ABSTRACT
The envelope of bacteria is a complex multilayered shield that ensures multiple essential functions, including protecting the cell from external assaults. Hence, bacterial cells have evolved intricate mechanisms called envelope stress response systems (ESRS) to monitor all kinds of perturbations affecting the integrity of their envelope and to mount an appropriate response to contain or repair the damage. In the model bacterium Escherichia coli, several ESRS are built around a two-component system, in which envelope stress triggers a phosphotransfer between a sensor protein in the inner membrane of the envelope and a response regulator in the cytoplasm. In this review, we focus on two major ESRS in E. coli, the Rcs and Cpx pathways, in which additional proteins not directly involved in the phosphotransfer modulate signal transduction. Both the Rcs and Cpx systems can be turned on by a lipoprotein anchored in the outer membrane, RcsF and NlpE, respectively, providing a molecular connection between the most exterior layer of the envelope and the ground control center in the cytoplasm. Here, we review how these two lipoproteins, which share a striking set of features while being distinct in several aspects, act as sentinels at the front line of the bacterium by sensing and transducing stress to the downstream components of the Rcs and Cpx systems.
KEYWORDS: Escherichia coli, cell envelope, lipoproteins, outer membrane, periplasm, stress response
INTRODUCTION
Bacterial cells are surrounded by an elaborate envelope that is essential for survival. Indeed, the envelope is the first line of defense of bacteria, offering a multilayered armor against harsh and ever-changing environmental conditions, while ensuring the selective import of nutrients from the surroundings (1). In diderm bacteria, the envelope is arranged in several chemically and functionally distinct strata (Fig. 1) (2). The first layer is the cytoplasmic membrane or inner membrane (IM), an impermeable phospholipid bilayer in which numerous proteins are present. The vast majority of IM proteins are inserted in the membrane by alpha-helical transmembrane segments. These proteins play important cellular functions, including the transfer of electrons across the respiratory complexes or the active transport of biomolecules and inorganic compounds across the membrane. A small number of lipoproteins, which are globular proteins anchored to a membrane by a lipid moiety, are also present in the IM (3). IM lipoproteins are oriented toward the periplasm, a viscous and oxidizing compartment that constitutes the second layer of the envelope. The periplasm is densely packed with soluble proteins (4) and contains a critically important structure called the cell wall, which is made of a continuous polymer of peptidoglycan (PG). The role of the cell wall is multiple: it resists turgor pressure and therefore preserves the osmotic stability of the cell; it also dictates the shape of the cell and underlies the fundamental processes of cell elongation and division (5). The third coat of the envelope, which separates the periplasm from the extracellular milieu, is the outer membrane (OM) (6). It differs from the IM in at least two aspects. First, the outer leaflet of the OM is composed of lipopolysaccharides (LPS), which are negatively charged molecules making the OM impermeable to hydrophobic compounds (7). Second, proteins that are embedded in the OM are not alpha-helical but fold instead into β-barrels with transmembrane domains consisting of amphipathic β-strands. Some of these β-barrels, called porins, function like pores through which small hydrophilic molecules can diffuse across the OM (8). In addition to β-barrels, the OM accommodates the majority of the lipoproteins encoded by bacterial genomes. In the model bacterium Escherichia coli, most OM lipoproteins are thought to be facing the periplasm (3), although, as described below, some OM lipoproteins “float in the most peculiar way” (akin to David Bowie's Major Tom). Thus, the envelope exhibits a sophisticated composition, ensuring that the bacterial cell is efficiently protected from toxic substances or the stressful conditions of the environment. Preserving the integrity of this multilayered shell is therefore a matter of life and death for Gram-negative bacteria.
FIG 1.
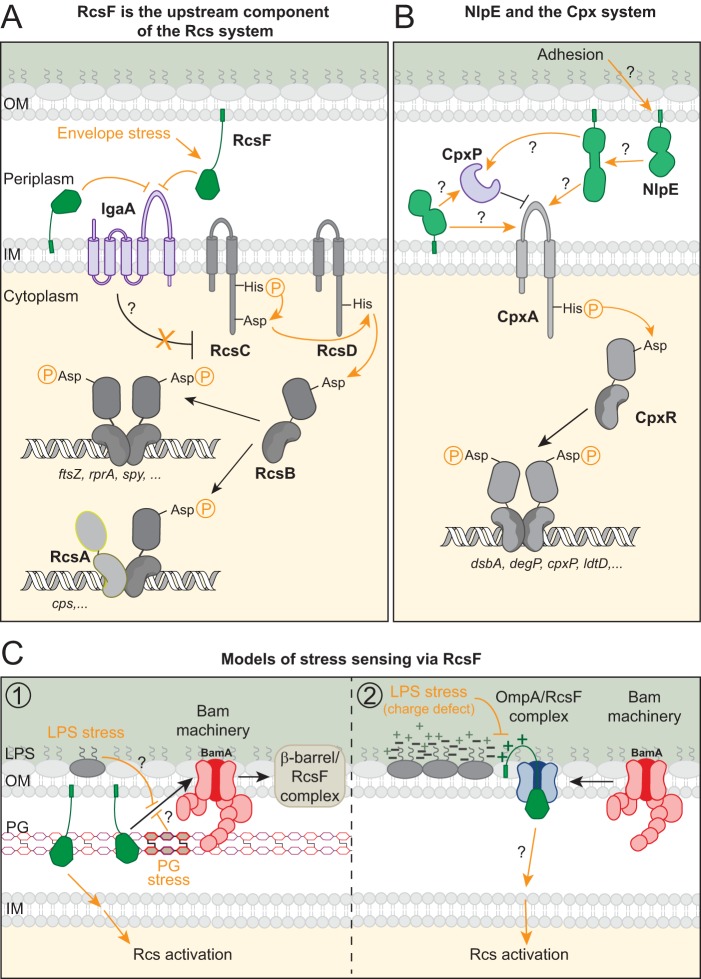
The lipoproteins RcsF and NlpE trigger the Rcs and Cpx envelope stress response pathways. Question marks indicate protein interactions or mechanisms that remain unknown. Drawings are not to scale. Orange lines or text indicate stress-related events. (A) Under nonstress conditions, RcsF is sequestered by β-barrel proteins (not shown; see panel C), and the IM protein IgaA inhibits the Rcs system (possibly via interaction with RcsC or RcsD). RcsF detects perturbations in the PG or in the OM via partially understood mechanisms (see panel C). Under stress, the signaling domain of RcsF interacts with IgaA, releasing the inhibition of IgaA on the Rcs system and triggering a phosphorelay between the sensor kinase RcsC, the phosphotransmitter RcsD, and the response regulator RcsB. Phosphorylated RcsB either homodimerizes or associates with RcsA to control the expression of a vast regulon. When retargeted to the IM, RcsF interacts with IgaA, activating Rcs. (B) Overexpression or mislocalization of NlpE at the IM triggers the Cpx phosphorelay, in which the sensor kinase CpxA autophosphorylates and transfers the phosphate to the response regulator CpxR. Whether NlpE interacts with CpxA, its periplasmic inhibitor CpxP, or other partners, is unknown. Adhesion to abiotic surfaces or to undifferentiated epithelial cells induce the Cpx response in an NlpE-dependent manner by an unknown mechanism. The crystal structure of NlpE reveals two globular domains and suggests that part of the protein unfolds upon adhesion, facilitating access to downstream Cpx components or intermediate proteins. (C, model 1) Under nonstressed conditions, RcsF interacts with BamA; the Bam machinery then mediates the assembly of RcsF with the major β-barrels OmpA/F/C. In complex with these proteins, part of RcsF is accessible to the cell surface and hence occluded from IgaA. In this model, drugs targeting LPS (polymyxin B) or PG (amdinocillin, A22) prevent newly synthesized RcsF from interacting with BamA by a still-unknown mechanism, leading to the accumulation of free RcsF (not associated with β-barrels) (16). This free RcsF has access to IgaA in the IM, triggering Rcs (16). (Model 2) It has been proposed that RcsF is threaded into β-barrels by BamA during β-barrel folding, with the lipid moiety of RcsF anchored in the outer leaflet of the OM and its signaling domain in the periplasm (41). Considering this topology, interactions between the positively charged residues of the RcsF linker and the negatively charged LPS molecules are likely and RcsF could sense charge alterations in LPS via these interactions (46). In this model, activation does not require new RcsF synthesis (46). How RcsF then activates Rcs while in complex with β-barrel proteins is unknown.
ENVELOPE STRESS AND HOW BACTERIA DEAL WITH IT
To protect their precious envelope, bacteria have evolved intricate signaling pathways that not only detect different kinds of envelope alterations but also turn on appropriate cellular responses to cope with stress and repair damage by controlling gene expression. These pathways, which are called envelope stress response systems (ESRS), have been studied extensively in E. coli (9, 10). It is important to emphasize at this stage that the generic term “envelope stress” encompasses innumerable conditions that, for instance, perturb the fine balance between PG synthesis and remodeling (therefore modifying the tightly regulated chemical architecture of the cell wall), alter membrane integrity, or prevent the correct folding and/or targeting of proteins within the envelope. Given the broad diversity of potential stresses, bacteria rely on several ESRS to sense and respond to the different kinds of damage that can affect their envelope. While each system can be elicited by more than one stimulus, it is generally considered that each ESRS is specialized to deal with a set of stimuli that fall into a common theme. In this review, we focus on two major ESRS in E. coli, the Rcs and Cpx systems, which are traditionally considered to cope with OM and cell wall defects, or with the accumulation of periplasmic proteins and IM stress, respectively.
VARIATIONS AROUND THE SAME THEME: THE Rcs AND THE Cpx SIGNAL TRANSDUCTION PATHWAYS
Both the Rcs and Cpx systems are built around a two-component system (TCS), a signaling strategy that is mostly present in prokaryotes but has also been reported in certain eukaryotes (11). In their simplest variation, bacterial TCS employ a sensor protein, often a histidine kinase that is either cytoplasmic or embedded in the IM, and a cognate response regulator in the cytoplasm. When activated, the sensor protein autophosphorylates on a histidine residue before transferring the phosphoryl group to an aspartate present in the receiver domain of the response regulator. Following phosphorylation, typical response regulators dimerize and bind DNA via a second domain to control the transcription of a series of target genes (11). The Cpx pathway is a classical TCS, as it involves the direct transfer of a phosphoryl group from the histidine kinase CpxA in the IM to the response regulator CpxR (Fig. 1B) (12, 13). In contrast, Rcs is significantly more complex than the TCS core, since a phosphorelay is required to transduce the signal between the sensor and the regulator. Here, the phosphoryl group on the histidine kinase domain of the IM sensor RcsC is first transferred to an aspartate present in a receiver domain on the same protein, before being handed over to a histidine residue on RcsD, another IM protein that acts as a phosphotransmitter between RcsC and the receiver domain of the response regulator RcsB (Fig. 1A) (14). Interestingly, the activities of both Rcs and Cpx are modulated by proteins that do not directly participate in the cascade of phosphoryl transfer reactions. In Rcs, RcsB can form heterodimers with other regulators (e.g., the cytoplasmic protein RcsA) to control a different set of genes than the RcsB homodimer, while the essential IM protein IgaA acts as a brake on the system, keeping its activity at check (see below) (15, 16). In Cpx, the periplasmic protein CpxP functions as a negative regulator of CpxA kinase activity (17, 18). Finally, both Rcs and Cpx can be triggered by an OM lipoprotein, RcsF and NlpE, respectively, providing a connection between the most outward layer of the envelope and the cytoplasm. Thus, via RcsF and NlpE, information can be transmitted through the envelope to the ground control center in the cytoplasm, which then deals with the problem by turning on or off an appropriate regulon.
Several extensive reviews describe the inputs and outputs of the Cpx and Rcs signaling pathways (13, 19–23). Here, we review the current knowledge on how the OM-anchored lipoproteins RcsF and NlpE function as sentinels at the front line of the bacterial cell by detecting stress and triggering the Rcs and Cpx systems, respectively. Although these two proteins are different in many aspects, they share a striking set of features that we will describe. Table 1 gives an overview of the large diversity of Rcs and Cpx-inducing cues and whether RcsF or NlpE are engaged in transducing these signals. Finally, we present outstanding questions that remain unsolved on the mechanisms of stress sensing by the Rcs and Cpx systems.
TABLE 1.
Inducing cues of the Rcs and Cpx envelope stress response systems
| System induced by stress | Cue type | Cues |
|---|---|---|
| Cpx | NlpE dependent | nlpE overexpression (32); artificial mislocalization of NlpE (37, 39); adhesion of stationary phase cells to hydrophobic glass (49, 50); adhesion of stationary phase EHEC cells to undifferentiated epithelial cells (50) |
| NlpE independent | Overproduction of PapE and PapG pilus subunits in the absence of cognate chaperone (59); alkaline pH (Cpx activation shown in reference 60 and NlpE independence from unshown data in reference 59); stationary phase growth (Cpx activation shown in references 59 and 61 and NlpE independence from unshown data in reference 59); lipid II accumulation (Cpx activation shown in reference 62 and NlpE independence from unshown data in reference 59); lack of phosphatidylglycerol or phosphatidylethanolamine (63, 64); EDTA (Cpx activation shown in reference 65 and NlpE independence from unshown data in reference 59); drugs targeting PG synthesis (mecillinam, cephalexin, and A22 [37]) | |
| Undetermined role of NlpE | Loss of 4 PBPs (involved in PG synthesis) (66); overproduction of type IV pilus subunit (67); CuSO4 (68); ΔdegP (69); high osmolarity (65, 70, 71); Curli overproduction and ompR mutation (71); IM stress due to lack of YidC, lack of protease FtsH or HtpX, or accumulation of IM substrates of FtsH like SecY or SecY mutations (72, 73); indole, ethanol, and dibucaine (65); mammalian PG recognition proteins and gentamicin (74); ΔcpxP (59) | |
| Rcs | RcsF dependent | rcsF overexpression (24); artificial mislocalization of RcsF (29); mutation in the LPS biosynthesis pathway (deep rough rfa mutants) (26); cationic antimicrobial peptides, including polymyxin B (29, 46); treatments targeting the PG (lysozyme [47], the β-lactams mecillinam and cefsulodin [48], and MreB inhibitor A22 [16]); defective lipoprotein sorting due to lack of the periplasmic chaperone LolA or dominant-negative LolA mutant (36) or to inhibition of lipoprotein signal peptidase LspA by globomycin 38; lack of the periplasmic chaperone SurA involved in OM biogenesis (30); low levels of BamA in a bamA101 strain (16); mutation (pgsA) in phospholipid biosynthesis leading to lack of phosphatidylglycerol and cardiolipin (28); lack of membrane-derived oligosaccharides (mdo mutations) (data not shown in reference 30); a set of multicopy inducers (from reference 25) (26); growth in excess zinc at high temperature (27) |
| RcsF independent | Overproduction of the IM protein DjlA (28), which acts as an Rcs inhibitor under native expression levels (75); overproduction of the putative lipoprotein YpdI (76) | |
| Undetermined role of RcsF | Loss of 4 PBPs (involved in PG synthesis) (66); growth on a solid surface (77) |
OM-ANCHORED LIPOPROTEINS TRIGGER THE Cpx AND Rcs SYSTEMS
Linking the lipoproteins RscF and NlpE to stress sensing in the envelope.
The implication of the RcsF and NlpE lipoproteins being involved in the Rcs and Cpx signaling pathways was uncovered in the 1990s when they appeared as hits in two independent multicopy suppressor screens. The first screen aimed to identify genes that phenocopied rcsB overexpression and could therefore act as regulators of RcsB. Like rcsB, rcsF overexpression was sufficient to restore the growth of an ftsZ-thermosensitive mutant strain and induced the secretion of colanic acid, a capsule exopolysaccharide that makes colonies mucoid. Because these rcsF-mediated phenotypes were rcsB dependent, these data showed that RcsF acts upstream of RcsB (24). It was only 10 years later that the team of Majdalani et al. demonstrated that RcsF in fact lies upstream of RcsC (25, 26). Importantly, RcsF is necessary to transduce most known Rcs-inducing cues to the system, including defects in cell wall and OM integrity (25, 27–31), establishing the critical position occupied by this lipoprotein in the Rcs envelope stress response pathway in E. coli. Although artificially increasing RcsF synthesis induces Rcs activity, the known physiological Rcs-inducing cues do not increase rcsF expression, suggesting that RcsF triggers the Rcs phosphorelay via protein-protein interactions (26). However, the molecular mechanism by which RcsF “detects” stress and activates the Rcs ESRS remained completely mysterious until very recently, as described in more detail below.
The connection between NlpE and Cpx was also discovered more than 20 years ago in a seminal work from the group of Thomas Silhavy, in which they screened for suppressors of the toxicity of the tripartite LamB-LacZ-PhoA fusion (32). When secreted in large amounts into the periplasm, this fusion forms disulfide-bonded aggregates whose accumulation is lethal (33). Activation of the Cpx regulatory system suppresses this toxicity (34), probably thanks to an increased synthesis of the periplasmic protease DegP (35), which degrades the LamB-LacZ-PhoA misfolded protein (34). The link between NlpE and Cpx became apparent when it was shown that NlpE overproduction rescued cells from the accumulation of the toxic aggregates in a manner that was fully dependent on cpxR (32). Consistently, the suppression of LamB-LacZ-PhoA toxicity conferred by NlpE overproduction was partially prevented by the inactivation of degP (32). Important to note, whereas NlpE overexpression clearly activates Cpx by a yet-unknown mechanism, the role of this lipoprotein in controlling Cpx activity when expressed at chromosomal levels is less clear (see below). This is in sharp contrast with RcsF, which is required to sense most Rcs-inducing cues.
Localization matters: rerouting RscF and NlpE stimulates their cognate stress response pathway.
While RcsF and NlpE are anchored in the OM, the closest Rcs and Cpx members are positioned in the IM. Recent data suggest that RcsF activates Rcs by interacting with the IM protein IgaA (see reference 16 and below), which most likely alleviates the inhibition that IgaA exerts on Rcs (Fig. 1A). The downstream interaction partner of NlpE remains unknown, although the IM histidine kinase CpxA appears to be a likely candidate (Fig. 1B). Thus, it has been proposed that both RcsF and NlpE are occluded from their cognate (verified or hypothetical) IM partner under normal conditions but become able to interact with them under stress. Supporting this, several pieces of evidence show that localization matters when it comes to Cpx and Rcs activation by NlpE and RcsF, respectively. For instance, accumulating either lipoprotein in the IM (by altering the lipoprotein maturation pathway [36] or mutating key residues in their targeting motif [29, 37, 38]) strongly induces the expression from specific reporters of the respective systems. Likewise, versions of RcsF that are soluble in the periplasm also trigger Rcs (29, 38). Importantly, the activating effect is independent of overexpression, as the rerouted proteins induced the signaling cascades even when expressed at physiological levels (16, 29, 37). Thus, it is tempting to speculate that when present in the IM, NlpE and RcsF trigger their respective stress responses because they gain free access to their downstream partner (29, 32, 37, 39). Actually, a recent study proposed that NlpE serves as a sentinel to alert the Cpx system in cells with a modified lipoprotein content and defective lipoprotein sorting between the IM and the OM (see below and reference 40), which further supports the importance of lipoprotein localization for function. In the following sections, we address the intriguing question of how stress can modulate the interaction between RcsF and its partners and examine various hypotheses concerning the role of NlpE in Cpx activation.
MECHANISM OF STRESS SENSING BY RcsF
RscF is exposed at the cell surface.
LPS alterations induce the Rcs system in an RcsF-dependent manner (Table 1). Therefore, it was hypothesized that in order to sense perturbations in LPS, RcsF could be exposed on the cell surface instead of having its membrane-fastened soluble domain “floating” in the periplasm like most lipoproteins (41). Four different methods by the Silhavy group and ours indeed demonstrated that RcsF is surface exposed: (i) RcsF was labeled by the NHS-LC-LC biotin probe, which is considered to be OM impermeable (41); (ii) biotinylated RcsF was partially cleaved by a protease added in the milieu (41); (iii) RcsF was detected on whole (nonfixed, nonpermeabilized) cells by an anti-RcsF antibody in dot blot assays (16, 41); and (iv) RcsF was detected on nonpermeabilized cells by quantitative immunofluorescence microscopy using an anti-RcsF antibody (16). The surface exposure of RcsF triggers two major questions: how does it get there, and which portion of the lipoprotein is facing the outside?
The ability of RcsF to form complexes with three major β-barrels (OmpA, OmpF, and OmpC) was discovered when looking for partners interacting with RcsF (16, 41). In these experiments, RcsF was pulled down after incubation of the cells with chemical cross-linkers, and the complexes were subjected to mass spectrometry for protein identification. OmpA, OmpC, and OmpF appeared as major partners of RcsF in one study (41), whereas the OmpA-RcsF complex was the most abundant in a second one (16). This discrepancy could be explained by differences in the RcsF expression levels (overexpression [41] versus native levels [16]) and in the efficiencies of the chemical cross-linkers used in these experiments. Nevertheless, both studies could demonstrate that RcsF interacts with abundant β-barrels, suggesting that these proteins could serve as vehicles allowing RcsF to reach the surface.
The topology of the complexes that RcsF forms with β-barrels was further dissected using site-specific photo-cross-linking of RcsF variants carrying the UV-cross-linkable unnatural amino acid p-benzoyl-l-phenylalanine (pBpa) at several positions in the protein (16, 41). Upon UV exposure, pBpa covalently binds to any residue in tight proximity (<3 Å), which allows the precise mapping of interaction interfaces between a protein of interest and its partners (42). Most RcsF residues that cross-linked to the β-barrels belonged to the linker domain of RcsF, an intrinsically disordered segment located at the N terminus of the protein (43, 44) (Fig. 2). Cross-linking was also observed when pBpa was inserted at a few sites in the folded globular domain of the protein (16, 41). Interestingly, a pBpa residue located at the beginning of the globular domain (G60) was found to cross-link with OmpF/C peptides that face the lumen of the β-barrels (41). In addition, when a Flag tag was inserted at various positions in RcsF, it could only be detected by an anti-Flag antibody when present in the linker domain or in the beginning of the globular domain (41). These results led to a model in which the lipid moiety of RcsF is anchored in the outer leaflet of the OM instead of the inner leaflet, and the N-terminal linker is exposed on the cell surface before being threaded through the lumen of β-barrel proteins. In this topology, most of the globular domain of RcsF is present inside the cell (41) (Fig. 1C, right). However, this model raises a number of questions that will have to be addressed in the future to fully elucidate how RcsF gains access to the surface. First, an anti-RcsF antibody, which specifically recognizes the globular domain of RcsF, could bind whole cells in two different assays, suggesting that at least part of the signaling domain of RcsF is facing the cell exterior (16). Second, it will be interesting to determine how the lumen of OmpF/C accommodates the rest of the globular domain of RcsF after residue G60, which is intriguing given the small size of the pore. Finally, it will also have to be established how OmpA and RcsF interact. Indeed, whereas OmpC and OmpF form large 16-stranded β-barrels with a central pore large enough to accommodate a disordered segment, such as the RcsF linker, the conformation adopted by OmpA is less clear. Although some reports suggest that OmpA may adopt a conformation similar to that of OmpC and OmpF, the predominant view is that this protein assumes a 2-domain structure, with an 8-stranded N-terminal β-barrel and a C-terminal periplasmic domain (45). In this conformation, the β-barrel does not have an open channel that could accommodate a polypeptide. Thus, more work will be required to clarify how OmpA and RcsF interact.
FIG 2.
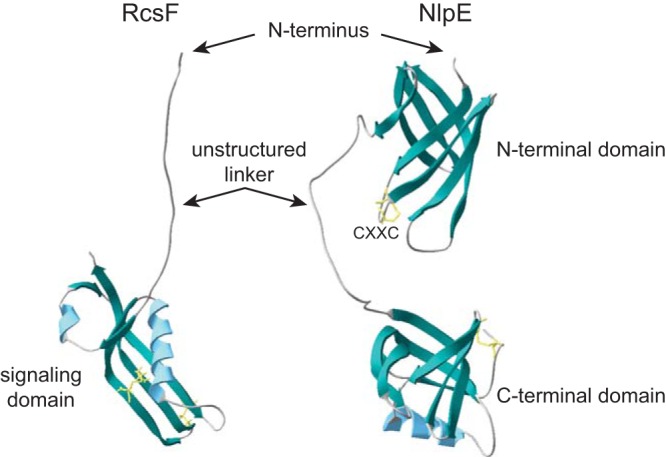
Structures of RcsF and NlpE. Ribbon representations of the protein structures of RcsF (left, PDB 2L8Y [44]) and NlpE (right, PDB 2Z4H [53]). Predicted β-sheets are in green, α-helices are in blue, and disulfide bonds are in yellow. The N terminus of the proteins (lacking the lipobox and signal peptide) and domain composition are indicated. RcsF harbors an unstructured linker region at the N terminus and a globular domain, which is sufficient for Rcs activation. Type I NlpE homologs have both the N-terminal and the C-terminal domains, separated by a region that is predicted to be unstructured and flexible, whereas type II homologs only carry the N-terminal domain. A highly conserved CXXC motif is present in the N-terminal domain of NlpE.
How are the RcsF-β-barrel complexes formed? BamA, the essential β-barrel of the multiprotein Bam complex that assembles β-barrels in the OM, was among the proteins that were cross-linked to RcsF (16, 41), suggesting that Bam promotes the assembly of complexes between RcsF and β-barrels. In this hypothesis, the BamA-RcsF complex would be an intermediate in the formation of the RcsF-OmpA, -OmpC, and -OmpF complexes during their assembly in the OM. Several lines of evidence support this model. First, less OmpA-RcsF complex was observed in strains depleted of BamA or lacking bamE, a gene encoding a nonessential lipoprotein of the Bam complex (OmpC and OmpF were not tested) (16, 41, 46). Overexpression of BamA alone, which cannot function when expressed without the other Bam components, had a similar impact (16). In addition, the OmpA-RcsF complex could be formed in vitro when denatured OmpA was allowed to refold in the presence of RcsF but not when RcsF was added to previously refolded OmpA (41). The direct role of Bam in this process remains, however, to be clearly established.
RscF uses its interaction with BamA and β-barrels to detect stress in the envelope.
RcsF activates the Rcs system in response to different types of envelope stress. These include defects in the LPS layer caused by cationic antimicrobial peptides (29) or mutations in the LPS biosynthesis pathway (26), and alterations in the peptidoglycan caused by exposure to lysozyme (47), β-lactam antibiotics (48), or an MreB inhibitor (16). In addition, decreased levels of BamA (16) or defects in the biosynthesis of certain phospholipids (28) also induce Rcs via RcsF. Both the complexity of the Rcs system and the large diversity of conditions leading to its activation have hampered the elucidation of the stress-sensing mechanism(s). Recent studies from the Silhavy lab (41) and from ours (16) have started, however, to shed some light into this.
In collaboration with the Typas lab, we reported that the interaction between RcsF and BamA is key in the ability of this lipoprotein to activate Rcs under stress (16). In particular, we found that stressing the cells with the cationic antimicrobial peptide polymyxin B or with the β-lactam antibiotic amdinocillin, which inhibits the essential transpeptidase penicillin-binding protein 2 (PBP2), decreases the levels of the BamA-RcsF complex. Our results suggested that this decrease was due to the inability of the newly synthesized RcsF to interact with BamA under stress. On the basis of these results, we proposed the following model for RcsF sensing (Fig. 1C, left). In the absence of stress, BamA continuously funnels RcsF to β-barrels (OmpA/C/F). In this process, at least portions of RcsF become exposed on the surface. When bound to BamA or OmpA/C/F, RcsF is prevented from interacting with IgaA and cannot activate Rcs. Stress conditions impair the ability of RcsF to interact with BamA and therefore to be funneled to OmpA/C/F. This keeps RcsF exposed to the periplasm, free to activate the Rcs cascade. Because a functional Bam machinery is required to funnel RcsF to OmpA/C/F, we proposed that RcsF senses envelope damages by monitoring the activity of the Bam complex (16). Further work will be required to understand how envelope stress prevents newly synthesized RcsF from interacting with BamA.
An alternative model has been proposed by Konovalova et al. to explain how RcsF activates Rcs in response to LPS defects (Fig. 1C, right). In this model, RcsF, when in complex with β-barrels, uses its positively charged surface-exposed N-terminal linker to directly sense perturbations in the LPS, detecting when the interactions that take place between LPS molecules are disturbed (41). This model is based on (i) the observation that exposure to polymyxin B turns on Rcs even when protein synthesis is blocked, therefore indicating that Rcs induction in response to LPS defects does not require new RcsF synthesis; (ii) the fact that LPS alterations do not induce Rcs in cells lacking ompA or defective in the assembly of OmpA-RcsF, indicating that the formation of this complex is necessary for stress sensing; and (iii) the finding that the positively charged amino acids in the RcsF linker are important for sensing LPS stress. Because LPS defects do not cause the disassembly of the OmpA-RcsF complex, it remains to be established how RcsF can interact both with OmpA in the OM and with IgaA in the IM under stress conditions.
Due to the intricacy of the Rcs system, the many factors involved, and the large variety of inputs that can trigger the cascade, our knowledge of its detailed functioning remains incomplete despite these recent major advances. The two models described above are not necessarily mutually exclusive and could, for instance, be used to sense different types of stress. Carefully dissecting the Rcs system, and in particular how RcsF interacts with its different partners with or without stress, is a prerequisite to understand its detailed functioning at the molecular level.
MECHANISM OF STRESS SENSING BY NlpE
Is NlpE sensing surface adhesion?
Most Cpx-inducing cues trigger the TCS independently of NlpE (Table 1), hinting to a major difference with RcsF on which most Rcs-inducing signals rely. Actually, there are only two reports of an NlpE-dependent activation of the Cpx pathway in E. coli other than overproduction (49), and both studies propose that NlpE is turning on Cpx upon adhesion of the bacterium to surfaces. In the first study published 15 years ago by Otto and Silhavy, the transcription of several Cpx reporters quickly increased in E. coli cells that attached to hydrophobic acid-cleaned glass beads compared to planktonic cells (49). For reasons that remain unknown, Cpx activation by adhesion occurred only in stationary-phase cells grown in rich medium. Interestingly, the activity of a Cpx reporter did not increase upon adhesion of cells lacking nlpE, cpxR, or cpxA, suggesting that NlpE is required to signal surface attachment to the TCS. Furthermore, it was proposed that the Cpx system and NlpE facilitate the adhesion of E. coli to another type of abiotic surface (hydrophobic quartz crystals). More recently, the potential role of NlpE in the adhesion-induced Cpx response was investigated in an enterohemorrhagic E. coli (EHEC) strain (50). Consistent with previous findings, Cpx activity increased in a nlpE-dependent manner when stationary-phase EHEC cells grown in rich medium adhered to hydrophobic, but not hydrophilic, glass beads (50). Interestingly, the Cpx system was also turned on when EHEC attached to intestinal epithelial cells, although to a lesser extent than on glass beads and in a manner that was partially dependent on NlpE (50). In this assay, Cpx was activated only when a concentrated bacterial inoculum was used, suggesting the existence of additional NlpE-independent factors that depend on cell density, in addition to the known stimulating effect of stationary phase on the Cpx TCS. Intriguingly, Cpx induction was NlpE dependent when EHEC cells were placed in contact with undifferentiated epithelial cells but not with differentiated or HeLa cells, suggesting that a specific feature on the cell surface is recognized by NlpE directly or indirectly, or that it modulates the NlpE-Cpx pathway. While both reports indicate that NlpE could be involved in sensing adhesion to specific surfaces and triggering the Cpx pathway, possible polar effects were not completely ruled out. Moreover, the physiological significance of E. coli adhering to the tested abiotic surfaces remains to be determined, and it is unknown if stationary phase and high cell density mimic physiological changes that occur in bacteria when they encounter epithelial cells or other surfaces in vivo. Since the Cpx response affects virulence in several species (reviewed in reference 21), it would be interesting to see if nlpE mutants are less virulent than wild-type cells in an in vivo infection model to validate the importance of NlpE in surface adhesion.
Finally, additional studies support a role for NlpE in biofilm formation (which is intimately linked to surface adhesion), although it is unknown if this effect goes through the Cpx system. For instance, it was found that nlpE deletion has a negative impact on the production of E. coli biofilms (51). Moreover, a multidrug-resistant clinical strain of Acinetobacter baumannii, which is more prone to biofilm formation than the wild type, has higher levels of NlpE in its envelope than the wild-type strain (52), raising the intriguing possibility of a connection between elevated levels of NlpE and surface adhesion and virulence. However, the NlpE homolog in A. baumannii lacks the C-terminal domain found in E. coli (Fig. 2), so it is possible that NlpE functions differently in these species. Hence, the physiological implication of NlpE in sensing surfaces and transmitting this information to the Cpx system awaits further investigation.
Hints on NlpE mode of action from its crystal structure.
Insights into the primary function of NlpE may eventually come from its structure, which was solved in 2007 using a soluble form of the protein (lacking the signal sequence and a short N-terminal stretch) (53) (Fig. 2). NlpE harbors two globular domains: an 8-stranded antiparallel β-barrel at the N terminus, which shares some similarity with the lipid-binding bacterial lipocalin Blc, and a C-terminal domain that resembles the OB (oligonucleotide/oligosaccharide) fold and is composed of a 5-stranded antiparallel β-barrel and one α-helix. Strikingly, a search for homologs in other species revealed two classes of NlpE: type I NlpE have both the N-terminal and C-terminal domains (as in E. coli, Fig. 2), while type II only carry the N-terminal domain (as in A. baumannii), suggesting the unexplored possibility that the C-terminal domain has been selected during evolution to modulate the function of the N-terminal domain of NlpE. Another intriguing feature of NlpE is the highly conserved CXXC in the N-terminal domain, a motif that usually performs redox or metal binding functions in other proteins (54, 55) but has an unknown role in NlpE. The redox state of the solution seems to influence the formation of a disulfide bond in the CXXC motif of purified NlpE, suggesting that this is redox sensitive (53), but this has never been shown in vivo. Two more cysteines are present in the C-terminal domain, where they form a structural disulfide bond (53). Interestingly, the NlpE crystal contained an asymmetric unit of two molecules arranged in a three-dimensional-domain-swapped dimer in which the N-terminal β-barrel of one NlpE molecule accommodates one strand from the C-terminal domain of the second molecule. It was also proposed that the linker region between the two domains, which appears unstructured and highly flexible in the monomer, contains a β-sheet in the domain-swapped dimer. However, NlpE is monomeric in solution (53); (A. Delhaye, G. Laloux, and J.-F. Collet, unpublished data), suggesting that it exists as a monomer in vivo as well and questioning the biological relevance of the swapped dimer. Altogether, these structural features led Hirano et al. to hypothesize that unfolding at the N terminus (upon metal binding by the CXXC motif, for example) may extend NlpE, allowing the protein to reach a partner, like CpxA, for instance, via its C-terminal domain while still being anchored to the OM (53). Alternatively, or in addition, modification of the relative orientation of the N- and C-terminal domains may trigger Cpx via an unknown mechanism. So far, these attractive models, including the putative contact between NlpE and CpxA, have not been verified experimentally. A systematic functional dissection of NlpE in vivo should reveal how this lipoprotein talks to the Cpx pathway.
NlpE, LIKE Cpx, CONTRIBUTES TO CELL SURVIVAL WHEN LIPOPROTEIN CONTENT AND TRAFFICKING ARE ALTERED
Very recently, the group of Silhavy challenged the traditional view that all components of the Lol system, which extracts lipoproteins from the IM and transports them to the OM, are essential. Indeed, they found that E. coli cells lacking both the periplasmic chaperone LolA, which escorts lipoproteins across the periplasm, and the OM lipoprotein LolB, which inserts lipoproteins in the OM, could survive, provided that the most abundant OM lipoprotein, Lpp, was absent, that the Rcs system was shut down, and that the Cpx response was constitutively on (40). This led to the interesting possibility that under certain conditions, a novel route becomes accessible upon Cpx activation to direct lipoproteins to the OM, thereby ensuring cell survival. Indeed, essential machineries, like Bam or Lpt, absolutely need OM lipoproteins to function (7, 56). Rcs-off Lpp-lacking LolB-depleted cells barely grew in the absence of cpxR (40), consistent with a protective role of the Cpx response when cells face Lol deficiency in this mutant background. Interestingly, survival of the Rcs-off Lpp-lacking LolB-depleted cells was similarly compromised when nlpE was missing (while cpxR was present), suggesting that NlpE could be solely responsible for signaling lipoprotein defects to the Cpx system (40). Assuming that NlpE remains stuck in the IM in the tested mutant strain, it was therefore hypothesized that the Cpx system can provide a beneficial response to lipoprotein trafficking problems by monitoring NlpE localization (40) (remember that NlpE mislocalization to the IM is known to activate the Cpx response [37, 39]). Although these findings could point to the first physiological role for NlpE as a sentinel for the Cpx two-component system (40), direct evidence showing that defaults in lipoprotein trafficking turn on the Cpx response, and that this is dependent on NlpE, remains to be provided. Also, whether NlpE signals lipoprotein-sorting defects to Cpx only in cells lacking Lpp and a functional Rcs or in other more physiological backgrounds, is unknown. If confirmed, such a new “sentinel” role of NlpE will trigger future work to uncover the underlying molecular mechanism leading to Cpx activation.
CONCLUSION AND UNSOLVED MYSTERIES
The remarkable similarities between RcsF and NlpE raise at least two attractive hypotheses. First, it may suggest that the modulation of signaling pathways by OM lipoproteins represent a paradigm to scan the status of all three envelope layers via a molecular connection between the most outward and inward shields of the cell. In the case of stress signaling, this eventually leads to the signal being transferred from outside the cell to the decision center in the cytoplasm. Following this idea, the case of RcsF and NlpE is reminiscent of other OM lipoproteins that take part into transenvelope complexes to fine-tune the activity of cell wall synthesis proteins occupying more inward positions (57, 58).
Second, we may envision that the molecular mechanism of Cpx activation by NlpE, which is still largely mysterious, shares one or more principles with the one used by RcsF to detect envelope stress and communicate with the Rcs components. Hence, it would be particularly interesting to determine if NlpE also interacts with β-barrels in the OM, if it is occluded under nonstress conditions by exposure at the cell surface, or if it associates directly or not with Cpx proteins, like CpxA or CpxP. However, it is also conceivable that NlpE and RcsF function in completely distinct ways, since NlpE seems to be dispensable under most known Cpx-activating conditions, whereas the opposite is true for RcsF. Thus, the different requirements of the Cpx and Rcs systems for an OM lipoprotein sentinel potentially reflect different molecular mechanisms of activation employed by NlpE and RcsF. Alternatively, we cannot exclude the possibility that an unknown category of Cpx-inducing stress remains to be discovered, which may lead to the identification of novel NlpE-dependent signals. Uncovering the mechanistic detail of Cpx activation by NlpE will certainly require extensive work, and this is even more true considering that the molecular nature of the signal(s) sensed by the IM sensor CpxA is still unknown. Moreover, we cannot exclude the possibility that NlpE plays another role in a pathway unrelated to Cpx.
Whereas recent discoveries on the stress-sensing mechanism of RcsF have greatly improved our understanding of how the Rcs pathway is functioning, several key questions remain unsolved. For instance, what are the dynamics and precise topology of RcsF surface exposure when in complex with large (OmpC/F) and small (OmpA) β-barrel proteins, with or without stress? How do stress conditions prevent RcsF from interacting with BamA? Is the mechanism used to sense LPS defects different from the one used to detect PG alterations? Does the newly synthesized RcsF play a different role in stress sensing than the “old” RcsF? Finally, future research should also reveal how Bam assembles complexes between RcsF and β-barrels and test whether this constitutes a novel mechanism to export lipoproteins on the surface of E. coli and other bacteria.
ACKNOWLEDGMENTS
We are grateful to the members of our lab for helpful comments and discussions. We apologize to all authors whose work could not be cited due to space limitations. The title of this review was inspired by David Bowie's song “Space Oddity.”
G.L. is a chargée de recherche and J.-F.C. is maître de recherche of the FRS-FNRS and an Investigator of the FRFS-WELBIO. This work was supported by grants from the WELBIO and the FRS-FNRS.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dufresne K, Paradis-Bleau C. 2015. Biology and assembly of the bacterial envelope. Adv Exp Med Biol 883:41–76. doi: 10.1007/978-3-319-23603-2_3. [DOI] [PubMed] [Google Scholar]
- 3.Szewczyk J, Collet JF. 2016. The journey of lipoproteins through the cell: one birthplace, multiple destinations. Adv Microb Physiol 69:1–50. doi: 10.1016/bs.ampbs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Goemans C, Denoncin K, Collet J-F. 2013. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta 1843:1517–1528. doi: 10.1016/j.bbamcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollauer SE, Sooreshjani MA, Noinaj N, Buchanan SK. 2015. Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc Lond B Biol Sci 370:20150023. doi: 10.1098/rstb.2015.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz N, Silhavy TJ. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol 8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 10.MacRitchie DM, Buelow DR, Price NL, Raivio TL. 2008. Two-component signaling and Gram negative envelope stress response systems Adv Exp Med Biol 631:80–110. doi: 10.1007/978-0-387-78885-2_6. [DOI] [PubMed] [Google Scholar]
- 11.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunke S, Keller R, Müller VS. 2011. Signal integration by the Cpx-envelope stress system. FEMS Microbiol Lett 326:12–22. doi: 10.1111/j.1574-6968.2011.02436.x. [DOI] [PubMed] [Google Scholar]
- 14.Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol 40:440–450. doi: 10.1046/j.1365-2958.2001.02393.x. [DOI] [PubMed] [Google Scholar]
- 15.Domínguez-Bernal G, Pucciarelli MG, Ramos-Morales F, García-Quintanilla M, Cano DA, Casadesús J, García-del Portillo F. 2004. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol Microbiol 53:1437–1449. doi: 10.1111/j.1365-2958.2004.04213.x. [DOI] [PubMed] [Google Scholar]
- 16.Cho S-H, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, Asmar A, Laloux G, Hov A-K, Leverrier P, Van der Henst C, Vertommen D, Typas A, Collet J-F. 2014. Detecting envelope stress by monitoring β-barrel assembly. Cell 159:1652–1664. doi: 10.1016/j.cell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 17.Raivio TL, Popkin DL, Silhavy TJ. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol 181:5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Keller R, Volkmer R, Krauss N, Scheerer P, Hunke S. 2011. Structural basis for two-component system inhibition and pilus sensing by the auxiliary CpxP protein. J Biol Chem 286:9805–9814. doi: 10.1074/jbc.M110.194092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raivio TL. 2014. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta 1843:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Grabowicz M, Silhavy TJ. 2017. Envelope stress responses: an interconnected safety net. Trends Biochem Sci 42:232–242. doi: 10.1016/j.tibs.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt SL, Raivio TL. 2011. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 22.Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol 5:1173–1184. doi: 10.2217/fmb.10.83. [DOI] [PubMed] [Google Scholar]
- 23.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 24.Gervais FG, Drapeau GR. 1992. Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J Bacteriol 174:8016–8022. doi: 10.1128/jb.174.24.8016-8022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majdalani N, Hernandez D, Gottesman S. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol 46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 26.Majdalani N, Heck M, Stout V, Gottesman S. 2005. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J Bacteriol 187:6770–6778. doi: 10.1128/JB.187.19.6770-6778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J Bacteriol 185:5735–5746. doi: 10.1128/JB.185.19.5735-5746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiba Y, Yokoyama Y, Aono Y, Kiuchi T, Kusaka J, Matsumoto K, Hara H. 2004. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J Bacteriol 186:6526–6535. doi: 10.1128/JB.186.19.6526-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farris C, Sanowar S, Bader MW, Pfuetzner R, Miller SI. 2010. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J Bacteriol 192:4894–4903. doi: 10.1128/JB.00505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castanié-Cornet M-P, Cam K, Jacq A. 2006. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J Bacteriol 188:4264–4270. doi: 10.1128/JB.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet 3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder WB, Silhavy TJ. 1995. Beta-galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J Bacteriol 177:953–963. doi: 10.1128/jb.177.4.953-963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cosma CL, Danese PN, Carlson JH, Silhavy TJ, Snyder WB. 1995. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol 18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- 35.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev 9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 36.Tao K, Narita S-I, Tokuda H. 2012. Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J Bacteriol 194:3643–3650. doi: 10.1128/JB.00553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delhaye A, Collet J-F, Laloux G. 2016. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7(1):e00047-16. doi: 10.1128/mBio.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiba Y, Miyagawa H, Nagahama H, Matsumoto K, Kondo D, Matsuoka S, Hara H. 2012. Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiology 158:1238–1248. doi: 10.1099/mic.0.056945-0. [DOI] [PubMed] [Google Scholar]
- 39.Miyadai H, Tanaka-Masuda K, Matsuyama S-I, Tokuda H. 2004. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem 279:39807–39813. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]
- 40.Grabowicz M, Silhavy TJ. 2017. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci U S A 114:4769–4774. doi: 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. 2014. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc Natl Acad Sci U S A 111:E4350–E4358. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin JW, Schultz PG. 2002. In vivo photocrosslinking with unnatural amino acid mutagenesis. Chembiochem 3:1135–1137. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Leverrier P, Declercq J-P, Denoncin K, Vertommen D, Hiniker A, Cho S-H, Collet J-F. 2011. Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J Biol Chem 286:16734–16742. doi: 10.1074/jbc.M111.224865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogov VV, Rogova NY, Bernhard F, Lohr F, Dotsch V. 2011. A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF. J Biol Chem 286:18775–18783. doi: 10.1074/jbc.M111.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reusch RN. 2012. Insights into the structure and assembly of Escherichia coli outer membrane protein A. FEBS J 279:894–909. doi: 10.1111/j.1742-4658.2012.08484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konovalova A, Mitchell AM, Silhavy TJ. 2016. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 5:5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callewaert L, Vanoirbeek KGA, Lurquin I, Michiels CW, Aertsen A. 2009. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J Bacteriol 191:1979–1981. doi: 10.1128/JB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otto K, Silhavy TJ. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc Natl Acad Sci U S A 99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu T, Ichimura K, Noda M. 2015. The surface sensor NlpE of enterohemorrhagic Escherichia coli contributes to regulation of the type III secretion system and flagella by the Cpx response to adhesion. Infect Immun 84:537–549. doi: 10.1128/IAI.00881-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JAJ, Molin S, Prensier G, Arbeille B, Ghigo J-M. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- 52.Siroy A, Cosette P, Seyer D, Lemaître-Guillier C, Vallenet D, Van Dorsselaer A, Boyer-Mariotte S, Jouenne T, Dé E. 2006. Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J Proteome Res 5:3385–3398. doi: 10.1021/pr060372s. [DOI] [PubMed] [Google Scholar]
- 53.Hirano Y, Hossain MM, Takeda K, Tokuda H, Miki K. 2007. Structural studies of the Cpx pathway activator NlpE on the outer membrane of Escherichia coli. Structure 15:963–976. doi: 10.1016/j.str.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Quan S, Schneider I, Pan J, Hacht Von A, Bardwell JCA. 2007. The CXXC motif is more than a redox rheostat. J Biol Chem 282:28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 55.Wee SK, Burns JL, DiChristina TJ. 2013. Identification of a molecular signature unique to metal-reducing Gammaproteobacteria. FEMS Microbiol Lett 350:90–99. doi: 10.1111/1574-6968.12304. [DOI] [PubMed] [Google Scholar]
- 56.Selkrig J, Leyton DL, Webb CT, Lithgow T. 2014. Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta 1843:1542–1550. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 57.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, Blaauwen Den T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer membrane proteins. Cell 143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DiGiuseppe PA, Silhavy TJ. 2003. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol 185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danese PN, Silhavy TJ. 1998. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 180:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Wulf P, Lin EC. 2000. Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. J Bacteriol 182:1423–1426. doi: 10.1128/JB.182.5.1423-1426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danese PN, Oliver GR, Barr K, Bowman GD, Rick PD, Silhavy TJ. 1998. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J Bacteriol 180:5875–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Itou A, Matsumoto K, Hara H. 2012. Activation of the Cpx phosphorelay signal transduction system in acidic phospholipid-deficient pgsA mutant cells of Escherichia coli. Biochem Biophys Res Commun 421:296–300. doi: 10.1016/j.bbrc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Mileykovskaya E, Dowhan W. 1997. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol 179:1029–1034. doi: 10.1128/jb.179.4.1029-1034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet 5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans KL, Kannan S, Li G, de Pedro MA, Young KD. 2013. Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli. J Bacteriol 195:4415–4424. doi: 10.1128/JB.00596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nevesinjac AZ, Raivio TL. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J Bacteriol 187:672–686. doi: 10.1128/JB.187.2.672-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto K, Ishihama A. 2005. Transcriptional response of Escherichia coli to external copper. Mol Microbiol 56:215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- 69.Isaac DD, Pinkner JS, Hultgren SJ, Silhavy TJ. 2005. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc Natl Acad Sci U S A 102:17775–17779. doi: 10.1073/pnas.0508936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jubelin G, Vianney A, Beloin C, Ghigo J-M, Lazzaroni J-C, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol 183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653–662. doi: 10.1046/j.1365-2443.2002.00554.x. [DOI] [PubMed] [Google Scholar]
- 73.Shimohata N, Nagamori S, Akiyama Y, Kaback HR, Ito K. 2007. SecY alterations that impair membrane protein folding and generate a membrane stress. J Cell Biol 176:307–317. doi: 10.1083/jcb.200611121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kashyap DR, Wang M, Liu L-H, Boons G-J, Gupta D, Dziarski R. 2011. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat Med 17:676–683. doi: 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shiba Y, Matsumoto K, Hara H. 2006. DjlA negatively regulates the Rcs signal transduction system in Escherichia coli. Genes Genet Syst 81:51–56. doi: 10.1266/ggs.81.51. [DOI] [PubMed] [Google Scholar]
- 76.Potrykus J, Wegrzyn G. 2004. The ypdI gene codes for a putative lipoprotein involved in the synthesis of colanic acid in Escherichia coli. FEMS Microbiol Lett 235:265–271. doi: 10.1111/j.1574-6968.2004.tb09598.x. [DOI] [PubMed] [Google Scholar]
- 77.Ferrières L, Clarke DJ. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol 50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]