Abstract
Virus-based gene therapy by CRISPR/Cas9-mediated genome editing and knockout may provide a new option for treatment of inherited and acquired ocular diseases of the retina. In support of this notion, we show that Streptococcus pyogenes (Sp) Cas9, delivered by lentiviral vectors (LVs), can be used in vivo to selectively ablate the vascular endothelial growth factor A (Vegfa) gene in mice. By generating LVs encoding SpCas9 targeted to Vegfa, and in parallel the fluorescent eGFP marker protein, we demonstrate robust knockout of Vegfa that leads to a significant reduction of VEGFA protein in transduced cells. Three of the designed single-guide RNAs (sgRNAs) induce in vitro indel formation at high frequencies (44%–93%). A single unilateral subretinal injection facilitates RPE-specific localization of the vector and disruption of Vegfa in isolated eGFP+ RPE cells obtained from mice five weeks after LV administration. Notably, sgRNA delivery results in the disruption of Vegfa with an in vivo indel formation efficacy of up to 84%. Sequencing of Vegfa-specific amplicons reveals formation of indels, including 4-bp deletions and 2-bp insertions. Taken together, our data demonstrate the capacity of lentivirus-delivered SpCas9 and sgRNAs as a developing therapeutic path in the treatment of ocular diseases, including age-related macular degeneration.
Keywords: SpCas9, lentiviral vector, retinal gene knockout, RPE cells, VEGFA, subretinal delivery
Introduction
Development of gene knockout and replacement strategies for treatment of retinal diseases relies on efficient delivery of genetic tools ablating or substituting genomic sequences.1 Recent studies have demonstrated efficient biallelic2 and allele-specific3 knockout in animal models of retinitis pigmentosa (RP),4 Leber’s congenital amaurosis (LCA),5 and age-related macular degeneration (AMD)6 following non-viral or viral delivery of CRISPR/Cas97, 8, 9 orthologs.
In autosomal-dominant RP, disease penetrance is conferred by a monoallelic gain of function mutation.10 Presently, 24 genes have been implicated in the etiology of autosomal dominant RP, with Rho variations representing the highest proportion of RP cases.11 The Rho gene encodes a G protein-coupled receptor that is the primary photon detector in rod photoreceptor cells.10 To develop a simplified therapeutic approach restoring retinal function Bakondi et al. used subretinal injection of naked plasmid DNA encoding single-guide RNA (sgRNA) molecules and Streptococcus pyogenes (Sp)Cas9, followed by electroporation for allele-specific targeting of the dominant RhoS334 mutation in heterozygous S334ter-3 rats.12 Using this strategy, allele-specific disruption of RhoS334 was demonstrated resulting in photoreceptor cell preservation, alleviation of retinal degeneration, and improved visual acuity. In another study, likewise based on non-viral delivery, Latella et al. targeted the Rho gene in a mutation-independent manner, using two sgRNAs to excise the region around the P23H mutation site,4 leading to proof-of-principle that SpCas9 knockout and subsequent gene replacement can be utilized for treatment of retinal diseases.
To further improve efficacy of gene knockout, viral vectors have been used to deliver Cas9 to the retina. Hung et al. targeted yellow fluorescent protein (YFP) in Thy1-YFP transgenic mice using an adeno-associated virus (AAV)-based SpCas9 delivery approach.13 Because of limited cargo capacity, the AAV is not able to deliver both SpCas9 and sgRNA in a single vector.14 Hence, in the study by Hung et al., sgRNA and Cas9 were delivered by co-transduction of separate AAV vectors. Similarly, sgRNA and SpCas9 delivered in separate AAVs were found to ameliorate the effects of a mutation introducing a cryptic splice site in the Cep290 gene, often affected in LCA10 patients.5 Even though gene knockout was demonstrated by the two-vector AAV system, this approach may be limited by the need for co-transduction.
To overcome the limitations related to the two-vector AAV system, other Cas9 orthologs have been investigated.15, 16 Because of its smaller size (∼3.2 kb), the Streptococcus aureus Cas9 (SaCas9)17 is suitable for combined all-in-one AAV-based vectors encoding both Cas9 and sgRNA. Although SaCas9 and SpCas9 have comparable endonuclease activities, they differ in requirements related to the protospacer-adjacent motif (PAM). Hence, SaCas9 requires a 5′-NNGRRT-3′ PAM sequence, which enables targeting of various genomic positions differing from sites containing the SpCas9 5′-NGG-3′ PAM.18, 19 Smaller Cas9 orthologs with alternative PAM preferences thus may be crucial to further improve efficacy and specificity of in vivo genome knockout and editing. Exploiting this strategy, a recent study showed efficient in vivo genome editing by Campylobacter jejuni Cas9 (CjCas9) targeting vascular endothelial growth factor A (Vegfa) following intravitreal delivery of an all-in-one AAV.6 Even though lentiviral vectors (LVs) have the capacity to deliver large Cas9 proteins together with sgRNAs and marker proteins, as well as a high transduction efficiency and specificity to target a range of different tissues of both dividing and non-dividing cells,20 only a few studies have demonstrated in vivo gene editing following lentiviral delivery of Cas9.21
In this study, we demonstrate for the first time in vivo gene knockout in the murine retina mediated by LVs encoding sgRNA and SpCas9 targeted to the Vegfa gene in the retinal pigment epithelium (RPE) cells. As dysregulation of the Vegfa gene, which promotes aberrant angiogenesis, is linked to exudative AMD22, 23, 24 as well as other retinal neovascular disorders, the presented data further support the notion that in vivo genome knockout with Cas9 may provide a new platform for the treatment of AMD.
Results
Design of sgRNAs Targeted to the Vegfa Gene
To induce efficient knockout of the murine Vegfa gene, different Vegfa-targeting sgRNAs were designed. The Vegfa gene consists of eight exons alternatively spliced to produce several VEGFA isoforms, which perform distinctive tasks in different compartments of the body. The predominant variant is VEGFA, which consists of exon 1–5, 7, and 8. All splice variants share exons 1–5. A large exon provides more sgRNA target sites, increasing the possibility of computing highly efficient sgRNAs. Exon 3 is the largest exon of the first five exons, and indel formation in the beginning of the gene would induce a frameshift leading to nonsense-mediated RNA decay (NMD).31, 32 On this basis, exon 3 of Vegfa was chosen for targeting.
MIT CRISPR Design and Broad Institute CRISPRko were used to assess off-target and on-target efficiency, respectively. CRISPRko internally ranked the designed sgRNAs for off-target scores. Four sgRNAs were designed to guide the SpCas9 by the two sgRNA prediction tools. Two sgRNAs were chosen based on optimal off-target scores (MIT CRISPR Design) combined with the highest possible on-target scores (sgRNA1 and 4) (Figure 1A). sgRNA2 and sgRNA3 were selected by the highest on-target scores (CRISPRko) (Table S1). A non-targeting control sgRNA was included and is designated here by sgRNA-irrelevant (Irr) (5′-ACGGAGGCTAAGCGTCGCAA-3′).
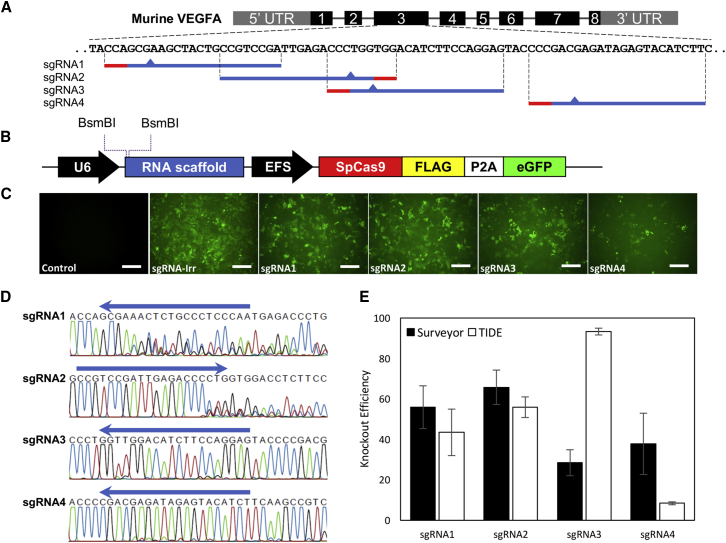
Figure 1.
Design and Validation of LV/Cas9-sgRNA Vectors In Vitro
(A) Murine Vegfa target sequence used for sgRNA design. Based on on- and off-target activities assessed by MIT CRISPR Design and Broad Institute CRISPRko, four sgRNAs were designed. Sequence of each sgRNA is indicated with subsequent PAM sequence (red) and supposed cut-site (triangle). (B) Schematics of the LV/Cas9-sgRNA multigenic expression vector. The lentiviral transfer vector incorporates a U6 promoter that drives the expression of the gRNA scaffold. Indicated BsmBI restriction sites were used for cloning of the sgRNA sequences. The EFS promoter drives expression of the SpCas9 transcript also encoding a FLAG tag, a P2A self-cleaving sequence, and eGFP. (C) Immunofluorescence analysis of eGFP expression five days post-transduction. HEK293-VEGFA cells transduced with 500 ng p24 of LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, LV/Cas9-sgRNA3, LV/Cas9-sgRN4, or LV/Cas9-sgRNA-Irr. Immunofluorescence analysis of eGFP indicates transduction efficiency after 5 days. A non-transduced control was included (control). (D) Sequencing of Vegfa gDNA following LV transduction. Five days post-transduction, gDNA was isolated from LV-transduced HEK293-VEGFA cells and used as template for PCR. Amplification of Vegfa using sequence-specific primers resulted in a 567-bp PCR product, which was sequenced using the forward PCR primer. The resultant chromatograms are shown. (E) Indel formation efficiency by Surveyor Nuclease Assay and TIDE analysis. The Vegfa gene was amplified using gDNA isolated from HEK293-VEGFA cells transduced with LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, LV/Cas9-sgRNA3, LVCas9-sgRNA4, LV/Cas9-sgRNA-Irr, or the non-transduced control (control). For surveyor nuclease assay, PCR products were reannealed to generate heteroduplexes followed by surveyor nuclease digestion and analysis by gel electrophoresis (black columns). Indel formation frequencies were calculated according to the Materials and Methods. Gel electrophoresis following surveyor nuclease digestion is presented in Figure S1A. Sequencing results from LV/Cas9-sgRNA1-, LV/Cas9-sgRNA2-, LV/Cas9-sgRNA3-, and LV/Cas9-sgRNA4-transduced HEK293-VEGFA cell samples were analyzed by TIDE in comparison to the LV/Cas9-sgRNA-Irr sample (white columns). Indel formation frequencies are shown as mean ± SD (n = 3). Scale bar, 200 μm. EFS, elongation factor 1α short; HEK293, human embryonic kidney cells; LV, lentiviral vector; PAM, protospacer-adjacent motif; sgRNA, single-guide RNA; TIDE, tracking of indels by decomposition; Vegfa, vascular endothelial growth factor A.
LV constructs, called LV/Cas9-sgRNA1-4 or LV/Cas9-sgRNA-Irr, were generated by cloning each sgRNA into the RNA scaffold induced by the U6 promoter in the lentiCRISPRv2 vector. In parallel, the LV constructs, under the control of the elongation factor 1α short (EFS) promoter, encode SpCas9. For easy visualization, the lentiCRISPRv2 vector contains a DNA sequence of a FLAG tag fused to the SpCas9 cDNA sequence followed by DNA sequences encoding a P2A self-cleaving peptide and the eGFP marker protein (Figure 1B). The resultant LV constructs were used in all further experiments.
In Vitro Validation of Gene Knockout
To validate efficiency of the designed sgRNAs, human embryonic kidney 293 (HEK293)-VEGFA cells stably expressing high levels of VEGFA were utilized. The HEK293-VEGFA cell line was transduced with 500 ng p24 LV/Cas9-sgRNA(1-4) or LV/Cas9-sgRNA-Irr. At day five, fluorescence analysis of eGFP expression showed high transduction levels for all LVs (Figure 1C), except for LV/Cas9-sgRNA4. As the SpCas9 and eGFP proteins were translated from a single transcript, their expression should be similar, meaning that robust eGFP expression indicated high SpCas9 expression and possibly high knockout efficiency. At day five, cells were harvested and analyzed by sequencing, tracking of indels by decomposition (TIDE) software, and the surveyor nuclease assay for indel formation.
DNA sequencing chromatograms obtained from analysis of cells transduced with LV/Cas9-sgRNA1 and LV/Cas9-sgRNA2 showed alternating sequences from the cut site (3-bp upstream of the PAM) indicating formation of different kinds of indels as result of double-stranded break formation by the SpCas9 and subsequent non-homologous end joining (Figure 1D). Low-frequency indel formation, in the form of small alterations from the cut site and forward, was observed in the chromatograms resulting from the analysis of HEK293-VEGFA cells transduced with LV/Cas9-sgRNA3 or LV/Cas9-sgRNA4. As expected, inspection of chromatograms obtained from cells transduced with LV/Cas9-sgRNA-Irr showed no changes compared to the Vegfa sequence (data not shown). By comparing the obtained sequencing data to the Vegfa sequence resulting from cells transduced with LV/Cas9-sgRNA-Irr, the TIDE software revealed high editing efficiency of 44% ± 12%, and 56% ± 5% for cells treated with LV/Cas9-sgRNA1 or LV/Cas9-sgRNA2, respectively. These efficiency scores are in accordance with the respective chromatograms.
The sequencing analysis of genomic DNA (gDNA) isolated from HEK293-VEGFA cells transduced with LV/Cas9-sgRNA3 or LV/Cas9-sgRNA4 revealed chromatograms showing low frequency alterations from the cut-site and forward. However, the TIDE scores significantly differed as editing efficiency for cells treated with LV/Cas9-sgRNA3 was 93% ± 2% compared to 8% ± 1% in the case of LV/Cas9-sgRNA4. Additionally, PCR amplicons of the Vegfa-target region were used for surveyor nuclease assay. Cell samples transduced with LV/Cas9-sgRNA1 or LV/Cas9-sgRNA2 scored similarly to the TIDE results with a 56% ± 11% and 66% ± 9% indel formation, respectively (Figure 1E). In contrast, indel formation efficiencies of 29% ± 6% and 38% ± 15% were observed for amplicons obtained from HEK293-VEGFA cells transduced with LV/Cas9-sgRNA3 or LV/Cas9-sgRNA4, respectively.
In summary, the LV/Cas9-sgRNA1 and LV/Cas9-sgRNA2 vectors repeatedly resulted in high indel formation using different analysis methods. LV/Cas9-sgRNA3 also provided high indel formation as judged by the TIDE analysis, but seemed mediocre at best based on the chromatogram and surveyor nuclease assay. An apparently low transduction efficiency based on eGFP fluorescence of the LV/Cas9-sgRNA4, resulting in low levels of indel formation, rendered this vector inappropriate for further studies. Hence, the in vitro validation demonstrated that three out of four sgRNAs delivered by LVs were able to induce indel formation with high efficiency and were thus relevant for in vivo validation.
Genomic Vegfa Knockout Effects on Protein Level
Based on LVs encoding effective Vegfa-targeted sgRNAs, we wanted to investigate whether indel formation affected the functional protein levels. Using the same experimental setup as described above, the VEGFA protein levels in HEK293-VEGFA cells transduced with either LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA-Irr were examined. As a negative control non-transduced cells were included. In accordance with the findings presented in Figure 1C, immunofluorescence analysis of eGFP at day five post transduction (Figure 2A) showed efficient transduction for all three LV/Cas9-sgRNAs. As the VEGFA protein exhibits its mitogenic activity on nearby endothelial cells after secretion, we assessed VEGFA protein content in both cell lysates and in the cultivation medium by means of western blotting (Figure 2B).
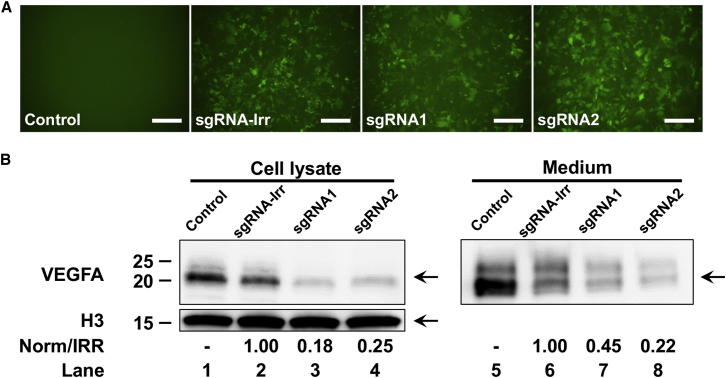
Figure 2.
In Vitro Validation of Functional Knockout
(A) Immunofluorescence analysis of eGFP expression. HEK293-VEGFA cells transduced with 500 ng p24 LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA-Irr evaluated for eGFP expression by fluorescence five days post-transduction. A non-transduced control (control) was included. (B) Western blot analysis of VEGFA following LV transduction of HEK293-VEGFA cells. Five days post-transduction media from all four samples were collected, and the cells were lysed for protein purification. 15 μg of total protein was used for LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, LV/Cas9-sgRNA-Irr, and the non-transduced control (control) (lanes 1–4), as well as 15 μL media for each sample (lanes 5–8). The cell lysates and media were subjected to electrophoresis and blotting. The resultant membranes were incubated with a rabbit anti-VEGFA antibody. A rabbit antibody against H3 was used as loading control for the cell lysate samples. VEGFA- and H3-specific bands were visualized using an ImageQuant LAS4000 digital imaging system. Quantification of VEGFA relative to IRR samples is indicated below the samples. The positions of VEGFA and H3 are indicated with arrows. Molecular sizes in kilodaltons are indicated on the left. The presented western blot is a representative of three independent experiments. Scale bars, 200 μm. H3, histone H3.
The western blot analysis revealed an immune-reactive band between ∼20 and ∼25 kDa,27 representing the VEGFA protein. VEGFA protein was detected in both cell lysates and medium from the transduced cells. In comparison to the cells transduced with LV/Cas9-sgRNA-Irr (Figure 2B, lane 2), the lysate obtained from cells transduced with LV/Cas9-sgRNA1 or LV/Cas9-sgRNA2 contained significantly lower levels of VEGFA protein (18%–25% of normal level) (Figure 2B, lanes 3 and 4). Analysis of the cell culture medium showed similar results as the samples obtained from the cell lysate. Hence, significant reduction of the VEGFA levels (22%–45% normal level) was obtained in medium from cells treated with LV/Cas9-sgRNA1 or LV/Cas9-sgRNA2 compared to the LV/Cas9-sgRNA-Irr sample (Figure 2B, compare lanes 6–8).
These results showed that both cell lysate and medium obtained from transduced cells encoding SpCas9 and either sgRNA1 or sgRNA2 resulted in significantly reduced levels of VEGFA protein compared to cells transduced with LV/Cas9-sgRNA-Irr, demonstrating efficient knockout of Vegfa when the cells were treated with SpCas9-sgRNAs targeting Vegfa. Thus, the ability of the Vegfa-specific sgRNAs to induce indel formation, validated by TIDE and surveyor nuclease assay (Figure 1E), directly translates into decreased VEGFA protein levels, thereby confirming the link between genomic and functional knockout.
SpCas9 Expression in RPE Cells following Subretinal Injections of LVs
As in vitro validation of LV-delivered sgRNAs demonstrated genomic and functional knockout of Vegfa, in vivo efficiency of the LVs was examined in mice following subretinal injections. To transduce the RPE cells, the primary production site of VEGFA in the retina, four groups of 5–13 C57BL/6J mice were subretinally injected with 100 ng p24 LV/Cas9-sgRNA-eGFP either guided by sgRNA1, sgRNA2, sgRNA3, or sgRNA-Irr (Figure 3A).
Figure 3.
In Vivo Validation of Lentiviral CRISPR/cas9 Targeting of RPE Cells
(A) Schematics of the experimental workflow for in vivo evaluation of genomic knockout. Subretinal injections of LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, LV/Cas9-sgRNA3, or LV/Cas9-sgRNA-Irr in C57BL/6J mice, 8–9 weeks of age at day 0. Funduscopic eGFP assessment at days 14, 21, 28, and 35 PI. At day 35, PI mice were euthanized, and their retinas were dissected followed by FACS-mediated isolation of eGFP+ RPE cells. Following PCR, amplification of gDNA Vegfa-specific amplicons was used for TIDE analysis and TOPO cloning. (B and C) Immunofluorescence assessment of eGFP expression in RPE following cryosectioning. At day 35 PI, non-injected control (control) mice (n = 3) and LV/Cas9-sgRNA1 (sgRNA1) subretinally injected C57BL/6J mice (n = 3) were euthanized, and the eyes were enucleated and fixated in PFA for cryosectioning. eGFP signals were evaluated following nuclear staining with DAPI. Scale bar, 20 μm. PFA, paraformaldehyde.
At day 35, post injection (p.i.) mice were euthanized and the eyes enucleated. To further investigate expression from the LVs, three eyes of LV/Cas9-sgRNA1 injected mice were dissected and cryosectioned. Three non-injected control eyes were used as negative controls. The RPE cells and the outer nuclear layer were analyzed by confocal laser scanning microscopy, revealing RPE specific eGFP expression in the LV/Cas9-sgRNA1 injected eyes (Figure 3C). As expected, no eGFP expression was observed in the RPE layer of the control eyes (Figure 3B). However, green fluorescent signal was observed outside the RPE layer, representing autofluorescence due to abundancy of fluorophors, including all-trans retinol, A2-PE and FAD.33, 34, 35 As the cDNA sequences encoding SpCas9 and eGFP were transcribed as a single mRNA, separated by a P2A self-cleaving peptide in the LV, the observed RPE-specific eGFP expression pattern indicated that SpCas9 was likewise produced in the RPE cell layer following subretinal administration of the LVs.
In Vivo Knockout Assessed in Isolated eGFP+ Cells following Fluorescence-Activated Cell Sorting
Our data showed that the eGFP expression was confined to the RPE cell layer (Figure 3C). As eGFP signals mark cells positive for transduction, we aimed at isolating eGFP+ RPE cells in order to validate the in vivo knockout efficiency induced by the SpCas9 and sgRNA1-, sgRNA2-, and sgRNA3-encoding LVs. Unilateral subretinal injections of 30 C57BL/6J mice divided into four groups receiving either LV/Cas9-sgRNA1 (n = 10), LV/Cas9-sgRNA2 (n = 5), LV/Cas9-sgRNA3 (n = 5), or LV/Cas9-sgRNA-Irr (n = 10) were performed. The experiment was conducted as delineated in Figure 3A. Five weeks p.i., the mice were euthanized and the eyes enucleated and dissected. As eGFP expression was observed solely in RPE cells in the histological examination, only the eyecup comprised of the sclera, choroid, and RPE cell layer was used. The cell suspensions from each group (five or ten mice) were then pooled and subjected to fluorescence-activated cell sorting (FACS) analysis. Even though the neuroretina was removed before FACS analysis, the results showed that the cell solution was not a uniform population of RPE cells, but rather a collection of positive cells primarily consisting of RPE cells (Figure 4A).
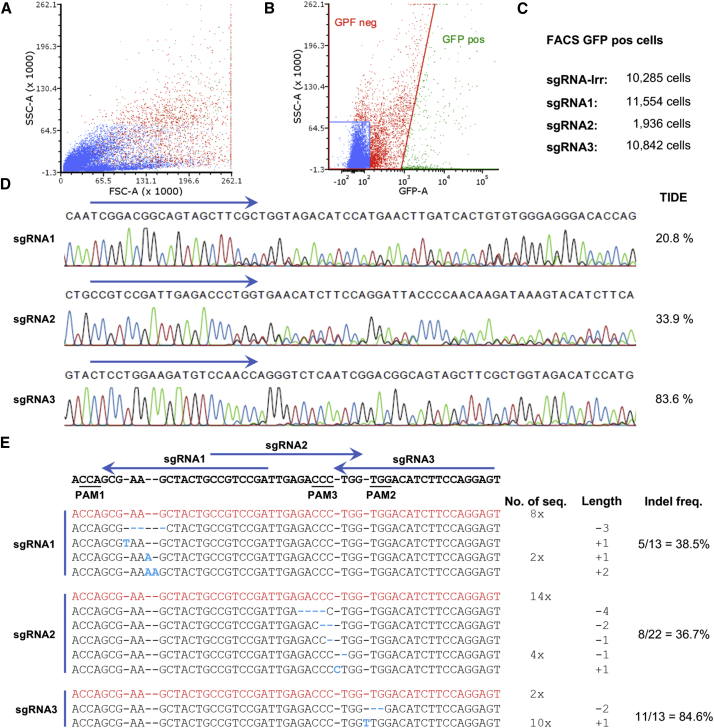
Figure 4.
In Vivo Gene Editing following FACS Analysis
(A–E) Following subretinal injection of LV/Cas9-sgRNA1 (n = 13), LV/Cas9-sgRNA2 (n = 5), LV/Cas9-sgRNA3 (n = 5), or LV/Cas9-sgRNA-Irr (n = 10) in C57BL/6J mice, animals were euthanized at day 35 PI, and the retinas were dissected. (A) FACS analysis of the resultant RPE cells. The RPE cells from all mice in each group were pooled before FACS analysis. Representative results from the FACS analysis (LV/Cas9-sgRNA3). No cells were excluded because of size (forward-scattered light [FSC]) or granularity (side-scattered light [SSC]). (B) Illustration of eGFP versus SSC FACS analysis (LV/Cas9-sgRNA3), including thresholds for eGFP+ and eGFP− cells. Because of these criteria, cells were marked as eGFP+ or eGFP−. (C) Numbers of FACS-mediated isolation of eGFP+ RPE cells from the four groups resulted in LV/Cas9-sgRNA-Irr 10,285 cells (n = 10), LV/Cas9-Cas9-sgRNA1 11,554 cells (n = 10), LV/Cas9-sgRNA2 1,936 cells (n = 5), and LV/Cas9-sgRNA3 10,842 cells (n = 5). (D) Sequencing chromatograms of PCR amplicons following administration of LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA3. Chromatograms were assessed for indels by TIDE in comparison to LV/Cas9-sgRNA-Irr. (E) Sequencing analysis of TOPO-cloned PCR amplicons obtained from FACS-isolated eGFP+ RPE cells. PCR amplicons following delivery of LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA3 were TOPO cloned into the pCR-Blunt-II-TOPO vector. Purified DNA samples were amplified by means of PCR and Vegfa exon 3-specific amplicons sequenced. The resultant sequences were designated with the number of sequences (1× by default), altered length, and overall indel formation frequency. The wild-type is shown in red; insertions and deletions are indicated in bold and cyan. Top: schematic overview of the three target sites in Vegfa exon 3. Blue arrows shown on top of the sequence represent the different Cas9-sgRNAs. PAM sequences are underlined and indicated beneath the sequence. PAM, protospacer-adjacent motif; FSC, forward-scattered light; SSC, side-scattered light.
eGFP+ cells were isolated according to the gating shown in Figure 4B, resulting in ∼10.000 positive cells from the groups subretinally injected with LV/Cas9-sgRNA1, LV/Cas9-sgRNA3 or LV/Cas9-sgRNA-Irr, while only ∼2.000 eGFP+ cells were obtained from the LV/Cas9-sgRNA2-transduced group (Figure 4C). A short region of the Vegfa gene was PCR-amplified from gDNA obtained from the isolated eGFP+ cell population using primers specific for exon three of Vegfa. To determine the indel formation efficiency, TIDE analysis and TOPO cloning were applied. Sequencing chromatograms showed specific alterations following the cut site in samples obtained from mice injected with LV/Cas9-sgRNA1 and LV/Cas9-sgRNA2, and TIDE analysis scored 20.8% and 33.9% indel formation, respectively, in comparison to the LV/Cas9-sgRNA-Irr sample (Figure 4D). The sequencing of DNA obtained from eGFP+ cells treated with LV/Cas9-sgRNA3 showed limited alterations in the chromatogram, but interestingly an extra A nucleotide was inserted after the cut site. Following TIDE analysis, the indel formation score was calculated to 83.6% in comparison to the LV/Cas9-sgRNA-Irr sample, of which 79.1% was a 1-bp insertion (Figure S1C), which is in accordance with the sequencing analysis. Hence, the scores obtained from the in vivo experiment were comparable to the in vitro scores for each of the investigated guide sequences (Figure 1E).
To investigate the nature and frequency of specific SpCas9-induced indels, TOPO-cloning was performed using Vegfa exon 3 PCR amplicons from gDNA of eGFP+ cells isolated from murine retinas by FACS (see Figure 3A). In the case of LV/Cas9-sgRNA1, 13 clones were sequenced showing three 1-bp insertions, one 2-bp insertion, and a 3-bp deletion, resulting in a 38.5% knockout efficiency (5/13). Twenty-two colonies of the LV/Cas9-sgRNA2 sample were sequenced, resulting in eight insertions and deletions (−4-bp to +1-bp) giving a 36.7% knockout efficiency (8/22). Notably, the data from the LV/Cas9-sgRNA3 sample show that out of 13 colonies, 10 had an identical 1-bp insertion and one colony had a 2-bp deletion, resulting in an 84.6% indel formation (11/13). The 1-bp insertion was identical to the A nucleotide identified in the chromatogram.
To investigate the impact of in vivo genomic knockout on the VEGFA protein level western blot analysis was pursued. As a maximum of 10,000 eGFP+ RPE cells was retrieved following FACS analysis (see above), the VEGFA level was assessed in 10,000 eGFP− cells. Since no VEGFA protein was detected in these cells (data not shown), most likely due to the limited number of isolated RPE cells, western blot analysis on FACS-sorted eGFP+ cells was not performed.
In summary, we demonstrated efficient in vivo knockout of the Vegfa gene in isolated eGFP+ RPE cells following subretinal injection of LVs encoding SpCas9 and different sgRNAs.
Discussion
Our study is the first to report LV-based retinal delivery of CRISPR/Cas9 generating genomic knockout indel formation. LVs have previously been applied for in vitro screening of various targets including Vegfa36, 37, 38, 39 but only sparsely for in vivo CRISPR/Cas9 delivery.21 In this study, we show (1) indel formation in Vegfa, (2) functional knockout of genomic Vegfa in vitro following LV transduction, (3) delivery of LVs expressing sgRNA with SpCas9 to the RPE cells following a subretinal injection in mice, and (4) in vivo targeting of Vegfa as result of LV transduction of Cas9 in combination with three different sgRNAs. These data collectively provide proof of principle for lentivirus-delivered CRISPR/Cas9 to the murine retina and subsequent Vegfa genomic knockout.
LVs expressing sgRNA1 and sgRNA2 provided a frameshift knockout indel formation in eGFP+ RPE cells. The indels identified by TOPO cloning predominantly caused frameshifts leading to premature termination codons (only one 3-bp deletion resulted from LV/Cas9-sgRNA1), which in the end resulted in synthesis of truncated VEGFA protein and/or NMD. LV/Cas9-sgRNA3 targeting of Vegfa in HEK293-VEGFA and retinal RPE cells primarily induced a unique 1-bp insertion as assessed by TIDE analysis (Figures S1B and S1C), as well as TOPO cloning. Surveyor Nuclease Assay of cells transduced with LV/Cas9-sgRNA3 revealed low frequency of indel formation. However, as this unique 1-bp insertion is the predominant variant in the sample (∼84%), reannealing primarily generates identical homoduplexes (containing the 1-bp insertion) and thereby results in a situation in which the production of surveyor nuclease digestion sites may be limited.
The applied polyclonal anti-VEGF antibody targets epitopes formed by a synthetic peptide corresponding to amino acid residues 50–150 of VEGFA. As the designed sgRNAs target Vegfa sequences encoding amino acid residues 45–72 of VEGFA, truncated protein (encoded by Vegfa cDNA) might be produced in HEK293-VEGFA cells, which are not recognized by the antibody. On the other hand, in the case of targeting genomic Vegfa in vivo, this issue may not be considered a major concern because LV/Cas9-sgRNA1-3 all result in frameshift mutations and introduction of premature termination codons before the last exon, thereby rendering the resultant transcripts prone to NMD31, 32 and hence resulting in limited synthesis of truncated VEGFA protein. However, it cannot be ruled out that fractions of truncated VEGFA are produced in the HEK293-VEGFA cells. Notably, our results clearly show almost undetectable levels of full-length VEGFA in HEK293-VEGFA cells following delivery of LV/Cas9-sgRNA1 or LV/Cas9-sgRNA2. Recently, genomic disruption of VEGFA in human RPE (ARPE-19) cells using LV-delivered CRISPR/Cas9 demonstrated indel formation in Vegfa at frequencies up to 37.0% with corresponding decreases in secreted VEGFA protein up to 41.2%.39 In the present study using HEK293-VEGFA cells LV-delivered CRISPR/Cas9 induced indel formation in Vegfa at frequencies up to 93% with corresponding decreases in secreted VEGFA protein up to 78%. The observed differences obtained in the two model systems may reflect variations in transduction efficiency.
As VEGFA dysregulation is the main contributor to AMD development, we targeted the primary site of VEGFA production: the RPE cells. Initially mice were inspected by in vivo fundoscopy at day 14, 21, 28 and 35 p.i. prior to euthanization and TIDE analysis of RPE cells isolated from dissected eyes. Unexpectedly, no eGFP+ cells were detected in the retina by fundoscopy and the TIDE analysis provided no indel formation at any of the investigated time points. Limited transduction efficacy, as a consequence of either low functional virus titer or complications following the subretinal injections, combined with a very short exposure time (66 ms) restricted by the Micron IV Retinal Imaging Microscope, may explain these findings. However, analysis of cryosections obtained from the 35 days p.i. group (LV/Cas9-sgRNA1) demonstrated eGFP expression in the RPE cells, and subsequently presence of LVs in the target cells. Hence, to improve the capability to detect indel formation in transduced cells following lentiviral delivery in vivo, FACS-mediated isolation of eGFP+ RPE cells was pursued. Notably, TIDE analysis and TOPO cloning performed on the FACS-sorted cells from retinas treated with LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA3 collectively revealed significant levels of indel formation in transduced cells.
Another recent study by Kim et al.6 has used the CRISPR/Cas system for gene knockout in the retina.6 In this study, the authors investigated the specificity of CjCas9 targeting the Rosa26, Vegfa and Hif1a genes. Following intravitreal injection of AAV expressing CjCas9, the three targets were evaluated by analysis of whole retinas and RPE cells, revealing indel formation frequencies ranging from 15%–60%, with ∼20% indel formation when targeting Vegfa. Furthermore, they observed ∼20% decrease in VEGFA protein levels as a result of genomic knockout. Our LV-based results further supported these findings, and LV/Cas9-sgRNAs cause indels with frequencies in line with the indel formation frequencies obtained by the CjCas9-AAV system, thereby paving the way for possible therapeutic application of CRISPR/Cas9 delivered by viral vectors for retinal diseases.
LVs have a larger cargo capacity compared to AAVs, which allows the expression of the larger Cas9 proteins, enabling targeting of more genomic sites. Multiple sgRNAs can be expressed for multigenic targeting or for excision of exact sequences, all in combination with appropriate marker proteins. The larger cargo capacity potentially brings endless possibilities to the experimental setup. When using dual AAVs for delivery, this versatility is not possible. A drawback of the dual AAV system is that it requires co-transduction to be functional, ultimately targeting fewer cells and wasting AAV particles on cells only transduced by one vector. Subsequently, increased amounts of AAV particles are needed to obtain efficacy, and as a consequence the risk of immuno- as well as genotoxicity will likewise increase.40 Still, AAV possesses high transduction efficiency and versatility of serotypes, making it a suitable delivery vehicle for Cas9 for a range of experimental setups. A toolbox with AAV-delivered SaCas9 and LV-delivered SpCas9 will increase the available genomic targets.
As LVs primarily transduce the RPE cells following subretinal injection,41 acquired and autosomal dominantly inherited retinal diseases affecting the RPE cells might be targets for LV-based CRISPR/Cas therapy. A number of retinal targets have been examined by CRISPR/Cas for elimination/reduction of a specific protein, as in our study, or mutation specific targeting of disease alleles. Targeting the disease allele would, if dominant, alleviate the condition, as the healthy allele would restore normal function of the protein.42 Bakondi et al.12 used this approach in a heterozygous S334ter-3 rat model of RP substituting a serine with a termination codon. By specifically targeting two SNPs, one in the PAM and one 10-bp upstream of the PAM in the sgRNA, indel formation solely in RhoS334 was detected, leaving Rhowild-type untouched to restore physiological conditions.
In conclusion, this study further supports the use of viral vectors for retinal gene therapy and emphasizes the possibility of adapting this system for clinical use in the future. We expand the CRISPR/Cas toolbox with LV delivery, which holds potential for RPE-specific delivery of CRISPR/Cas systems.
Materials and Methods
sgRNA Design and Construct Generation
sgRNAs targeting the exonic regions of the murine Vegfa gene were designed. The MIT CRISPR Design19 and Broad Institute CRISPRko25 software algorithms were used to estimate on-target and off-target gene knockout efficiency. A non-targeting sgRNA, designated sgRNA-Irr, was used as a control (5′-ACGGAGGCTAAGCGTCGCAA-3′). sgRNA-Irr was obtained from a Human GeCKOv2 CRISPR knockout pooled library, and BLAST analysis showed no targeting in the murine genome. Human GeCKOv2 CRISPR knockout pooled library was a gift from Feng Zhang (Addgene Pooled Library # 1000000048, #1000000049).26 Four Vegfa targeting sgRNAs (1–4) were designed: sgRNA1: 5′-TCGGACGGCAGTAGCTTCGC-3′, sgRNA2: 5′-CCGTCCGATTGAGACCCTGG-3′, sgRNA3: 5′-CTCCTGGAAGATGTCCACCA-3′, and sgRNA4: 5′-AAGATGTACTCTATCTCGTC-3′. LentiCRISPRv2-egfp was constructed by replacing a puromycin-resistant component in the lentiCRISPRv2 with an eGFP gene using Gibson Assembly. LentiCRISPRv2 was a gift from Feng Zhang (addgene plasmid #52961).26 The sgRNAs were cloned into the BsmBI sites of the lentiCRISPRv2-egfp plasmid. Expression of sgRNA was under the control of a U6 promoter. In parallel, the lentiCRISPRv2 plasmid, under the control of the EFS promoter, encodes SpCas9 fused to the FLAG tag and a P2A self-cleaving peptide followed by the eGFP marker protein.26 The resultant LV constructs were designated LV/Cas9-sgRNA1-4 or LV/Cas9-sgRNA-Irr.
Stable VEGFA-Expressing Cell Line
The murine Vegfa sequence (GenBank AAH61468.1, coding part of BC061468.1) was cloned into the NotI sites of the pT2/CMV vector. Human embryonic kidney (HEK293) cells were stably transfected with the pT2/CMV-VEGFA construct and evaluated by immunostaining and qPCR (data not shown) as previously shown.27 Stable VEGFA-expressing HEK293 cells are designated by HEK293-VEGFA from this point onward.
Cell Culture, LV Production, and Transductions
HEK293-T (catalog number CRL-1573; American Type Culture Collection) and HEK293-VEGFA27 cells were maintained in DMEM (Lonza, Schweiz), supplemented with 10% fetal calf serum (Sigma-Aldrich, Broendby, Denmark), 0.29 mg/mL glutamine (Sigma-Aldrich), 0.06 mg/mL penicillin (FarmaPlus, Oslo, Norway), and 0.1 mg/mL streptomycin (Sigma-Aldrich). Cells were cultured in tissue culture flasks or 6-well plates (both Sarstedt, Nürnbrecht, Germany) at 37°C with 5% (v/v) CO2. LVs were produced using calcium phosphate triple transfection as previously described.28 For in vitro validation, 200,000 HEK293-VEGFA cells were seeded in 6-well plates. The following day, the medium was replaced by fresh DMEM containing 8 μg/mL polybrene (Sigma-Aldrich) before transducing the cells with 500 ng p24 of LV/Cas9-sgRNA(1-4) or LV/Cas9-sgRNA-Irr. One day post-transduction, medium was replaced by fresh DMEM, and 5 days post-transduction, cells were harvested for genomic DNA (gDNA) purification.
Indel Analysis: TIDE and Surveyor Nuclease Assay
DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) was utilized for gDNA isolation from LV/Cas9-sgRNA1-4- or LV/Cas9-sgRNA-Irr-transduced HEK293-VEGFA cells following the manufacturer’s protocol and quantified using a spectrophotometer (NanoDrop, BioNordika, Herlev, Denmark). For TIDE analysis (https://tide-calculator.nki.nl/), two primer sets were utilized: one set for the amplification of the whole-Vegfa gene of HEK293-VEGFA and another exon 3-specific sequence for in vivo knockout analysis. The sequences were amplified by PCR using the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Hvidovre, Denmark). PCR products were purified by MiniElute PCR purification (QIAGEN) and subsequently sequenced (GATC, Copenhagen). For the surveyor nuclease assay, 200 ng purified PCR product was applied for duplex formation by denaturation and subsequent heteroduplex formation (95°C for 10 min, ramped down to 25°C at 5°C/min, and 25°C for 1 min). The surveyor nuclease assay (Surveyor Mutation Detection Kit, Integrated DNA Technologies, Leuven, Belgium) was performed according to the manufacturer’s protocol and incubated for 1 hr at 42°C. A 2% TAE agarose gel was used for visualizing the PCR product, and ImageJ (https://imagej.nih.gov/ij/; National Institute of Health, Bethesda, MD, USA) was used for quantification of the DNA band intensities. Indel formation was analyzed using the following equation: indel% = 100 ⋅ (1 – (1 – fcut)1/2), where fcut is the fraction of total cleaved DNA.29 Primer sequences are shown in Table S2.
Western Blotting
Western blot analysis was performed on extracts from HEK293-VEGFA cells transduced with either 500 ng p24 LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA-Irr. Non-transduced cells were used as control. 5 days post-transduction, medium and cells were harvested. Medium was filtered (22 μm), and 15 μL was used for western blotting as previously described.28 In brief, 200 μL RIPA lysis buffer (VWR, Soeborg, Denmark) with cOmplete Mini (Roche Applied Science, Hvidovre, Denmark) was added to cells in 6-well plates and incubated on ice for 15 min. The lysates were collected and after 30 min of centrifugation at 13,000 rpm, the supernatant was isolated, and 15 μg of total protein was used for western blotting. Cell lysates and cell medium were electrophoresed on a Mini-PROTEAN TGX 4%–15% gel (Bio-Rad, Copenhagen, Denmark). The gel was run at 200 V for 30 min in 1× TGS buffer (Bio-Rad). Proteins were blotted to polyvinylidene fluoride membranes (Bio-Rad) using a Trans-Blot Turbo Transfer system (Bio-Rad). Membranes were blocked for 1 hr in Tris-buffered saline-tween20 (TBS-T) containing 5% (w/v) skim-milk powder (BD Difco, Franklin Lakes, NJ, USA) and incubated at 4°C overnight (ON) with rabbit anti-VEGFA antibody (ab46154; Abcam, Cambridge, UK). Membranes were washed for 3 to 5 min in TBS-T before incubation with horseradish-peroxidase (HRP)-conjugated goat-anti-rabbit antibody (DAKO, Glostrup, Denmark) for 1 hr at room temperature (RT). Bound antibodies were visualized with Clarity Western ECL Blotting substrate (Bio-Rad) on an ImageQuant LAS4000 digital imaging system (GE Healthcare, Cleveland, OH). Anti-histone H3 antibody (ab1791; Abcam) was used as a loading control for 1 hr incubation followed by 3 to 5 min TBS-T wash and goat-anti-rabbit HRP-conjugated antibody for 1 hr before visualization.
Animals
All animal experiments were approved by the Danish Animal Inspectorate (Case # 2015-15-0201-00691). Male C57BL/6J mice, 8–9 weeks of age, were purchased from Janvier Labs (Le Genest-Saint-Isle, France). The mice were kept on a 12 hr/12 hr light/dark cycle at the Animal Facilities, Department of Biomedicine, Aarhus University. A combination of ketamine 60–100 mg/kg (Ketador, Richter Pharma AG, Wels, Austria) and medetomidine hydrochloride 0.5–1 mg/kg (Cepetor, ScanVet Animal Health A/S, Fredensborg, Denmark) were used as anesthesia prior to subretinal injections or in vivo fundoscopy. A drop of 1% tropicamide solution (Mydriacyl, Alcon Nordic A/S, Copenhagen, Denmark) and 10% phenylephrine hydrochloride (Metaoxedrin, Skanderborg Apotek, Skanderborg, Denmark) were used for dilation of pupils. Eyes were lubricated prior to subretinal injection and fundoscopy with a carbomer eye-gel (Viscotears, 2 mg/g, Alcon Nordic A/S). Atipamezole hydrochloride (Antisedan, Orion Pharma, Espoo, Finland) 0.5–1 mg/kg was used to bring the mice out of sedation, and mice were kept warm until mobile, before being transferred back into their cages. In the period one day prior to and three days after subretinal injection, the mice were treated with the NSAID carprofen 5 mg/150 mL (Norodyl, ScanVet Animal Health A/S) via their drinking water. In addition, to ensure adequate analgesia mice received subcutaneous injection of carprofen (5 mg/kg) 24 hr prior to and immediately after suretinal injection.
Subretinal Injections
A total of 36 C57BL/6J male mice, 8–9 weeks of age, were divided into five groups; LV/Cas9-sgRNA1 (13 mice, 10 for FACS-sorting and 3 for cryosectioning), LV/Cas9-sgRNA2 (5 mice), LV/Cas9-sgRNA3 (5 mice), LV/Cas9-sgRNA-Irr (10 mice), and 3 mice as non-injected controls for cryosectioning. Subretinal injections of 100 ng p24 in a total volume of 2 μL of LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, LV/Cas9-sgRNA3, or LV/Cas9-sgRNA-Irr were performed in accordance with the method described by Bemelmans et al.30
Cryosectioning and Visualization of eGFP
Cryosectioning and histological analysis were performed as previously described.28 In brief, mice were euthanized by cervical dislocation, and eyes were fixated in 4% paraformaldehyde (PFA) for 5 min at RT. The lens and cornea were removed, and the eyes were fixated for two hours at RT. The eyes were washed in PBS and kept in a 30% sucrose solution ON at 4°C. The eyes were frozen on dry ice in optimal cutting temperature compound (Tissue Tek; Sakura Finetek, AV Alphen aan den Rijn, the Netherlands) and stained with DAPI (Sigma-Aldrich). Cells were visualized using a confocal laser scanning microscope (LSM 710, Zeiss, Jena, Germany).
Retinal Dissection and FACS
Mice were euthanized, and the eyes were enucleated and dissected immediately. The lens, cornea, and neuroretina were removed, and the eyecup was placed in 1% trypsin (Sigma-Aldrich) for 1 hr at 37°C. Cells in the eyecup were rinsed in Hank’s balanced salt solution (Sigma-Aldrich), and the cell population was placed in 1% trypsin for 1 hr to obtain a single-cell solution. The solution was sorted on a FACSAria III high-speed cell sorter (BD Biosciences, San Jose, CA, USA) for sorting of eGFP+ cells. During the sorting, no cells were excluded due to aggregation, size, or debris. The only sorting criterion was eGFP expression.
TOPO Cloning
Exon 3-specific amplicons of the Vegfa gene generated by PCR using gDNA obtained from eGFP+ FACS-sorted RPE cells from mice treated with LV/Cas9-sgRNA1, LV/Cas9-sgRNA2, or LV/Cas9-sgRNA3 were TOPO cloned into the pCR-Blunt-II-TOPO (Invitrogen, Taastrup, Denmark) vector and transformed in E. coli. Colonies were analyzed by PCR amplification using genomic primers for the Vegfa exon 3 region. Purified PCR products were sequenced (GATC Biotech, Constance, Germany).
Author Contributions
Writing – Original Draft, A.H. and T.J.C.; Writing – Reviewing and Editing, A.H. and T.J.C.; Supervision, T.J.C., J.G.M., and A.L.A.; Investigation, A.H., A.L.A., J.N.E.B., Y.C., and E.A.T.; Methodology, T.J.C., A.L.A., T.B., and J.G.M.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
The authors would like to thank Tina Hindkjaer, Kamilla Zahll Hornbek, Christian Knudsen, and Karen Kathrine Brøndum for their excellent technical support. Cell sorting was performed at the FACS Core Facility, Aarhus University, Denmark. This work was supported by the Faculty of Health Sciences (PhD scholarship to A.H.), the Danish Council for Independent Research (4183-00017B to T.J.C.), the Gene Therapy Initiative Aarhus (GTI-Aarhus) funded by the Lundbeck Foundation (R126-2012-12456 to T.J.C. and J.G.M.), the Danish Eye Foundation (to T.J.C.), Aase og Ejnar Danielsen’s Foundation (to T.J.C.), Knud and Edith Eriksen’s Foundation (to T.J.C.), Maskinfabrikant Jochum Jensen og hustru Mette Marie Jensen f. Poulsens Mindelegat (to T.J.C.), the Riisfort Foundation (to T.J.C.), and Svend Helge Schrøder og hustru Ketty Lydia Larsen Schrøders fund (to T.J.C.).
Footnotes
Supplemental Information includes one figure and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.08.016.
Supplemental Information
References
- 1.Trapani I., Toriello E., de Simone S., Colella P., Iodice C., Polishchuk E.V., Sommella A., Colecchi L., Rossi S., Simonelli F. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum. Mol. Genet. 2015;24:6811–6825. doi: 10.1093/hmg/ddv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S., Sengel C., Emerson M.M., Cepko C.L. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev. Cell. 2014;30:513–527. doi: 10.1016/j.devcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latella M.C., Di Salvo M.T., Cocchiarella F., Benati D., Grisendi G., Comitato A., Marigo V., Recchia A. In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol. Ther. Nucleic Acids. 2016;5:e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan G.X., Barry E., Yu D., Lukason M., Cheng S.H., Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol. Ther. 2017;25:331–341. doi: 10.1016/j.ymthe.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson J.H., Wensel T.G. The nature of dominant mutations of rhodopsin and implications for gene therapy. Mol. Neurobiol. 2003;28:149–158. doi: 10.1385/MN:28:2:149. [DOI] [PubMed] [Google Scholar]
- 11.Rossmiller B., Mao H., Lewin A.S. Gene therapy in animal models of autosomal dominant retinitis pigmentosa. Mol. Vis. 2012;18:2479–2496. [PMC free article] [PubMed] [Google Scholar]
- 12.Bakondi B., Lv W., Lu B., Jones M.K., Tsai Y., Kim K.J., Levy R., Akhtar A.A., Breunig J.J., Svendsen C.N., Wang S. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol. Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung S.S.C., Chrysostomou V., Li F., Lim J.K.H., Wang J.H., Powell J.E., Tu L., Daniszewski M., Lo C., Wong R.C. AAV-mediated CRISPR/Cas gene editing of retinal cells in vivo. Invest. Ophthalmol. Vis. Sci. 2016;57:3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 21.Blasco R.B., Karaca E., Ambrogio C., Cheong T.C., Karayol E., Minero V.G., Voena C., Chiarle R. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 22.Miller J.W., Adamis A.P., Shima D.T., D’Amore P.A., Moulton R.S., O’Reilly M.S., Folkman J., Dvorak H.F., Brown L.F., Berse B. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am. J. Pathol. 1994;145:574–584. [PMC free article] [PubMed] [Google Scholar]
- 23.Askou A.L. Development of gene therapy for treatment of age-related macular degeneration. Acta Ophthalmol. 2014;92(Thesis3):1–38. doi: 10.1111/aos.12452. [DOI] [PubMed] [Google Scholar]
- 24.Corydon T.J. Antiangiogenic eye gene therapy. Hum. Gene Ther. 2015;26:525–537. doi: 10.1089/hum.2015.064. [DOI] [PubMed] [Google Scholar]
- 25.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pihlmann M., Askou A.L., Aagaard L., Bruun G.H., Svalgaard J.D., Holm-Nielsen M.H., Dagnaes-Hansen F., Bek T., Mikkelsen J.G., Jensen T.G., Corydon T.J. Adeno-associated virus-delivered polycistronic microRNA-clusters for knockdown of vascular endothelial growth factor in vivo. J. Gene Med. 2012;14:328–338. doi: 10.1002/jgm.2623. [DOI] [PubMed] [Google Scholar]
- 28.Askou A.L., Aagaard L., Kostic C., Arsenijevic Y., Hollensen A.K., Bek T., Jensen T.G., Mikkelsen J.G., Corydon T.J. Multigenic lentiviral vectors for combined and tissue-specific expression of miRNA- and protein-based antiangiogenic factors. Mol. Ther. Methods Clin. Dev. 2015;2:14064. doi: 10.1038/mtm.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 30.Bemelmans A.P., Kostic C., Crippa S.V., Hauswirth W.W., Lem J., Munier F.L., Seeliger M.W., Wenzel A., Arsenijevic Y. Lentiviral gene transfer of RPE65 rescues survival and function of cones in a mouse model of Leber congenital amaurosis. PLoS Med. 2006;3:e347. doi: 10.1371/journal.pmed.0030347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzmiak H.A., Maquat L.E. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Kim S.R., Nakanishi K., Itagaki Y., Sparrow J.R. Photooxidation of A2-PE, a photoreceptor outer segment fluorophore, and protection by lutein and zeaxanthin. Exp. Eye Res. 2006;82:828–839. doi: 10.1016/j.exer.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L.-L., Qu J.-L., Chen D.-N., Niu H.-B. Layered-resolved autofluorescence imaging of photoreceptors using two-photon exicitation. J. Biomed. Sci. Eng. 2009;2:363–365. [Google Scholar]
- 35.Chen C., Tsina E., Cornwall M.C., Crouch R.K., Vijayaraghavan S., Koutalos Y. Reduction of all-trans retinal to all-trans retinol in the outer segments of frog and mouse rod photoreceptors. Biophys. J. 2005;88:2278–2287. doi: 10.1529/biophysj.104.054254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 39.Yiu G., Tieu E., Nguyen A.T., Wong B., Smit-McBride Z. Genomic disruption of VEGF-A expression in human retinal pigment epithelial cells using CRISPR-Cas9 endonuclease. Invest. Ophthalmol. Vis. Sci. 2016;57:5490–5497. doi: 10.1167/iovs.16-20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandler R.J., Sands M.S., Venditti C.P. Recombinant adeno-associated viral integration and genotoxicity: insights from animal models. Hum. Gene Ther. 2017;28:314–322. doi: 10.1089/hum.2017.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balaggan K.S., Ali R.R. Ocular gene delivery using lentiviral vectors. Gene Ther. 2012;19:145–153. doi: 10.1038/gt.2011.153. [DOI] [PubMed] [Google Scholar]
- 42.Monteys A.M., Ebanks S.A., Keiser M.S., Davidson B.L. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol. Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.