Abstract
Water lilies are not only highly favored aquatic ornamental plants with cultural and economic importance but they also occupy a critical evolutionary space that is crucial for understanding the origin and early evolutionary trajectory of flowering plants. The birth and rapid radiation of flowering plants has interested many scientists and was considered ‘an abominable mystery’ by Charles Darwin. In searching for the angiosperm evolutionary origin and its underlying mechanisms, the genome of Amborella has shed some light on the molecular features of one of the basal angiosperm lineages; however, little is known regarding the genetics and genomics of another basal angiosperm lineage, namely, the water lily. In this study, we reviewed current molecular research and note that water lily research has entered the genomic era. We propose that the genome of the water lily is critical for studying the contentious relationship of basal angiosperms and Darwin’s ‘abominable mystery’. Four pantropical water lilies, especially the recently sequenced Nymphaea colorata, have characteristics such as small size, rapid growth rate and numerous seeds and can act as the best model for understanding the origin of angiosperms. The water lily genome is also valuable for revealing the genetics of ornamental traits and will largely accelerate the molecular breeding of water lilies.
Introduction
Ornamentals, cultural symbols and economic value
Water lilies are beautiful aquatic flowering plants that are distributed worldwide. These plants are found in the aquatic section of nearly every botanic garden because of their highly valued ornamental features. The almost full spectrum of petal colors range from black to white, making water lilies the most diversely colored flowering plants (Figure 1). The lovely cup-like flower shapes and floating leaves such as the famous Victoria and Amazon water lilies are also favored ornamental characteristics. In Bangladesh and Sri Lanka, water lilies were chosen as the national flower because they are regarded as a symbol of truth, purity, and discipline.
Figure 1.
Water lilies are ornamental plants with beautiful flowers and leaves: (a) Nymphaea ‘Hermine’, (b) N. ‘Marliacea Chromatella’, (c) N. ‘Wanvisa’, (d) N. ‘Gigantea Hybrid1’, (e) N. colorata, (f) N. ‘Muang Wiboonlak’, (g) N. ‘Piyalarp’, (h) N. ‘Agkee Sri Non’, (i) leaf ornamental Victoria water lily.
Beyond being beautiful ornamental plants, water lilies have been utilized as an ingredient in many products, including beneficial and cosmetic substances, soap, perfume, hand cream, flower tea bags, and traditional medicine.1 In Asian countries, some water lilies such as Brasenia schreberi, Euryale ferox, Nymphaea spp. are traditional vegetables with edible parts including young leaves, stems, and seeds. Several Nymphaea species have also been used to purify heavy metal-contaminated water and soap-polluted wastewater.2
Critical evolutionary place
In taxonomy, plants categorized in the Order Nymphaeales share the common name water lily.3 Water lilies are divided into three families: Hydatellaceae, Cabombaceae, and Nymphaeaceae.4 The family Nymphaeaceae has the most species of the three families and consists of six genera: Barclaya, Euryale, Nuphar, Nymphaea, Ondinea, and Victoria.4,5 Floral organs differ greatly among each family in the order Nymphaeales. In the genus Nymphaea, flowers are composed of 4 sepals, 50 to 70 petals, 30 to 40 carpels, and 120 to 250 stamens. These characteristics are often regarded as the most primitive angiosperm floral characteristics, as seen in various ancestral flowering plant fossils.6
In the tree of plant life, basal angiosperms consisting of three orders Nymphaeales, Amborellales, and Austrobaileyales, have long been regarded as the basal branches of angiosperms using both molecular phylogenetic and developmental classifications.7,8 Although multiple lines of evidence support Amborella as the basal-most angiosperm,7,9–11 the water lily-basal or Amrorella-water lily co-basal theories cannot yet be ruled out.12–15 The genomic sequences of the water lily may be critical in resolving the early evolution of angiosperms, because among all basal angiosperms only the genome of Amborella is currently known.
Limited genetic and genomic analysis of water lilies
Despite the importance of water lilies in phylogenetic research and as an aquatic ornamental plant, limited genetic and genomic information is available. Previous chromosome number and size studies have provided the karyotype background of approximately 65 water lily species 16,17 (Table 1). Only two homologs of INO genes,18 two reference genes for expression studies,19 six floral organ identity genes,20 and ABC model genes 21 have been cloned (Table 1). Genetic markers, such as the matK genes 4 and inter-simple sequence repeats 22 have been applied in DNA barcoding of the water lily germplasm.
Table 1. Available molecular research on water lilies.
| Taxon name | Chromosome (n) | Genome size (Mb) | Genetic study | Transcriptome | |
|---|---|---|---|---|---|
| Nymphaeaceae | Nymphaea alba L. | 42 [1] | 1950 [6] | INO gene [8], ITS2+matK [9] | |
| Nymphaea amazonum Mart. & Zucc. | 9 [1], ~18 [2] | 821.52 [2] | ITS2+matK [9] | ||
| Nymphaea ampla (Salisb.) DC. | 14 [2] | 772.62 [2] | |||
| Nymphaea atrans S. W. L. Jacobs | 42 [1] | 1408.32 [2] | |||
| Nymphaea bissetii hort. | 42 [1] | ||||
| Nymphaea caerulea Savigny | 14 [1] | ITS2+matK [9] | |||
| Nymphaea carpentariae ‘Julia Leu’ | ~42 [2] | 1447.44 [2] | |||
| Nymphaea candida C. Presl | 56 [1] | 1936.44 [2] | |||
| Nymphaea capensis Thunb. | 14 [1] | trnH-psbA, rpoC1 [10] | |||
| Nymphaea colorata Peter | 14 [2] | 489 [2] | INO gene [8] | ||
| Nymphaea conardii Wiersema | 9 [1] | ||||
| Nymphaea daubeniana Hort. ex O. Thomas. | 28 [1] | ||||
| Nymphaea dentatamagnifica Bisset. | 42 [1] | ||||
| Nymphaea gardneriana Planch. | 14 [1] | ||||
| Nymphaea gigantea Hook. | 112 [1] | 2709.06 [2] | |||
| Nymphaea heudelotii Planch. | 14 [1] | ||||
| Nymphaea immutabilis S. W. L. Jacobs | 42 [1] | 1408.32 [2] | |||
| Nymphaea jamesoniana Planch. | 14 [1] | ITS2+matK [9] | |||
| Nymphaea japono-koreana Nakai | 56 [1] | ||||
| Nymphaea lasiophylla Mart. & Zucc. | 9 [1] | ||||
| Nymphaea lingulata Wiersema | 9 [1] | ||||
| Nymphaea lotus L. | 28 [1] | 1779.96/1682.16 [2] | ITS2+matK [9], trnH-psbA, rpoC1 [10] | ||
| Nymphaea mexicana Zucc. | 28 [1] | 586.80 [2] | |||
| Nymphaea micrantha Guill. & Perr. | 14 [2] | 889.98 [2] | ITS2+matK [9] | ||
| Nymphaea minuta | 14 [2] | 449.88 [2] | |||
| Nymphaea nouchali Burm. | 38 [1], 42 [2] | 1193.16 [2] | ITS2+matK [9], trnH-psbA, rpoC1 [10] | ||
| Nymphaea nouchali var. caerulea (Savigny) Verdc. | 14 [1] | 567.24 [2] | |||
| Nymphaea novogranatensis Wiersema | 14 [1] | ITS2+matK [9] | |||
| Nymphaea odorata Aiton | 28 [1], 56 [2] | 1574.58 [2] | ITS2+matK [9] | ||
| Nymphaea oxypetala Planch. | 42 [1] | ITS2+matK [9] | |||
| Nymphaea pubescens Willd. | 28 [2] | 1975.56 [2] | |||
| Nymphaea prolifera Wiersema | 9 [1] | ||||
| Nymphaea pubescens Willd. | 12 [1] | ITS2+matK [9] | |||
| Nymphaea rubra Roxb. ex Andrews | 42 [1] | ITS2+matK [9] | |||
| Nymphaea rudgeana G. Mey. | 21 [1] | 792.18 [2] | |||
| Nymphaea stellata var. versicolor (Sims) Hook. & Thomson | 28 [1] | ||||
| Nymphaea sturtevantii J. N. Gerard | 28 [1] | ||||
| Nymphaea tenerinervia Casp. | 10 [1] | ||||
| Nymphaea tetragona Georgi | 42 [1] | ITS2+matK [9] | |||
| Nymphaea tetragona subsp. Leibergii / Nymphaea leibergii | 56 [1] | ||||
| Nymphaea thermarum E. Fisch. | 14 [1] | 498.78 [2] | |||
| Nymphaea tuberosa / Nymphaea odorata subsp. Tuberosa | 42 [1] | ||||
| Nymphaea violacea | 56 [2] | 1770.18 [2] | |||
| Euryale ferox | 29 [1] | 870.42 [2] | ITS2+matK [9] | ||
| Nuphar advena (Aiton) W. T. Aiton | 17 [1] | 2709.06/2718.84 [2] | ITS2+matK [9] | y [11] | |
| Nuphar lutea (L.) Sm. | 17 [1] | 2875.32 [2] | ITS2+matK [9] | ||
| Nuphar microphylla (Pers.) Fernald | 17 [1] | ITS2+matK [9] | |||
| Nuphar polysepalum Engelm. | 17 [1] | 3080.70/3070.92 [2] | ITS2+matK [9] | ||
| Nuphar pumila (Timm) DC. | 17 [1] | ITS2+matK [9] | |||
| Nuphar variegata Durand | 17 [1] | ITS2+matK [9] | |||
| Nuphar × spenneriana Gaudin | 17 [1] | 2581.92 [2] | |||
| Nuphar intermedia Ledeb. | 17 [1] | ||||
| Nuphar japonica DC. | 17 [1] | 2699.28 [2] | ITS2+matK [9] | ||
| Nuphar subintegerrima Makino | 17 [1] | ITS2+matK [9] | |||
| Barclaya longifolia | 17 [1] | ITS2+matK [9] | |||
| Victoria cruziana | 12 [1] | 4009.80 [2] | ITS2+matK [9] | ||
| Victoria yamasu | 12 [1] | ||||
| Victoria amazonica | 10 [1] | 4557.48 [2] | ITS2+matK [9] | ||
| Victoria ‘Longwood Hybrid’ | 11 [2] | 4303.20 [2] | ITS2+matK [9] | ||
| Cabombaceae | Cabomba caroliniana | 52 [2] | 3471.9 [2] | ITS2+matK [9] | |
| Brasenia schreberi | 36 [3], 40 [1], [2] | 1193.16 [2] | ITS2+matK [9] | ||
| Hydatellaceae | Trithuria submersa | 28 [1] | ~2680 [7] | ITS2+matK [9] | y [12] |
| Trithuria inconspicua | 12 [1] | ITS2+matK [9] | |||
| Trithuria konkanensis | 20 [4] | ||||
| Trithuria australis | 7 [5] | ITS2+matK [9] |
At the omics level, genome-wide expressed sequence tags (ESTs) were generated in 2006 from the yellow water lily Nuphar advena for genome duplication analysis.23 Later, the transcriptomes from seven tissues/organs were sequenced and analyzed from the same species24 (Table 1). Recently, the transcriptome of six samples from two coloring stages of the beautiful blue water lily Nymphaea ‘King of Siam’ were sequenced, together with metabolic analysis, to reveal the blue flower’s formation.25 So far, no water lily genome has been reported.
The water lily holds the key to Darwin’s abominable mystery
The origin and rapid massive expansion of flowering plants in a relatively short geological time, which resulted in most current-day flora, fascinated Charles Darwin, who called it an ‘abominable mystery’ and the ‘most perplexing phenomenon’, beyond which there was ‘nothing... more extraordinary’.26 Over the 137 years since this expansion was proposed by Charles Darwin in 1879, evolutionary biologists have long attempted to reconstruct the early history of angiosperms. One of the most critical questions to solving this mystery is to determine which lineage is the most basal angiosperm. So far, there have been several hypotheses.
From ANITA to ANA basal hypothesis to Amborella and water lily co-basal hypothesis
In 1999, relying on molecular phylogenetic methods, several groups proposed that the Amborella, Nymphaeales, and Illiciales-Trimeniaceae-Austrobaileya (ANITA) clade is the extant basal angiosperm8,27,28 (Figure 2). However, these phylogenetic trees were all based on a single gene or a few genes, mainly from chloroplasts.28 In 2005, based on several plastid, mitochondrial, and nuclear genes, researchers proposed that the Amborella, Nymphaeaceae, and Austrobaileyales (ANA) clade (Figure 2) were the basal sister clades to all other angiosperms.29 This classification sets either Amborella or Amborella and Nymphaeales as the sister to all other angiosperms.29 However, it was not clear which was the most basal angiosperm. Recent releases of new genome sequences has greatly improved phylogenomic or phylotranscriptomic analysis for species tree reconstruction.30 A phylogenetic analysis of 61 plastid genes first reported Nymphaeales and the Amborella, the extant relatives, as the most basal lineage of flowering plants.31 This was later supported by two phylotranscriptomic analyses.9,10 In the last few years, phylogentists have attempted to resolve which angiosperm is the most basal.
Figure 2.
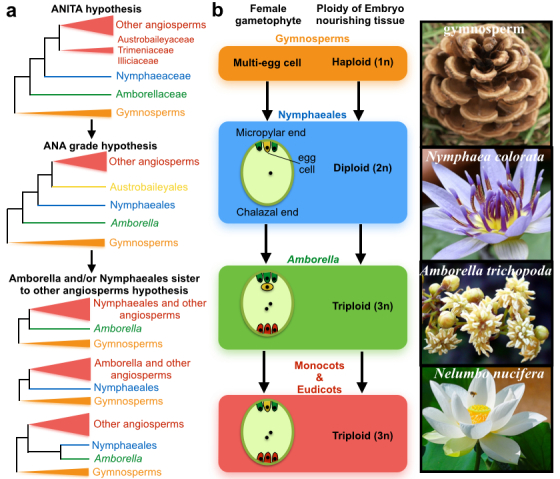
Phylogenetic uncertainty among Amborella, water lily, and other angiosperms. (a) Hypothesized phylogenetic relationships of basal angiosperms. (b) Developmental evidence suggests water lily as the most basal angiosperm.
Amborella as the most basal angiosperm
Unlike single gene-based phylogenetics, when using three mitochondrial genes, one chloroplast gene and one nuclear gene, an early phylogenetic analysis placed Amborella, and not water lilies, as the most basal angiosperm branch9 (Figure 2). This species tree topology is well supported by two recent phylotranscriptomic analyses9,10 using nuclear genes and one phylogenomic analysis using plastid and mitochondria genes.
Water lilies and Amborella as the basal sister to all other angiosperms
In other studies, Amborella and water lilies have been thought to form sister groups that both represent the first lineage to all other angiosperms. Relying on both nuclear and plastid genes, Xi and colleagues in 2014 placed Amborella and water lilies as sister groups using the coalescent-based phylogenetic method, and these sister groups serve as the most basal angiosperm clade32 (Figure 2).
Water lilies as the most basal angiosperms
There is still evidence to support water lilies as the most basal angiosperm. Relying on concatenation-based phylogenetic analysis of the whole chloroplast coding genes and using the transversion of the third position of the codon, researchers found that the water lily was the earliest branch of all extant angiosperms.13 A comparison of the female gametophyte and the embryo-nourishing tissue ploidy also suggested that Amborella was an exception in the ANITA group, which contained triploid endosperm and nine cells in the embryo sac and is thereby closest to monocots and eudicots13,33 (Figure 2). In addition, water lilies contain fewer stomatal modifications from the ancestral angiosperm stomata, whereas Amborella exhibited extensive modifications of stomata.34 In addition, the first known fossil flower of a water lily is from the early cretaceous period, approximately 125–115 million years ago.35 Another Jurassic fossil with flowers and other above-ground organs including the archaefructus is also placed within Nymphaeales.6
Phylogenetic signals hold the key for basal angiosperm phylogeny
A major concern in phylogenomics is the selection of the best phylogenetic signals, which are now generally regarded to be low/single-copy nuclear genes36 that should fulfill two important criteria: high neutrality and low saturation.13 For the selected genes, position 1 and position 2 codons lack synonymous mutation rates and suffer extremely low neutrality.13 Researchers found that position 3 transversion rates are suitable for both shallow and deep phylogenetic tree constructions.13 Based on this position 3 transversion, most single-gene-based trees placed the water lily as the most basal lineage of angiosperms.13 For species tree construction for angiosperms, we suggest the utilization of both protein sequences and nucleotide sequences as a more accurate method for land plant species tree construction.10 Most importantly, phylogenetic signals for species tree reconstruction should be genome-wide and contain a large number of signals but not rare genes or a limited number of signals.
Concatenation VS coalescent methods
In recent years, phylogenomics have relied on both the concatenation method and coalescent method.14,32 Although the coalescent method is theoretically sound to explain incomplete lineage sorting, both theories and applications show that concatenation could yield misleading results when highly conflicting gene trees exist, due to incomplete lineage sorting. These two methods have been under heated debate regarding the effects of tree estimation error in phylogenomics.37–41 Strong phylogenetic signals are still needed for more accurate species tree inference.42
Until recently, our understanding of plant phylogeny has largely depended on studies of plastid, mitochondrial, and ribosomal genes. However, recently, using large-scale comparisons of dozens of genes comprising thousands of DNA bases, phylogenomics have reshuffled most of our long-established trees of life, such as trees for eukaryotic life,43 bird life,44 fish life,45 and major nodes of eudicots of plants.46 The availability of the Amborella genome21 has shed light on basal angiosperm tree construction, but definite resolution of basal angiosperm phylogeny has not been resolved.
Pantropical water lilies could serve as the model for studying basal angiosperms
To understand basal angiosperm evolution and the radiation of angiosperms, a good model species is needed. Among all basal angiosperms, the enormous genome size for Austrobaileyales, ~7050 Mb,47 is a major challenge for genome decoding and genetic experiments. A slow growth rate and the woodiness of Austrobaileyale plants and Amborella may also be challenging due to the difficulty of producing experimental materials. In the water lily order, Cabomba displays multiple features as a model for basal angiosperms, such as small size and rapid vegetative growth, but its large genome size, 3290 Mb,47 excludes it from gene functional studies, as large genomes usually harbor redundant gene copies, are highly heterogenetic and thereby not appropriate for gene functional studies. Although Trithuria species grow into small herbs, their genomes are still too large for genetic studies. Luckily, four pantropical diploid (2n=28) water lilies (or subgenus Brachyceras) may be good choices, as they have the smallest genomes, N. caerulea=567.24 Mb, N. colorata=489 Mb, N. minuta=449.88 Mb, N. thermarum=498.78 Mb.17 The native habitats of all four water lilies are in Africa, and all are annual plants (Table 2). N. caerulea and N. colorata are famous ornamental water lilies and have been widely used to breed new cultivars. N. minuta and N. thermarum are minute water lilies with thumb-sized flowers. Unlike hardy water lilies (a in Figure 1), these four tropical water lilies are easy to cultivate and maintain hundreds of plants in a single green house; it is easy to trigger flowering via temperature control (below 18 °C). All four water lilies can produce hundreds of seeds in a single flower (Figure 3) and can be used to generate a large mutant library. These plants are also easy to self-pollinate in nature to generate pure lines, and can also easily be cross-pollinated. They have a relatively short life cycle of approximately three months from seed to seed in tropical regions. In addition, N. thermarum has recently been well studied for its potential as a model system for basal angiosperms.48 These characteristics make these four water lilies the best candidates for genome sequencing and the best model for functional studies.
Table 2. Characteristics of four pantropical water lilies.
| Species | Genome size | Chromosome | Classification | Distribution | Flowers in diameter |
|---|---|---|---|---|---|
| Nymphaea caerulea | 567.24 Mb | 2n=28 | Nymphaea, Brachyceras | East Africa | 10–15 cm |
| Nymphaea colorata | 489 Mb | 2n=28 | Nymphaea, Brachyceras | tropical East Africa | 11–14 cm |
| Nymphaea minuta | 449.88 Mb | 2n=28 | Nymphaea, Brachyceras | Madagascar | 2 cm |
| Nymphaea thermarum | 498.78 Mb | 2n=28 | Nymphaea, Brachyceras | Rwanda, Africa | 10–15 cm |
The four listed genome sizes and chromosome counts have been reported.17
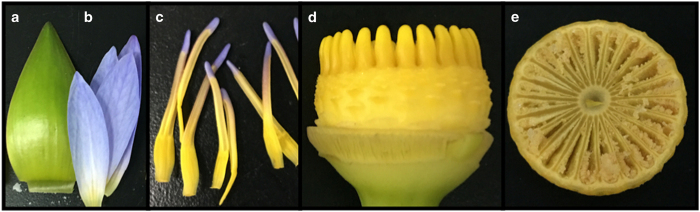
Figure 3.
Floral organs of a typical tropical water lily. (a) petal, (b) sepal, (c) stamen, (d) carpels on the receptacle, (e) numerous young seeds.
The water lily genome for basic evolutionary research and applied horticulture
Based on the advantages of pantropical water lilies, we launched a genome-sequencing project of N. colorata using a third-generation single-molecule real-time sequencing method. We produced half of the reads >20 kb, and this has facilitated the assembly of complex repeating sequences and GC-rich regions that are usually highly fragmented or even unassembled in next-generation sequencing projects.49 We have annotated the genes and other key DNA elements using multiple tools. The future reference water lily genome will provide genomic information for reconstructing the karyotype of an angiosperm ancestor, with the species trees of basal angiosperms and early massive radiation of angiosperms. This availability of the water lily reference genome will greatly help us to understand Charles Darwin’s ‘abominable mystery’, the early evolution trajectory of angiosperms, the aquatic life style of angiosperms, the evolution of a 4-celled embryo sac and diploid endosperm, the comparative analyses of genes and other elements such as conserved non-coding elements and telomeres. The water lily genome is also needed to revisit the age of angiosperms and whether they evolved 0.1 billion-years ago50 or 0.2 billion years ago.51 The reference genome will also provide genetic information for breeders and geneticists. Currently, only seven aquatic plants have their genomes decoded, and only two aquatic ornamental plants, the water lily and the sacred lotus, have sequenced genomes (Table 3). The similar appearance of the water lily and the lotus does not actually indicate a tight relationship; the former is a basal angiosperm and the latter is a eudicot. Thus, the genome of the water lily will serve as a template to accelerate genomic studies of other aquatic ornamentals.
Table 3. Sequenced aquatic plants.
| Scieitific name | Common name | Classification | Ornamental plant | Habitate | Genome size |
|---|---|---|---|---|---|
| Nymphaea spp. | Water lilies | Basal angiosperms | Partially yes | Floating leaf type | ~400 Mb–2.7 Gb |
| Nelumbo nucifera | Sacred lotus | Basal eudicot | Yes | Emerged plant | 929 Mb |
| Utricularia gibba | Floating bladderwort | Eudicot, Lentibulariaceae | No | Floating plant | 82 Mb |
| Zostera marina | Seagrass | Monocot, Zosteraceae | No | Submergent plant | 202.3 Mb |
| Spirodela polyrhiza | Duckweed | Monocot, Araceae | No | Free-floating | 158 Mb |
| Zizania latifolia | Manchurian wildrice | Monocot, Poaceae | No | Wetland plant | 586 Mb |
| Oryza sativa | Rice | Monocot, Poaceae | No | Wetland plant | 420 Mb |
The genome of Nymphaea colorata was sequenced recently by our team. Other genomes have been sequenced and are publicly available.
Conclusions
Upgrading sequencing technologies and bioinformatics tools have provided high-resolution genomic details, showing great potential for understanding the large questions in biology (including Darwin’s famous abominable mystery), and are valuable resources for molecular breeding. Although the genetics and genomics of water lilies are incipient, four pantropical water lilies, especially N. colorata, show great potential as a model system to study basal angiosperms; their genomes will greatly enhance our current knowledge, including Charles Darwin’s abominable mystery, the early evolutionary trajectory of angiosperms, the aquatic life style of angiosperms, and molecular breeding.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81502437) and a start-up fund from the Fujian Agriculture and Forestry University to LZ We acknowledge Dr Xianxian Yu from Xuchang University for providing the N. colorata picture in Figure 1.
Footnotes
The authors declare no conflict of interest.
References
- Raja MKMM, Sethiya NK, Mishra SH. A comprehensive review on Nymphaea stellata: a traditionally used bitter. J Adv Pharm Technol Res 2010; 1: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani M, Sawada A, Oyabu T, Seiryo K. Ability of water lilies to purify water polluted by soap and their application in domestic sewage disposal facilities. Sens Mater 2006; 18: 91–101. [Google Scholar]
- Friedman WE. Hydatellaceae are water lilies with gymnospermous tendencies. Nature 2008; 453: 94–97. [DOI] [PubMed] [Google Scholar]
- Biswal DK, Debnath M, Kumar S, Tandon P. Phylogenetic reconstruction in the Order Nymphaeales: ITS2 secondary structure analysis and in silico testing of maturase k (matK) as a potential marker for DNA bar coding. BMC Bioinformatics 2012; 13: S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Les DH, Schneider EL, Padgett DJ, Soltis PS, Soltis DE, Zanis M. Phylogeny, classification and floral evolution of water lilies (Nymphaeaceae; Nymphaeales): a synthesis of non-molecular rbcL, matK, and 18S rDNA data. Syst Bot 1999; 24: 28–46. [Google Scholar]
- Gandolfo MA, Nixon KC, Crepet WL. Cretaceous flowers of Nymphaeaceae and implications for complex insect entrapment pollination mechanisms in early angiosperms. Proc Natl Acad Sci USA 2004; 101: 8056–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D, Soltis P, Nickrent D et al. Angiosperm phylogeny inferred from 18S ribosomal DNA sequences. Ann Miss Bot Gard 2015; 71: 607–630. [Google Scholar]
- Qiu Y, Lee J, Bernasconi-quadroni F, Zimmer EA, Chen Z, Savolainen V. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 1999; 402: 404–407. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergencetimes. Nat Commun 2014; 5: 4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 2014; 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew BT, Ruhfel BR, Smith SA et al. Another look at the root of the angiosperms reveals a familiar tale. Syst Biol 2014; 63: 368–382. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J, Raubeson LA, Cui L et al. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Bio Evol 2005; 22: 1948–1963. [DOI] [PubMed] [Google Scholar]
- Yang X, Tuskan GA, Tschaplinski TJ, Cheng Z-M. Third-codon transversion rate-based Nymphaea basal angiosperm phylogeny -- concordance with developmental evidence. Nat Preced 2007, 1–20.
- Simmons MP, Gatesy J. Coalescence vs. concatenation: sophisticated analyses vs. first principles applied to rooting the angiosperms. Mol Phylogenet Evol 2015; 91: 98–122. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Nikiforova SV, Cavalieri D, Pindo DM, Lockhart P. The root of flowering plants and total evidence. Syst Biol 2015; 64: 879–891. [DOI] [PubMed] [Google Scholar]
- Diao Y, Chen L, Yang G et al. Nuclear DNA C-values in 12 species in Nymphaeales. Caryologia 2006; 59: 25–30. [Google Scholar]
- Pellicer J, Kelly LJ, Magdalena C, Leitch IJ. Insights into the dynamics of genome size and chromosome evolution in the early diverging angiosperm lineage Nymphaeales (water lilies). Genome 2013; 56: 437–449. [DOI] [PubMed] [Google Scholar]
- Yamada T, Ito M, Kato M. Expression pattern of INNER NO OUTER homologue in Nymphaea (water lily family, Nymphaeaceae). Dev Genes Evol 2003; 213: 510–513. [DOI] [PubMed] [Google Scholar]
- Luo H, Chen S, Wan H, Chen F, Gu C, Liu Z. Candidate reference genes for gene expression studies in water lily. Anal Biochem 2010; 404: 100–102. [DOI] [PubMed] [Google Scholar]
- Luo H, Chen S, Jiang J et al. The expression of floral organ identity genes in contrasting water lily cultivars. Plant Cell Rep 2011; 30: 1909–1918. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science 2013; 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Chaveerach A, Tanee T, Sudmoon R. Molecular identification and barcodes for the genus. Nymphaea. Acta Biol Hungarica 2011; 62: 328–340. [DOI] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-mack JH et al. Widespread genome duplications throughout the history of flowering plants. Genome Res 2006; 16: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MJ, Chanderbali AS, Altman NS, Soltis PS, Soltis DE. Evolutionary trends in the floral transcriptome: Insights from one of the basalmost angiosperms, the water lily Nuphar advena (Nymphaeaceae). Plant J 2010; 64: 687–698. [DOI] [PubMed] [Google Scholar]
- Wu Q, Wu J, Li S-S et al. Transcriptome sequencing and metabolite analysis for revealing the blue flower formation in waterlily. BMC Genomics 2016; 17: 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman WE. The meaning of Darwin’s ‘abominable mystery’. Am J Bot 2009; 96: 5–21. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 1999; 402: 402–404. [DOI] [PubMed] [Google Scholar]
- Kuzoff RK, Gasser CS. Recent progress in reconstructing angiosperm phylogeny. Trends Plant Sci 2000; 5: 330–336. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Dombrovska O, Lee J et al. Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes. Int J Plant Sci 2005; 166: 815–842. [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 2005; 6: 361–375. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Nikiforova SV, Biggs PJ et al. The evolutionary root of flowering plants. Syst Biol 2013; 62: 50–61. [DOI] [PubMed] [Google Scholar]
- Xi Z, Liu L, Rest JS, Davis CC. Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst Biol 2014; 63: 919–932. [DOI] [PubMed] [Google Scholar]
- Friedman WE. Embryological evidence for developmental lability during early angiosperm evolution. Nature 2006; 441: 337–340. [DOI] [PubMed] [Google Scholar]
- Carpenter KJ. Stomatal architecture and evolution in basal angiosperms. Am J Bot 2005; 92: 1596–1615. [DOI] [PubMed] [Google Scholar]
- Friis E, Pedersen K, Crane PR. Fossil evidence of water lilies (Nymphaeales) in the early cretaceous. Nature 2001; 410: 357–360. [DOI] [PubMed] [Google Scholar]
- Sang T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit Rev Biochem Mol Biol 2002; 37: 121–147. [DOI] [PubMed] [Google Scholar]
- Betancur-R R, Naylor GJP, Ortí G. Conserved genes, sampling error, and phylogenomic inference. Syst Biol 2014; 0: 1–6. [DOI] [PubMed] [Google Scholar]
- Mirarab S, Bayzid MS, Boussau B, Warnow T. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 2014; 346: 1250463. [DOI] [PubMed] [Google Scholar]
- Liu L, Edwards SV. Comment on ‘Statistical binning enables an accurate coalescent-based estimation of the avian tree’. Science 2014; 350: 171. [DOI] [PubMed] [Google Scholar]
- Springer MS, Gatesy J. The gene tree delusion. Mol Phytolgenet Evol 2016; 94: 1–33. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Xi Z, Janke A et al. Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics. Mol Phytolgenet Evol 2016; 94: 447–462. [DOI] [PubMed] [Google Scholar]
- Salichos L, Rokas A. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 2013; 497: 327–331. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One 2007; 2: e790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum RO, Berv JS, Dornburg A et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015; 526: 569–573. [DOI] [PubMed] [Google Scholar]
- Ricardo B-R, Broughton RE, Wiley EO et al. The tree of life and a new classification of bony fishes. PLOS Curr Tree Life 2013, 1–52. [DOI] [PMC free article] [PubMed]
- Zeng L, Zhang N, Zhang Q, Endress PK, Huang J, Ma H. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol 2017; 214: 1338–1354. [DOI] [PubMed] [Google Scholar]
- Vialette-Guiraud ACM, Alaux M, Legeai F et al. Cabomba as a model for studies of early angiosperm evolution. Ann Bot 2011; 108: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povilus RA, Losada JM, Friedman WE. Floral biology and ovule and seed ontogeny of Nymphaea thermarum, a water lily at the brink of extinction with potential as a model system for basal angiosperms. Ann Bot 2015; 115: 211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, Bryant D, Edger PP et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 2015; 527: 508–511. [DOI] [PubMed] [Google Scholar]
- Buggs RJA. The deepening of Darwin’s abominable mystery. Nat Eco Evol 2017; 1: 169. [DOI] [PubMed] [Google Scholar]
- Herendeen PS, Friis EM, Pedersen KR, Crane PR. Palaeobotanical redux: revisiting the age of the angiosperms. Nat Plants 2017; 3: 17015. [DOI] [PubMed] [Google Scholar]
- Gaikward SP, Yadav SR. Further morphotaxonomical contribution to the understanding of the family Hydatellaceae. J Swamy Bot 2003; 20: 1–10. [Google Scholar]
- Iles WJD, Rudall PJ, Sokoloff DD et al. Molecular phylogenetics of Hydatellaceae (Nymphaeales): sexual-system homoplasy and a new sectional classification. Am J Bot 2012; 99: 663–676. [DOI] [PubMed] [Google Scholar]
- Kynast RG, Joseph JA, Pellicer J, Ramsay MM, Rudall PJ. Chromosome behavior at the base of the angiosperm radiation: karyology of Trithuria submersa (Hydatellaceae, Nymphaeales). Am J Bot 2014; 101: 1447–1455. [DOI] [PubMed] [Google Scholar]
- Marques I, Montgomery SA, Barker MS et al. Transcriptome-derived evidence supports recent polyploidization and a major phylogeographic division in Trithuria submersa (Hydatellaceae, Nymphaeales). New Phytol 2016; 210: 310–323. [DOI] [PubMed] [Google Scholar]