Abstract
Current therapies against hepatitis B virus (HBV) do not reliably cure chronic infection, necessitating new therapeutic approaches. The T cell response can clear HBV during acute infection, and the adoptive transfer of antiviral T cells during bone marrow transplantation can cure patients of chronic HBV infection. To redirect T cells to HBV-infected hepatocytes, we delivered plasmids encoding bispecific antibodies directed against the viral surface antigen (HBsAg) and CD3, expressed on almost all T cells, directly into the liver using hydrodynamic tail vein injection. We found a significant reduction in HBV-driven reporter gene expression (184-fold) in a mouse model of acute infection, which was 30-fold lower than an antibody only recognizing HBsAg. While bispecific antibodies triggered, in part, antigen-independent T cell activation, antibody production within hepatocytes was non-cytotoxic. We next tested the bispecific antibodies in a different HBV mouse model, which closely mimics the transcriptional template for HBV, covalently closed circular DNA (cccDNA). We found that the antiviral effect was noncytopathic, mediating a 495-fold reduction in HBsAg levels at day 4. At day 33, bispecific antibody-treated mice exhibited 35-fold higher host HBsAg immunoglobulin G (IgG) antibody production versus untreated groups. Thus, gene therapy with HBsAg/CD3-bispecific antibodies represents a promising therapeutic strategy for patients with HBV.
Keywords: hepatitis B virus, viral hepatitis, bispecific antibody, T cell, gene therapy, immunotherapy, liver
Introduction
Hepatitis B virus (HBV) is a partially double-stranded DNA virus with tropism to the liver, infecting over 300 million people chronically worldwide and causing cirrhosis and liver cancer in a significant number of these patients.1 Once chronically infected, very few HBV patients develop antibodies (Abs) against and clear hepatitis B surface antigen (HBsAg), which serves as a clinical biomarker for functional cure.2 There is no effective treatment for chronic HBV patients; a 5-year treatment course with entecavir, a reverse transcriptase inhibitor, results in HBsAg seroconversion in only 1.4% of patients.3 These antiviral inhibitors suppress serum HBV DNA levels but have no effect on covalently closed circular DNA (cccDNA), the episomal transcriptional template of HBV. This molecule is very stable once formed in the hepatocyte, and cccDNA has been shown to persist for years.4 Pegylated interferon (IFN)-α is also approved for HBV therapy but has shown efficacy only in a minority of patients and is not well tolerated.5
In patients who clear HBV during the acute infection, the CD8-positive T cell response is crucial.6 This immune response is, in part, noncytopathic, relying primarily on secreted cytokines, IFN-γ and tumor necrosis factor (TNF)-α, to mediate cccDNA degradation.7 However, the frequency of HBV-specific T cells is low in chronically infected HBV patients,8 and their functionality is impaired.9 Given the paucity of antiviral T cells in the host, T cells have been redirected to attack HBV-infected hepatocytes using chimeric antigen receptors (CARs) specific for HBsAg.10 Redirected T cells were shown to reduce cccDNA from infected primary hepatocytes in vitro10 and mediated transient viral reduction in an HBV transgenic mouse model.11 While CAR-T cells represent a potential therapy against HBV, T cell products currently have to be produced for each patient individually, limiting their potential utility as a readily available therapeutic. To develop an “off-the-shelf product” to redirect T cells to HBsAg-positive hepatocytes, we investigated the use of bispecific antibodies that recognize HBsAg and CD3, which is expressed on almost all T cells.
Bispecific antibodies targeting CD3 to direct T cells to cell surface antigens were originally reported over 30 years ago12, 13 and have shown promising antitumor activity in numerous preclinical models. However, only blinatumomab, a bispecific Ab that targets CD3 and CD19, expressed on B cell malignancies, has received Food and Drug Administration (FDA) approval so far.14 Current bispecific Ab approaches are challenged by a complicated manufacturing process, short half-lives requiring continuous infusions, and side effects secondary to systemic T cell activation.15 These hurdles could be overcome through in situ expression of bispecific Abs from DNA or RNA templates in patient tissues, but there have been few reports on such strategies.16, 17, 18 The liver absorbs major fractions of gene therapy vectors, nanoparticles, or liposomes, allowing gene constructs to be delivered more readily than in any other organ. Expression of bispecific Abs in the liver should have several advantages compared to the passive infusion of recombinant proteins for treating HBV. Local expression should result in increased Ab concentrations in the liver, before being diluted in the circulation. Moreover, soluble HBsAg in the serum of HBV patients can reduce efficacy by neutralizing a substantial fraction of infused Abs,19 and the formed HBsAg/Ab immune complexes carry the risk of immune-complex disorders in HBV patients.20
Herein, we have developed an approach to express in situ a bispecific Ab to redirect T cells to HBsAg. Our results in transfection-based murine models of HBV suggest a rapid reduction of the virus in a predominately noncytopathic manner.
Results
Hydrodynamic Tail Vein Injection of a Plasmid Expressing HBsAg-Specific Antibodies Results in the Production of Functional Antibody In Vivo
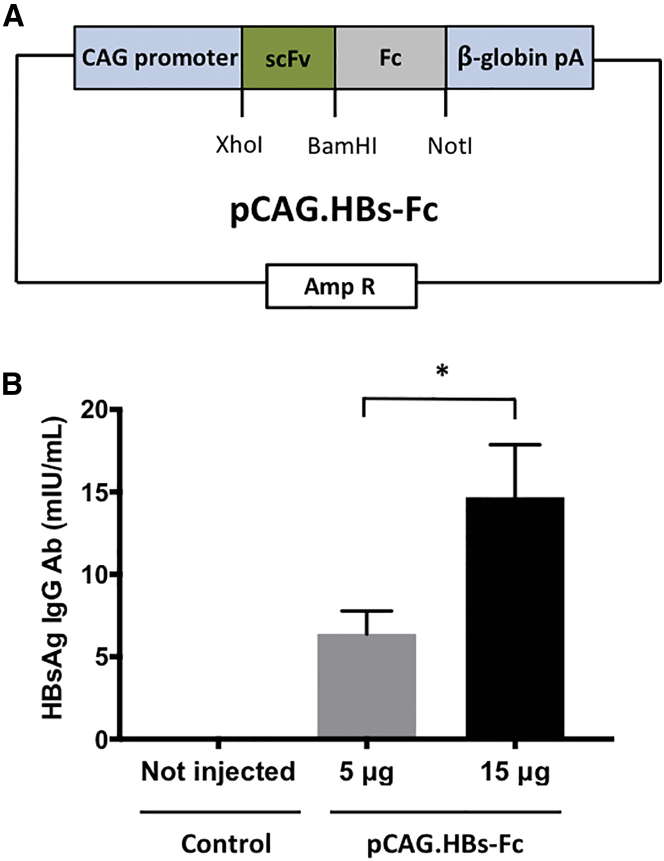
To evaluate the feasibility of expressing functional HBsAg-specific Ab in vivo, we cloned a minigene encoding a HBsAg-specific Ab (HBs-Fc), consisting of the immunoglobulin heavy-chain leader peptide, a single chain variable fragment (scFv) derived from the HBsAg-specific Ab 19.79.5,19, 21, 22 and the Fc domain of human immunoglobulin G1 (IgG1), into the expression plasmid pCAG (pCAG.HBs-Fc; Figure 1A). Hydrodynamic tail vein (HTV) injection was employed to deliver plasmids into the liver, wherein a large volume bolus (10% fluid-body volume) is injected with plasmid DNA,23 resulting in specific delivery into hepatocytes by punching holes into cell membranes.24 5 or 15 μg pCAG.HBs-Fc was injected via HTV injection into immunocompetent mice, and the plasma concentration of HBs-Fc was measured 4 days post-injection by ELISA. Mean HBs-Fc concentrations were 6.4 mIU/mL for 5 μg and 14.7 mIU/mL for 15 μg injected plasmid (Figure 1B). Thus, HTV injection of pCAG.HBs-Fc results in significant, dose-dependent production of HBsAg-specific Abs in vivo.
Figure 1.
HTV Injection of pCAG.HBs-Fc Results in HBs-Fc Antibody Expression In Vivo
(A) Scheme of pCAG.HBs-Fc. (B) Serum was collected 4 days post-HTV injection of pCAG.HBs-Fc. The serum concentration of HBs-Fc was determined by HBsAg IgG Ab ELISA (mean ± SEM, n = 4, *p < 0.05).
HBs-Fc Gene Delivery Has Antiviral Activity In Vivo
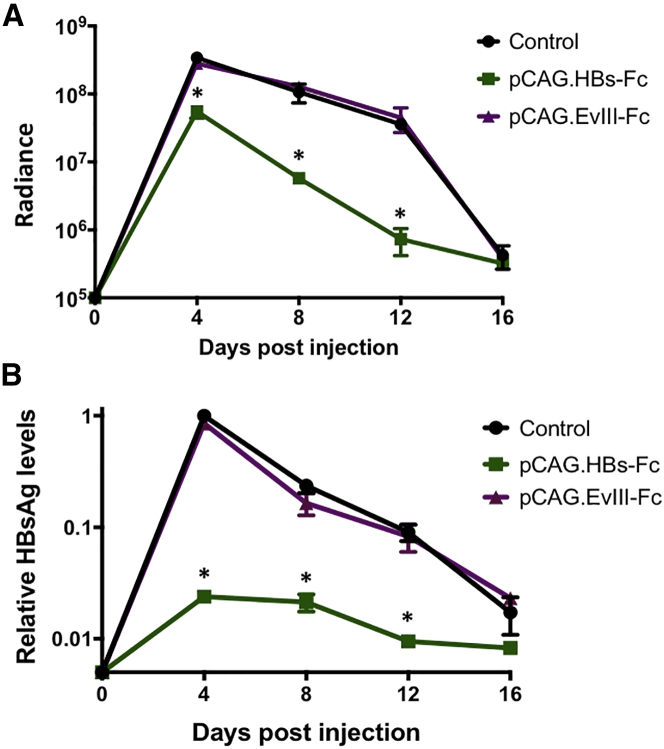
To evaluate if in vivo expression of HBs-Fc has antiviral activity, we adapted a murine model that allows for measuring the clearance of HBV using non-invasive bioluminescence imaging, which correlated with serum HBsAg and HBV DNA levels.25 Bioluminescence of a reporter gene has previously been shown to be a sensitive readout for CD8 T cell responses in the liver against a co-delivered antigen gene.26, 27 Briefly, we generated a plasmid encoding the HBV genome and a GFP-2A-firefly luciferase (GFP-2A-ffLuc) expression cassette, both under the transcriptional control of identical HBV core promoters (pHBV-ffLuc; Figure S1A). HTV injection of pHBV-ffLuc into NOD scid gamma (NSG) mice resulted in luciferase expression in the liver as judged by bioluminescence imaging, confirming the functionality of pHBV-ffLuc (Figures S1B and S1C). The introduction of HBV plasmid DNA by HTV injection into immunocompetent mice results in immune clearance over 2 weeks, in a process resembling acute HBV infection.28 To evaluate if expression of HBs-Fc induces clearance of HBV, pHBV-ffLuc was co-injected with pCAG.HBs-Fc, a pCAG plasmid encoding an Ab specific for an irrelevant antigen29 (EGFRvIII; pCAG.EvIII-Fc), or a control plasmid. Keeping the total amount of DNA injected consistent, co-injection of 5 μg pHBV-ffLuc with 15 μg pCAG.HBs-Fc resulted in a significantly lower luciferase signal in comparison to co-injection with 15 μg pCAG.EvIII-Fc or control plasmid (Figure 2A). In addition, pHBV-ffLuc/pCAG.HBs-Fc co-injected mice had significantly lower levels of serum HBsAg (Figure 2B) compared to the other treatment groups, indicating that HBs-Fc has antiviral activity in vivo.
Figure 2.
HBs-Fc Gene Delivery into Hepatocytes Has Anti-HBV Activity In Vivo
(A) Immunocompetent mice were co-injected by HTV injection with 5 μg pHBV-ffLuc and 15 μg pCAG.HBs-Fc, pCAG.EvIII-Fc, or control plasmid. Quantitative bioluminescence imaging data (radiance = photons/sec/cm2/sr) for all mice are shown (mean ± SEM, n = 3, *p < 0.05). (B) HBsAg levels were determined by ELISA. Data were normalized to the day 4 HBsAg level of the pHBV-ffLuc/control plasmid group (mean ± SEM, n = 3, *p < 0.05).
Including an Anti-CD3 Domain in HBs-Fc Enhances the Antiviral Activity In Vivo
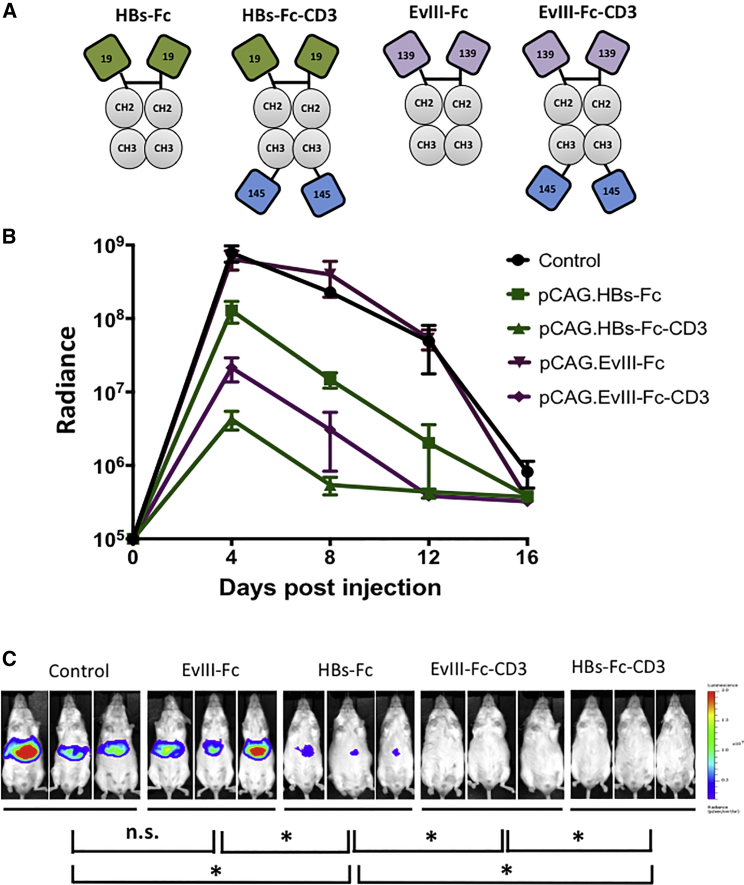
Having established that pCAG.HBs-Fc has anti-HBV activity in vivo, we next determined if inclusion of an scFv specific for murine CD3, which activates T cells, further enhances its antiviral activity. We generated pCAG expression plasmids encoding HBs-Fc-CD3 or EvIII-Fc-CD3 bispecific Abs by inserting the murine CD3-binding scFv from monoclonal antibody (mAb) 145-2C1130, 31 at the C terminus of HBs-Fc or EvIII-Fc, respectively (pCAG.HBs-Fc-CD3; pCAG.EvIII-Fc-CD3; Figure 3A).
Figure 3.
Addition of CD3 Binding Domain Enhances Anti-HBV Activity of HBs-Fc
(A) Scheme of Ab constructs (green: HBsAg-specific scFv derived from mAb 19.79.5 [19]; purple: EvIII-specific scFv derived from mAb 139 [139]; gray: IgG1-Fc [CH2, CH3]; and blue: murine CD3-specific scFv derived from mAb 145-2C11 [145]). (B) Immunocompetent mice were co-injected by HTV injection with 15 μg pCAG.HBs-Fc-CD3, pCAG.EvIII-Fc-CD3, pCAG.HBs-Fc, pCAG-EvIII-Fc, or control plasmid and 5 μg pHBV-ffLuc. Quantitative bioluminescence imaging data (radiance = photons/sec/cm2/sr) for all mice are shown (mean ± SEM, n = 3). (C) Mouse images for different constructs are shown for day 4 post-injection (n = 3, *p < 0.05) using the identical exposure time. n.s., not significant.
Keeping the total amount of DNA injected consistent, 5 μg pHBV-ffLuc was injected by HTV injection in combination with 15 μg control plasmid or plasmids encoding the respective Ab. In this experiment, we compared pCAG.HBs-Fc, pCAG.HBs-Fc-CD3, EvIII-Fc, and pCAG.EvIII-Fc-CD3. Inclusion of a CD3-specific scFv enhanced the antiviral activity of pCAG.HBs-Fc 30-fold (p < 0.05) as judged by bioluminescence imaging (Figure 3B). Representative bioluminescence images of mice at day 4 post-injection are shown, and there was a significant difference between pCAG.EvIII-Fc and pCAG.EvIII-Fc-CD3 at this time point (p < 0.05), indicating that bispecific Abs induce unspecific T cell activation (Figure 3C). Unspecific T cell activation was confirmed with a pCAG plasmid encoding an Ab specific for the irrelevant antigen EphA2 and CD3 (pCAG.EphA2-Fc-CD3; Figure S2).32
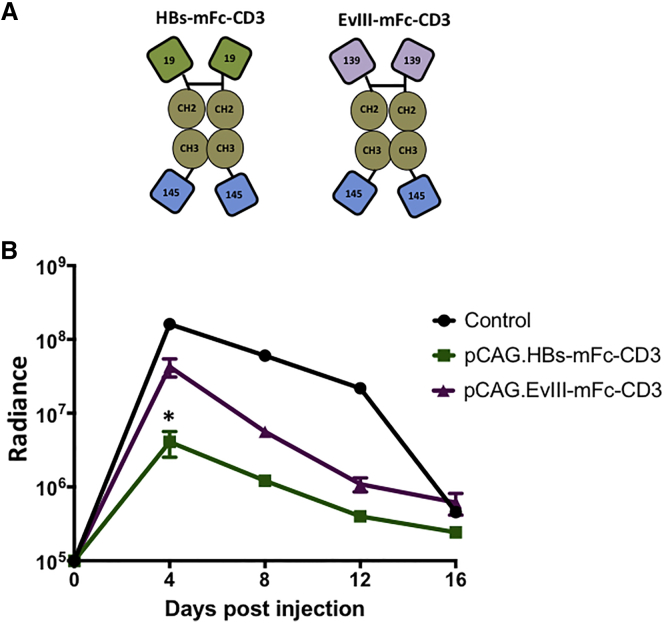
To evaluate the contribution of Fc receptor-mediated phagocytosis or cell killing to the observed antiviral activity of our bispecific Abs, we replaced the wild-type human IgG1 Fc domain in HBs-Fc-CD3 and EvIII-Fc-CD3 with a mutated human IgG4 Fc (mFc) domain that does not bind to Fc receptors33 (HBs-mFc-CD3; EvIII-mFc-CD3) (Figure 4A). We compared the antiviral activity of pCAG.HBs-mFc-CD3 to pCAG.EvIII-mFc-CD3 in our model. Both bispecific Abs had antiviral activity as judged by bioluminescence imaging (Figure 4B). HBs-mFc-CD3 had significantly greater antiviral activity at day 4 (10-fold) than EvIII-mFc-CD3 (Figure 4B), while as shown previously using the non-mutated Fc (Figure 3B), the HBs-Fc-CD3 had only 5-fold greater activity than the corresponding EvIII-Fc-CD3 at the same time point. We confirmed this observation with additional replicates (Figure S3) and thus selected HBs-mFc-CD3 and EvIII-mFc-CD3 as a control for our subsequent experiments.
Figure 4.
Fc Receptor Binding Does Not Contribute to the Anti-HBV Activity of HBs-Fc-CD3
(A) Scheme of Ab constructs (green: HBsAg-specific scFv derived from mAb 19.79.5 [19]; purple: EvIII-specific scFv derived from mAb 139 [139]; brown: IgG4-Fc with mutated Fc receptor binding sites [CH2, CH3]; and blue: murine CD3-specific scFv derived from mAb 145-2C11 [145]). (B) Immunocompetent mice were co-injected by HTV injection with 15 μg pCAG.HBs-mFc-CD3, pCAG.EvIII-mFc-CD3, or control plasmid with 5 μg pHBV-ffLuc. Quantitative bioluminescence imaging data (radiance = photons/sec/cm2/sr) for all mice are shown (mean ± SEM, n = 3). pCAG.HBs-mFc-CD3 had significantly greater anti-HBV activity than pCAG-EvIII-mFc-CD3 (*p < 0.05).
Bispecific Antibodies Act Early after Injection and in a CD3-Dependent Manner in the HBV Model
We sought to explore the kinetics of HBs-mFc-CD3 action over the course of acute clearance. We co-injected 5 μg pHBV-ffLuc and 15 μg pCAG.HBs-mFc-CD3 or control, measuring bioluminescence each day for the first 4 days. We found that a 13-fold difference between control and HBs-mFc-CD3 treatment had already occurred at day 1 post-injection (Figure S4A), with a similar difference maintained subsequently. This matches the published kinetics of gene expression after HTV injection, which peaks at 8 hr post-injection,23 and suggests that ongoing bispecific Ab production and/or antiviral T cell activation after day 1 is minimal.
We also investigated the requirements for T cell signaling by substituting a different moiety for T cell stimulation, replacing the CD3 targeting portion. For this purpose, we utilized the extracellular domain of the mouse CD80 protein, which can interact with CD28 expressed on T cells. We cloned this portion at the N terminus in order to most closely resemble its natural orientation. We found that the injected pCAG.CD80-mFc-HBs plasmid had significantly higher bioluminescence (28-fold higher) compared to the pCAG.HBs-mFc-CD3 plasmid (Figure S4B), indicating that CD3 activation of T cells is critical for the observed antiviral activity.
In Vivo Expression of HBs-mFc-CD3 Does Not Result in Hepatocyte Toxicity
Having demonstrated that unspecific T cell activation contributes to the antiviral activity of HBs-mFc-CD3, we next explored unspecific hepatotoxicity triggered by HBs-mFc-CD3 expression. We first evaluated liver transaminase elevation that might result from bispecific Ab expression, knowing that the HTV procedure itself results in transient hepatocyte injury and serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) increases.34 No difference between the conditions was observed at day 4, with AST levels still mildly elevated at this time point post-injection (Figure S5A). Since it is difficult to separate bulk damage from the HTV procedure versus direct transgene toxicity by examining transaminases alone, we sought to more precisely study toxicity in individual transgene-modified hepatocytes. Toward this goal, we developed a new hepatotoxicity assay based on bioluminescence imaging to evaluate whether transfected hepatocytes persisted. Transgenic ROSA26-LoxP-STOP-LoxP-ffLuc (Rosa-Luc) mice were injected by HTV injection with pCMV-nuclear localization sequence (NLS)-Cre and pCAG.HBs-mFc-CD3 or control plasmids. The injection results in co-delivery of plasmids to the same hepatocytes; expressed Cre recombinase in transfected cells of Rosa-Luc mice induces ffLuc expression, and the resulting bioluminescence signal correlates with the number of viable, transfected hepatocytes in vivo (details in the Materials and Methods). HBs-mFc-CD3 expression did not reduce the bioluminescence signal on day 4 or 8 post-injection versus control mice (Figure S5B), indicating that HBs-mFc-CD3 is non-toxic in hepatocytes. These findings were confirmed using standard histological examination (H&E staining) of liver sections from mice co-injected with pHBV-ffLuc and control or pCAG.HBs-mFc-CD3 on day 4 post-injection (Figure S5C). Likewise, no histomorphological changes indicating toxicity were noted when transfecting previously used Ab constructs (Figure S6). These histological stains also did not demonstrate a notable increase in lymphocyte levels among the different test constructs at day 4, indicating that additional T cells were not recruiting into the liver, but rather tissue-resident T cells in the liver were likely activated (Figure S6).
In Vivo Expression of HBs-mFc-CD3 Has Antiviral Activity in a Recombinant cccDNA HBV Mouse Model
Finally, we wanted to explore the ability of HBs-mFc-CD3 to induce cccDNA clearance in vivo, which more closely mimics HBV transcriptional templates in human cells than plasmids carrying the HBV genome. We adapted previously reported HBV murine models that utilize recombinases to generate a recombinant cccDNA-like (rcccDNA) molecule lacking bacterial DNA.35, 36 In our system, we constructed a floxed HBV genome with an NLS-Cre recombinase (containing an internal intron) cassette driven by a cytomegalovirus (CMV) promoter in cis on the same plasmid (pCLX; Figure S7A; R.L.K, M.B., X.L., F.P.P., B. Bissig-Choisat, C. Whitten-Bauer, B.L. Slagle, U. Garaigorta, K.D.B., unpublished data). In the floxed, unexcised state without Cre recombinase, there is no detection of HBV antigens, since viral transcripts and/or proteins are interrupted by the LoxP sequences preventing expression (Figures S7B and S7C). After HTV injection of pCLX, Cre expression and resultant rcccDNA formation yields a high level of HBsAg production and HBV core expression 1 week post-injection, demonstrating the functionality of our model (Figures S7B and S7C). When pCLX is introduced into Rosa-Luc mice, the Cre recombinase also induces ffLuc expression. Thus, bioluminescence imaging serves as a non-invasive means to monitor viable HBV transfected cells and, as in our previous Rosa-Luc experiments, can be used to monitor hepatotoxicity of our bispecific Ab therapy.
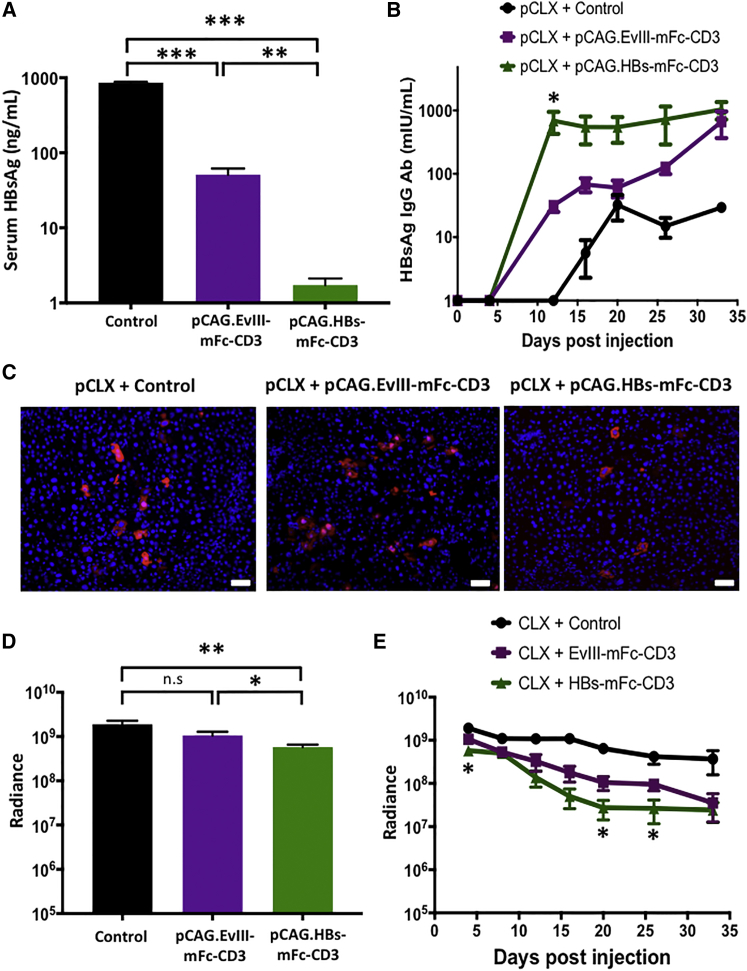
To determine the antiviral activity and safety of HBs-mFc-CD3 in our rcccDNA model, we co-injected pCLX with pCAG.HBs-mFc-CD3, pCAG.EvIII-mFc-CD3, or control plasmid. On day 4 post-injection, we measured 495-fold and 30-fold lower HBsAg levels in pCAG.HBs-mFc-CD3-injected mice compared to mice receiving control plasmid and pCAG.EvIII-mFc-CD3, respectively (Figure 5A). pCAG.HBs-mFc-CD3 therapy not only reduced HBsAg levels, but it also induced HBsAg Abs earlier and at higher levels than pCAG.EvIII-mFc-CD3 and controls (Figure 5B). The antiviral activity of pCAG.HBs-mFc-CD3 therapy was also confirmed by immunofluorescence in a subset of animals on day 4 post-injection, showing less HBV core expression within individual hepatocytes compared to the pCAG.EvIII-mFc-CD3 and control groups (Figure 5C). We used bioluminescence imaging to determine the safety of pCAG.HBs-mFc-CD3 therapy. On day 4 post-injection, there was a 3.3-fold reduction in the bioluminescence signal in pCAG.HBs-mFc-CD3-injected mice compared to mice receiving control plasmid (Figure 5D), and long-term follow-up revealed a further decline in the bioluminescence signal of pCAG.HBs-mFc-CD3-injected mice. At days 4, 20, and 26 post-injection, there were significant differences between pCAG.HBs-mFc-CD3- and pCAG.EvIII-mFc-CD3-injected mice (p < 0.05, Figure 5E).
Figure 5.
In Vivo Expression of HBs-mFc-CD3 in a Recombinant cccDNA Model of HBV Has Antiviral Effects and Induces Endogenous HBsAg Antibodies
Rosa-Luc mice were co-injected by HTV injection with 5 μg pCLX and 15 μg pCAG.HBs-mFc-CD3, pCAG.EvIII-mFc-CD3, or control plasmid (n = 4). (A) HBsAg serum levels were determined by ELISA at day 4 post-injection (mean ± SEM, n = 4, **p < 0.005, ***p < 0.0001). (B) The development of host HBsAg IgG Abs was determined by ELISA at the indicated time points (mean ± SEM, n = 4, *p < 0.05). (C) Tissue was harvested from mice at day 4 post-injection, and HBV core protein (red) expression was assessed by immunofluorescence (blue = DAPI; scale bars represent 50 μm). (D and E) Quantitative bioluminescence imaging data (radiance = photons/sec/cm2/sr) for all mice is shown on (D) day 4 post-injection and (E) during long-term follow-up (mean ± SEM, *p < 0.05, **p < 0.01). n.s., not significant.
Discussion
Here we developed a novel therapeutic strategy using bispecific Abs for HBV immunotherapy. We describe the production of bispecific Abs in the liver leading to local T cell activation, modulating the immune response in the organ. This differs from recombinant protein strategies, which do not specifically accumulate at tissue sites unless additional targeting moieties are included, or from using cell-based carriers for delivery.32 Our strategy utilizes the recruitment of naive T cells against HBV, as opposed to relying on exhausted, dysfunctional HBV-specific T cells8, 9 to mediate antiviral effects.
We demonstrated that Abs can be expressed in hepatocytes and retain their functionality. As expected, the HBs-Fc Ab was specific and effectively mediated an antiviral effect, similar to the parent monoclonal Ab,19, 22 while the addition of CD3-specific scFv led to an even more potent antiviral response. This antiviral effect peaked at day 1 post-injection, and we were unable to detect significant levels of bispecific antibodies in the serum at day 4, likely due to activation and/or engagement with mouse T cells preventing high-level accumulation. Delivering co-stimulation through the CD80 ectodomain instead of CD3-specific scFv did not elicit high-level antiviral responses. Interestingly, the bispecific Ab targeting CD3 and an unrelated antigen had significant anti-HBV activity in vivo, indicating that T cell activation occurred independent of HBsAg binding. Fc receptor cross-linking did not play a role in enhancing antiviral effects or in facilitating T cell activation,37 since the construct with mutated Fc receptor binding sites, HBs-mFc-CD3, had similar anti-HBV activity to HBs-Fc-CD3. Antigen-independent secretion of IFN-γ has been observed previously in vitro with the bispecific Ab format used in this study.38 In addition, antigen-independent T cell activation by bispecific Abs secreted from cell lines has also been reported.39 While the exact mechanism of antigen-independent T cell activation by bispecific Abs is not completely understood, self-aggregation, resulting in cross-linking of CD3, is the most likely explanation.
In this study, we utilized a novel procedure for dual monitoring of hepatocyte viability and HBV markers, adapting recombination methods to generate cccDNA-like molecules similar to previous reports,35, 36 while also leveraging recombination of host cell reporter genes. Our study was able to replicate other reports that initial T cell effects against HBV in the liver are noncytopathic,7 since the HBsAg levels were reduced to a much larger extent in the bispecific Ab-treated groups compared to bioluminescent radiance, reflecting hepatocyte viability. Beyond activating T cells, HBs-mFc-CD3 may have also reduced serum HBsAg levels in the rcccDNA model via the ability of Abs to block HBsAg secretion from hepatocytes through their engagement with the neonatal Fc receptor.40 A later decrease in radiance in groups treated with bispecific Abs was observed, similar to the pattern of final clearance of infected cells in chimpanzees.6 In conjunction with the higher levels of HBsAg IgG Ab production, this indicates that bispecific Abs increased the host adaptive immune response versus the control group.
Our study differs from previous gene therapy reports expressing an individual cytokine, IFN-γ, in the liver to yield antiviral activity.41, 42 When tested in a surrogate woodchuck model, adenoviral delivery of IFN-γ and TNF-α genes to the liver did not result in an immediate reduction in the woodchuck hepatitis virus (WHV), but rather the later adaptive response against adenovirus by T cells decreased WHV viral loads, suggesting that expressing individual cytokines alone may not be sufficient.43 Thus, activating T cells through CD3, which results in the production of not only IFN-γ but also other proinflammatory cytokines such as TNF-α and granulocyte macrophage colony-stimulating factor (GM-CSF),44 might be more effective. Our targeted approach to HBV is also distinct from the infusion of a recombinant T cell receptor (TCR)-like Ab to deliver IFN-α to HBV-infected hepatocytes45 and from gene therapy with a Apo-A1/IFN-α fusion protein.46 In addition, our strategy targeting HBsAg and CD3 format differs from previous protein-based bispecific Ab strategies targeting two different epitopes on HBsAg,47, 48 as well as a strategy targeting HBx and CD3 for hepatocellular carcinoma.49
The current study is limited in relying on HTV injection, which results in efficient co-delivery of plasmids to the same cells50 but cannot be applied to patients. To overcome this limitation, we are planning to explore clinical translatable liver gene delivery systems such as adeno-associated virus (AAV)51 or mRNA nanoparticles.52, 53 In a recent study, mRNA nanoparticles effectively targeted the liver in mice and systemically secreted bispecific antibodies to engage claudin-6 and CD3, triggering T cell cytotoxicity against subcutaneously injected tumors.18 This strategy could be applied to our approach to treat in situ liver disease. Another limitation is that HTV injection results in an acute model of HBV, rather than a chronic HBV mouse model, which can be established in immunocompetent mice with AAV54 or low-dose adenoviral55 delivery of HBV genomes. Future work will seek to treat in chronic HBV models with translatable delivery systems. In these studies, it will be important to evaluate the kinetics and expression of bispecific antibodies from these other vector systems. We also plan to evaluate the exact cytokine profile induced by bispecific antibodies in the liver, as well as the effects on intrahepatic viral markers.
In conclusion, we developed a novel therapeutic approach to target HBV infection by expressing bispecific Abs in hepatocytes, leading to a local and predominantly noncytopathic immune response against HBV. Our approach may be of value for treating not only HBV but also other viral liver diseases.
Materials and Methods
Plasmid Constructs
Generation of Ab Constructs
A codon-optimized minigene was synthesized by Integrated DNA Technologies (IDTDNA; Coralville, IA) containing the immunoglobulin heavy-chain leader peptide (MDWIWRILFLVGAATGAHS), the HBs-specific mAb 19.79.521 heavy chain, a glycine (G) serine (S) linker [(G4S)3], and the 19.79.5 light chain flanked by 5′ XhoI and 3′ BamHI sites. The human IgG1 Fc domain was PCR amplified from a plasmid encoding a CAR containing an IgG1 Fc hinge with PCR primers possessing 5′ BamHI and 3′ NotI. The minigene and PCR product was cloned into pCAG by three-way ligation to create pCAG-HBs-Fc. To create the EvIII-Fc control plasmid, the 139 scFv specific for EGFRvIII29 was PCR cloned from pSFG.139-CD3-I-mOrange with PCR primers containing 5′ XhoI and 3′ BamHI sites. The three-way ligation reaction was performed to create pCAG-EvIII-Fc. Cloning was confirmed by sequencing (Lone Star Labs, Houston, TX).
Generation of Bispecific Ab Constructs
The 145-2C1130 scFv specific for murine CD3 was PCR amplified from pRV2011.145-2C11-1D3-I-Thy1.1 with 5′ EcoRV and 3′ NotI sites. Four-way ligation was performed with 5′ XhoI-leader-HBs or EvIII scFv-3′ BamHI, 5′ BamHI-Fc-3′ EcoRV, 5′ EcoRV-145-2C11 scFv-3′ NotI, and pCAG digested with XhoI and NotI to generate pCAG.HBs-Fc-CD3 and pCAG.EvIII-Fc-CD3. pCAG.HBs-mFc-CD3 and pCAG.EvIII-mFc-CD3 were generated in a similar fashion except a codon-optimized minigene (IDTDNA, Coralville, IA) encoding the human IgG4 Fc with mutated Fc binding sites33 and flanking 5′ BamHI and 3′ EcoRV sites was used instead of 5′ BamHI-Fc-3′ EcoRV. pCAG.CD80-mFc-HBs was generated by synthesizing the extracellular domain including the leader sequence of murine CD80 (B7.1) protein between 5′ XhoI and 3′ BamHI sites. Separately, PCR amplification of HBs scFv added 5′ EcoRV and 3′ NotI sites. The four-way ligation reaction with 5′-XhoI-CD80-3′ BamHI, 5′ BamHI-mFc-3′ EcoRV, and 5′ EcoRV-HBs-3′ NotI and pCAG digested with XhoI and NotI was performed to generate pCAG.CD80-mFc-HBs. Cloning was confirmed by sequencing (Lone Star Labs, Houston, TX).
Generation of pCAG
The pCAG vector was constructed from the pCIG vector (containing the hybrid promoter CMV enhancer chicken beta actin [CAG] promoter, rabbit beta globin 3′UTR, polyadenylation sequence, internal ribosome entry site (IRES)-NLS-GFP, and SV40 origin of replication) by removing IRES-NLS-GFP, leaving XhoI and NotI sites for inserting transgenes (Figure 1A).
Generation of pHBV-ffLuc
pHBV-ffLuc was generated by inserting a GFP-2A-ffLuc expression cassette by PCR cloning (provided by Dr. Inder Verma, Salk Institute, San Diego, CA) into the SmaI and SacI sites of pSP65ayw1.3 (provided by Dr. Stefan Wieland, University of Basel, Switzerland). pSP65ayw1.3 encodes an over-length HBV genome from genotype D, subtype ayw, GenBank V01460. The SmaI site is located at the 3′ end of the over-length HBV genome and the PCR primers for GFP-2A-ffLuc subcloning were designed so that GFP-2A-ffLuc translation is in frame with core protein translation at the 3′ end of the HBV genome (see Figure S8 for more sequence information). Cloning was confirmed by sequencing (Lone Star Labs, Houston, TX).
Bioluminescence Imaging
The IVIS system (Xenogen Corp., Alameda, CA) was used for bioluminescence imaging. Mice were anesthetized with isoflurane and injected intraperitoneally with 200 μL of 7.5 mg/mL luciferin solution (GoldBio, Olivette, MO). Luciferin was allowed to circulate for 10 min post-injection, and mice were placed ventral side up and imaged promptly thereafter. Luminescence signals were quantified using Living Image 4.2 software (Caliper Life Sciences, Hopkinton, MA) with a region of interest (ROI) circling the area over the liver.
HBsAg ELISA
HBsAg levels were determined as previously described.56 Briefly, serum HBsAg levels were evaluated with commercially sold ELISA kits according to the manufacturer’s instructions (International Immuno Diagnostics, Foster City, CA). Quantification of serum HBsAg was made by comparing serial dilutions of known standards (Alpha Diagnostic International, San Antonio, TX). HBsAg levels were reported in nanograms per milliliter, consistent with the standards utilized. The conversion ratio to international units per milliliter is not provided by the manufacturer, but many kits have conversions of 1 or 10 ng/mL to 1 IU/mL HBsAg as approximate guidelines.57
HBsAg IgG Antibody ELISA
Serum HBsAg IgG Ab levels were quantified by ELISA according to the manufacturer’s instructions (Alpha Diagnostic International, San Antonio, TX). HBsAg IgG Ab levels were reported as milli-international units per milliliter. The ELISA assay can detect both human and mouse immunoglobulin.
Transaminase Analysis
Serum ALT and AST were measured by the Comparative Pathology Laboratory (Baylor College of Medicine, Houston, TX) using the COBAS INTEGRA 400 plus analyzer (Roche Diagnostics, Indianapolis, IN).
Animal Experiments
All animal experiments followed a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. For all experiments using immunocompetent mice, the Rosa-Luc strain from Jackson Labs [FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J] was utilized, in which the expression of the firefly luciferase (Luc) gene is blocked by a loxP-flanked STOP fragment placed between the Luc sequence and the Gt(ROSA)26Sor promoter. Even though most experiments did not utilize the endogenous luciferase reporter, using Rosa-Luc mice for all experiments reduced the risk of inter-strain variability. Rosa-Luc mice, aged 6–10 weeks, were selected for HTV injection. pCMV-Gaussia luciferase (Thermo Fisher, Waltham, MA) was used as a filler or control plasmid so that an equal amount of DNA was injected in each group of mice. Plasmid DNA was diluted into 0.9% normal saline solution to a total volume equaling 10% of murine body weight. Mice were placed under a heat lamp for 5–10 min to dilate the lateral tail veins, and injection was performed over 4–6 s.58 Mice were bled retro-orbitally, and serum was collected after centrifugation for 30 min at 2.3 × g. Serum was stored at −80°C until further use for HBsAg ELISA and HBsAg IgG ELISA quantification.
Histology
Frozen tissue slides from livers were fixed with 3% paraformaldehyde (PFA) for 15 min and stained for HBV core overnight at 4°C in PBS-T buffer (PBS 1× containing 0.5% BSA and 0.2% of Triton-100) using the primary Ab, rabbit anti-hepatitis B virus core antigen (Dako/Agilent, Santa Clara, CA). The primary Ab was washed with PBS 1×, and slides were incubated with Alexa Fluor secondary antibodies (Molecular Probes, Eugene, OR) in PBS-T buffer. Vectashield plus DAPI (Vector Labs, Burlingame, CA) was used for slide mounting. In other experiments, liver tissue was fixed in 4% PFA overnight, and serial sections of paraffin-embedded liver were stained with H&E.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, CA). Data measurements are presented as the mean ± SEM. Mean differences were tested using appropriate tests, including unpaired, parametric, one-tailed t tests. The significance level used was p < 0.05, unless otherwise specified.
Author Contributions
R.L.K., S.G., and K.B. designed the experiments; R.L.K., T.S., and F.P.P. performed in vivo experiments; M.B. did immunostaining; X.L. did ELISA. All authors read and approved the final manuscript.
Conflicts of Interest
R.L.K., T.S., S.G., and K.B. have a patent application filed covering the work in this manuscript.
Acknowledgments
We thank Beatrice Bissig-Choisat (Baylor College of Medicine) for help with animal experiments, and Stefan Wieland (University of Basel) for providing pSP65-HBVayw1.3. K.B. is supported by National Institute of Allergy and Infectious Diseases (NIAID) grant R01AI094409, National Heart, Lung and Blood Institute (NHLBI) grant R01HL134510, the Texas Hepatocellular Carcinoma Consortium (THCCC) (CPRIT no. RP150587), and the Diana Helis Henry and Adrienne Helis Malvin Medical Research Foundations. R.L.K. and T.S. were supported by NIH grant T32DK060445. T.S. was supported by NIH grant 5T32HL092332. The Dan L. Duncan Cancer Center is supported by NIH grant P30CA125123. The Integrated Microscopy Core at the Texas Medical Center Digestive Disease Center is supported by NIH grant P30DK56338.
Footnotes
Supplemental Information includes eight figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.08.006.
Contributor Information
Stephen Gottschalk, Email: smg@bcm.edu.
Karl-Dimiter Bissig, Email: bissig@bcm.edu.
Supplemental Information
References
- 1.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142 doi: 10.1053/j.gastro.2011.12.061. 1264.e1–1273.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J., Yang H.-I., Lee M.-H., Lu S.-N., Jen C.-L., Wang L.-Y., You S.L., Iloeje U.H., Chen C.J., REVEAL-HBV Study Group Incidence and determinants of spontaneous hepatitis B surface antigen seroclearance: a community-based follow-up study. Gastroenterology. 2010;139:474–482. doi: 10.1053/j.gastro.2010.04.048. [DOI] [PubMed] [Google Scholar]
- 3.Chang T.-T., Lai C.-L., Kew Yoon S., Lee S.S., Coelho H.S.M., Carrilho F.J., Poordad F., Halota W., Horsmans Y., Tsai N. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:422–430. doi: 10.1002/hep.23327. [DOI] [PubMed] [Google Scholar]
- 4.Werle-Lapostolle B., Bowden S., Locarnini S., Wursthorn K., Petersen J., Lau G., Trepo C., Marcellin P., Goodman Z., Delaney W.E., 4th Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Perrillo R. Benefits and risks of interferon therapy for hepatitis B. Hepatology. 2009;49(Suppl):S103–S111. doi: 10.1002/hep.22956. [DOI] [PubMed] [Google Scholar]
- 6.Thimme R., Wieland S., Steiger C., Ghrayeb J., Reimann K.A., Purcell R.H., Chisari F.V. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia Y., Stadler D., Lucifora J., Reisinger F., Webb D., Hösel M., Michler T., Wisskirchen K., Cheng X., Zhang K. Interferon-γ and tumor necrosis factor-α produced by T cells reduce the HBV persistence form, cccDNA, without cytolysis. Gastroenterology. 2016;150:194–205. doi: 10.1053/j.gastro.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 8.Boni C., Fisicaro P., Valdatta C., Amadei B., Di Vincenzo P., Giuberti T., Laccabue D., Zerbini A., Cavalli A., Missale G. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 2007;81:4215–4225. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J.-J., Wong D.K., Wahed A.S., Lee W.M., Feld J.J., Terrault N., Khalili M., Sterling R.K., Kowdley K.V., Bzowej N., Hepatitis B Research Network Hepatitis B virus--specific and global T-cell dysfunction in chronic hepatitis B. Gastroenterology. 2016;150:684–695. doi: 10.1053/j.gastro.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohne F., Chmielewski M., Ebert G., Wiegmann K., Kürschner T., Schulze A., Urban S., Krönke M., Abken H., Protzer U. T cells redirected against hepatitis B virus surface proteins eliminate infected hepatocytes. Gastroenterology. 2008;134:239–247. doi: 10.1053/j.gastro.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Krebs K., Böttinger N., Huang L.R., Chmielewski M., Arzberger S., Gasteiger G., Jäger C., Schmitt E., Bohne F., Aichler M. T cells expressing a chimeric antigen receptor that binds hepatitis B virus envelope proteins control virus replication in mice. Gastroenterology. 2013;145:456–465. doi: 10.1053/j.gastro.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 12.Staerz U.D., Kanagawa O., Bevan M.J. Hybrid antibodies can target sites for attack by T cells. Nature. 1985;314:628–631. doi: 10.1038/314628a0. [DOI] [PubMed] [Google Scholar]
- 13.Staerz U.D., Bevan M.J. Hybrid hybridoma producing a bispecific monoclonal antibody that can focus effector T-cell activity. Proc. Natl. Acad. Sci. USA. 1986;83:1453–1457. doi: 10.1073/pnas.83.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przepiorka D., Ko C.-W., Deisseroth A., Yancey C.L., Candau-Chacon R., Chiu H.-J., Gehrke B.J., Gomez-Broughton C., Kane R.C., Kirshner S. FDA approval: blinatumomab. Clin. Cancer Res. 2015;21:4035–4039. doi: 10.1158/1078-0432.CCR-15-0612. [DOI] [PubMed] [Google Scholar]
- 15.Mau-Sørensen M., Dittrich C., Dienstmann R., Lassen U., Büchler W., Martinius H., Tabernero J. A phase I trial of intravenous catumaxomab: a bispecific monoclonal antibody targeting EpCAM and the T cell coreceptor CD3. Cancer Chemother. Pharmacol. 2015;75:1065–1073. doi: 10.1007/s00280-015-2728-5. [DOI] [PubMed] [Google Scholar]
- 16.Compte M., Nuñez-Prado N., Sanz L., Alvarez-Vallina L. In vivo secretion of bispecific antibodies recruiting lymphocytic effector cells. Antibodies. 2013;2:415–425. [Google Scholar]
- 17.Pang X., Ma F., Zhang P., Zhong Y., Zhang J., Wang T., Zheng G., Hou X., Zhao J., He C., Chen Z.Y. Treatment of human B-cell lymphomas using minicircle DNA vector expressing anti-CD3/CD20 in a mouse model. Hum. Gene Ther. 2017;28:216–225. doi: 10.1089/hum.2016.122. [DOI] [PubMed] [Google Scholar]
- 18.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 19.Galun E., Eren R., Safadi R., Ashour Y., Terrault N., Keeffe E.B., Matot E., Mizrachi S., Terkieltaub D., Zohar M. Clinical evaluation (phase I) of a combination of two human monoclonal antibodies to HBV: safety and antiviral properties. Hepatology. 2002;35:673–679. doi: 10.1053/jhep.2002.31867. [DOI] [PubMed] [Google Scholar]
- 20.van Nunen A.B., Baumann M., Manns M.P., Reichen J., Spengler U., Marschner J.P., de Man R.A., International Study Group Efficacy and safety of an intravenous monoclonal anti-HBs in chronic hepatitis B patients. Liver. 2001;21:207–212. doi: 10.1034/j.1600-0676.2001.021003207.x. [DOI] [PubMed] [Google Scholar]
- 21.Eren R., Lubin I., Terkieltaub D., Ben-Moshe O., Zauberman A., Uhlmann R., Tzahor T., Moss S., Ilan E., Shouval D. Human monoclonal antibodies specific to hepatitis B virus generated in a human/mouse radiation chimera: the Trimera system. Immunology. 1998;93:154–161. doi: 10.1046/j.1365-2567.1998.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eren R., Ilan E., Nussbaum O., Lubin I., Terkieltaub D., Arazi Y., Ben-Moshe O., Kitchinzky A., Berr S., Gopher J. Preclinical evaluation of two human anti-hepatitis B virus (HBV) monoclonal antibodies in the HBV-Trimera mouse model and in HBV chronic carrier chimpanzees. Hepatology. 2000;32:588–596. doi: 10.1053/jhep.2000.9632. [DOI] [PubMed] [Google Scholar]
- 23.Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G., Gao X., Song Y.K., Vollmer R., Stolz D.B., Gasiorowski J.Z., Dean D.A., Liu D. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11:675–682. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang S.-Q., Du J., Yan H., Zhou Q.-Q., Zhou Y., Yuan Z.-N., Yan S.D., Fu Q.X., Wang X.H., Jia S.Z., Peng J.C. A mouse model for studying the clearance of hepatitis B virus in vivo using a luciferase reporter. PLoS ONE. 2013;8:e60005. doi: 10.1371/journal.pone.0060005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stabenow D., Frings M., Trück C., Gärtner K., Förster I., Kurts C., Tüting T., Odenthal M., Dienes H.P., Cederbrant K. Bioluminescence imaging allows measuring CD8 T cell function in the liver. Hepatology. 2010;51:1430–1437. doi: 10.1002/hep.23575. [DOI] [PubMed] [Google Scholar]
- 27.Rai U., Huang J., Mishra S., Li X., Shiratsuchi T., Tsuji M. A new method to determine antigen-specific CD8+ T cell activity in vivo by hydrodynamic injection. Biomolecules. 2012;2:23–33. doi: 10.3390/biom2010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang P.L., Althage A., Chung J., Chisari F.V. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan R.A., Johnson L.A., Davis J.L., Zheng Z., Woolard K.D., Reap E.A., Feldman S.A., Chinnasamy N., Kuan C.T., Song H. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum. Gene Ther. 2012;23:1043–1053. doi: 10.1089/hum.2012.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leo O., Foo M., Sachs D.H., Samelson L.E., Bluestone J.A. Identification of a monoclonal antibody specific for a murine T3 polypeptide. Proc. Natl. Acad. Sci. USA. 1987;84:1374–1378. doi: 10.1073/pnas.84.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jost C.R., Titus J.A., Kurucz I., Segal D.M. A single-chain bispecific Fv2 molecule produced in mammalian cells redirects lysis by activated CTL. Mol. Immunol. 1996;33:211–219. doi: 10.1016/0161-5890(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 32.Iwahori K., Kakarla S., Velasquez M.P., Yu F., Yi Z., Gerken C., Song X.T., Gottschalk S. Engager T cells: a new class of antigen-specific T cells that redirect bystander T cells. Mol. Ther. 2015;23:171–178. doi: 10.1038/mt.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudecek M., Sommermeyer D., Kosasih P.L., Silva-Benedict A., Liu L., Rader C., Jensen M.C., Riddell S.R. The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol. Res. 2015;3:125–135. doi: 10.1158/2326-6066.CIR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonamassa B., Hai L., Liu D. Hydrodynamic gene delivery and its applications in pharmaceutical research. Pharm. Res. 2011;28:694–701. doi: 10.1007/s11095-010-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi Z., Li G., Hu H., Yang C., Zhang X., Leng Q., Xie Y., Yu D., Zhang X., Gao Y. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J. Virol. 2014;88:8045–8056. doi: 10.1128/JVI.01024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo X., Chen P., Hou X., Xu W., Wang D., Wang T.-Y., Zhang L., Zheng G., Gao Z.L., He C.Y. The recombined cccDNA produced using minicircle technology mimicked HBV genome in structure and function closely. Sci. Rep. 2016;6:25552. doi: 10.1038/srep25552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinnooy Kan E.A., Wright S.D., Welte K., Wang C.Y. Fc receptors on monocytes cause OKT3-treated lymphocytes to internalize T3 and to secrete IL-2. Cell. Immunol. 1986;98:181–187. doi: 10.1016/0008-8749(86)90278-9. [DOI] [PubMed] [Google Scholar]
- 38.Kuo S.R., Wong L., Liu J.S. Engineering a CD123xCD3 bispecific scFv immunofusion for the treatment of leukemia and elimination of leukemia stem cells. Protein Eng. Des. Sel. 2012;25:561–569. doi: 10.1093/protein/gzs040. [DOI] [PubMed] [Google Scholar]
- 39.Compte M., Álvarez-Cienfuegos A., Nuñez-Prado N., Sainz-Pastor N., Blanco-Toribio A., Pescador N., Sanz L., Alvarez-Vallina L. Functional comparison of single-chain and two-chain anti-CD3-based bispecific antibodies in gene immunotherapy applications. OncoImmunology. 2014;3:e28810. doi: 10.4161/onci.28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann A.U., Phillips S., Levine I., Ijaz S., Dahari H., Eren R., Dagan S., Naoumov N.V. Novel mechanism of antibodies to hepatitis B virus in blocking viral particle release from cells. Hepatology. 2010;52:875–885. doi: 10.1002/hep.23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumortier J., Schönig K., Oberwinkler H., Löw R., Giese T., Bujard H., Schirmacher P., Protzer U. Liver-specific expression of interferon gamma following adenoviral gene transfer controls hepatitis B virus replication in mice. Gene Ther. 2005;12:668–677. doi: 10.1038/sj.gt.3302449. [DOI] [PubMed] [Google Scholar]
- 42.Shin E.-C., Protzer U., Untergasser A., Feinstone S.M., Rice C.M., Hasselschwert D., Rehermann B. Liver-directed gamma interferon gene delivery in chronic hepatitis C. J. Virol. 2005;79:13412–13420. doi: 10.1128/JVI.79.21.13412-13420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y., Cullen J.M., Aldrich C.E., Saputelli J., Miller D., Seeger C., Mason W.S., Jilbert A.R. Adenovirus-based gene therapy during clevudine treatment of woodchucks chronically infected with woodchuck hepatitis virus. Virology. 2004;327:26–40. doi: 10.1016/j.virol.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Sen M., Wankowski D.M., Garlie N.K., Siebenlist R.E., Van Epps D., LeFever A.V., Lum L.G. Use of anti-CD3 x anti-HER2/neu bispecific antibody for redirecting cytotoxicity of activated T cells toward HER2/neu+ tumors. J. Hematother. Stem Cell Res. 2001;10:247–260. doi: 10.1089/15258160151134944. [DOI] [PubMed] [Google Scholar]
- 45.Ji C., Sastry K.S.R., Tiefenthaler G., Cano J., Tang T., Ho Z.Z., Teoh D., Bohini S., Chen A., Sankuratri S. Targeted delivery of interferon-α to hepatitis B virus-infected cells using T-cell receptor-like antibodies. Hepatology. 2012;56:2027–2038. doi: 10.1002/hep.25875. [DOI] [PubMed] [Google Scholar]
- 46.Berraondo P., Di Scala M., Korolowicz K., Thampi L.M., Otano I., Suarez L., Fioravanti J., Aranda F., Ardaiz N., Yang J. Liver-directed gene therapy of chronic hepadnavirus infection using interferon alpha tethered to apolipoprotein A-I. J. Hepatol. 2015;63:329–336. doi: 10.1016/j.jhep.2015.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park S.S., Ryu C.J., Kang Y.J., Kashmiri S.V., Hong H.J. Generation and characterization of a novel tetravalent bispecific antibody that binds to hepatitis B virus surface antigens. Mol. Immunol. 2000;37:1123–1130. doi: 10.1016/s0161-5890(01)00027-x. [DOI] [PubMed] [Google Scholar]
- 48.Tan W., Meng Y., Li H., Chen Y., Han S., Zeng J., Huang A., Li B., Zhang Y., Guo Y. A bispecific antibody against two different epitopes on hepatitis B surface antigen has potent hepatitis B virus neutralizing activity. MAbs. 2013;5:946–955. doi: 10.4161/mabs.26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y., Tang Z., Liu K., Ye S., Li J., Huang Z., Wang D., Segal D. Preparation and application of anti-HBx/anti-CD3 bispecific monoclonal antibody (BsAb) retargeting effector cells for lysis of human hepatoma xenografts in nude mice. Oncol. Rep. 1996;3:637–644. doi: 10.3892/or.3.4.637. [DOI] [PubMed] [Google Scholar]
- 50.Sebestyén M.G., Budker V.G., Budker T., Subbotin V.M., Zhang G., Monahan S.D., Lewis D.L., Wong S.C., Hagstrom J.E., Wolff J.A. Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. Hepatocyte uptake of various molecules. J. Gene Med. 2006;8:852–873. doi: 10.1002/jgm.921. [DOI] [PubMed] [Google Scholar]
- 51.Nathwani A.C., Tuddenham E.G.D., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dion S., Bourgine M., Godon O., Levillayer F., Michel M.-L. Adeno-associated virus-mediated gene transfer leads to persistent hepatitis B virus replication in mice expressing HLA-A2 and HLA-DR1 molecules. J. Virol. 2013;87:5554–5563. doi: 10.1128/JVI.03134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang L.R., Gäbel Y.A., Graf S., Arzberger S., Kurts C., Heikenwalder M., Knolle P.A., Protzer U. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology. 2012;142:1447–1450. doi: 10.1053/j.gastro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 56.Billioud G., Kruse R.L., Carrillo M., Whitten-Bauer C., Gao D., Kim A., Chen L., McCaleb M.L., Crosby J.R., Hamatake R. In vivo reduction of hepatitis B virus antigenemia and viremia by antisense oligonucleotides. J. Hepatol. 2016;64:781–789. doi: 10.1016/j.jhep.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 57.Locarnini S., Bowden S. Hepatitis B surface antigen quantification: not what it seems on the surface. Hepatology. 2012;56:411–414. doi: 10.1002/hep.25732. [DOI] [PubMed] [Google Scholar]
- 58.Kovacsics D., Raper J. Transient expression of proteins by hydrodynamic gene delivery in mice. J. Vis. Exp. 2014;87:51481. doi: 10.3791/51481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.