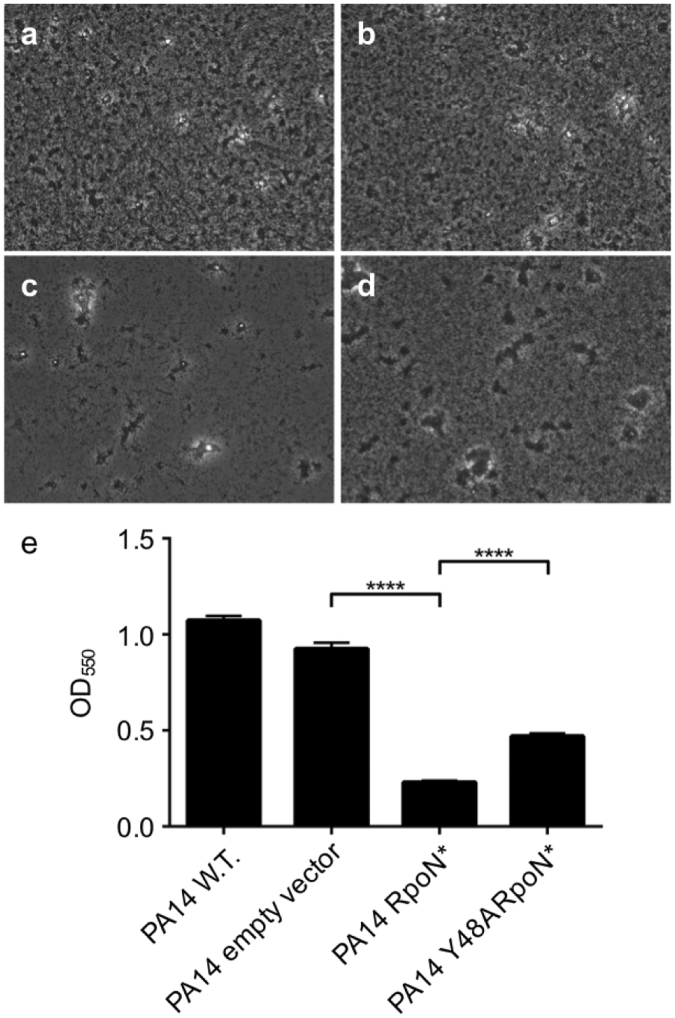
Biofilm formation is a major obstacle in treating many bacterial infections. Microarray analysis revealed that RpoN* reduced transcription of genes involved in biofilm formation, including cdrA, paL1, and pprB. Additionally, a recent study on sigma-factor regulons in P. aeruginosa PA14 identified the biofilm-related pel genes as having a RpoN binding site24. Thus, we sought to confirm if RpoN* could hinder biofilm formation in vitro using both an air-liquid interface (ALI) assay and a microtiter-dish based assay. Wild type P. aeruginosa PA14 was used as a positive control. The test conditions were P. aeruginosa wild-type (empty vector), RpoN*, or the point mutant Y48A RpoN*. If RpoN* reduced expression of genes involved in biofilm formation, we expected to see diminished formation of biofilms in vitro. As expected, wild-type P. aeruginosa PA14 formed dense biofilms (Fig. 5a,b,e), with or without the empty vector plasmid. Conversely, P. aeruginosa expressing RpoN* diminished biofilm formation in both the ALI (Fig. 5c) and microtiter-dish assays (Fig. 5e; Student’s t-test p ≤ 0.0001). Expression of the Y48A RpoN* also reduced biofilm formation, but to a lesser extent than RpoN* (Fig. 5d,e; Student’s t-test p ≤ 0.0001). These results are consistent with RpoN* acting as an antagonist of biofilm formation in P. aeruginosa.
Figure 5.
RpoN* decreases biofilm formation in vitro. (a–d) Air-Liquid Interface (ALI) biofilm formation assay conducted in well bottoms of 6-well microtiter plates for 24 h and visualized by phase contrast microscopy at 200x. Strains used: P. aeruginosa PA14 wild type (a), empty vector (b), RpoN* (c), and Y48A* point mutant (d). Images shown are representative fields of view from two independent experiments with three technical replicates for each strain. (e) Biofilms formed in 96-well microtiter plates were stained with crystal violet, solubilized, and quantified at OD550 (n = 28). Data presented as mean ± SEM. Student’s t test performed (****p ≤ 0.0001).
RpoN* increases C. elegans survival in paralytic killing assay
Phenotypic assays and microarray analysis indicated that expression of RpoN* could repress production of virulence factors in P. aeruginosa. Thus, the effects of the roadblock were next evaluated with a P. aeruginosa–C. elegans infection model. C. elegans is an efficient in vivo model for evaluating P. aeruginosa virulence, as C. elegans has innate defenses to protect from bacteria, some of which have homologs in the human innate system35. Additionally, C. elegans is often used for screening antimicrobials36. Thus, this model is a valuable tool to study the effects of RpoN* in P. aeruginosa virulence. Each infection model, or killing strategy, involves different virulence factors, with varied types of media used to promote each specific killing strategy37,38. This is in accordance with other Gram-negative bacteria, where choice of media is known to affect gene expression39.
A paralytic killing model, where lethal paralysis is mediated by hydrogen cyanide production by P. aeruginosa, was one of the infection assays used37,40. Some of the genes found to be associated with paralytic killing, particularly hcnC, soxA, prpC, and PA074540, were affected by RpoN* in the microarray analysis. The test conditions were wild-type P. aeruginosa expressing RpoN* or the point mutant Y48A in RpoN* from an IPTG-inducible promoter. If RpoN* reduced gene expression of virulence factors, we expected to see increased C. elegans survival compared to the positive control. As expected, the positive control caused C. elegans paralysis and killing, while both negative controls had no effect on C. elegans survival (Fig. 6a). Expression of RpoN* or the Y48A point mutant significantly increased C. elegans survival compared to the wild type strain (Mantel Log Rank Test, p ≤ 0.0001). Furthermore, expression of RpoN* increased C. elegans survival significantly more compared to the Y48A point mutant (Mantel-Cox Log-Rank Test, p ≤ 0.0001). Thus, RpoN* expression improved worm survival in a C. elegans–P. aeruginosa infection model mediated by cyanide production.
Figure 6.
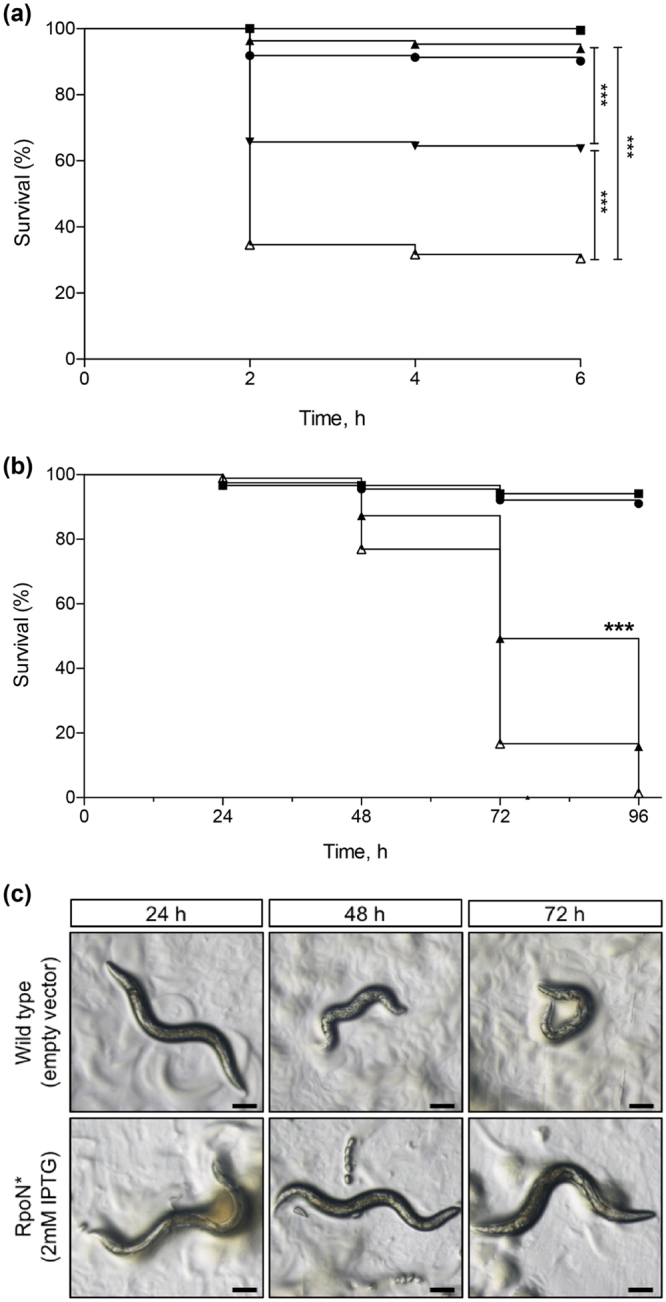
RpoN* increases C. elegans survival in C. elegans – P. aeruginosa infection assays. (a) Brain-heart infusion media with bacto-agar was supplemented with 30 mg/L gentamicin and at least 1 mM IPTG. All paralytic killing assays were conducted at room temperature (22–24 °C), and scored every 2 h. Strains used: E. coli (▪, n = 195), P. aeruginosa ΔlasR (⚫, n = 197), wild-type P. aeruginosa PAO1 (Manoil strain) (empty vector, Δ, n = 205), RpoN* (▴, n = 301), and Y48A* point mutant (▾, n = 376). (b) Slow killing agar (0.35% bactopeptone, 2% bactoagar) was supplemented with 30 mg/L gentamicin and at least 1 mM IPTG, when necessary. Slow killing assays were conducted at 20 °C and scored every 24 h. Strains used: E. coli (▪, n = 90), P. aeruginosa ΔlasR (⚫, n = 90), wild-type P. aeruginosa PA19660 Xen5 (empty vector, Δ, n = 171), RpoN* (▴, n = 160). (c) Appearance of worms fed on P. aeruginosa wild type (upper panels) or RpoN* (lower panels). All images taken on an Olympus stereoscope (mag. 4.5x). Scale bars are 100 μm. Points on the Kaplan-Meier survival curves represent the combined survival of three or more separate assays. Mantel-Cox log-rank tests performed to analyze curves (***p ≤ 0.0001).
RpoN* prolongs C. elegans survival in a slow killing model
Multiple P. aeruginosa–C. elegans infection assays can be used to evaluate pathogenicity of the bacteria. The paralytic killing assay revealed that RpoN* improved C. elegans survival in an infection assay mediated by cyanide production. While the paralytic killing assay mimics infection conditions similar to a cystic fibrosis lung, other assays model alternate forms of P. aeruginosa infections41. Thus, we also evaluated the effects of RpoN* in a slow killing assay, where death is facilitated by the establishment and proliferation of an infection38, and is mediated by lasR, gacA, lemA, and ptsP 38. Wild-type P. aeruginosa PA19660 (empty vector) was the positive, virulent control. P. aeruginosa ΔlasR:gent R and E. coli, both lacking a vector, were negative, avirulent controls. The test condition was wild-type P. aeruginosa expressing RpoN* from an IPTG-inducible promoter. If RpoN* affected genes associated with P. aeruginosa pathogenicity, we expected improved C. elegans survival. As expected, the negative controls did not affect C. elegans survival (Fig. 6b). The positive control diminished C. elegans survival, with an LT50 of 72 h (Fig. 6b). C. elegans exposed to the wild-type strain appeared unhealthy as early as 48 h (Fig. 6c, top panels). RpoN* expression significantly improved C. elegans survival compared to wild type and increased the LT50 to 96 h (Fig. 6b; Mantel-Cox Log-Rank Test, p ≤ 0.0001). Furthermore, C. elegans appeared healthier on P. aeruginosa lawns expressing RpoN* (Fig. 6c, bottom panels). While it was expected that RpoN* would reduce P. aeruginosa virulence, there are several possible factors why RpoN* expression did not have a larger effect on P. aeruginosa survival in the slow killing assay. C. elegans often require higher concentrations of compounds due to selective uptake in the intestine and limited permeability of the cuticle36. In fact, a previous study reported that the levels of drug absorbed and metabolized by C. elegans was time and dose dependent, and that the presence of live bacteria decreased drug concentration in media42. Thus, it is possible that concentrations of gentamicin and IPTG were not sustained at adequate levels in the C. elegans gut to maintain selection and expression of the roadblock. A liquid assay would help alleviate this problem, as C. elegans would be suspended in media for the duration of the experiment43, which would allow gentamicin and IPTG to be pumped into the C. elegans gut as it feeds, helping to maintain selection and expression.
Conclusion
Here, we demonstrated that the RpoN molecular roadblock altered gene expression and reduced virulence of P. aeruginosa. More genes were altered by RpoN* regulation than those with an RpoN consensus promoter, as the roadblock reduced expression of various regulatory proteins, subsequently reducing expression of their downstream targets. We also showed that in the absence of native RpoN, the molecular roadblock altered gene transcription, significantly reducing motility and other virulence-associated phenotypes. In these instances, RpoN* provides clues to the nuanced levels of transcriptional regulation within bacteria for genes controlled by multiple promoters. When native RpoN is absent, these interactions became clearer with notable decreases in gene expression of specific genes and reduction in specific virulence phenotypes. The molecular roadblock is a DNA binding agent that is cis-acting directly at RpoN consensus promoters. We propose that, regardless of the presence of native RpoN, the molecular roadblock alters transcription, reducing virulence in P. aeruginosa (Fig. 3c). Although mutations may arise to evade the molecular roadblock, the numerous RpoN binding sites within the P. aeruginosa genome precludes resistance. The effectiveness of the roadblock to reduce gene expression and virulence, along with the lower likelihood of developing resistance, suggests targeting RpoN consensus promoters is a powerful strategy to combat pathogenic P. aeruginosa.
Future studies will help us to further understand the effects and mechanism of action of the molecular roadblock. For instance, RpoN has been linked to P. aeruginosa tolerance of several antibiotics, specifically carbapenems, quinolones, and tobramycin44–46. The molecular roadblock reduced expression of mex family genes, including mexH, which are associated with multidrug efflux pumps. Additionally, the roadblock affected expression of rpoS and quorum sensing-related genes that regulate tolerance of P. aeruginosa to ofloxacin47. Thus, studies are needed to address synergistic effects between the roadblock and antibiotics. Additionally, the RpoN-box25,27,48, which facilitates DNA binding, as well as the RpoN consensus promoter sites49,50 are conserved among both Gram-positive and Gram-negative bacteria. Therefore, studies are also needed to evaluate potential broad-spectrum applications of the molecular roadblock. Unfortunately, the roadblock is not druggable in its current form, limiting its application, but its binding sites in the genome should serve as new targets for drug discovery. Finding small molecules with similar cis-acting function is crucial for development into a new antimicrobial. Targeting RpoN at its consensus promoters in a similar manner to the molecular roadblock may be an effective and desirable strategy to treat P. aeruginosa or other bacterial infections while minimizing the development of resistance.
Materials and Methods
Bacteria and Nematodes
P. aeruginosa PAO1 was provided by C. Manoil37 and D. Haas16. P. aeruginosa PA14 was provided by F. Ausubel51. P. aeruginosa PA19660 Xen5 was purchased from PerkinElmer. The lecA::luxΔlasR P. aeruginosa PAO1 mutant was provided by S.P. Diggle52. The P. aeruginosa PAO1 ΔrpoN mutant was provided by D. Haas16. E. coli OP50 was provided by D. Pruyne (Upstate Medical University). Bacteria were grown overnight in Lennox Broth (Difco or Fisher BioReagents) at 37 °C with shaking, glycerol was added to 10%, and stocks were stored frozen at −80 °C. Caenorhabditis elegans N2 was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN), and maintained on E. coli (OP50) on nematode growth media agar (NGM) at 20 °C53. To create a synchronized population, hermaphroditic, gravid adults were transferred by wire pick to fresh NGM plates with E. coli for 2–4 hours to lay eggs, and then removed. Eggs were grown to the young adult stage at 20 °C54.
Plasmids
Plasmids and oligonucleotides used in this study are given in Supplementary Table S1. Plasmids were maintained in E. coli Top10 (Invitrogen). For plasmid and marker selection, the following antibiotics were used: ampicillin, 100 mg/L E. coli; kanamycin, 50 mg/L E. coli or 500 mg/L P. aeruginosa; gentamicin, 20 mg/L E. coli or 30 mg/L P. aeruginosa. For induction of gene expression, isopropyl β-D-1-thiogalactopyranoside (IPTG) was used, up to concentrations of 2 mM.
Construction of RpoN*
A tightly regulated, IPTG-inducible expression vector for P. aeruginosa was constructed by cloning the region encoding LacIQ and the trc-promoter of pTrc99a (Pharmacia Biotech, Sweden) into the Xba I and Sph I sites of the broad-host range plasmid pBBR1MCS-5 to yield pBRL32055. EcoR I and Sac I sites of the multiple cloning region in the pTrc99a fragment were removed by site-directed mutagenesis using QuikchangeTM (Stratagene) and oligonucleotides BL331.f/BL331.r (Supplementary Table S1) to give pBRL344.
A gene (rpoN*) encoding the last 60 amino acids of RpoN of A. aeolicus was synthesized and codon optimized for expression in E. coli by DNA 2.0 (Menlo Park, California). Nde I and Sac I restriction sites were engineered into the 5′ and 3′ ends of rpoN*, respectively. The rpoN* gene was then subcloned from pJ201:42178 into the Nde I/Sac I sites of the E. coli expression pET-vector pKH22 to give pBRL32756. pBRL327 was then digested with Xba I and Sac I to liberate the rpoN* gene with an upstream ribosome binding site. This fragment was cloned into the Xba I/Sac I sites of pBRL344 to yield pBRL348. To attenuate the rpoN* in pBRL348, the tyrosine at position 48 was changed to alanine using QuickchangeTM (Stratagene, Santa Clara, California) and oligonucleotides BL330.f/BL330.r to give pBRL34934.
Transformation
Plasmids were introduced to P. aeruginosa through electroporation prior to all experiments, as previously described57. Transformed bacteria were selected for on brain heart infusion (BHI) or LB agar, and individual colonies were picked for each assay.
RNA isolation and microarray analysis
RNA isolation and microarray analysis were done in quadruplicate. P. aeruginosa PAO1 or P. aeruginosa ΔrpoN were transformed with either pBRL344 or pBRL348 were grown in LB supplemented with 30 mg/L gentamicin for 24 h at 37 °C and 200 rpm. Fresh LB supplemented with 30 mg/L gentamicin was inoculated with 0.5% (v/v) of the LB-grown seed culture. The inoculated cultures were then grown at 37 °C and 200 rpm to an optical density at 600 nm (OD600) of 0.2. Cultures were treated with IPTG at a final concentration of 1 mM, and induced cultures were grown for an additional 2 h. For P. aeruginosa ΔrpoN, media was supplemented with 1 mM glutamine. RNA was purified using a Qiagen RNeasy kit with addition of RNAprotect® reagent and on-column DNase digestion58,59. RNA samples were analyzed for quality with a Bioanalyzer (Agilent) and for DNA contamination by PCR. Microarray studies were carried out at the Microarray Core Facility at SUNY Upstate Medical University (Syracuse, NY). Microarray experiments were performed as indicated in the Affymetrix GeneChip® Expression Analysis Technical Manual (Pub. 702232, Rev. 3) and by established protocol60. Data was processed with Affymetrix software for quality control, calculating signal intensities, and indicating presence of a gene. The RMA method was used to normalize data (GeneTraffic software, Stratagene, La Jolla). Additional statistical analysis was performed with the MultiExperiment Viewer (MeV v4.6.2) to identify genes with significant differences in intensities. Microarray data was deposited in Gene Expression Omnibus and is accessible through GEO Series accession number GSE3563261.
Phenotyping Assays
Assays to measure swimming and twitching62, protease63, elastase64,65, pyoverdine and pyocyanin66, and rhamnolipids67,68 were conducted according to standard protocols. Freshly transformed P. aeruginosa PA19660 Xen5, PAO1 wild type,or PAO1 ΔrpoN, and lecA::luxΔlasR P. aeruginosa PAO1 were grown on the appropriate media with 30 µg/mL gentamicin with or without 2 mM IPTG. Plates for motility, protease and elastase assays were incubated for 24–48 h at 37 °C and photographs were taken and bioluminescence was measured by IVIS (PerkinElmer). For the motility assays, the colony diameter was measured across the point of inoculation to the edges of the bacterial patch using Living Image software (PerkinElmer). Elastase, pyoverdine and pyocyanin assays in liquid broth were grown at 37 °C, 200 rpm for the appropriate time and absorbance measured at 459 nm, 408 nm, and 520 nm, respectively. For PAO1 and PA19660, media was not supplemented with additional nutrients as it was unnecessary for adequate growth. For the ΔrpoN mutant, media was supplemented with 1 mM glutamine.
Biofilm Assay
Freshly transformed P. aeruginosa PA14 were inoculated in LB broth with 30 mg/L gentamicin and grown overnight at 37 °C. Cultures were diluted 1:3 in LB containing 30 mg/L gentamicin and 1 mM IPTG, grown for 3 h at 37 °C, and subcultured 1:50 in M63 minimal media with 0.4% arginine, 1 mM MgSO4, 30 mg/L gentamicin, and 1 mM IPTG. Air-liquid interface (ALI) and microtiter dish biofilm formation assays were conducted by standard protocol69–71. For the ALI assay, biofilms were visualized by phase contrast at 200x on a Leica AF6000 microscope. For the microtiter dish assay, biofilms were stained with crystal violet, extracted in ethanol and absorbance at 550 nm was measured on a Synergy H1 Multi-Mode reader (Biotek).
Paralytic Killing Assay
Freshly transformed P. aeruginosa PAO1 were swabbed onto BHI agar with 30 mg/L gentamicin and at least 1 mM IPTG, when appropriate. For lecA::luxΔlasR P. aeruginosa PAO1 and E. coli (OP50), bacteria were grown from frozen stocks overnight at 37 °C with shaking in BHI broth containing or lacking 30 mg/L gentamicin, respectively. These overnight cultures were diluted 1:100 in BHI and 170 μL was spread on BHI agar with or without antibiotics, per standard protocol37. All BHI agar plates contained 1.7% BactoAgar in 60-mm petri plates. Plates were incubated at 37 °C for 24 h.
Slow Killing Assay
Freshly transformed P. aeruginosa PA19660 Xen5 was swabbed onto modified NGM (0.35% peptone)38 with 30 mg/L gentamicin and when applicable, at least 1 mM IPTG. For lecA::luxΔlasR P. aeruginosa PAO1 and E. coli (OP50), fresh bacteria plates were used to inoculate LB with or without 30 mg/L gentamicin, respectively, and grown overnight at 37 °C with shaking. A 30 µL volume of overnight culture was spread on modified NGM with or without antibiotics, per standard protocol38. All modified NGM plates were incubated at 37 °C for 24 h, then at room temperature (22–26 °C) for an additional 24 h.
Statistics
Data were analyzed using Excel and GraphPad Prism with a significance of p ≤ 0.05 (Microsoft, Washington; GraphPad Software Inc., California).
Data Availability
The datasets generated during the current study are deposited in the Gene Expression Omnibus and is accessible through GEO Series accession number GSE35632 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35632). Otherwise, the datasets generated for and analyzed during the current study are either included in this article (and its supplementary files) or is available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by the NIH (CTN: 2R15GM104880), by The Hill Collaboration (JFM and CTN: 58482), by Cystic Fibrosis Canada (TFM: 2613) and by the Natural Sciences and Engineering Research Council of Canada (TFM: 341358-2010). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author Contributions
M.G.L. wrote the manuscript. C.T.N. and J.F.M. conceived the study. M.G.L., B.R.L., C.W.H., L.B.-P.G., and T.F.M. conducted the experiments. M.G.L., C.W.H., L.B.-P.G., and T.F.M. generated the figures. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12667-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bessa LJ, Fazii P, Di Giulio M, Cellini L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: some remarks about wound infection. Int Wound J. 2015;12:47–52. doi: 10.1111/iwj.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnihotri N, Gupta V, Joshi RM. Aerobic bacterial isolates from burn wound infections and their antibiograms–a five-year study. Burns. 2004;30:241–243. doi: 10.1016/j.burns.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Edlich RF, et al. Developing an organized approach in the Food and Drug Administration to ban dangerous devices that can injure the patient and health care worker. J Emerg Med. 2012;42:454–456. doi: 10.1016/j.jemermed.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Registry, C. F. F. P. 2015 Annual Data Report. (2016).
- 5.Porras-Gomez M, Vega-Baudrit J, Nunez-Corrales S. Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches. Journal of Biomaterials and Nanobiotechnology. 2012;3:9. doi: 10.4236/jbnb.2012.324053. [DOI] [Google Scholar]
- 6.Tacconelli, E. & Magrini, N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organization, 1–7 (2017).
- 7.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 9.Hung DT. Anti-virulence appraoches to antimicrobial therapy. The FASEB Journal. 2007;21:A94–A94. [Google Scholar]
- 10.Sarkar S, et al. Comprehensive analysis of type 1 fimbriae regulation in fimB-null strains from the multidrug resistant Escherichia coli ST131 clone. Mol Microbiol. 2016;101:1069–1087. doi: 10.1111/mmi.13442. [DOI] [PubMed] [Google Scholar]
- 11.Dickey, S. W., Cheung, G. Y. C. & Otto, M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov, 10.1038/nrd.2017.23 (2017). [DOI] [PubMed]
- 12.Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Totten PA, Lara JC, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damron FH, et al. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol. 2012;194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heurlier K, Denervaud V, Pessi G, Reimmann C, Haas D. Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. Journal of bacteriology. 2003;185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson LS, Webb JS, Rice SA, Kjelleberg S. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2003;220:187–195. doi: 10.1016/S0378-1097(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 18.Boucher JC, Schurr MJ, Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- 19.Caiazza NC, O’Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrickson EL, Plotnikova J, Mahajan-Miklos S, Rahme LG, Ausubel FM. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J Bacteriol. 2001;183:7126–7134. doi: 10.1128/JB.183.24.7126-7134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinbaum RL, et al. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 2012;8:e1002813. doi: 10.1371/journal.ppat.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz S, et al. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 2015;11:e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor M, et al. The RpoN-box motif of the RNA polymerase sigma factor sigma N plays a role in promoter recognition. Mol Microbiol. 1996;22:1045–1054. doi: 10.1046/j.1365-2958.1996.01547.x. [DOI] [PubMed] [Google Scholar]
- 26.Bose D, et al. Organization of an activator-bound RNA polymerase holoenzyme. Mol Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doucleff M, Pelton JG, Lee PS, Nixon BT, Wemmer DE. Structural basis of DNA recognition by the alternative sigma-factor, sigma54. Journal of molecular biology. 2007;369:1070–1078. doi: 10.1016/j.jmb.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrick MJ. In a class of its own–the RNA polymerase sigma factor sigma 54 (sigma N) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 29.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond JH, Dolben EF, Smith TJ, Bhuju S, Hogan DA. Links between Anr and quorum sensing in Pseudomonas aeruginosa biofilms. Journal of bacteriology. 2015;197:2810–2820. doi: 10.1128/JB.00182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampel KJ, et al. Characterization of the GbdR regulon in Pseudomonas aeruginosa. J Bacteriol. 2014;196:7–15. doi: 10.1128/JB.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Gralla JD. Roles for the C-terminal region of sigma 54 in transcriptional silencing and DNA binding. J Biol Chem. 2001;276:8979–8986. doi: 10.1074/jbc.M009587200. [DOI] [PubMed] [Google Scholar]
- 35.Schulenburg H, Hoeppner MP, Weiner J, 3rd, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 2008;213:237–250. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 36.O’Reilly LP, Luke CJ, Perlmutter DH, Silverman GA, Pak SCC. elegans in high-throughput drug discovery. Adv Drug Deliv Rev. 2014;69-70:247–253. doi: 10.1016/j.addr.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darby C, Cosma CL, Thomas JH, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blair JM, Richmond GE, Bailey AM, Ivens A. & Piddock, L. J. Choice of bacterial growth medium alters the transcriptome and phenotype of Salmonella enterica Serovar Typhimurium. PLoS One. 2013;8:e63912. doi: 10.1371/journal.pone.0063912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallagher LA, Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Utari PD, Quax WJ. Caenorhabditis elegans reveals novel Pseudomonas aeruginosa virulence mechanism. Trends Microbiol. 2013;21:315–316. doi: 10.1016/j.tim.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Zheng SQ, Ding AJ, Li GP, Wu GS, Luo HR. Drug absorption efficiency in Caenorhbditis elegans delivered by different methods. PLoS One. 2013;8:e56877. doi: 10.1371/journal.pone.0056877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mozhei OI, et al. [Evaluating the mitochondrial dna copy number in leukocytes and adipocytes from metabolic syndrome patients: pilot study] Mol Biol (Mosk) 2014;48:677–681. [PubMed] [Google Scholar]
- 44.Viducic D, et al. rpoN gene of Pseudomonas aeruginosa alters its susceptibility to quinolones and carbapenems. Antimicrob Agents Chemother. 2007;51:1455–1462. doi: 10.1128/AAC.00348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. RpoN Modulates Carbapenem Tolerance in Pseudomonas aeruginosa through Pseudomonas Quinolone Signal and PqsE. Antimicrob Agents Chemother. 2016;60:5752–5764. doi: 10.1128/AAC.00260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. RpoN Promotes Pseudomonas aeruginosa Survival in the Presence of Tobramycin. Front Microbiol. 2017;8:839. doi: 10.3389/fmicb.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kayama S, et al. The role of rpoS gene and quorum-sensing system in ofloxacin tolerance in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;298:184–192. doi: 10.1111/j.1574-6968.2009.01717.x. [DOI] [PubMed] [Google Scholar]
- 48.Burrows PC, Severinov K, Ishihama A, Buck M, Wigneshweraraj SR. Mapping sigma 54-RNA polymerase interactions at the -24 consensus promoter element. J Biol Chem. 2003;278:29728–29743. doi: 10.1074/jbc.M303596200. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Gralla JD. Multiple in vivo roles for the -12-region elements of sigma 54 promoters. J Bacteriol. 1998;180:5626–5631. doi: 10.1128/jb.180.21.5626-5631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrios H, Valderrama B, Morett E. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic acids research. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 52.Fletcher MP, et al. A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environmental Microbiology. 2007;9:2683–2693. doi: 10.1111/j.1462-2920.2007.01380.x. [DOI] [PubMed] [Google Scholar]
- 53.Wood, W. B. The Nematode Caenorhabditis elegans: Introduction to C. elegans biology. (Cold Spring Harbor Laboratory Press, 1988).
- 54.Byerly L, Cassada RC, Russell RL. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev Biol. 1976;51:23–33. doi: 10.1016/0012-1606(76)90119-6. [DOI] [PubMed] [Google Scholar]
- 55.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 56.Lundgren BR, Boddy CN. Sialic acid and N-acyl sialic acid analog production by fermentation of metabolically and genetically engineered Escherichia coli. Org Biomol Chem. 2007;5:1903–1909. doi: 10.1039/b703519e. [DOI] [PubMed] [Google Scholar]
- 57.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 58.Lundgren BR, et al. Gene PA2449 is essential for glycine metabolism and pyocyanin biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2013;195:2087–2100. doi: 10.1128/JB.02205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lundgren BR, et al. Genetic analysis of the assimilation of C5-dicarboxylic acids in Pseudomonas aeruginosa PAO1. J Bacteriol. 2014;196:2543–2551. doi: 10.1128/JB.01615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Middleton FA, Carrierfenster K, Mooney SM, Youngentob SL. Gestational ethanol exposure alters the behavioral response to ethanol odor and the expression of neurotransmission genes in the olfactory bulb of adolescent rats. Brain Res. 2009;1252:105–116. doi: 10.1016/j.brainres.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Toole GA, et al. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 63.Kulasekara, H. D. Protocols: Pseudomonas Phenotyping, http://miller-lab.net/MillerLab/protocols/all-about-phenotyping/ (2012).
- 64.Rust L, Messing CR, Iglewski BH. Elastase assays. Methods Enzymol. 1994;235:554–562. doi: 10.1016/0076-6879(94)35170-8. [DOI] [PubMed] [Google Scholar]
- 65.Ohman DE, Cryz SJ, Iglewski BH. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Essar DW, Eberly L, Han CY, Crawford IP. DNA sequences and characterization of four early genes of the tryptophan pathway in Pseudomonas aeruginosa. J Bacteriol. 1990;172:853–866. doi: 10.1128/jb.172.2.853-866.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siegmund I, Wagner F. New method for detecting rhamnolipids excreted byPseudomonas species during growth on mineral agar. Biotechnology Techniques. 1991;5:265–268. doi: 10.1007/BF02438660. [DOI] [Google Scholar]
- 68.Pinzon NM, Ju L-K. Improved detection of rhamnolipid production using agar plates containing methylene blue and cetyl trimethylammonium bromide. Biotechnology letters. 2009;31:1583–1588. doi: 10.1007/s10529-009-0049-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhang L, et al. Identification of genes involved in Pseudomonas aeruginosa biofilm-specific resistance to antibiotics. PLoS One. 2013;8:e61625. doi: 10.1371/journal.pone.0061625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merritt, J. H., Kadouri, D. E. & O’Toole, G. A. In Current Protocols in Microbiology (John Wiley & Sons, Inc., 2005).
- 71.O’Toole, G. A. Microtiter dish biofilm formation assay. J Vis Exp, 10.3791/2437 (2011). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are deposited in the Gene Expression Omnibus and is accessible through GEO Series accession number GSE35632 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35632). Otherwise, the datasets generated for and analyzed during the current study are either included in this article (and its supplementary files) or is available from the corresponding author on reasonable request.