ABSTRACT
The aim of this study was to characterize the plasmids carrying antimicrobial resistance (AMR) determinants in multiple Salmonella serotypes recovered from the commercial swine farm environment after manure application on land. Manure and soil samples were collected on day 0 before and after manure application on six farms in North Carolina, and sequential soil samples were recollected on days 7, 14, and 21 from the same plots. All environmental samples were processed for Salmonella, and their plasmid contents were further characterized. A total of 14 isolates including Salmonella enterica serotypes Johannesburg (n = 2), Ohio (n = 2), Rissen (n = 1), Typhimurium var5− (n = 5), Worthington (n = 3), and 4,12:i:− (n = 1), representing different farms, were selected for plasmid analysis. Antimicrobial susceptibility testing was done by broth microdilution against a panel of 14 antimicrobials on the 14 confirmed transconjugants after conjugation assays. The plasmids were isolated by modified alkaline lysis, and PCRs were performed on purified plasmid DNA to identify the AMR determinants and the plasmid replicon types. The plasmids were sequenced for further analysis and to compare profiles and create phylogenetic trees. A class 1 integron with an ANT(2″)-Ia-aadA2 cassette was detected in the 50-kb IncN plasmids identified in S. Worthington isolates. We identified 100-kb and 90-kb IncI1 plasmids in S. Johannesburg and S. Rissen isolates carrying the blaCMY-2 and tet(A) genes, respectively. An identical 95-kb IncF plasmid was widely disseminated among the different serotypes and across different farms. Our study provides evidence on the importance of horizontal dissemination of resistance determinants through plasmids of multiple Salmonella serotypes distributed across commercial swine farms after manure application.
IMPORTANCE The horizontal gene transfer of antimicrobial resistance (AMR) determinants located on plasmids is considered to be the main reason for the rapid proliferation and spread of drug resistance. The deposition of manure generated in swine production systems into the environment is identified as a potential source of AMR dissemination. In this study, AMR gene-carrying plasmids were detected in multiple Salmonella serotypes across different commercial swine farms in North Carolina. The plasmid profiles were characterized based on Salmonella serotype donors and incompatibility (Inc) groups. We found that different Inc plasmids showed evidence of AMR gene transfer in multiple Salmonella serotypes. We detected an identical 95-kb plasmid that was widely distributed across swine farms in North Carolina. These conjugable resistance plasmids were able to persist on land after swine manure application. Our study provides strong evidence of AMR determinant dissemination present in plasmids of multiple Salmonella serotypes in the environment after manure application.
KEYWORDS: Salmonella, plasmid, antimicrobial resistance, horizontal gene transfer, environment, environmental microbiology, swine farm
INTRODUCTION
The emergence of antimicrobial resistance (AMR) in bacterial pathogens has threatened the sustainability of an effective global public health response to infectious diseases (1, 2). There are major gaps in our understanding of AMR transmission within agricultural sites and the potential impacts on humans, animals, and the environment due to a lack of studies conducted on actual commercial food animal farms (3–5). A number of studies have documented the abundance of AMR pathogens associated with livestock production due to the intensive use of antimicrobials in animal husbandry practices for therapeutic and nontherapeutic purposes (6–9). However, there is limited knowledge about the effect of manure application on the spread of AMR pathogens and AMR genes by means of horizontal gene transfer (HGT), such as by plasmids, transposons, and integrons, in the environment (4, 10, 11). Exposure of bacterial pathogens to antimicrobials in the environment increases the evolution of resistance and has an influence on the abundance, distribution, and transfer of AMR genes into different bacterial species (9, 12). We recently reported the dissemination of AMR Salmonella isolates in manure from commercial swine farms that were able to persist on land for at least 21 days after manure application, and it was clearly observed that Salmonella bacteria were rarely present in the soil before the land application (13). Given the potential risk of disseminating AMR Salmonella bacteria into the environment during manure application, we further characterized the plasmids that were detected in the multiple Salmonella serotypes isolated in our previous study.
The dissemination of undesirable AMR genes in Gram-negative pathogenic bacteria has been mainly regarded as the acquisition of multiple plasmid-located AMR genes by HGT (14, 15). Conjugation is considered the main mode of HGT of AMR genes among the Enterobacteriaceae family and helps to increase bacterial genetic diversity (16, 17). Plasmids conferring resistance have been identified as hindering antimicrobial therapy, including the use of extended-spectrum cephalosporins and fluoroquinolones, which are regarded as drugs of choice for bacterial infection in human clinical cases (14, 18, 19). Studies from several parts of the world have demonstrated the distribution of plasmids harboring extended-spectrum β-lactamase (ESBL) genes (blaCTX, blaSHV, blaCMY, and blaTEM) or ampC and plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, and qnrS) in Escherichia coli and Salmonella among animal, human, and environmental sources (16, 20–22). The presence of plasmid-mediated transfer of a recently identified mobile colistin resistance gene (mcr-1) is another example of the threat posed to public health (23, 24). The comparative analysis of mcr-1-containing plasmids maintained in the Enterobacteriaceae family revealed that they are disseminated in a broad host range, including human, animal, and food sources, and are now being reported from different countries worldwide (25–27). Plasmids that confer resistance in carbapenem-resistant Enterobacteriaceae (CRE) pose an urgent threat to public health with their global expansion (28, 29). Mollenkopf et al. (30) reported that the CRE carrying blaIMP-27 plasmids were recovered from the environment of a swine production area in the United States. The farm environment is considered a potential reservoir of AMR Salmonella strains that probably exchange AMR determinants with humans and animals by plasmid horizontal transfer (13, 22, 31, 32).
The objective of this study was to determine and characterize the resistance plasmid profiles isolated from multiple AMR Salmonella serotypes recovered in manure and environmental samples after land application of manure on commercial swine farms in North Carolina. To address this, we performed antimicrobial susceptibility testing (AST), plasmid replicon typing, conjugation assays, and plasmid sequencing to fully understand the role of these plasmids in transferring AMR determinants in the environment.
RESULTS
Salmonella serotypes and plasmid characterization.
A total of 14 different Salmonella serotypes isolated from commercial swine farms in North Carolina were selected to determine whether the AMR genes were located on transmissible plasmids. We also wanted to find out whether dissemination of AMR Salmonella bacteria through manure application assists in the transmission of genes via plasmids to other susceptible bacterial populations. Salmonella isolates collected from the swine farm environment after manure application were selected from each farm based on type of sample (lagoon and soil), sampling day, serotype, and resistance phenotype (Table 1). All 14 Salmonella donors harbored at least one large plasmid larger than 40 kb in size, and their plasmid profiles were dependent on farm origin and donor Salmonella serotype. PCR-based replicon typing (PBRT) revealed four plasmid replicons (FI, FII, I1, and N) among the 14 isolates carrying plasmids (Table 1). IncN plasmids (n = 3) of 50 kb in size were found in Salmonella enterica serotype Worthington isolates from both lagoon and soil samples in North Carolina farm 1 (NCF1). In NCF3, 100-kb IncI1 (n = 2) plasmids were isolated from S. enterica serotype Johannesburg while S. enterica serotype Typhimurium var5− was the predominant serotype at this farm and carried IncFII plasmids (n = 4) of 95 kb in size. Furthermore, IncFII plasmids were also found in S. Typhimurium var5− from NCF5 and 4,12:i:− from NCF6. A single S. enterica serotype Rissen isolate from a lagoon sample in NCF6 carried an IncI1 plasmid of 90 kb in size. The heterogeneous IncF group was the predominant replicon type detected in this study. Within the IncF group, we detected the subgroups FIA, FIB, FIC, FIIA, and Frep, with IncFIC and Frep being the most prevalent subgroups. The IncFI plasmid group found in 10 Salmonella isolates (Table 1) was determined to consist of small plasmids (less than 40 kb in size each). However, the plasmids identified in our study were represented by more than one replicon family in each isolate.
TABLE 1.
Conjugative resistance plasmid content of 14 environmental isolates harboring AMR genes recovered from Salmonella donor isolates after manure application on commercial swine farms in North Carolina
|
Salmonella donor isolate |
Plasmid |
|||||||
|---|---|---|---|---|---|---|---|---|
| Farm and sourcea | Day of sampling | Serotype | Name | Size (kb) | Inc groupb | ST (Inc group)c | Resistance pattern (MIC [μg/ml])d | AMR gene(s) |
| NCF1 | ||||||||
| Lagoon | 0 | Worthington | pS6 | 50 | N (50 kb), FI | ST5 (N) | FIS (>256), GEN (16), STR (64), TET (>32) | sul1, aadA2, tet(A) |
| Soil | 0 | Worthington | pS7 | 50 | N (50 kb), FI | ST5 (N) | FIS (>256), STR (64), TET (>32) | sul1, aadA2, tet(A) |
| Soil | 7 | Worthington | pS8 | 50 | N (50 kb), FI | ST5 (N) | FIS (>256), STR (64), TET (>32) | sul1, aadA2, tet(A) |
| NCF3 | ||||||||
| Lagoon | 0 | Johannesburg | pS9 | 100, 95 | I1 (100 kb), FI (95 kb) | ST12 (I1) | AMP (>32), AUG2 (32/16), AXO (16), FOX(32) | blaCMY-2 |
| Soil | 7 | Johannesburg | pS10 | 95 | I1, FI (95 kb) | AMP (>32), AUG2 (32/16), AXO (16), FOX (32) | blaCMY-2 | |
| Lagoon | 0 | Typhimurium var5− | pS12 | 95 | FII, FI | AMP (>32), FIS (>256) | sul1 | |
| Soil | 7 | Typhimurium var5− | pS13 | 95 | FII, FI | AMP(>32), FIS(>256) | sul1 | |
| Soil | 14 | Typhimurium var5− | pS14 | 95 | FII | AMP (>32), FIS (>256) | ||
| Soil | 21 | Typhimurium var5− | pS15 | 95 | FII | AMP (>32), FIS (>256) | ||
| NCF6 | ||||||||
| Lagoon | 0 | Rissen | pS20 | 90 | I1 (90 kb), FI | ST155 (I1) | TET(>32) | tet(A), tet(B) |
| Soil | 0 | 4,12:i:− | pS24 | 95 | FII, FI | AMP (>32), FIS (>256), STR (>64), TET (>32) | blaTEM, sul2, aadA | |
| NCF5 | ||||||||
| Lagoon | 0 | Typhimurium var5− | pS27 | 95 | FII, FI | AMP (>32), CHL (>32), FIS (>256), STR (32), TET (32) | sul1, aadA2 | |
| Lagoon | 0 | Ohio | pS28 | 40 | FI | TET (>32) | tet(A) | |
| Soil | 0 | Ohio | pS29 | 40 | FI | TET (>32) | tet(A) | |
NCF, North Carolina farm.
Incompatibility group based on PBRT (45).
Sequence type (ST) based on pMLST (https://pubmlst.org/plasmid/) (41).
Nalidixic acid (NAL) resistance was not detected in the plasmid isolated from the transconjugant. MIC ranges of the drugs are as follows: amoxicillin-clavulanic acid (AUG2), 1/0.5 to 32/16 μg/ml (breakpoint, ≥32/16 μg/ml); ampicillin (AMP), 1 to 32 μg/ml (breakpoint, ≥32 μg/ml); cefoxitin (FOX), 0.5 to 32 μg/ml (breakpoint, ≥32 μg/ml); ceftriaxone (AXO), 0.25 to 64 μg/ml (breakpoint, ≥4 μg/ml); chloramphenicol (CHL), 2 to 32 μg/ml (breakpoint, ≥32 μg/ml); gentamicin (GEN), 0.25 to 16 μg/ml (breakpoint, ≥16 μg/ml); streptomycin (STR), 32 to 64 μg/ml (breakpoint, ≥32 μg/ml); sulfisoxazole (FIS), 16 to 256 μg/ml (breakpoint, ≥512 μg/ml); and tetracycline (TET) 4 to 32 μg/ml (breakpoint, ≥16 μg/ml).
Antimicrobial resistance phenotypes.
To determine the AMR phenotypes and MICs for all 14 nalidixic acid-resistant (NALr) E. coli confirmed transconjugants and the 14 AMR Salmonella donor isolates from the environmental source, we conducted antimicrobial susceptibility testing using broth microdilution. The results of transconjugant AST correlated with the AMR profiles and the MICs for the Salmonella donor isolates, confirming the successful transfer of plasmids from the donors to the recipient strains (Table 2). NALr was detected in all 14 transconjugants since the NALr E. coli JM109 strain was used as a recipient for plasmid transfer. Five out of 14 plasmids were considered multidrug resistant (MDR; resistant to more than three classes of antimicrobials) including plasmids pS6 (S. Worthington donor), pS9 and pS10 (S. Johannesburg donor), pS24 (S. enterica 4,12:i:− donor), and pS27 (S. Typhimurium var5− donor) (Table 1). The plasmid pS6 showed resistance to sulfisoxazole (FIS), gentamicin (GEN), streptomycin (STR), and tetracycline (TET), while plasmids pS7 and pS8 had the MDR pattern FIS-STR-TET. These three transconjugants were successfully transferred to the recipient E. coli from S. Worthington donors recovered from NCF1, but only transconjugant pS6 had a 100% AMR profile that matched that of the donor isolate. Two plasmids, pS9 and pS10, were isolated from transconjugants of S. Johannesburg on NCF3 representing identical MDR patterns, with resistance to ampicillin (AMP), amoxicillin-clavulanic acid (AUG2), ceftriaxone (AXO), and cefoxitin (FOX). However, the ceftiofur (XNL) resistance represented in S. Johannesburg isolates was not detected in the transconjugants (Table 2). The plasmid pS27 from S. Typhimurium var5− recovered from NCF5 showed resistance to AMP, chloramphenicol (CHL), FIS, STR, and TET. Plasmids pS12, pS13, pS14, and pS15 isolated from transconjugants of S. Typhimurium var5− on farm 3 had the resistance pattern AMP-FIS. The plasmid from NCF6, pS24 with the MDR pattern AMP-FIS-STR-TET, was isolated from an S. enterica 4,12:i:− transconjugant. Another plasmid from farm 6 (pS20) from S. Rissen was resistant to only TET. All the transconjugants with AMP resistance were selected on Luria-Bertani (LB) plates with AMP and NAL as the markers, while the rest of the transconjugants were selected on NAL and TET marker LB plates.
TABLE 2.
Antimicrobial susceptibilities with MICs of AMR environmental Salmonella isolates and corresponding E. coli transconjugants
| Salmonella isolate or transconjuganta | MIC (μg/ml)b |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AUG2 | AXO | AZI | CHL | CIP | FIS | FOX | GEN | NAL | STR | SXT | XNL | TET | |
| S6 | <1 | <1/0.5 | <0.25 | 4 | 8 | <0.015 | >256 | 4 | 16 | 2 | 64 | <0.12/2.38 | 1 | >32 |
| TC-S6 | <1 | 2/1 | <0.25 | 2 | 8 | 0.06 | >256 | 4 | 16 | >32 | 64 | <0.12/2.38 | 0.5 | >32 |
| S7 | <1 | <1/0.5 | <0.25 | 4 | 8 | <0.015 | >256 | 4 | 16 | 2 | >64 | <0.12/2.38 | 0.5 | >32 |
| TC-S7 | 2 | 2/1 | <0.25 | 2 | 8 | 0.12 | >256 | 4 | 8 | >32 | 64 | <0.12/2.38 | 0.5 | >32 |
| S8 | <1 | <1/0.5 | <0.25 | 4 | 8 | <0.015 | >256 | 4 | >16 | 2 | 64 | <0.12/2.38 | 0.5 | >32 |
| TC-S8 | 2 | 2/1 | <0.25 | 2 | 8 | 0.12 | >256 | 2 | 8 | >32 | 64 | <0.12/2.38 | 0.5 | >32 |
| S9 | >32 | 32/16 | 16 | 8 | 8 | 0.03 | 256 | 32 | 0.5 | 4 | 4 | <0.12/2.38 | >8 | <4 |
| TC-S9 | >32 | 32/16 | 16 | 2 | 8 | 0.12 | <16 | 32 | <0.25 | >32 | 4 | <0.12/2.38 | 4 | <4 |
| S10 | >32 | 32/16 | 16 | 8 | 8 | 0.03 | 256 | 32 | 0.5 | 4 | 8 | <0.12/2.38 | >8 | <4 |
| TC-S10 | >32 | 32/16 | 16 | 2 | 8 | 0.25 | <16 | >32 | 0.5 | >32 | 4 | <0.12/2.38 | 4 | <4 |
| S12 | >32 | 8/4 | <0.25 | 4 | 8 | <0.015 | >256 | 2 | 0.5 | 4 | 8 | <0.12/2.38 | 0.5 | <4 |
| TC-S12 | >32 | 8/4 | <0.25 | 4 | 8 | 0.12 | >256 | 4 | 0.5 | >32 | 8 | <0.12/2.38 | 1 | <4 |
| S13 | >32 | 8/4 | <0.25 | 4 | 8 | <0.015 | >256 | 2 | 0.5 | 4 | 8 | 0.25/4.75 | 0.5 | <4 |
| TC-S13 | >32 | 8/4 | <0.25 | 4 | 8 | 0.12 | >256 | 4 | 0.5 | >32 | 8 | <0.12/2.38 | 1 | <4 |
| S14 | >32 | 8/4 | <0.25 | 4 | 8 | <0.015 | >256 | 2 | 0.5 | 4 | 8 | <0.12/2.38 | 1 | <4 |
| TC-S14 | >32 | 8/4 | <0.25 | 4 | 8 | 0.12 | >256 | 2 | 0.5 | >32 | 8 | <0.12/2.38 | 1 | <4 |
| S15 | >32 | <1/0.5 | <0.25 | 4 | 8 | 0.25 | >256 | 2 | 0.5 | 4 | 8 | <0.12/2.38 | 1 | <4 |
| TC-S15 | >32 | 8/4 | <0.25 | 4 | 8 | 0.25 | >256 | 2 | 0.5 | >32 | 16 | 0.25/4.75 | 1 | <4 |
| S20 | <1 | <1/0.5 | <0.25 | 8 | 8 | 0.03 | 64 | 4 | 0.5 | 4 | 4 | <0.12/2.38 | 1 | >32 |
| TC-S20 | 2 | 2/1 | <0.25 | 4 | 8 | 0.12 | <16 | 4 | <0.25 | >32 | 4 | <0.12/2.38 | <0.12 | >32 |
| S24 | >32 | 4/2 | <0.25 | 8 | 8 | 0.03 | >256 | 2 | 0.5 | 8 | >64 | <0.12/2.38 | 1 | >32 |
| TC-S24 | >32 | 8/4 | <0.25 | 4 | 8 | 0.12 | >256 | 2 | 0.5 | >32 | >64 | <0.12/2.38 | 0.5 | >32 |
| S27 | >32 | 32/16 | 8 | 8 | >32 | <0.015 | >256 | 16 | 0.5 | 4 | >64 | <0.12/2.38 | 8 | >32 |
| TC-S27 | >32 | 8/4 | <0.25 | 4 | >32 | 0.12 | >256 | 2 | <0.25 | >32 | 32 | <0.12/2.38 | 0.5 | 32 |
| S28 | <1 | <1/0.5 | <0.25 | 8 | 8 | <0.015 | 64 | 2 | <0.25 | 2 | 4 | <0.12/2.38 | 1 | >32 |
| TC-S28 | 2 | 2/1 | <0.25 | 8 | 8 | 0.12 | <16 | 8 | <0.25 | >32 | 4 | <0.12/2.38 | 0.5 | >32 |
| S29 | <1 | <1/0.5 | <0.25 | 4 | 8 | <0.015 | 64 | 2 | 0.5 | 4 | 8 | <0.12/2.38 | 1 | >32 |
| TC-S29 | 2 | 2/1 | <0.25 | 4 | 8 | 0.12 | <16 | 2 | <0.25 | >32 | <2 | <0.12/2.38 | 0.5 | >32 |
E. coli transconjugants are indicated by designations beginning with “TC.” Salmonella isolate designations begin with the letter “S.”
MIC ranges of the drugs are as follows: amoxicillin-clavulanic acid (AUG2), 1/0.5 to 32/16 μg/ml (breakpoint, ≥32/16 μg/ml); ampicillin (AMP), 1 to 32 μg/ml (breakpoint, ≥32 μg/ml); azithromycin (AZI), 0.12 to 16 μg/ml (breakpoint, ≥32 μg/ml); cefoxitin (FOX), 0.5 to 32 μg/ml (breakpoint, ≥32 μg/ml); ceftiofur (XNL), 0.12 to 8 μg/ml (breakpoint, ≥8 μg/ml); ceftriaxone (AXO), 0.25 to 64 μg/ml (breakpoint, ≥4 μg/ml); chloramphenicol (CHL), 2 to 32 μg/ml (breakpoint, ≥32 μg/ml); ciprofloxacin (CIP), 0.015 ± 4 μg/ml (breakpoint, ≥4 μg/ml); gentamicin (GEN), 0.25 to 16 μg/ml (breakpoint, ≥16 μg/ml); nalidixic acid (NAL), 0.5 to 32 μg/ml (breakpoint, ≥32 μg/ml); streptomycin (STR), 32 to 64 μg/ml (breakpoint, ≥32 μg/ml); sulfisoxazole (FIS), 16 to 256 μg/ml (breakpoint, ≥512 μg/ml); trimethoprim-sulfamethoxazole (SXT), 0.12/2.38 to 4/76 μg/ml (breakpoint, ≥4/76 μg/ml); and tetracycline (TET), 4 to 32 μg/ml (breakpoint, ≥16 μg/ml). Boldface indicates resistance of the Salmonella isolate or transconjugant to the antimicrobial.
Determination of antimicrobial resistance genes.
Following the conjugation experiment and AST, 14 AMR-encoding genes were tested using a PCR-based method (Table 3). Only eight of these marker genes, including blaCMY-2, blaTEM, sul1, sul2, aadA, aadA2, tet(A), and tet(B), were detected in plasmids. The blaCMY-2 gene was detected in a 100-kb IncI1 plasmid (pS9). The blaTEM gene was found in an IncFII plasmid (pS27). We detected tet(A) or tet(B) in plasmids that encoded tetracycline resistance. In plasmids carrying streptomycin resistance, aadA1, and aadA2 were found. The sul1 gene was the most prevalent among plasmids which were resistant to the antimicrobial sulfisoxazole. Plasmids pS14 and pS15 did not test positive for any AMR genes which were tested in this study. The resistance genotypes of all 14 plasmids are tabulated in Table 1.
TABLE 3.
Primers used for PCR detection of resistance genes
| Gene | Forward oligonucleotide sequence (5′ to 3′) | Reverse oligonucleotide sequence (5′ to 3′) | Expected size (bp) | Reference |
|---|---|---|---|---|
| blaCMY-2 | GACAGCCTCTTTCTCCACA | TGGAACGAAGGCTACGTA | 1015 | 76 |
| blaPSE-1 | TTTGGTTCCGCGCTATCTG | TACTCCGAGCACCAAATCCG | 150 | 77 |
| blaTEM | GCACGAGTGGGTTACATCGA | GGTCCTCCGATCGTTGTCAG | 860 | 78 |
| aadA | GTGGATGGCGGCCTGAAGCC | AATGCCCAGTCGGCAGCG | 528 | 79 |
| aadA2 | CGGTGACCATCGAAATTTCG | CTATAGCGCGGAGCGTCTCGC | 250 | 80 |
| strA | CCTGGTGATAACGGCAATTC | CCAATCGCAGATAGAAGGC | 548 | 79 |
| strB | ATCGTCAAGGGATTGAAACC | GGATCGTAGAACATATTGGC | 509 | 79 |
| sul1 | CGGACGCGAGGCCTGTATC | GGGTGCGGACGTAGTCAGC | 591 | 75 |
| sul2 | GCGCTCAAGGCAGATGGCATT | GCGTTTGATACCGGCACCCGT | 285 | 78 |
| cmlA | TGGACCGCTATCGGACCG | CGCAAGACACTTGGGCTGC | 641 | 75 |
| tet(A) | GCTACATCCTGCTTGCCTTC | CATAGATCGCCGTGAAGAGG | 210 | 40 |
| tet(B) | TTGGTTAGGGGCAAGTTTTG | GTAATGGGCCAATAACACCG | 659 | 40 |
| tet(C) | CTTGAGAGCCTTCAACCCAG | ATGGTCGTCATCTACCTGCC | 418 | 40 |
| tet(G) | CAGCTTTCGGATTCTTACGG | GATTGGTGAGGCTCGTTAGC | 844 | 40 |
Plasmid sequencing and analysis.
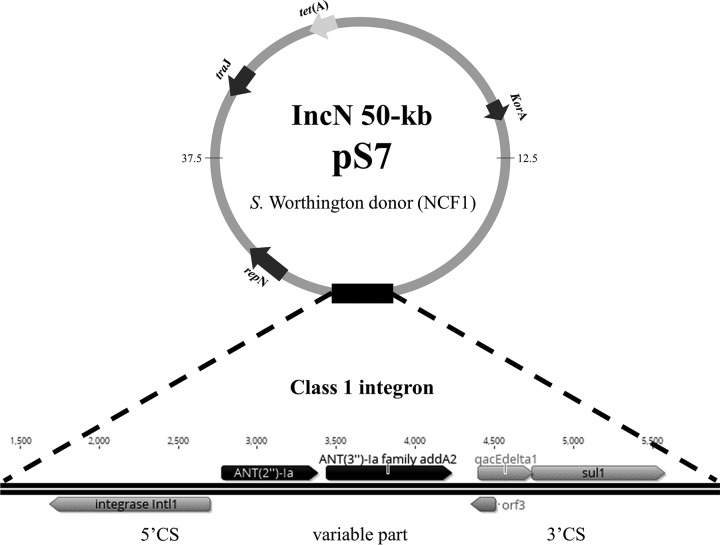
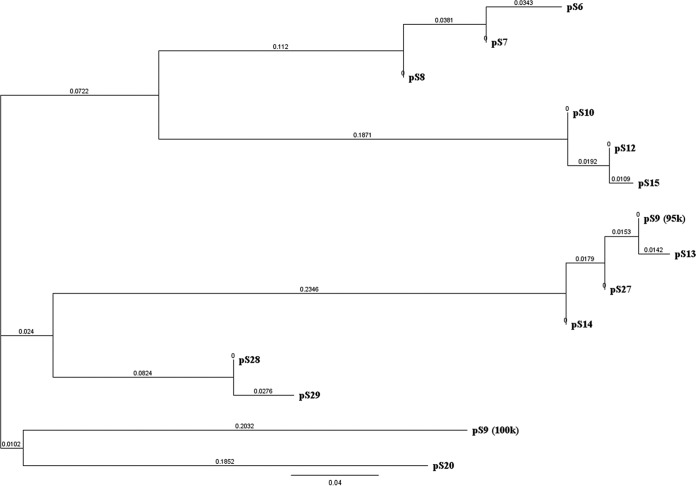
The incompatibility (Inc) group and resistance genes of plasmids were confirmed using sequencing (Table 1). Plasmid sequencing was able to identify the replicon families of each individual plasmid. A blastn comparison revealed that 95-kb IncF plasmids from different farms and serotypes (pS9, pS10, pS12, pS13, pS14, pS15, and pS27) (Table 1) were identical to another fully sequenced plasmid, pSTY1-H2662 previously isolated from S. Typhimurium from human stool (GenBank accession number CP014980) (33). A class 1 integron was identified in plasmids pS6 to pS8 isolated from S. Worthington using in silico analysis. This integron was comprised of a 5′ conserved segment (CS), variable part, and 3′ conserved segment (Fig. 1, pS7). The unusual variable part contained an ANT(2″)-Ia-aadA2 gene cassette, which is responsible for aminoglycoside resistance, while the sul1 gene was always found in the 3′ CS responsible for sulfonamide resistance. In addition, plasmid sequence analysis revealed the presence of VirB-family type IV secretion systems (T4SS) in all 14 plasmids, together with multiple tra genes, including traC, traF, traG, traI, traJ, traO, and traU. The evolutionary tree of 14 plasmid sequences was created using Geneious R10 software (Fig. 2). At 70% similarity, the plasmids from the same Salmonella donor were clustered together, including pS6, pS7, and pS8 (from S. Worthington) and pS28 and pS29 (from S. enterica serotype Ohio). The plasmids with distinct sizes, the 100-kb pS9 and 90-kb pS20, were separated from the other group. Plasmid pS24 was not included in the analysis because of the incomplete sequencing output. The plasmid multilocus sequence typing (pMLST) database revealed that three 50-kb IncN plasmids isolated from S. Worthington belonged to sequence type 5 (ST5). The IncI1 plasmid (pS9) isolated from S. Johannesburg was assigned to ST12 and clonal complex 12 (CC-12); another IncI1 plasmid (pS20) isolated from S. Rissen was typed as ST155, but the clonal complex was not defined (Table 1).
FIG 1.
Schematic representation of a class 1 integron in 50-kb IncN plasmid pS7: in the 5′conserved segment, the intl1 integrase gene; in the variable region, ANT(2″)-Ia, producing the aminoglycoside resistance enzyme, and aadA2, an ANT(3″)-Ia family aadA2 gene producing streptomycin resistance; in the 3′ conserved segment, qacEΔ1, a partially deleted gene that encodes quaternary ammonium compound resistance, sul1, producing sulfonamide resistance, and orf3, of unknown function, on the gene cassette recognized by the integrase. Arrows indicate the direction of the coding sequence.
FIG 2.
Phylogenetic diversity for sequences of 14 plasmids acquired from environmental Salmonella isolates. Evolutionary distances between plasmids were computed using a neighbor-joining algorithm. The distance was obtained from pairwise alignments with 70% similarity and no outgroup. The plasmid label names relate to data in Table 1. Phylogenetic analyses were conducted in Geneious R10.
DISCUSSION
The aim of the study was to characterize the plasmids identified in different AMR Salmonella serotypes isolated from a commercial swine farm environment after land application of manure. We also wanted to determine the role of plasmids in the dissemination of AMR genes to other potential bacterial recipients in the environment. The results potentially addressed the key role played by plasmids in the horizontal gene transfer that leads to the rapid proliferation of AMR genes in the environment. It is important to stress that our study was conducted at commercial swine farms and not at a research station in North Carolina, which is one of the top two leading pork-producing states in the United States. The Salmonella serotypes carrying multiple plasmids are common in the Enterobacteriaceae family (34). However, we focused on large (defined as being ≥40 kb in size) plasmids which are abundant in E. coli and Salmonella and comprise important pools of adaptive and transferable genetic information, especially AMR-corresponding genes, in these bacteria (34, 35). The large plasmids, in the range of 40 to 200 kb, have been suggested to be the necessary markers for extended-spectrum β-lactamases (ESBL), β-lactamase-encoding genes, and plasmid-mediated quinolone resistance (PMQR) (14, 16, 36). In our study, 5 out of 14 plasmids that we detected were 95 kb in size and were isolated from the S. Typhimurium var5− serotype (n = 5). The plasmid profiles of these five isolates were similar although they were recovered from different farms and at different time points, indicating the persistence of this plasmid in this serotype in the environment after manure deposition. The results correlated with those of a previous study that reported that Salmonella plasmids were conserved and primarily serotype specific, including those of S. Typhimurium and S. enterica serotype Heidelberg, and that they tended to persist for a long period in the environment (34). These plasmids were in contrast to E. coli plasmids which were more variable and not specific to particular strains (22, 34). The pS24 plasmid isolated from S. enterica 4,12:i:− had a profile similar to that of S. Typhimurium plasmids, and the parent strain was also isolated from a different swine farm environment. During the last decade, S. enterica 1,4,12:i:−, 1,4,[5],12:i:−, and 4,12:i;− have emerged around the world and have frequently been isolated from human, animal, agricultural production, and environmental sources (37–39). These serotypes are believed to be a mosaic variant of S. Typhimurium and are related to plasmid-mediated colistin resistance encoded by the mcr-1 gene (37, 39, 81). We detected one S. Rissen plasmid of approximately 90 kb that carried a tetracycline resistance marker. This is in comparison to our previous report where we identified from a farm environment in North Carolina a 90- to 100-kb plasmid in a tetracycline-resistant S. Rissen isolate carrying the tet(A) gene (31). This serotype is not common in the U.S. agricultural system and was identified for the first time in North Carolina swine farms in 2009 (42).
Typing of plasmid incompatibility (Inc), the inability of two plasmids of the same family to coexist in the same host cell, classifies plasmids based on their stability during conjugation (43, 82). This classification helps to categorize plasmids into clusters and relies on their phylogenetic relatedness, distribution in the host cells and environment, and their evolutionary origin (43, 44). Currently, 27 Inc groups are identified among the Enterobacteriaceae family (43, 45). On the basis of the PCR-based replicon typing (PBRT) method, 18 Inc groups were detected in our study. We used total plasmid DNA from each isolate in conducting PBRT, so the results did not differentiate individual plasmids in multiplasmid isolates. Most of the isolates were positive for more than one replicon family either because the isolates contained multiple plasmids from different incompatibility groups or because a single plasmid carried replication or partitioning genes from more than one incompatibility group. However, we were able to identify the exact replicon families after assessing the plasmid sequencing data (Table 1). We did not differentiate the heterogeneous IncF plasmids into individual groups because of their partitioning of replication genes (34), and the small (<40 kb in size) plasmids were not characterized in this study.
Particular plasmid Inc families, including IncN, IncI1, and IncF, are more frequently associated with the dissemination of AMR genes (14). These three plasmid Inc families have been associated with specific Salmonella serotypes and geographic farm areas in our study (34). The IncN family was detected in S. Worthington, which was consistently isolated from NCF1, while IncI1 was detected in S. Johannesburg isolated from NCF3. Both families are associated with large plasmids related to MDR phenotypes (Table 1). The IncF family was detected in multiple serotypes and farms (NCF3, -5, and -6). These findings are in accordance with those of previous studies that found that IncF and IncI1 are the most prevalent replicon types distributed among the Enterobacteraceae (14, 34). The IncI and IncF plasmids generally recovered from E. coli and Salmonella from human and animal sources are considered the source of several ESBL genes (14, 20, 23).
The IncFI group including FIA, FIB, and FIC, together with the IncFIIA subtype, was the most frequently detected replicon type in this study. All 14 Salmonella isolates carried at least one IncF plasmid. Our result supports the view that the IncF (both FI and FII) family was well adapted and commonly distributed in E. coli and Salmonella (14, 15, 34, 46). Wang et al. (14) reported that IncFIIA was detected only in S. enterica serotype Typhimurium, which correlates with our findings; however, we also detected the FIIA type in the S. enterica serotype 4,12:i:−. IncF family plasmids have been reported to contribute to the spread of AMR in Enterobacteriaceae and have been associated with specific genes conferring resistance to aminoglycosides, β-lactams, and quinolones (43, 46, 47).
Conjugative plasmids of the IncI1 replicon type were usually associated with multiple resistance compounds, especially extended-spectrum cephalosporinases of both the CTX-M and CMY types (47–49). The IncI1 plasmids carrying TEM-52 have been identified in E. coli and Salmonella cultured from humans and from chicken and turkey products in the European Union (50–52). The blaCMY-IncI1 plasmids linked to poultry, ground beef, and tomato sources have been identified to be responsible for ceftriaxone-resistant Salmonella outbreaks in the United States during 2011 and 2012 (18). Reports indicated that Salmonella enterica serotypes Heidelberg, Infantis, Typhimurium, and Newport were associated with IncI plasmids carrying the blaCMY gene. Similar to results of our study, IncI plasmids carrying the blaCMY gene were identified in a ceftriaxone-resistant S. enterica serotype Johannesburg isolate from a commercial swine farm environment sampled in our study.
IncN plasmids are the major vehicles for the dissemination of PMQR and ESBL genes, including blaCTX-M (22, 53, 54). In contrast to results of our study, IncN plasmids were identified in S. Worthington transconjugants and exhibited resistance to sulfisoxazole, streptomycin, and tetracycline but not to quinolones and ampicillin. Thus, characterization based on plasmid profiling and the corresponding Inc group using the PBRT technique is an essential tool for plasmid epidemiological surveillance, enhancing discrimination between Salmonella serotypes and tracing the spread of AMR genes (14, 16).
Multiple MDR-coding genes were found in plasmids. We detected plasmids carrying sul1 and sul2 genes conferring sulfisoxazole resistance, while plasmids with streptomycin resistance carried the aadA and aadA2 genes. Similarly, the tet(A) and tet(B) genes were found in plasmids in Salmonella strains that were resistant to tetracycline. β-Lactamase-encoding (bla) genes, including blaTEM and blaCMY, were detected in the plasmids which encoded the resistance to ampicillin and cephalosporin group antimicrobials. Several mechanisms are available for bla genes to support HGT between bacteria, thereby ensuring the spread of these markers to new hosts and the environment (14, 55). The heavy use of specific antimicrobials such as tetracycline plays a key role in plasmid dissemination and allows for the selection and enrichment of bacteria with multidrug-resistant plasmids (22, 56, 57).
The class 1 integron with an ANT(2″)-Ia-aadA2 gene cassette was detected in plasmids pS6 to pS8 retrieved from S. Worthington (Fig. 1, pS7). The integron had an unusual organization, with an ANT(2″)-Ia gene cassette which is responsible for resistance against gentamicin (58). The gentamicin resistance was not identified in pS7 but in Salmonella isolate S7 (pS7 donor) and pS6 (Tables 1 and 2). After BLAST analysis at NCBI, pS6 to pS8 showed genetic relatedness to a Klebsiella pneumoniae MDR IncN plasmid reported from Japan (59). However, the K. pneumoniae plasmid harbored different resistance genes than those we detected in the Salmonella serotypes from our study. The integrons are able to locate on either a chromosome or a mobile genetic element such as a plasmid (60). Several studies have stated that the integrons harboring aadA or a variant of aadA genes are common among Salmonella species (10, 61–63). The variable parts of integrons might be composed of variants of aad, dfr, or bla genes that contribute to aminoglycoside, sulfonamide, and cephalosporin resistance, respectively (10, 61). S. enterica serotype Worthington detected in our study is commonly found in poultry, poultry products, and the environment in several parts of the world and harbors integrons either on the chromosome or plasmids (62, 64–66). The presence of genetic elements such as integrons, transposons, and plasmids has consequently been associated with multidrug resistance phenotypes among Salmonella isolates (10). Our study reports an emerging multidrug-resistant clone isolated from Salmonella serotypes in a commercial swine farm environment carrying a large conjugative plasmid with an ANT(2″)-Ia-aadA2 gene cassette located on an integron.
Though the Salmonella plasmids were transferred to an E. coli JM109 recipient under laboratory conditions, the presence of VirB-family type IV secretion systems (T4SS) and tra genes in our study confirms that HGT by conjugation is likely to occur in the environment. The T4SS in Gram-negative bacteria functionally encompass the conjugation system and the effector translocators for interbacterial transfer of AMR genes, virulence determinants, and genes encoding other traits beneficial to the host (67). IncN plasmids (pS6 to pS8) and IncI1 plasmids (pS9 [100-kb] and pS20) employed TraJ, which the has ability to conjugate, and the conjugation process could be stimulated approximately 100-fold, demonstrating functional conservation of a significant regulatory feature of F-like conjugation modules (68).
The phylogenetic tree of 14 plasmids (Fig. 2) at 70% similarity suggested that the plasmids analyzed in our study were clustered based on the Salmonella donor serotypes, such as the S. Worthington cluster (pS6 to pS8) and S. Ohio cluster (pS28 and pS29). Within three Inc groups (IncI1, IncN, and IncF), the phylogenetic analysis also suggested the existence of an Inc group that is serotype specific (34). Based on the pMLST database, all IncN (pS6 to pS8) plasmids which were specific to S. enterica serotype Worthington belonged to the same ST5. These results were in accordance with the BLAST output for individual plasmids and the Salmonella clustering done by pulsed-field gel electrophoresis (PFGE) in our previous study (13).
Our study demonstrated that identical plasmids were recovered from different Salmonella serotypes isolated either from the same or different farm environments. Our findings provide evidence of a single, large 95-kb IncF plasmid being distributed across the swine production systems in North Carolina among different serotypes of Salmonella. In addition, we found that AMR plasmids were able to persist in the swine farm environment after manure application for a minimum period of 21 days (final sampling time point). The AMR determinants on these plasmids were transferable among Salmonella serotypes, which underlined the fact that manure deposition enriches the environmental resistome. We recommend conducting longitudinal studies on commercial food animal farms to determine the role of manure deposition on the environmental dissemination of AMR genes.
MATERIALS AND METHODS
Salmonella serotype selection.
A total of 168 AMR Salmonella isolates from commercial swine farm environments in North Carolina during 2013 to 2015 were tested for their plasmid components. The details of farm distribution, waste management systems, sample collection, and Salmonella isolation were described in a previous study (13). Briefly, manure samples from a lagoon and soil samples before and after manure spray application were collected on the first day (day 0) of the farm visit. The subsequent soil samples were collected on day 7, day 14, and day 21 from the same plots as on day 0. The serotyping, antimicrobial susceptibility testing (AST), and pulsed-field gel electrophoresis (PFGE) were performed for phenotypic and genotypic characterization of the Salmonella strains. The Salmonella isolates selected for plasmid characterization were chosen based on their temporal and spatial relationships, AMR profiles, AMR determinants, and PFGE fingerprint profiles. Based on the above criteria, a total of 14 isolates were finally selected for plasmid analysis and sequencing (Table 1). All isolates were maintained at −80°C in brucella broth (Difco, Becton-Dickinson, USA) until further characterization.
Conjugation experiments.
Conjugation experiments were conducted to evaluate intra- and interserovar transmission of AMR genes among AMR Salmonella serotypes. Fourteen AMR Salmonella isolates were selected to serve as donor strains, and the nalidixic acid-resistant (NALr) Escherichia coli JM109 strain was used as a recipient strain. A heat shock assay modified from Zeng et al. (69) was utilized for performing conjugation experiments. In brief, a loopful of overnight culture of the donor strain was gently mixed in Luria-Bertani (LB) broth (Difco, Becton-Dickinson, USA) with E. coli JM109. The donor and recipient DNA mixtures were kept on ice for 20 to 30 min, given heat shock in a water bath at 42°C for 30 to 60 s, and moved back on ice for 2 min. We added 250 to 1,000 μl of LB broth and incubated the culture mix at 37°C in a shaking incubator for 45 to 60 min. The culture mixtures were transferred to selective LB plates (Criterion; Hardy Diagnostics, USA) containing nalidixic acid (50 μg/ml) and one of the antimicrobials, depending on the resistance profile of the donor strain, and incubated at 37°C overnight. Transconjugants were confirmed on nontyphoidal Salmonella chromogenic plates (CHROMagar, Paris, France) and xylose lactose tergitol (XLT4) agar plates (Criterion; Hardy Diagnostics, USA). The antimicrobials and the concentrations used are as follows: ampicillin, 100 μg/ml; nalidixic acid, 50 μg/ml; and tetracycline, 20 μg/ml.
Antimicrobial susceptibility testing.
The transconjugant AMR and MIC profiles were determined by the broth microdilution method using a Gram-negative Sensititre (CMV3AGNF) plate (Trek Diagnostic Systems, OH). The panel of 14 antimicrobials tested include amoxicillin-clavulanic acid (AUG2; 1/0.5 to 32/16 μg/ml), ampicillin (AMP; 1 to 32 μg/ml), azithromycin (AZI; 0.12 to 16 μg/ml), cefoxitin (FOX; 0.5 to 32 μg/ml), ceftiofur (XNL; 0.12 to 8 μg/ml), ceftriaxone (AXO; 0.25 to 64 μg/ml), chloramphenicol (CHL; 2 to 32 μg/ml), ciprofloxacin (CIP; 0.015 to 4 μg/ml), gentamicin (GEN; 0.25 to 16 μg/ml), nalidixic acid (NAL; 0.5 to 32 μg/ml), streptomycin (STR; 2 to 64 μg/ml), sulfisoxazole (FIS; 16 to 256 μg/ml), trimethoprim-sulfamethoxazole (SXT; 0.12/2.38 to 4/76 μg/ml), and tetracycline (TET; 4 to 32 μg/ml). The MICs were determined, and breakpoints were interpreted based on the Clinical and Laboratory Standards Institute standards (CLSI) for broth microdilution (70, 71) and the National Antimicrobial Resistance Monitoring System (NARMS) (72). E. coli ATCC 25922 was used as a quality control strain. The transconjugants with MICs in the intermediate level were categorized as susceptible to avoid overestimation of resistance. The transconjugants with resistance to three or more classes of antimicrobials were classified as multidrug resistant (MDR).
Plasmid isolation.
Plasmid DNA was isolated from the confirmed transconjugant (NALr E. coli JM109) cultures by the modified alkali lysis method described by Sambrook et al. (73), which is suitable for the isolation of both large and small plasmids. The purified DNA concentrations of the plasmid extracts were calculated by measuring the absorbance at 260 and 280 nm using a NanoDrop ND-2000 Spectrophotometer (NanoDrop; Wilmington, DE) and Qubit, version 3.0, fluorometer (Invitrogen, Carlsbad, CA) to ensure that there was adequate plasmid DNA for sequencing. The plasmid DNA was stored frozen at −20°C until required.
PCR amplification of resistance genes.
The presence of resistance genes on plasmids of specific AMR Salmonella phenotypes was detected using PCR (31, 74). Overall, genes encoding resistance to ampicillin and cephalosporin (blaPSE-1, blaTEM, and blaCMY-2), chloramphenicol (cmlA), streptomycin (aadA1, aadA2, strA, and strB), sulfisoxazole (sul1 and sul2), and tetracycline [tet(A), tet(B), tet(C), and tet(G)] were tested. Template plasmid DNAs were extracted by the modified alkali lysis method mentioned above. The primers, amplicon sizes, and references used to detect the presence of the selected AMR genes are listed in Table 3. The PCR conditions for all resistance genes, except the cmlA and sul1 genes, included an initial denaturation at 95°C for 4 min, followed by 30 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 54°C, extension for 1 min at 72°C, and a final extension at 72°C for 7 min. For the cmlA and sul1 genes, the PCR conditions used have been described previously (75). Briefly, an initial denaturation at 94°C for 5 min was followed by 30 cycles of denaturation for 45 s at 94°C, annealing for 45 s at 57°C, extension for 1 min at 72°C, and a final extension at 72°C for 5 min. Salmonella enterica isolates carrying resistance genes and characterized in earlier studies were used as positive controls (31).
Plasmid PCR-based replicon typing (PBRT).
Single and multiplex PCRs were run to identify different incompatibility (Inc) groups, including FIA, FIB, FIC, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA. The primers and PCR running conditions have been described in a previous study (45). The purified plasmid DNA from the modified alkali lysis method was used as the template DNA. PCR running conditions used for the five multiplex PCRs and three single PCRs included an initial denaturation for 5 min at 94°C, followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 30 s at 60°C, and elongation for 1 min at 72°C, with a final extension of 5 min at 72°C. The single PCRs for Frep were performed under the same amplification conditions but with an annealing temperature of 52°C. The PCR products were electrophoresed on a 1.5% agarose gel in Tris-acetate-EDTA (TAE) buffer and UV visualized by staining with ethidium bromide.
Plasmid sequencing, assembly, and annotation.
Isolated plasmid DNA libraries were prepared for sequencing using a Nextera XT kit (Illumina, San Diego, CA). Multiplexed sequencing of these libraries was done with a single run on an Illumina MiSeq using 2-by-250- or 2-by-300-bp paired-end reads (MiSeq reagent kit, version 3). Following demultiplexing, sequences were analyzed using CLC Genomic Workbench 10 (Qiagen, Valencia, CA). For analyzing plasmid content, de novo assembly of unused reads into new contigs was applied. The initially assembled contigs were analyzed using the National Center for Biotechnology Information's Basic Local Alignment Search Tool (BLAST). In addition, individual sequence reads were mapped back to the assembled plasmids to confirm that there were continuous overlapping reads over the entire length of the assembled plasmid. Following completion of plasmid assembly, the plasmid sequences were run through a BLAST search individually and compared to GenBank sequences. The open reading frame (ORF) of each gene in plasmid contigs was identified, and the particular genes of interest were annotated using Geneious R10 software (BioMatters, New Zealand). Manual trimming and editing of terminally redundant contig ends generated circular plasmid genomes. The complete plasmid sequences were visualized using plasmid mapping in the CLC Workbench and deposited in the GenBank under prospective accession numbers.
Comparative genotypic analysis.
To further characterize the plasmids and compare their profiles, we mapped the PCR primers described by Carattoli et al. (45) to the assembled plasmid sequences with a BLAST search configured for short reads. Based on the annotations and BLAST output, the plasmids were assessed for the presence of known AMR genes, plasmid transfer (tra) genes, and mobile genetic elements, including class I integrons and transposons. The assembled plasmid sequences submitted to a BLAST search were compared to previously sequenced plasmids in GenBank. We identified 14 plasmid sequences and analyzed them for variation using the Geneious R10 software (BioMatters, New Zealand) global alignment with 70% similarity to construct neighbor-joining trees using the Tamura-Nei genetic distance model. In addition, all 14 plasmid sequences were typed by pMLST as previously described (41) and assigned to STs according to the plasmid MLST database (https://pubmlst.org/plasmid/) for ST prevalence analysis.
Accession number(s).
The sequencing output of the 14 Salmonella plasmids was submitted to the National Center for Biotechnology Information (NCBI) under the BioProject accession number PRJNA293224. Individual plasmid sequence reads have been deposited in the Sequence Read Archive (SRA) as BioSample numbers SAMN07345795 to SAMN07345807.
ACKNOWLEDGMENTS
We gratefully acknowledge the swine producers in the state of North Carolina for allowing access to their farms for collecting the manure and environmental samples. This study would not have been possible without their cooperation and support. We thank Joy Horovitz, who assisted in the plasmid sequencing.
This project was funded by the National Pork Board grant (project number 556678) and the College of Veterinary Medicine, North Carolina State University.
REFERENCES
- 1.Lammie SL, Hughes JM. 2016. Antimicrobial resistance, food safety, and one health: the need for convergence. Annu Rev Food Sci Technol 7:287–312. doi: 10.1146/annurev-food-041715-033251. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO). 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Singer AC, Shaw H, Rhodes V, Hart A. 2016. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front Microbiol 7:1728. doi: 10.3389/fmicb.2016.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thanner S, Drissner D, Walsh F. 2016. Antimicrobial resistance in agriculture. mBio 7:e02227-15. doi: 10.1128/mBio.02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO). 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Ferri M, Ranucci E, Romagnoli P, Giaccone V. 2017. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr 57:2857–2876. doi: 10.1080/10408398.2015.1077192. [DOI] [PubMed] [Google Scholar]
- 7.Grace D. 2015. Review of evidence on antimicrobial resistance and animal agriculture in developing countries. Evidence on Demand, UK. Department for International Development, London, United Kingdom. [Google Scholar]
- 8.Diarra MS, Malouin F. 2014. Antibiotics in Canadian poultry productions and anticipated alternatives. Front Microbiol 5:282. doi: 10.3389/fmicb.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landers TF, Cohen B, Wittum TE, Larson EL. 2012. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep 127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopes GV, Michael GB, Cardoso M, Schwarz S. 2016. Antimicrobial resistance and class 1 integron-associated gene cassettes in Salmonella enterica serovar Typhimurium isolated from pigs at slaughter and abattoir environment. Vet Microbiol 194:84–92. doi: 10.1016/j.vetmic.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 11.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley RL, Collignon P, Larsson DG, McEwen SA, Li XZ, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E. 2013. The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 13.Pornsukarom S, Thakur S. 2016. Assessing the impact of manure application in commercial swine farms on the transmission of antimicrobial resistant Salmonella in the environment. PLoS One 11:e0164621. doi: 10.1371/journal.pone.0164621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Stephan R, Karczmarczyk M, Yan Q, Hächler H, Fanning S. 2013. Molecular characterization of blaESBL-harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front Microbiol 4:188. doi: 10.3389/fmicb.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Fang T, Zhou X, Zhang D, Shi X, Shi C. 2016. IncHI2 plasmids are predominant in antibiotic-resistant Salmonella isolates. Front Microbiol 7:1566. doi: 10.3389/fmicb.2016.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, de Schaetzen MA, Van Huffel X, Imberechts H, Dierick K, Daube G. 2013. Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health 10:2643–2669. doi: 10.3390/ijerph10072643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folster JP, Grass JE, Bicknese A, Taylor J, Friedman CR, Whichard JM. 2017. Characterization of resistance genes and plasmids from outbreaks and illness clusters caused by Salmonella resistant to ceftriaxone in the United States, 2011–2012. Microb Drug Resist 23:188–193. doi: 10.1089/mdr.2016.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seiffert SN, Hilty M, Perreten V, Endimiani A. 2013. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Updat 16:22–45. doi: 10.1016/j.drup.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 20.McCollister B, Kotter CV, Frank DN, Washburn T, Jobling MG. 2016. Whole-genome sequencing identifies in vivo acquisition of a blaCTX-M-27-carrying IncFII transmissible plasmid as the cause of ceftriaxone treatment failure for an invasive Salmonella enterica serovar Typhimurium infection. Antimicrob Agents Chemother 60:7224–7235. doi: 10.1128/AAC.01649-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Accogli M, Fortini D, Giufrè M, Graziani C, Dolejska M, Carattoli A, Cerquetti M. 2013. IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clin Microbiol Infect 19:E238–E240. doi: 10.1111/1469-0691.12128. [DOI] [PubMed] [Google Scholar]
- 22.Dolejska M, Villa L, Hasman H, Hansen L, Carattoli A. 2013. Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J Antimicrob Chemother 68:333–339. doi: 10.1093/jac/dks387. [DOI] [PubMed] [Google Scholar]
- 23.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 24.Falagas ME, Karageorgopoulos DE, Nordmann P. 2011. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol 6:653–666. doi: 10.2217/fmb.11.49. [DOI] [PubMed] [Google Scholar]
- 25.Gao R, Hu Y, Li Z, Sun J, Wang Q, Lin J, Ye H, Liu F, Srinivas S, Li D, Zhu B. 2016. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog 12:e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 27.Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martínez R, Florez-Cuadrado D, Campos MJ, García M, Píriz S, Sáez JL, Domínguez L. 2016. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci 105:134–135. doi: 10.1016/j.rvsc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 28.CDC. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control, Atlanta, GA: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 29.Nordmann P, Poirel L, Toleman MA, Walsh TR. 2011. Does broad-spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J Antimicrob Chemother 66:689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 30.Mollenkopf DF, Stull JW, Mathys DA, Bowman AS, Feicht SM, Grooters SV, Daniels JB, Wittum TE. 2017. Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob Agents Chemother 61:e01298-16. doi: 10.1128/AAC.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keelara S, Thakur S. 2014. Dissemination of plasmid-encoded AmpC β-lactamases in antimicrobial resistant Salmonella serotypes originating from humans, pigs and the swine environment. Vet Microbiol 173:76–83. doi: 10.1016/j.vetmic.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Lynne AM, David DE, Tang H, Xu J, Nayak R, Kaldhone P, Logue CM, Foley SL. 2012. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS One 7:e51160. doi: 10.1371/journal.pone.0051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen SV, Harhay DM, Bono JL, Smith TP, Fields PI, Dinsmore BA, Santovenia M, Kelley CM, Wang R, Bosilevac JM, Harhay GP. 2016. Complete, closed genome sequences of 10 Salmonella enterica subsp. enterica serovar Typhimurium strains isolated from human and bovine sources. Genome Announc 4:e01212-16. doi: 10.1128/genomeA.01212-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams LE, Wireman J, Hilliard VC, Summers AO. 2013. Large plasmids of Escherichia coli and Salmonella encode highly diverse arrays of accessory genes on common replicon families. Plasmid 69:36–48. doi: 10.1016/j.plasmid.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Norman A, Hansen LH, Sorensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci 364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colobatiu L, Tabaran A, Flonta M, Oniga O, Mirel S, Mihaiu M. 2015. First description of plasmid-mediated quinolone resistance determinants and β-lactamase encoding genes in non-typhoidal Salmonella isolated from humans, one companion animal and food in Romania. Gut Pathog 7:16. doi: 10.1186/s13099-015-0063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cartwright EJ, Nguyen T, Melluso C, Ayers T, Lane C, Hodges A, Li X, Quammen J, Yendell SJ, Adams J, Mitchell J. 2016. A multistate investigation of antibiotic-resistant Salmonella enterica serotype I 4,[5],12:i:− infections as part of an international outbreak associated with frozen feeder rodents. Zoonoses Public Health 63:62–71. doi: 10.1111/zph.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henry I, Chemaly M, Granier S, Lalande F, Courtillon C, Salvat G, Cardinale E. 2015. Epidemiological analysis of Salmonella enterica serovar Typhimurium and serovar 1,4,[5],12:i:− isolates determined by pulsed field gel electrophoresis and antibiotic susceptibility: comparison of isolates from broiler chickens, humans and the environment in Reunion Island. Open Vet Sci J 9:10–18. doi: 10.2174/1874318801509010010. [DOI] [Google Scholar]
- 39.Ido N, Lee KI, Iwabuchi K, Izumiya H, Uchida I, Kusumoto M, Iwata T, Ohnishi M, Akiba M. 2014. Characteristics of Salmonella enterica serovar 4,[5],12:i:− as a monophasic variant of serovar Typhimurium. PLoS One 9:e104380. doi: 10.1371/journal.pone.0104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng LK, Mulvey MR, Martin I, Peters GA, Johnson W. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother 43:3018–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keelara S, Scott HM, Morrow WM, Gebreyes WA, Correa M, Nayak R, Stefanova R, Thakur S. 2013. Longitudinal study of distributions of similar antimicrobial-resistant Salmonella serovars in pigs and their environment in two distinct swine production systems. Appl Environ Microbiol 79:5167–5178. doi: 10.1128/AEM.01419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carattoli A. 2011. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol 301:654–658. doi: 10.1016/j.ijmm.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 44.DeNap JC, Hergenrother PJ. 2005. Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem 3:959–966. doi: 10.1039/b500182j. [DOI] [PubMed] [Google Scholar]
- 45.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Phan MD, Forde BM, Peters KM, Sarkar S, Hancock S, Stanton-Cook M, Zakour NL, Upton M, Beatson SA, Schembri MA. 2015. Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS One 10:e0122369. doi: 10.1371/journal.pone.0122369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcadé G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J Antimicrob Chemother 63:67–71. doi: 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- 48.Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. 2015. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid 80:118–126. doi: 10.1016/j.plasmid.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Önnberg A, Söderquist B, Persson K, Mölling P. 2014. Characterization of CTX-M-producing Escherichia coli by repetitive sequence-based PCR and real-time PCR-based replicon typing of CTX-M-15 plasmids. APMIS 122:1136–1143. doi: 10.1111/apm.12270. [DOI] [PubMed] [Google Scholar]
- 50.Olsen RH, Bisgaard M, Löhren U, Robineau B. 2014. Christensen H. Extended-spectrum β-lactamase-producing Escherichia coli isolated from poultry: a review of current problems, illustrated with some laboratory findings. Avian Pathol 43:199–208. doi: 10.1080/03079457.2014.907866. [DOI] [PubMed] [Google Scholar]
- 51.Randall LP, Clouting C, Horton RA, Coldham NG, Wu G, Clifton-Hadley FA, Davies RH, Teale CJ. 2010. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother 66:86–95. doi: 10.1093/jac/dkq396. [DOI] [PubMed] [Google Scholar]
- 52.Cloeckaert A, Praud K, Doublet B, Bertini A, Carattoli A, Butaye P, Imberechts H, Bertrand S, Collard JM, Arlet G, Weill FX. 2007. Dissemination of an extended-spectrum-β-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob Agents Chemother 51:1872–1875. doi: 10.1128/AAC.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flach CF, Johnning A, Nilsson I, Smalla K, Kristiansson E, Larsson DJ. 2015. Isolation of novel IncA/C and IncN fluoroquinolone resistance plasmids from an antibiotic-polluted lake. J Antimicrob Chemother 70:2709–2717. doi: 10.1093/jac/dkv167. [DOI] [PubMed] [Google Scholar]
- 54.Hammerum AM, Larsen J, Andersen VD, Lester CH, Skovgaard Skytte TS, Hansen F, Olsen SS, Mordhorst H, Skov RL, Aarestrup FM, Agersø Y. 2014. Characterization of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third-or fourth-generation cephalosporins. J Antimicrob Chemother 69:2650–2657. doi: 10.1093/jac/dku180. [DOI] [PubMed] [Google Scholar]
- 55.Chen J, Jin M, Qiu ZG, Guo C, Chen ZL, Shen ZQ, Wang XW, Li JW. 2012. A survey of drug resistance bla genes originating from synthetic plasmid vectors in six Chinese rivers. Environ Sci Technol 46:13448–13454. doi: 10.1021/es302760s. [DOI] [PubMed] [Google Scholar]
- 56.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5:e01918-14. doi: 10.1128/mBio.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahube TO, Yost CK. 2012. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application. J Appl Microbiol 112:1123–1133. doi: 10.1111/j.1365-2672.2012.05301.x. [DOI] [PubMed] [Google Scholar]
- 58.Hirsch DR, Cox G, D'Erasmo MP, Shakya T, Meck C, Mohd N, Wright GD, Murelli RP. 2014. Inhibition of the ANT(2″)-Ia resistance enzyme and rescue of aminoglycoside antibiotic activity by synthetic α-hydroxytropolones. Bioorg Med Chem Lett 24:4943–4947. doi: 10.1016/j.bmcl.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kayama S, Shigemoto N, Kuwahara R, Oshima K, Hirakawa H, Hisatsune J, Jové T, Nishio H, Yamasaki K, Wada Y, Ueshimo T. 2015. Complete nucleotide sequence of the IncN plasmid encoding IMP-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob Agents Chemother 59:1356–1359. doi: 10.1128/AAC.04759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stalder T, Barraud O, Casellas M, Dagot C, Ploy MC. 2012. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol 3:119. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng X, Zhang Z, Li K, Wang Y, Xia X, Wang X, Xi M, Meng J, Cui S, Yang B. 2017. Antibiotic susceptibility and molecular screening of class I integron in Salmonella Isolates recovered from retail raw chicken carcasses in China. Microb Drug Resist 23:230–235. doi: 10.1089/mdr.2015.0359. [DOI] [PubMed] [Google Scholar]
- 62.Sinwat N, Angkittitrakul S, Chuanchuen R. 2015. Characterization of antimicrobial resistance in Salmonella enterica isolated from pork, chicken meat, and humans in northeastern Thailand. Foodborne Pathog Dis 12:759–765. doi: 10.1089/fpd.2015.1946. [DOI] [PubMed] [Google Scholar]
- 63.Lopes GV, Michael GB, Cardoso M, Schwarz S. 2014. Identification and characterization of Salmonella enterica subsp. enterica serovar Derby isolates carrying a new aadA26 gene cassette in a class 1 integron obtained at pig slaughterhouses. FEMS Microbiol Lett 356:71–78. doi: 10.1111/1574-6968.12473. [DOI] [PubMed] [Google Scholar]
- 64.Sanad YM, Johnson K, Park SH, Han J, Deck J, Foley SL, Kenney B, Ricke S, Nayak R. 2016. Molecular characterization of Salmonella enterica serovars isolated from a turkey production facility in the absence of selective antimicrobial pressure. Foodborne Pathog Dis 13:80–87. doi: 10.1089/fpd.2015.2002. [DOI] [PubMed] [Google Scholar]
- 65.Mattiello SP, Drescher G, Barth VC, Ferreira CA, Oliveira SD. 2015. Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Antonie Van Leeuwenhoek 108:1227–1238. doi: 10.1007/s10482-015-0577-1. [DOI] [PubMed] [Google Scholar]
- 66.Pande VV, Gole VC, McWhorter AR, Abraham S, Chousalkar KK. 2015. Antimicrobial resistance of non-typhoidal Salmonella isolates from egg layer flocks and egg shells. Int J Food Microbiol 203:23–26. doi: 10.1016/j.ijfoodmicro.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 67.Christie PJ. 4 August 2016, posting date The mosaic type IV secretion systems. EcoSal Plus 2016 doi: 10.1128/ecosalplus.ESP-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rahube TO, Viana LS, Koraimann G, Yost CK. 2014. Characterization and comparative analysis of antibiotic resistance plasmids isolated from a wastewater treatment plant. Front Microbiol 5:558. doi: 10.3389/fmicb.2014.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng X, Ardeshna D, Lin J. 2015. Heat shock-enhanced conjugation efficiency in standard Campylobacter jejuni strains. Appl Environ Microbiol 81:4546–4552. doi: 10.1128/AEM.00346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.CLSI. 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed CLSI supplement VET01S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 71.CLSI. 2013. Performance standards for antimicrobial disc susceptibility test; approved standard, 10th ed CLSI document M02-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 72.Food and Drug Administration. 2011. National antimicrobial resistance monitoring system—enteric bacteria (NARMS): 2009 executive report. U.S. Department of Health and Human Services, Food and Drub Administration, Silver Spring, MD: http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM268954.pdf. [Google Scholar]
- 73.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 74.Beutlich J, Jahn S, Malorny B, Hauser E, Hühn S, Schroeter A, Rodicio MR, Appel B, Threlfall J, Mevius D, Helmuth R. 2011. Antimicrobial resistance and virulence determinants in European Salmonella genomic island 1-positive Salmonella enterica isolates from different origins. Appl Environ Microbiol 77:5655–5664. doi: 10.1128/AEM.00425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chuanchuen R, Padungtod P. 2009. Antimicrobial resistance genes in Salmonella enterica isolates from poultry and swine in Thailand. J Vet Med Sci 71:1349–1355. doi: 10.1292/jvms.001349. [DOI] [PubMed] [Google Scholar]
- 76.White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, McDermott PF, McDermott S, Wagner DD, Meng J. 2001. The isolation of antibiotic-resistant Salmonella from retail ground meats. N Engl J Med 345:1147–1154. doi: 10.1056/NEJMoa010315. [DOI] [PubMed] [Google Scholar]
- 77.Carlson SA, Bolton LF, Briggs CE, Hurd HS, Sharma VK, Fedorka-Cray PJ, Jones BD. 1999. Detection of multi resistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol Cell Probes 13:213–222. doi: 10.1006/mcpr.1999.0240. [DOI] [PubMed] [Google Scholar]
- 78.Aarestrup FM, Lertworapreecha M, Evans MC, Bangtrakulnonth A, Chalermchaikit T, Hendriksen RS, Wegener HC. 2003. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J Antimicrob Chemother 52:715–718. doi: 10.1093/jac/dkg426. [DOI] [PubMed] [Google Scholar]
- 79.Madsen L, Aarestrup FM, Olsen JE. 2000. Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet Microbiol 75:73–82. doi: 10.1016/S0378-1135(00)00207-8. [DOI] [PubMed] [Google Scholar]
- 80.Gebreyes WA, Thakur S. 2005. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob Agents Chemother 49:503–511. doi: 10.1128/AAC.49.2.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campos J, Cristino L, Peixe L, Antunes P. 2016. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1, 4,[5],12:i:− and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill 21:30270. doi: 10.2807/1560-7917.ES.2016.21.26.30270. [DOI] [PubMed] [Google Scholar]
- 82.Datta N, Hedges RW. 1971. Compatibility groups among fi− R factors. Nature 234:222–223. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]