Abstract
Long noncoding RNAs (lncRNAs) are emerging as important regulators during tumorigenesis by serving as competing endogenous RNAs (ceRNAs). In this study, the qRT-PCR results indicated that the lncRNA protein disulfide isomerase family A member 3 pseudogene 1 (PDIA3P) was overexpressed in oral squamous cell carcinoma (OSCC) and decreased the survival rate of OSCC patients. CCK-8 and clonal colony formation assays were used to detect the effects of PDIA3P on proliferation. Results revealed that silencing PDIA3P by small interfering RNA (siRNA) inhibited OSCC cell proliferation and repressed tumor growth and reduced the expression of proliferation antigen Ki-67 in vivo. Furthermore, the interaction between PDIA3P and miRNAs was then analyzed by qRT-PCR and luciferase reporter gene assay. We found that PDIA3P negatively regulated miR-185-5p in OSCC cells. Simultaneously, we found that silencing PDIA3P by siRNA suppressed proliferation via miR-185-5p in OSCC cells. Moreover, silencing PDIA3P by siRNA inhibited CCND2 protein (no influence on mRNA levels) expression via miR-185-5p in OSCC cells, and CCND2 facilitated cell proliferation of SCC4 and SCC15 cells induced by sh-PDIA3P#1. Therefore, our study demonstrated that PDIA3P may be a therapeutic target for the treatment of OSCC.
Keywords: protein disulfide isomerase family A member 3 pseudogene 1, PDIA3P, miR-185-5p, cyclin D2, oral squamous cell carcinoma, OSCC, tumorigenesis
Introduction
Oral squamous cell carcinoma (OSCC), an epithelial neoplasm found in 80%–90% of head and neck cancer,1, 2 is among the ten most frequent human malignancies and mostly arises in the context of alcohol abuse, smoking, and human papillomavirus (HPV) infection.3 OSCC lesions are histopathologically characterized by the presence of a different degree of keratin production, squamous differentiation, mitotic activity, invasive growth and metastasis, and nuclear pleomorphisms. No promising progress in the treatment of OSCC has been made over the past decades.4, 5 Although several advances have been obtained in its treatment, OSCC prognosis still remains poor, with a 5-year survival ratio of nearly 50%.1 It has been reported that the tumorigenesis and progression for OSCC are an intricate process involved in multiple genetic and epigenetic alterations.6, 7 Therefore, elucidation of the molecular mechanisms referred in the initiation and oncogenesis of OSCC is of great importance to improve treatment outcome.
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nt in length, which are involved in multiple biological functions, such as gene transcription regulation,8, 9 RNA processing,10 modulation of apoptosis and invasion,11 miRNA host genes,12 chromatin modification,13 and marker of cell fate,14 etc. Dysregulation of lncRNAs has occurred in multiple human diseases, including neurological disorders15 and cancers. Additionally, lncRNAs can not only function as a decoy for splicing factors or as antisense transcripts resulting in splicing malfunctioning,16, 17 but also serve as competing endogenous RNAs (ceRNAs) in several tumors.18, 19, 20 Several studies have implied that lncRNAs, including HOTAIR21 and GTF2IRD2P1,22 are related to OSCC tumorigenesis. Nevertheless, the detailed clinical significance and molecular mechanisms of lncRNAs in OSCC neoplasia and development still remain largely unclear.
Protein disulfide isomerase family A member 3 pseudogene 1 (PDIA3P1, NR_002305) is a 2099-bp lncRNA that maps to chromosome 1q21.1 (https://www.ncbi.nlm.nih.gov/gene/?term=PDIA3P). Recently, Zhang et al.22 have reported that PDIA3P is overexpressed in OSCC patients, whereas until now, the relationship between PDIA3P expression and OSCC oncogenesis has not been explored yet. Thus, the functions and underlying molecular mechanisms for PDIA3P in OSCC still need to be further elaborated.
In the present study, we demonstrated that PDIA3P was upregulated in OSCC tissues. PDIA3P upregulated the miR-185-5p target gene CCND2 by competitively spongeing miR-185-5p and then promoted OSCC cell proliferation. PDIA3P may function as a part of the ceRNA network.
Results
PDIA3P Is Overexpressed in OSCCs and Suppresses the Survival Rate of OSCC Patients
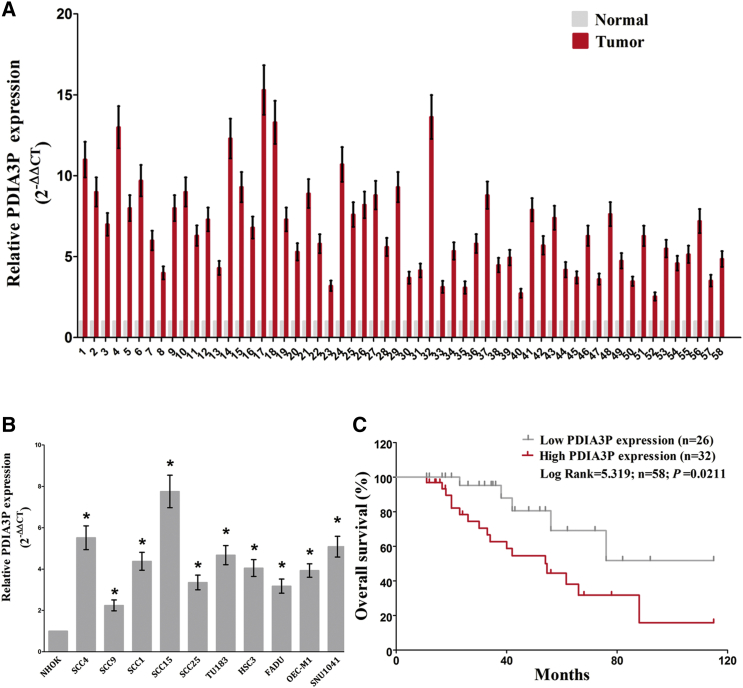
We randomly collected OSCC tissues and paired noncancerous tissues from 58 patients. The RNA expression level of PDIA3P was measured by qRT-PCR and found to be increased in OSCC tissues compared with corresponding noncancerous tissues (Figure 1A). Moreover, we found that the RNA expression level of PDIA3P was upregulated in OSCC cell lines (SCC4, SCC9, SCC1, SCC25, TU183, HSU3, FADU, OEC-M1, SNU1041, and SCC15) compared with a normal human oral keratinocytes cell line NHOK (p < 0.05), and PDIA3P expression was higher in SCC4 and SCC15 cells than in other OSCC cells (Figure 1B). Hence, the following experiments were performed using SCC4 and SCC15 cells. Results also indicated that PDIA3P reduced the survival rate of OSCC patients (p < 0.05) (Figure 1C). These results demonstrated that higher PDIA3P expression was related to poorer prognosis, and overexpression of PDIA3P might be crucial for OSCC progression.
Figure 1.
Relative PDIA3P Expression in OSCC Tissues and Cell Lines and Its Clinical Significance
(A) Relative expression of PDIA3P expression in OSCC tissues (n = 58) and in paired adjacent normal tissues (n = 58). PDIA3P expression was examined by qPCR and normalized to GAPDH expression (shown as 2−ΔΔCT). (B) Relative expression of PDIA3P expression in OSCC cell lines and normal lung epidermal cell. (C) The Kaplan-Meier survival analysis indicated that PDIA3P high expression (red line, n = 32) has a worse overall survival compared to the low expression subgroup (gray line, n = 26). *p < 0.05. Mean ± SEM is shown. Statistical analysis was conducted using Student’s t test.
PDIA3P Promotes OSCC Cell Growth In Vitro
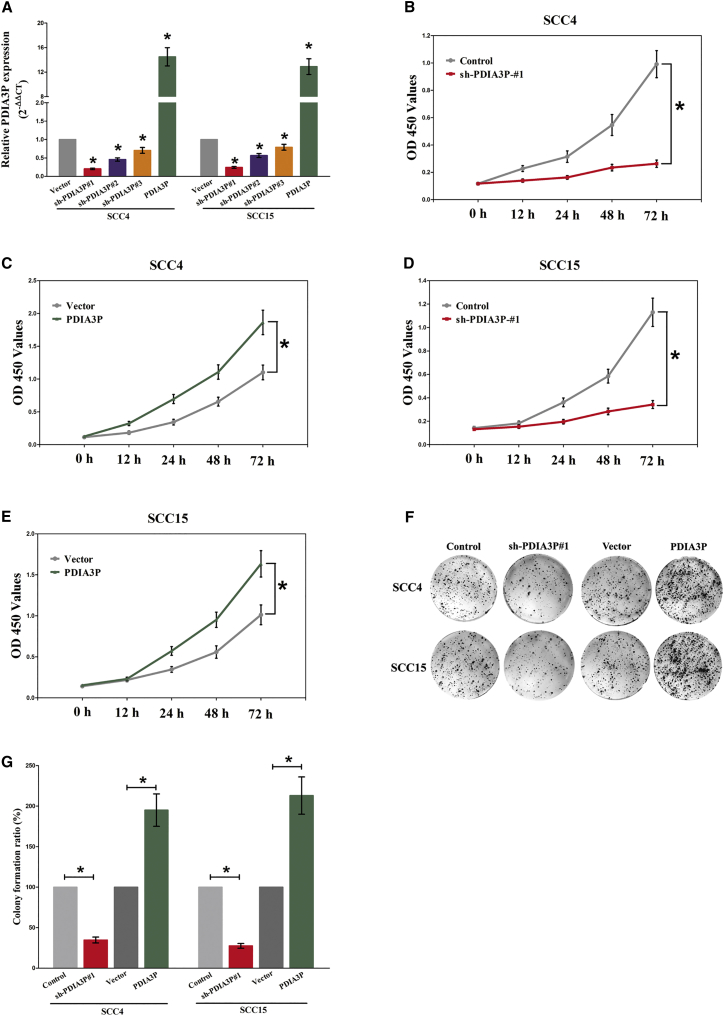
We also studied the impact of PDIA3P on OSCC cell proliferation. To this end, SCC4 and SCC15 cells were transfected with control short hairpin RNA (shRNA) or shRNAs against PDIA3P, i.e., sh-PDIA3P#1, sh-PDIA3P#2, and sh-PDIA3P#3, and pcDNA3.1-CT-GFP-PDIA3P. The qRT-PCR results indicated that sh-PDIA3P#1 effectively knocked down PDIA3P (Figure 2A). Thus, SCC4 and SCC15 cells were transfected with control or sh-PDIA3P#1 or control or pcDNA3.1-CT-GFP-PDIA3P. The CCK-8 results showed that silencing PDIA3P by RNAi inhibited SCC4 and SCC15 cell proliferation (p < 0.05) (Figures 2B–2E). Moreover, the clonal colony forming assay results also showed that silencing PDIA3P by RNAi inhibited SCC4 and SCC15 cell proliferation (p < 0.05) (Figures 2F and 2G). However, overexpressed PDIA3P facilitated cell growth of SCC4 and SCC15 cells (Figures 2B–2G), which clearly revealed that PDIA3P markedly promoted cell growth in OSCC cells.
Figure 2.
PDIA3P Promotes Tumor OSCC Cell Growth In Vitro
(A) Relative PDIA3P expression after transfection with control, sh-PDIA3P#1, sh-PDIA3P#2, sh-PDIA3P#2, or pcDNA3.1-CT-GFP-PDIA3P. (B–E) CCK-8 assays of SCC4 (B and C) and SCC15 (D and E) cells after transfection. (F) Shown are representative photomicrographs of colony formation assay after transfection for 14 days. (G) Statistics of colony formation assay after transfection for 14 days. Assays were performed in triplicate. *p < 0.05. Mean ± SEM is shown. Statistical analysis was conducted using Student’s t test (CCK-8) and one-way ANOVA test (colony formation assay).
PDIA3P Knockdown Suppresses OSCC Cell Growth In Vivo
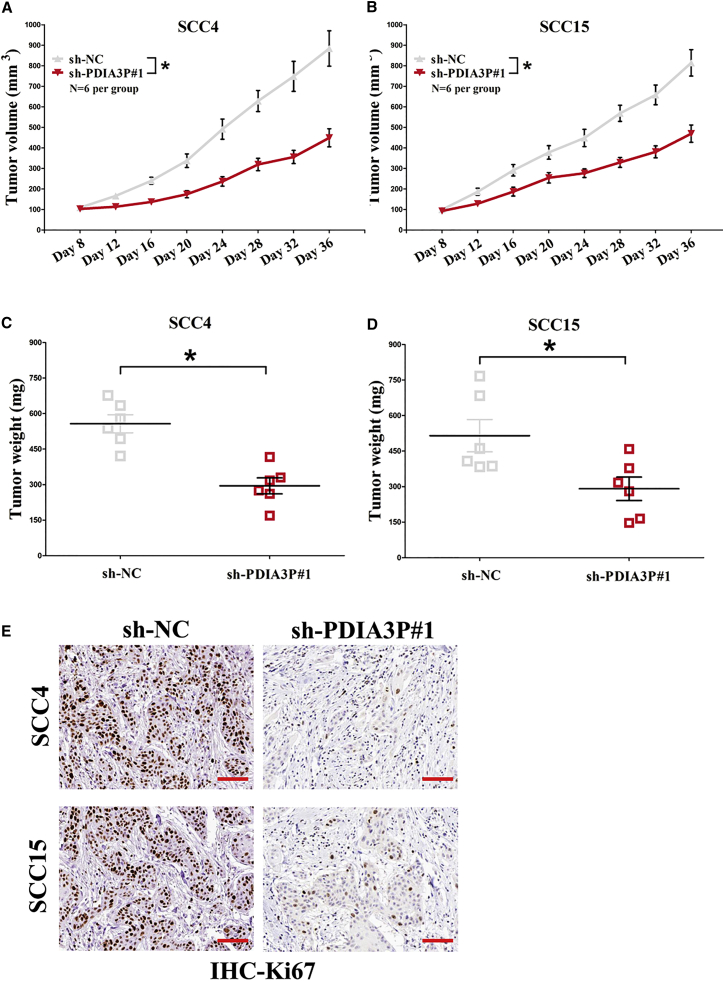
To further explore whether silencing PDIA3P could be involved in affecting tumor growth in vivo, SCC4 and SCC15 cells transfected with sh-NC or sh-PDIA3P#1 were inoculated into male nude mice. Results demonstrated that the tumor size in the sh-PDIA3P#1 group was smaller compared with the sh-NC group 36 days after injection (Figures 3A and 3B). The tumor weight of the sh-PDIA3P#1 group was also significantly less than that of the sh-NC group (Figures 3C and 3D). Additionally, immunohistochemistry (IHC) analysis identified that the tumors isolated from SCC4/sh-PDIA3P#1 and SCC15/sh-PDIA3P#1 cells revealed fewer Ki-67-staining areas than those from control cells (Figure 3E). Hence, we confirmed that silencing PDIA3P could repress tumor growth in vivo.
Figure 3.
PDIA3P Knockdown Suppresses NPC Cell Growth In Vivo
(A and B) Tumor volume in nude mice: SSC4 (A) and SCC15 (B). (C and D) Tumor weight in nude mice: SSC4 (C) and SCC15 (D). Each group contained six mice (n = 6). (E) Representative images of Ki-67 staining. Scale bar, 100 μm. Assays were performed in triplicate. *p < 0.05. Mean ± SEM is shown. Statistical analysis was conducted using Student’s t test.
PDIA3P Functions as a ceRNA for miR-185-5p in OSCC
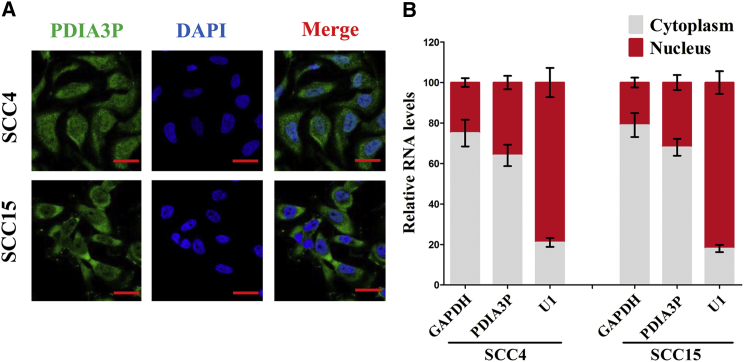
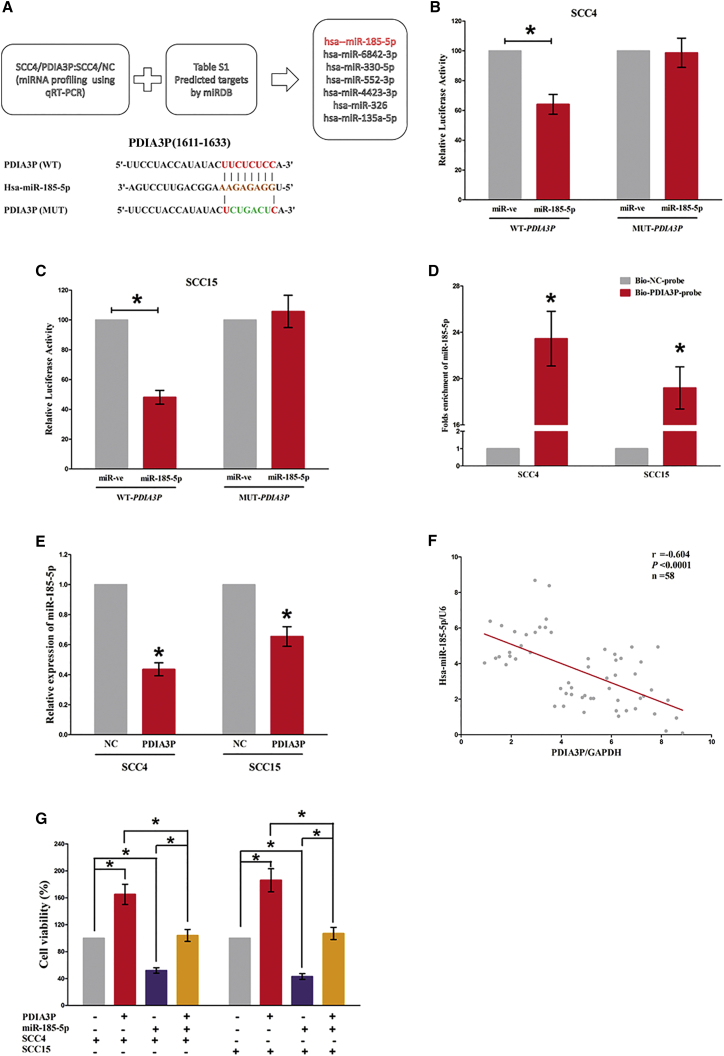
To explore the molecular mechanisms of PDIA3P involved in OSCC cells, we first analyzed the distribution of PDIA3P in SCC4 and SCC15 cells. We found that PDIA3P is distributed in both the cytoplasm and nucleus, but the ratio of PDIA3P in the cytoplasm is higher than that in the nucleus (Figures 4A and 4B). To investigate the interaction between PDIA3P and miRNAs, we predicted the miRNAs that may interact with PDIA3P using miRDB (Table S1). SCC4 cells were then transfected with control pcDNA3.1-CT-GFP or pcDNA3.1-CT-GFP-PDIA3P. Furthermore, qRT-PCR was used to screen differentially expressed miRNAs associated with PDIA3P (Figure S1). We selected 7 miRNAs: miR-185-5p, miR-6842-3p, miR-330-5p, miR-552-3p, miR-4423-3p, miR-326, and miR-135a-5p (Figure 5A). Among these seven miRNAs, miR-185-5p reduced the most; thus, it was chosen for further study. Moreover, we also performed dual-luciferase reporter assay to further verify if PDIA3P was a functional target of miR-185-5p. Our results demonstrated miR-185-5p may reduce the luciferase activity of pmirGLO-PDIA3P-WT, whereas it may not affect the luciferase activity of pmirGLO-PDIA3P-MUT (Figures 5B and 5C). Moreover, to find out whether PDIA3P could pull down miR-185-5p, we applied a biotin-avidin pull-down assay using a biotin-labeled-specific PDIA3P probe, and results indicated miR-185-5p was pulled down by PDIA3P (Figure 5D). Additionally, PDIA3P overexpression suppressed miR-185-5p expression both in SCC4 and SCC15 cells (Figure 5E). Furthermore, we assessed the association between PDIA3P RNA and miR-185-5p expression in 58 OSCC tissues as well, and results indicated the expression of PDIA3P RNA and miR-185-5p showed a remarkably negative correlation, as analyzed by Pearson correlation analysis (r = −0.604, p < 0.0001) (Figure 5F). These results revealed miR-185-5p could directly bind to PDIA3P at the miRNA recognition site.
Figure 4.
PDIA3P Locates Mainly in the Cytoplasm of OSCC Cells
(A) Fluorescence in situ hybridization (FISH) was used to detect the location of PDIA3P in OSCC cells. (B) Relative PDIA3P levels in SCC4 and SCC15 cell cytoplasm or nucleus were detected by qRT-PCR. GAPDH was used as cytoplasm control and U1 was used as nuclear control. Assays were performed in triplicate. *p < 0.05. Mean ± SEM is shown. Statistical analysis was conducted using Student’s t test.
Figure 5.
PDIA3P Is a Direct Target of miR-185-5p
(A) Up-screen of the candidate miRNAs that target PDIA3P predicted by miRDB and qPCR verification down-sequence alignment of miR-185-5p with the putative binding sites within the wild-type or mutant regions of PDIA3P. (B and C) The luciferase report assay demonstrated that overexpression of miR-185-5p could reduce the intensity of fluorescence in SCC4 (B) and SCC15 (C) cells transfected with the PDIA3P-WT vector, whereas it had no effect on the PDIA3P-MUT vector. (D) Detection of targeted miRNAs using qRT-PCR in the sample pulled down by biotinylated PDIA3P probe. (E) Detection of miR-185-5p using qRT-PCR in the PDIA3P overexpression SCC4 and SCC15 cell lines compared with the control group. (F) The correlation between PDIA3P mRNA and miR-185-5p expression in 58 OSCC tissues. (G) Upregulated miR-185-5p in SCC4 and SCC15 cells, which stably overexpressed PDIA3P, largely reversed the favorable effects of PDIA3P on cell proliferation. Assays were performed in triplicate. *p < 0.05. Mean ± SEM is shown. Statistical analysis was conducted using Student’s t test.
In addition, we also conducted Trypan blue staining assay to explore the interaction between miR-185-5p and PDIA3P on OSCC cell growth, and results demonstrated miR-185-5p suppressed cell growth both in SCC4 and SCC15 cells, whereas with co-transfection of miR-185-5p and pcDNA3.1-CT-GFP-PDIA3P, the growth-inhibitory role of miR-185-5p was reversed but the growth-expedited role of PDIA3P was also hampered (Figure 5G). These data demonstrated that PDIA3P facilitated cell growth via functioning as a ceRNA for miR-185-5p in OSCC cell lines.
PDIA3P Facilitates Cell Proliferation Partially via Spongeing miR-185-5p and Then Activating Cyclin D2
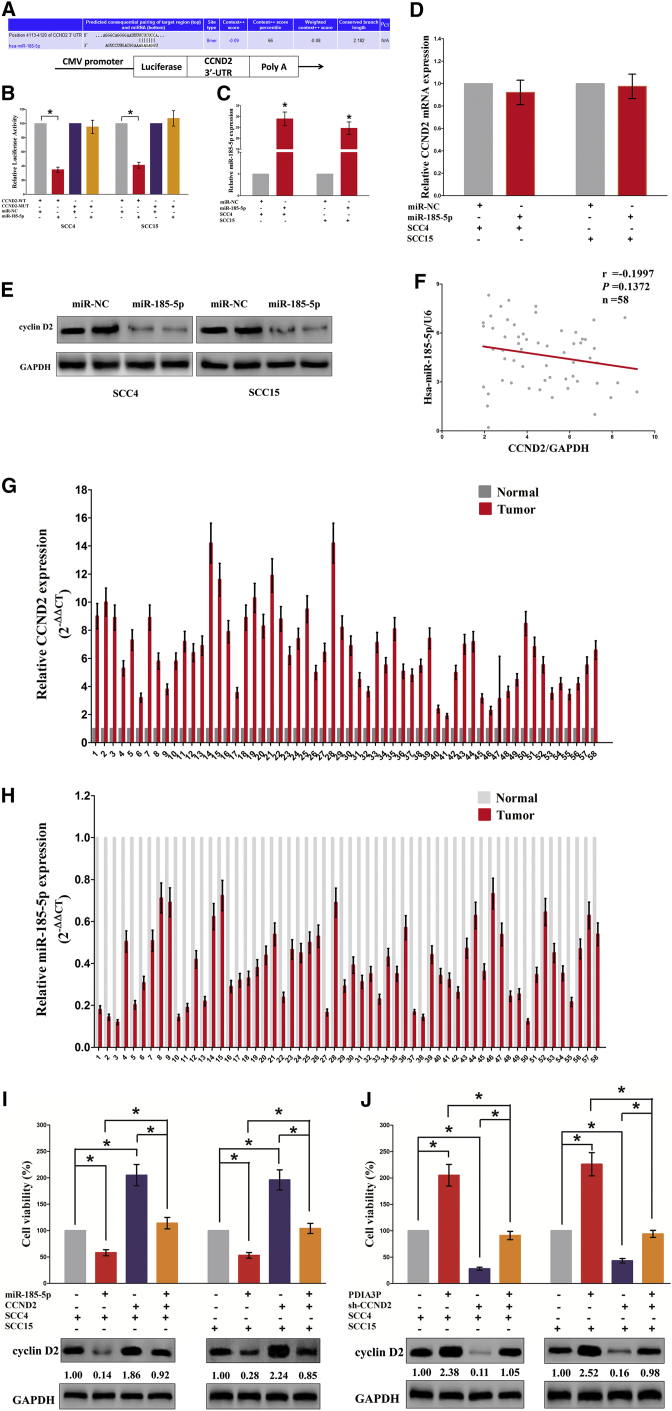
To study the role of miR-185-5p in the mechanisms and functions of PDIA3P, we analyzed direct targets of miR-185-5p using TargetScan and found that cyclin D2 (CCND2) may be related to miR-185-5p (Figure 6A). Then, we performed luciferase reporter assays to validate whether CCND2 expression was really influenced by miR-185-5p, and results revealed miR-185-5p reduced luciferase activity in both SCC4 cells and SCC15 cells that transfected with a wild-type (WT) CCND2 3′ UTR reporter plasmid, but no significant inhibition was observed in that transfected with a mutant (MUT) CCND2 3′ UTR reporter plasmid (Figure 6B). We also found that transfection with miR-185-5p significantly upregulated the miR-185-5p expression in SCC4 and SCC15 cells (Figure 6C). Moreover, miR-185-5p decreased the protein expression but had no influence on the mRNA expression for CCND2 in SCC4 and SCC15 cells (Figures 6D and 6E). Additionally, our results also demonstrated the mRNA of CCND2 expression had no significant correction with the expression of miR-185-5p in OSCC samples (r = −0.1997, p = 0.1372) (Figure 6F). Further, our results also demonstrated that CCND2 was highly expressed, whereas miR-185-5p was lowly expressed in OSCC tissues (Figures 6G and 6H). We then explored the role and mechanism of miR-185-5p on OSCC cell growth. Results of Trypan blue staining revealed miR-185-5p treatment suppressed cell growth, and pcDNA3.1-CT-GFP-CCND2 (with a full 3′ UTR mRNA for CCND2) treatment facilitated cell growth in SCC4 and SCC15 cells (Figures 6I and 6J). However, when SCC4 and SCC15 cells were treated with miR-185-5p plus pcDNA3.1-CT-GFP-CCND2, the advantageous role of CCND2 on cell growth was reversed by miR-185-5p, and the growth-inhibitory effect of miR-185-5p was inversed by CCND2 overexpression (Figures 6I and 6J). These data demonstrated that miR-185-5p suppressed cell growth by directly targeting 3′ UTR of CCND2 mRNA. Additionally, pcDNA3.1-CT-GFP-PDIA3P treatment reversed the growth inhibitory effect of sh-CCND2 in SCC4 and SCC15 cells (Figures 6I and 6J), which demonstrated that PDIA3P facilitated cell growth partially through upregulation of CCND2. Furthermore, we next explored the role of PDIA3P and miR-185-5p on the protein expression of CCND2. Results demonstrated that both miR-185-5p and sh-CCND2 treatment suppressed protein expression of CCND2, whereas both pcDNA3.1-CT-GFP-PDIA3P and pcDNA3.1-CT-GFP-CCND2 treatment significantly enhanced protein expression of CCND2 in SCC4 and SCC15 cells (Figures 6I and 6J), respectively. However, when SCC4 and SCC15 cells were treated with pcDNA3.1-CT-GFP-PDIA3P plus sh-CCND2, the beneficial role of PDIA3P on protein expression of CCND2 was suppressed by knockdown of CCND2, and the negative effect of sh-CCND2 was alleviated by overexpression of PDIA3P (Figures 6I and 6J). These findings suggest that the oncogenic roles of PDIA3P are at least partially mediated by miR-185-5p-CCND2 axis in OSCC.
Figure 6.
PDIA3P’s Oncogenic Activity Is in Part through Negative Regulation of miRNA-185-5p and Then Activation of Cyclin D2 in OSCC Cells
(A) The 3′ UTR of CCND2 harbors one miR-185-5p cognate site. (B) Relative luciferase activity of reporter plasmids carrying wild-type or mutant CCND2 3′ UTR in SCC4 and SCC15 cells co-transfected with miR-NC or miR-185-5p. (C) Relative miR-185-5p expression after transfection with miR-NC and miR-185-5p. (D) Relative CCND2 mRNA expression after transfection with miR-NC and miR-185-5p. (E) Relative cyclin D2 protein expression after transfection with miR-NC and miR-185-5p. (F) The relationship between CCND2 mRNA expression and miR-185-5p expression in OSCC tissues. (G and H) mRNA expression of CCND2 (G) and miR-185-5p (H) in OSCC tissues. (I and J) Up-statistical analysis of Trypan blue staining: SSC4 (I) and SCC15 (J). Down-protein expression of cyclin D2 in vector, miR-185-5p, pcDNA3.1-CT-GFP-CCND2, miR-185-5p plus pcDNA3.1-CT-GFP-CCND2, pcDNA3.1-CT-GFP-PDIA3P, sh-CCND2, or pcDNA3.1-CT-GFP-PDIA3P plus sh-CCND2-treated SCC4 and SCC15 cells. Assays were performed in triplicate. *p < 0.05. Mean ± SEM is shown. Statistical analysis was conducted using the one-way ANOVA test.
Discussion
lncRNAs can regulate protein-coding genes, transcription, and post-transcription, and play crucial roles in biological processes.9, 18, 23, 24 Many studies have shown that lncRNAs acted as oncogenes or tumor-suppressive genes to affect tumorigenesis, metastasis, prognosis, or diagnosis.10, 19, 20 Until now, the effects and mechanisms of lncRNAs on OSCC tumorigenesis and progression are still largely unclear. In the current study, we discovered that the lncRNA PDIA3P expression was higher in OSCC tissues than in corresponding noncancerous tissues. Concurrently, PDIA3P was associated with reduced survival in OSCC patients. We also proved that PDIA3P facilitated cell proliferation in OSCC cells. We also discovered that PDIA3P was a direct target for miR-185-5p. PDIA3P exerted its oncogenic role on OSCC in large part due to its effects to act as a ceRNA for miR-185-5p and subsequently activated the CCND2 signaling pathway. Therefore, we speculated that PDIA3P may play a vital role in the development and progression of OSCC.
To our knowledge, our present work is the first one to directly investigate the association between PDIA3P expression and OSCC. Herein, we found PDIA3P was markedly higher expressed in OSCC tissues than that of their counterparts. We also found that higher expressed PDIA3P corrected with poorer OSCC prognosis, as confirmed by Kaplan-Meier analysis, suggesting that PDIA3P levels might be used as a prognostic biomarker to assist in distinguishing patients with higher risks of OSCC progression. In addition, overexpressed PDIA3P significantly promoted OSCC cell growth in vitro, whereas silencing PDIA3P repressed it. Collectively, our data imply that PDIA3P may function as an oncogene and be favorable to OSCC tumorigenesis and progression.
An increasing number of studies have shown that miRNAs played important roles in regulating biological processes, including cell proliferation, metastasis, apoptosis, angiogenesis, and inflammation, in a variety of tumors.18, 25, 26, 27, 28, 29, 30 Specifically, many studies have indicated that various miRNAs, such as miR-455-5p,25 miR-211,31 miR-375,26 miR-19a/b,27 miR-204,32 and miR-187,33 were related to the development of OSCC. lncRNAs can function as ceRNA sponges for miRNAs to regulate the degradation of miRNA targets.18, 19, 20 A handle of lncRNAs have been reported, such as GAS5,28, 34 NEAT1 [1520906528], XIST,29 and CCAT1.20 In our study, we selected 7 miRNAs associated with PDIA3P, namely, miR-185-5p, miR-6842-3p, miR-330-5p, miR-552-3p, miR-4423-3p, miR-326, and miR-135a-5p, using bioinformatic prediction and qRT-PCR verification. We further validated the RNA expression levels of miRNAs and found that miR-185-5p was lowly expressed in OSCC tissues. We then found that PDIA3P negatively regulated miR-185-5p in OSCC cells, and PDIA3P facilitated cell proliferation via miR-185-5p in OSCC cells.
Having a revelation of the essential effects of miR-185-5p on the suppression OSCC progression, we searched for the potential mechanisms that participated in its function. It has been reported that miR-185 is a novel tumor suppressor that negatively modulates the Wnt/β-catenin pathway in human colorectal cancer.30 miR-185-5p regulates cell function Kruppel-like factor 2 (KLF2) in atherosclerosis-related vascular dysfunction.35 Zhu et al.36 also found that miR-185 inhibited cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Six2. But among all of the predicted target genes for miR-185-5p, we discovered CCND2 functioned as a critical effector of miR-185-5p. CCND2 plays an essential role in regulating progression from the G1-phase to the S-phase by forming complexes in the cytoplasm that will enter the nucleus and destroy cell-cycle suppressive retinoblastoma protein.37 A previous study has indicated that overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/CCND2.37 We confirmed that CCND2 was highly expressed in OSCC tissues, and overexpressed CCND2 facilitated cell proliferation in OSCC cell lines, whereas silencing CCND2 expression suppressed it. Moreover, we predicted CCND2 as a direct target of miR-185-5p at its 3′ UTR mRNA by bioinformatics, which was confirmed by luciferase reporter assays. Our results also demonstrated that miR-185-5p regulated CCND2 at the post-transcription level. Additionally, our data also revealed miR-185-5p could function as a tumor suppressor on OSCC by directly targeting CCND2. Furthermore, our data also revealed that PDIA3P’s oncogenic roles are in part through spongeing miR-185-5p and then activating CCND2.
In summary, our results indicated that lncRNA PDIA3P was overexpressed in OSCC and decreased the survival rate of OSCC patients. Moreover, PDIA3P facilitated cell proliferation in OSCC cells via miR-185-5p. A qRT-PCR was then used to screen differentially expressed miRNAs associated with PDIA3P. Specifically, PDIA3P negatively regulated miR-185-5p in OSCC cells, and silencing PDIA3P by shRNA suppressed CCND2 protein expression via miR-185-5p in OSCC cells. Our study demonstrated that PDIA3P may be a therapeutic target for the treatment of OSCC.
Materials and Methods
Ethical Statement and Tissue Collection
58 patients diagnosed as OSCC were involved in this study. All histologic diagnoses were performed by the pathology department in the Department of Oncology, Wuhan Pu-Ai Hospital, Tongji Medical College, Huazhong University of Science and Technology. Normal oral mucosal tissues were obtained from non-tumor adjacent tissues or patients with reshaping of gingival tissues. Informed consent was obtained from all subjects. All experimental protocols were approved by the Ethics Committee in Wuhan University and Huazhong University of Science and Technology. None of these patients received any pre-operative chemotherapy or radiotherapy.
Cell Lines and Plasmids
Ten OSCC cell lines (SCC4, SCC9, SCC1, SCC25, TU183, HSU3, FADU, OEC-M1, SNU1041, and SCC15) and the NHOK cell lines were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were routinely cultured as previously described.38 All cells were cultured at 37°C in a 5% CO2 atmosphere and maintained in 10% fetal bovine serum (FBS) (Kibbutz BeitHaemek, Israel) within 3 months of resuscitation from the frozen aliquots, with lower than 20 passages for each experiment. Plasmid pcDNA3.1-CT-GFP-CCND2 (NM_001759, with full length of 3′ UTR) and pcDNA3.1-CT-GFP-PDIA3P were prepared by us. RNAi sequences are shown in the Supplemental Information.
Cell Transfection and Stable Cell Lines
Cells were transfected with DNA plasmids using TransFast transfection reagent Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.39 For screening stable cell lines, 48 hr after transfection, cells were plated in the selective medium containing G418 (1,000–2,000 μg/mL, Invitrogen, UK) for the next 4 weeks or so, and the selective media were replaced every 3 days.
Western Blot Analysis
Western blot was performed using the protocol described previously.40, 41, 42 Briefly, the logarithmically growing cells were washed twice with ice-cold PBS (Hyclone) and lysed in a RIPA lysis buffer. Cells lysates were centrifuged at 12,000 × g for 20 min at 4°C after sonication on ice, and the supernatant was separated. After being boiled for 5–10 min in the presence of 2-mercaptoethanol, samples containing cell proteins were separated on a 10% SDS-PAGE and transferred onto a nitrocellulose membranes, then blocked in 10% dry milk-TBST (20 mM Tris-HCl [pH 7.6], 127 mM NaCl, and 0.1% Tween 20) for 1 hr at 37°C. Following three washes in Tris-HCl, pH 7.5, with 0.1% Tween 20, the blots were incubated with 0.2 μg/mL of antibody (appropriate dilution) overnight at 4°C. Following three washes, membranes were then incubated with secondary antibody for 60 min at 37°C or 4°C overnight in TBST. Signals were visualized by enhanced chemiluminescence (ECL). The following primary antibodies were used: rabbit anti-CCND2 (Santa Cruz, CA, USA) and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz, CA, USA).
qRT-PCR
RNA isolation and qRT-PCR was performed as described previously.43, 44 Total RNA was extracted from cells using Trizol reagent (Sangon Biotech) according to the manufacturer’s instructions. For mRNAs and lncRNA quantification, RNA was reverse transcribed to cDNA using the PrimeScript RT reagent Kit with gDNA Eraser (Takara). Quantitative real-time PCR was performed using cDNA primers specific for mRNA or lncRNA. The gene GAPDH was used as an internal control. For miRNA quantification, reverse transcription was performed using the Mir-X miRNA First Strand Synthesis Kit (Takara). miRNA-specific 5′ primers and the mRQ 3′ primer was used during quantitative real-time PCR. The gene U6 was used as an internal control. Primer sequences are provided in Table S2. All the real-time PCR reactions were performed using Takara’s SYBR Premix Ex Taq II (Tli RNaseH Plus) in Applied Biosystems 7300 Fast Real-Time PCR System. The 2-ΔΔCT method was used for quantification, and fold change for target genes was normalized by internal control.
Colony Formation Assay
The treated SCC4 and SCC15 cells were cultured for 14 days, and the colonies were then fixed with methanol and dyed with Giemsa dye solution before counting the colony-forming units.
Luciferase Reporter Assays
Luciferase reporter assays were conducted as previously described.45, 46, 47, 48
CCK-8 Assay
Cell growth was measured using the cell proliferation reagent WST-8 (Roche Biochemicals, Mannheim, Germany). After plating cells in 96-well microtiter plates (Corning Costar, Corning, NY) at 1.0 × 103/well, 10 μL of CCK-8 was added to each well at the time of harvest, according to the manufacturer’s instructions. 1 hr after adding CCK-8, cellular viability was determined by measuring the absorbance of the converted dye at 450 nm.
Trypan Blue Staining
Cell viability was assessed using the Trypan blue (Lonza, Basel, Switzerland) exclusion method. The SCC4 and SCC15 cells were seeded in 24-well culture plates at a density of 3 × 105 cells per well, and the cells were then transfected with Vector, miR-185-5p, pcDNA3.1-CT-GFP-CCND2, miR-185-5p plus pcDNA3.1-CT-GFP-CCND2, pcDNA3.1-CT-GFP-PDIA3P, sh-PDIA3P, or pcDNA3.1-CT-GFP-PDIA3P plus sh-PDIA3P. Each cell suspension was mixed with an equal volume of 0.4% Trypan blue solution, and the living cells were quantified using a hemocytometer. The cells were also counted using a microscope. The data are representative of three independent experiments performed on different days.
RNA Pull-Down Assays
PDIA3P transcripts were transcribed using T7 RNA polymerase (Ambio life) in vitro, and then by using the RNeasy Plus Mini Kit (QIAGEN) and treating with DNase I (QIAGEN). Purified RNAs were biotin labeled with the Biotin RNA Labeling Mix (Ambio life). Positive control, negative control, and biotinylated RNAs were mixed and incubated with SCC4 and SCC15 cell lysates. Then, magnetic beads were added to each binding reaction and incubated at room temperature. Finally, the beads were washed, and the eluted proteins were detected by western blot analysis.
Statistical Analysis
The results were analyzed using GraphPad Prism (GraphPad, La Jolla, CA, USA) and the SPSS 23.0 software (SPSS, Chicago, IL, USA). The significance of differences was assessed using Student’s t test or a one-way ANOVA. All data are expressed as mean ± SEM; p < 0.05 indicates a significant difference.
Author Contributions
Participated in research design: C.S., G.L., S.L., L.Z., and D.L.; Conducted experiments: C.S., S.L., and G.L.; Contributed new reagents or analytic tools: C.S., S.L., G.L., F.G., T.B., L.Z., and Y.F.; Performed data analysis: C.S., S.L., and G.L.; Wrote or contributed to the writing of the manuscript: C.S., G.L., S.L., L.Z., and D.L.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81271943) to D.L., the plan for the Scientific and Technological Innovation Team of High-tech Industries of Wuhan Municipal Science and Technology Bureau (No. 2015070504020219) to D.L., the Fundamental Research Funds for the Central Universities (No. 2015305020202) to C.S., and National Postdoctoral Program for Innovative Talents (No. BX201700178) to C.S.
Footnotes
Supplemental Information includes one figure and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.08.015.
Contributor Information
Cheng-Cao Sun, Email: chengcaosun@whu.edu.cn.
Qing-Ming Wu, Email: wuhe9224@sina.com.
De-Jia Li, Email: lodjlwhu@sina.com.
Supplemental Information
References
- 1.Vojtechova Z., Sabol I., Salakova M., Turek L., Grega M., Smahelova J., Vencalek O., Lukesova E., Klozar J., Tachezy R. Analysis of the integration of human papillomaviruses in head and neck tumours in relation to patients’ prognosis. Int. J. Cancer. 2016;138:386–395. doi: 10.1002/ijc.29712. [DOI] [PubMed] [Google Scholar]
- 2.Thompson L. World Health Organization classification of tumours: pathology and genetics of head and neck tumours. Ear Nose Throat J. 2006;85:74. [PubMed] [Google Scholar]
- 3.Zaravinos A. An updated overview of HPV-associated head and neck carcinomas. Oncotarget. 2014;5:3956–3969. doi: 10.18632/oncotarget.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter K.D., Parkinson E.K., Harrison P.R. Profiling early head and neck cancer. Nat. Rev. Cancer. 2005;5:127–135. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- 5.Yalniz Z., Demokan S., Suoglu Y., Ulusan M., Dalay N. Simultaneous methylation profiling of tumor suppressor genes in head and neck cancer. DNA Cell Biol. 2011;30:17–24. doi: 10.1089/dna.2010.1090. [DOI] [PubMed] [Google Scholar]
- 6.Wu M.J., Jan C.I., Tsay Y.G., Yu Y.H., Huang C.Y., Lin S.C., Liu C.J., Chen Y.S., Lo J.F., Yu C.C. Elimination of head and neck cancer initiating cells through targeting glucose regulated protein78 signaling. Mol. Cancer. 2010;9:283. doi: 10.1186/1476-4598-9-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 8.Gómez-Navarro N., Jordán-Pla A., Estruch F., E Pérez-Ortín J. Defects in the NC2 repressor affect both canonical and non-coding RNA polymerase II transcription initiation in yeast. BMC Genomics. 2016;17:183. doi: 10.1186/s12864-016-2536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich J.A., Kugel J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 10.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaitan D., Dinger M.E., Mazar J., Crawford J., Smith M.A., Mattick J.S., Perera R.J. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 12.Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin. Cell Dev. Biol. 2011;22:343–350. doi: 10.1016/j.semcdb.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Ginger M.R., Shore A.N., Contreras A., Rijnkels M., Miller J., Gonzalez-Rimbau M.F., Rosen J.M. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. USA. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan Z., Zheng D., Qing H. Regulatory roles of long non-coding RNAs in the central nervous system and associated neurodegenerative diseases. Front. Cell. Neurosci. 2017;11:175. doi: 10.3389/fncel.2017.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltran M., Puig I., Peña C., García J.M., Alvarez A.B., Peña R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C., Li S., Zhang F., Xi Y., Wang L., Bi Y., Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W.M., Zhu X., Wang W.M., Lu Y.F., Hu B.G., Wang H., Liang W.C., Wang S.S., Ko C.H., Waye M.M. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J. Hepatol. 2015;63:886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Ma M.Z., Chu B.F., Zhang Y., Weng M.Z., Qin Y.Y., Gong W., Quan Z.W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y., Zhang L., Zhang L., Wang Y., Li H., Ren X., Wei F., Yu W., Liu T., Wang X. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int. J. Oncol. 2015;46:2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S., Tian L., Ma P., Sun Q., Zhang K., GuanchaoWang, Liu H., Xu B. Potential role of differentially expressed lncRNAs in the pathogenesis of oral squamous cell carcinoma. Arch. Oral Biol. 2015;60:1581–1587. doi: 10.1016/j.archoralbio.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Sun C.C., Li S.J., Li G., Hua R.X., Zhou X.H., Li D.J. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang J., Sun C.C., Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem. Biophys. Res. Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 25.Cheng C.M., Shiah S.G., Huang C.C., Hsiao J.R., Chang J.Y. Up-regulation of miR-455-5p by the TGF-β-SMAD signalling axis promotes the proliferation of oral squamous cancer cells by targeting UBE2B. J. Pathol. 2016;240:38–49. doi: 10.1002/path.4752. [DOI] [PubMed] [Google Scholar]
- 26.Harrandah A.M., Fitzpatrick S.G., Smith M.H., Wang D., Cohen D.M., Chan E.K. MicroRNA-375 as a biomarker for malignant transformation in oral lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016;122:743–752.e1. doi: 10.1016/j.oooo.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 27.Christopher A.F., Gupta M., Bansal P. Micronome revealed miR-19a/b as key regulator of SOCS3 during cancer related inflammation of oral squamous cell carcinoma. Gene. 2016;594:30–40. doi: 10.1016/j.gene.2016.08.044. [DOI] [PubMed] [Google Scholar]
- 28.Tani H., Torimura M., Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song P., Ye L.F., Zhang C., Peng T., Zhou X.H. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 30.Dong-Xu W., Jia L., Su-Juan Z. MicroRNA-185 is a novel tumor suppressor by negatively modulating the Wnt/β-catenin pathway in human colorectal cancer. Indian J. Cancer. 2015;52(Suppl 3):E182–E185. doi: 10.4103/0019-509X.186576. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y.F., Yang C.C., Kao S.Y., Liu C.J., Lin S.C., Chang K.W. MicroRNA-211 enhances the oncogenicity of carcinogen-induced oral carcinoma by repressing TCF12 and increasing antioxidant activity. Cancer Res. 2016;76:4872–4886. doi: 10.1158/0008-5472.CAN-15-1664. [DOI] [PubMed] [Google Scholar]
- 32.Yu C.C., Chen P.N., Peng C.Y., Yu C.H., Chou M.Y. Suppression of miR-204 enables oral squamous cell carcinomas to promote cancer stemness, EMT traits, and lymph node metastasis. Oncotarget. 2016;7:20180–20192. doi: 10.18632/oncotarget.7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin S.C., Kao S.Y., Chang J.C., Liu Y.C., Yu E.H., Tseng S.H., Liu C.J., Chang K.W. Up-regulation of miR-187 modulates the advances of oral carcinoma by targeting BARX2 tumor suppressor. Oncotarget. 2016;7:61355–61365. doi: 10.18632/oncotarget.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shan K., Jiang Q., Wang X.Q., Wang Y.N., Yang H., Yao M.D., Liu C., Li X.M., Yao J., Liu B. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016;7:e2248. doi: 10.1038/cddis.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu S.M., Chen C.M., Jiang Z.Y., Yuan B., Ji M., Wu F.H., Jin J. MicroRNA-185 inhibits cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Six2. Eur. Rev. Med. Pharmacol. Sci. 2016;20:1712–1719. [PubMed] [Google Scholar]
- 37.Zeng Q., Tao X., Huang F., Wu T., Wang J., Jiang X., Kuang Z., Cheng B. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int. J. Mol. Med. 2016;37:1274–1280. doi: 10.3892/ijmm.2016.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen Y.C., Shiah S.G., Chu H.C., Hsu Y.M., Hsiao J.R., Chang J.Y., Hung W.C., Liao C.T., Cheng A.J., Lu Y.C. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol. Cancer. 2014;13:6. doi: 10.1186/1476-4598-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gui X., Li H., Li T., Pu H., Lu D. Long noncoding RNA CUDR regulates HULC and β-catenin to govern human liver stem cell malignant Differentiation. Mol. Ther. 2015;23:1843–1853. doi: 10.1038/mt.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun C., Yang C., Xue R., Li S., Zhang T., Pan L., Ma X., Wang L., Li D. Sulforaphane alleviates muscular dystrophy in mdx mice by activation of Nrf2. J. Appl. Physiol. (1985) 2015;118:224–237. doi: 10.1152/japplphysiol.00744.2014. [DOI] [PubMed] [Google Scholar]
- 41.Sun C.C., Li S.J., Yang C.L., Xue R.L., Xi Y.Y., Wang L., Zhao Q.L., Li D.J. Sulforaphane attenuates muscle inflammation in dystrophin-deficient mdx mice via NF-E2-related factor 2 (Nrf2)-mediated inhibition of NF-κB signaling pathway. J. Biol. Chem. 2015;290:17784–17795. doi: 10.1074/jbc.M115.655019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Sun C., Li S., Li D. Sulforaphane mitigates muscle fibrosis in mdx mice via Nrf2-mediated inhibition of TGF-beta/Smad signaling. J. Appl. Physiol. (1985) 2016;120:377–390. doi: 10.1152/japplphysiol.00721.2015. [DOI] [PubMed] [Google Scholar]
- 43.Sun C., Liu Z., Li S., Yang C., Xue R., Xi Y., Wang L., Wang S., He Q., Huang J. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6:25533–25574. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun C., Sang M., Li S., Sun X., Yang C., Xi Y., Wang L., Zhang F., Bi Y., Fu Y. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6:39756–39792. doi: 10.18632/oncotarget.5476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Sun C.C., Li S.J., Li D.J. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7:35960–35978. doi: 10.18632/oncotarget.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C., Huang C., Li S., Yang C., Xi Y., Wang L., Zhang F., Fu Y., Li D. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7:8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun C.C., Li S.J., Zhang F., Pan J.Y., Wang L., Yang C.L., Xi Y.Y., Li J. Hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. Oncotarget. 2016;7:21510–21526. doi: 10.18632/oncotarget.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun C., Li S., Yang C., Xi Y., Wang L., Zhang F., Li D. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem. Biophys. Res. Commun. 2016;471:82–88. doi: 10.1016/j.bbrc.2016.01.175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.