Abstract
Myogenic progenitor/stem cells retain their skeletal muscle differentiation potential by maintaining myogenic transcription factors such as MyoD. However, the mechanism of how MyoD expression is maintained in proliferative progenitor cells has not been elucidated. Here, we found that MyoD expression was reduced at the mRNA level by cell cycle arrest in S and G2 phases, which in turn led to the absence of skeletal muscle differentiation. The reduction of MyoD mRNA was correlated with the reduced expression of factors regulating RNA metabolism, including methyltransferase like 3 (Mettl3), which induces N6-methyladenosine (m6A) modifications of RNA. Knockdown of Mettl3 revealed that MyoD RNA was specifically downregulated and that this was caused by a decrease in processed, but not unprocessed, mRNA. Potential m6A modification sites were profiled by m6A sequencing and identified within the 5′ untranslated region (UTR) of MyoD mRNA. Deletion of the 5′ UTR revealed that it has a role in MyoD mRNA processing. These data showed that Mettl3 is required for MyoD mRNA expression in proliferative myoblasts.
Keywords: MyoD, Mettl3, HuR, m6A-sequencing, RNA processing, splicing
1. Introduction
Stem cells are able to self-renew and to multiply rapidly while maintaining their pluripotency, resulting in histogenesis [1,2]. The differentiation process from stem cells to various tissues has previously been elucidated by studies that characterized the factors or conditions required for differentiation into each organ or tissue [3–9]; by contrast, it is unclear how the differentiation potential is maintained in proliferative cells. In the case of skeletal muscle, which accounts for approximately 40% and 30% of adult male and female body weights, respectively [10], it is reasonable to assume that the differentiation of stem cells and progenitors requires multiple rounds of cell division to attain a high muscular density. However, the means by which the myogenic potential is maintained in proliferative skeletal muscle stem cells and progenitors remains to be determined.
The identification of MyoD and the MyoD family of myogenic regulatory factors (MRFs) has revealed that the myogenic potential is determined by the expression of transcription factors that influence myogenic gene expression. Conversely, the loss of MyoD and/or MRFs in skeletal myoblasts, both in vitro in C2C12 cells [11,12] and in vivo, suppresses skeletal myogenesis [13,14]. During differentiation, together with Pbx homeodomain protein, MyoD binds to the regulatory regions of myogenic genes, including Myog, Mylpf, Myh3, Tnni and Ryr1 [15], which triggers chromatin remodelling by recruiting the SWI/SNF chromatin remodelling factor Brg1 for myogenic gene transcription [16,17]. Before differentiation, MyoD labels myogenic gene loci by incorporating the variant histone H3.3 with chromodomain helicase DNA-binding domain 2 (Chd2), without activating transcription [8]. Thus, during proliferative phases, MyoD is critical for myoblast differentiation and the inheritance of differentiation potency.
Transcription factor expression is destabilized by RNA modifications that influence the differentiation potential of stem cells [18,19]. In embryonic stem (ES) cells, methyltransferase like 3 (Mettl3) [20] induces an N6-methyladenosine (m6A) modification in Nanog RNA, which is required for pluripotency and differentiation [19]. m6A modifications have been reported to affect RNA function by various mechanisms, including splicing, stabilization/destabilization [18,21], nuclear export [22] and translation efficiency [23,24]. RNA modifications or stabilization may also be crucial for skeletal muscle differentiation because MyoD mRNA has a short half-life of approximately 90 min [25,26]. The RNA-binding protein HuR has been reported to stabilize MyoD mRNA and to be necessary for terminal skeletal muscle differentiation [25,27]. It was also shown that MyoD mRNA levels are quite low in G0-arrested cells but increase upon re-entry into the cell cycle [28], implying that MyoD mRNA could be stabilized during the cell cycle. However, the underlying mechanisms, as well as the factors required for the maintenance of MyoD expression during proliferation, remain to be clarified.
Here, we explored the maintenance of MyoD mRNA levels in proliferative myoblasts. We found that cell cycle arrest reduced MyoD mRNA expression, thus suppressing myogenic differentiation, and that Mettl3 stabilized MyoD mRNA by promoting mRNA processing in skeletal myoblasts. Our results suggest that m6A modification by Mettl3 stabilizes MyoD mRNA levels for skeletal muscle differentiation.
2. Results
2.1. Cell cycle arrest in S and G2 phases reduces MyoD mRNA levels and inhibits myoblast differentiation
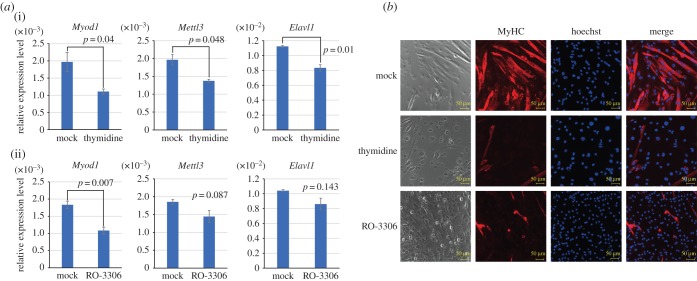
During proliferative phases, MyoD binding to target genes such as myogenic genes is required for skeletal muscle differentiation [8,13,29], suggesting that the maintenance of MyoD expression during cell cycle progression could be critical for differentiation. Because MyoD mRNA levels were reported to be low following cell cycle arrest at G0 [28], we hypothesized that cell cycle arrest may cause MyoD mRNA instability. C2C12 cells, a mouse myoblast cell line with both self-renewal and differentiation potential, were arrested either in the S phase by thymidine or in the G2 phase by the Cdk1 inhibitor RO-3306, and MyoD mRNA levels were analysed by quantitative reverse transcription PCR (qRT-PCR). Cell cycle arrest was confirmed by measuring the population in each cell cycle phase after exposure to thymidine or RO-3306 (electronic supplementary material, figure S1a). We observed cell cycle re-entry and the proliferation of cells after removal of the inhibitors (electronic supplementary material, figure S1b), confirming that the drug treatments did not induce cell death under our experimental conditions. qRT-PCR analysis showed that MyoD mRNA levels were significantly reduced after cell cycle arrest in both S and G2 phases in the growth state (p = 0.04 and 0.007, respectively; figure 1a(i)). On the other hand, mRNA levels of other skeletal muscle-specific transcription factors (Pax7 and Srf) were not substantially affected by cell cycle arrest in the proliferative state (electronic supplementary material, figure S2a).
Figure 1.
MyoD mRNA expression is reduced upon cell cycle arrest. (a) mRNA levels of Myod1, Mettl3 and Elavl1 in C2C12 cells treated with thymidine or RO-3306 for 48 h. (i) Cells treated with thymidine were compared with non-treated cells. (ii) Cells treated with RO-3306 were compared with DMSO-treated cells. (b) Morphology of C2C12 cells treated with DMSO (labelled as Mock), thymidine and RO-3306. RO-3306 was used as a Cdk1 inhibitor. Cells were immunostained for anti-MyHC and Hoechst at 72 h after the induction of differentiation. MyHC, myosin heavy chain.
Next, we examined the skeletal muscle differentiation of arrested C2C12 cells. Cells were cultured with 2% horse serum (HS) media for 48 h to induce differentiation. We found that myotube formation was diminished in thymidine- and RO-3306-treated cells but not in mock-treated cells (figure 1b). qRT-PCR analysis revealed that mRNA levels of MyoD as well as those of the skeletal muscle-specific genes Myog and Acta1 were decreased by cell cycle arrest in the differentiated state (electronic supplementary material, figure S2b). Taken together, these results suggest that during proliferative phases, cell cycle arrest leads to a reduction of MyoD mRNA expression that is required for skeletal muscle differentiation.
2.2. Cell cycle arrest affects mRNA levels of HuR and Mettl3
To examine the mechanism by which MyoD mRNA expression is decreased upon cell cycle arrest, we focused on two pathways potentially involved in RNA metabolism. One was the HuR (also known as Elavl1)-mediated stabilization of RNA, which involves HuR binding to AU-rich elements of MyoD mRNA in the early stages of skeletal myoblast differentiation [25,27,30]. The other was the m6A modification of RNA introduced by Mettl3 [20], which is also important for RNA stabilization. qRT-PCR analysis showed that Elavl1 and Mettl3 mRNA levels were significantly reduced by cell cycle arrest using thymidine (both p < 0.05; figure 1a(i)), but the effect was relatively limited following treatment with RO-3306 (p = 0.087 and 0.143, respectively; figure 1a(ii)). These results suggest that the decreased expression of HuR and/or Mettl3 may account for the decline in MyoD mRNA expression upon cell cycle arrest.
2.3. Knockdown of Mettl3, but not of HuR, downregulates MyoD mRNA levels in skeletal myoblasts
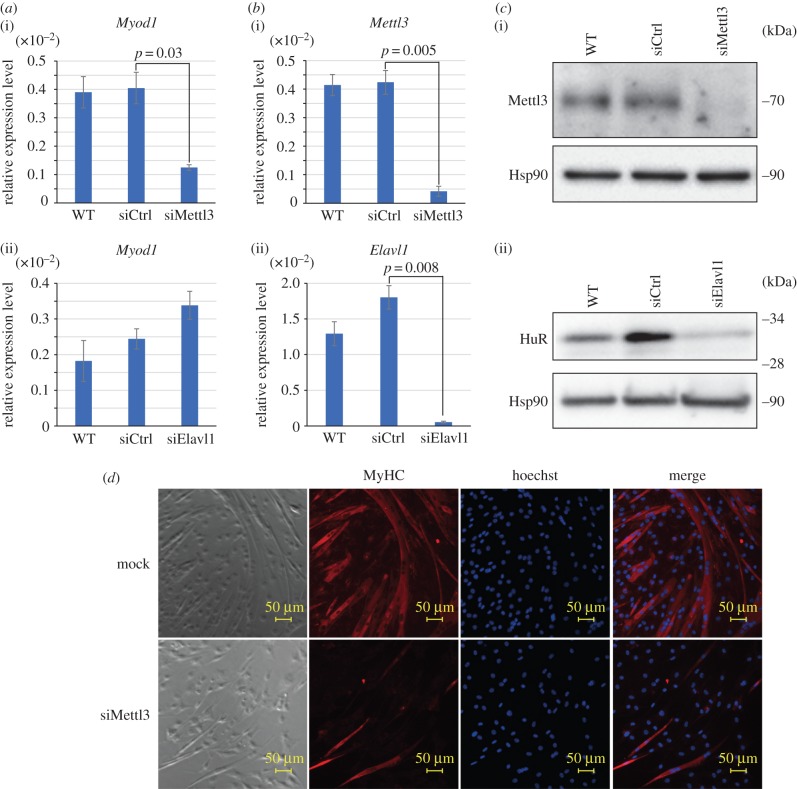
Next, to determine if HuR and/or Mettl3 regulate MyoD mRNA expression, we carried out small interfering (si)RNA-mediated knockdown of Mettl3 and HuR. qRT-PCR showed that MyoD mRNA levels were significantly reduced in Mettl3 knockdown cells (p = 0.03; figure 2a(i)), but not in HuR knockdown cells (figure 2a(ii)). Both siRNAs suppressed the expression of target genes at the mRNA (figure 2b) as well as protein level (figure 2c), validating the knockdown efficiency. These data suggest that Mettl3, but not HuR, is implicated in maintaining MyoD mRNA levels. Mettl3-targeted siRNA treatment also suppressed myotube formation (figure 2d).
Figure 2.
Knockdown of Mettl3, but not HuR, downregulates MyoD mRNA levels and suppresses skeletal muscle differentiation. (a) qRT-PCR analysis of Myod1 in (i) Mettl3 and (ii) HuR knockdown cells. (b) qRT-PCR analysis to monitor (i) Mettl3 and (ii) HuR knockdown efficiency. (c) Western blot analysis to monitor (i) Mettl3 and (ii) HuR knockdown efficiency. Hsp90 was used as a loading control. (d) Morphology of C2C12 cells with transfected Mettl3 siRNA. Cells were immunostained for anti-MyHC and Hoechst at 72 h after the induction of differentiation.
2.4. Knockdown of Mettl3 markedly downregulates processed, but not unprocessed, MyoD mRNA levels
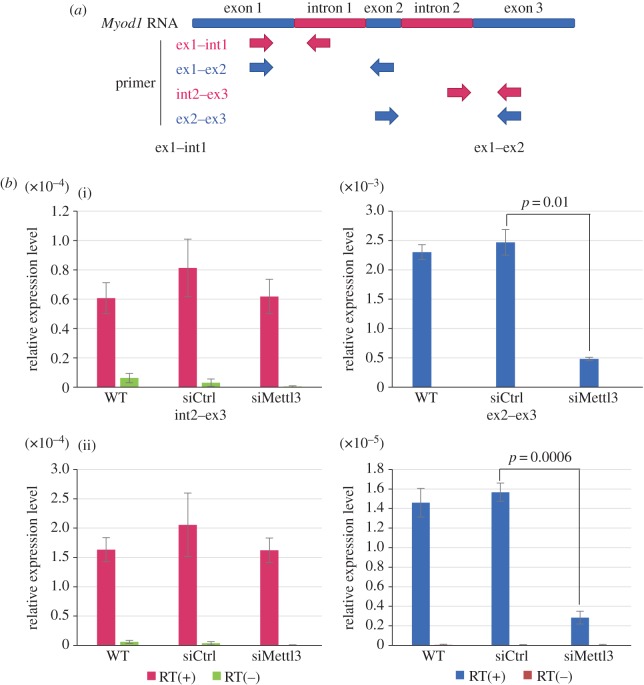
Because m6A modification by Mettl3 has been proposed to regulate RNA metabolism such as splicing [31–35], we examined the processing of MyoD pre-mRNA for the generation of mature MyoD mRNA. To investigate whether Mettl3 affects MyoD RNA splicing, two primer pairs were designed to anneal between two of the three MyoD exons and another two pairs to include both an intron and an exon (figure 3a). qRT-PCR analysis showed that in Mettl3 knockdown cells, fewer PCR products containing only exons were obtained (figure 3b(ii)), whereas there was no effect on the amount of PCR products containing an intron (figure 3b(i)). The length of PCR products amplified with exonic primer pairs was the correct splice size, and non-spliced RNA was not amplified (electronic supplementary material, figure S3a). Taken together, this indicates that knockdown of Mettl3 significantly decreased the amount of processed MyoD mRNA without affecting that of unprocessed MyoD RNA.
Figure 3.
Knockdown of Mettl3 downregulates processed, but not unprocessed, MyoD mRNA levels. (a) Schematic of Myod1 RNA which consists of three exons and two introns. Four primer pairs were designed: one with a forward primer in exon 1 and a reverse primer in intron 1, one with a forward primer in exon 1 and a reverse primer in exon 2, one with a forward primer in intron 2 and a reverse primer in exon 3, and one with a forward primer in exon 2 and a reverse primer in exon 3. ex: exon, int: intron. (b) qRT-PCR analysis of Myod1 in Mettl3 knockdown C2C12 cells using the above four primer pairs. To confirm the absence of DNA contamination, each sample was also analysed without reverse transcriptase. RT: reverse transcriptase. Error bars show ± s.d. (n = 3). p-value versus control siRNA.
To investigate whether the m6A reading process is associated with the maintenance of Myod1 mRNA, we focused on the representative m6A readers YTHDF2, YTHDC1, HNRNPA2B1 and HNRNPC [18,24,31–36], which were previously shown to be involved in RNA stability and processing [34,35]. We performed siRNA-mediated knockdown of these four factors and found that MyoD mRNA expression and myotube formation were only suppressed by Ythdf2 knockdown (electronic supplementary material, figure S3b,c). Knockdown of Ythdf2, however, led to a decrease of both processed and unprocessed MyoD RNA (electronic supplementary material, figure S3d), unlike the case of Mettl3 knockdown.
2.5. m6A modifications of MyoD mRNA are primarily enriched in the 5′ untranslated region
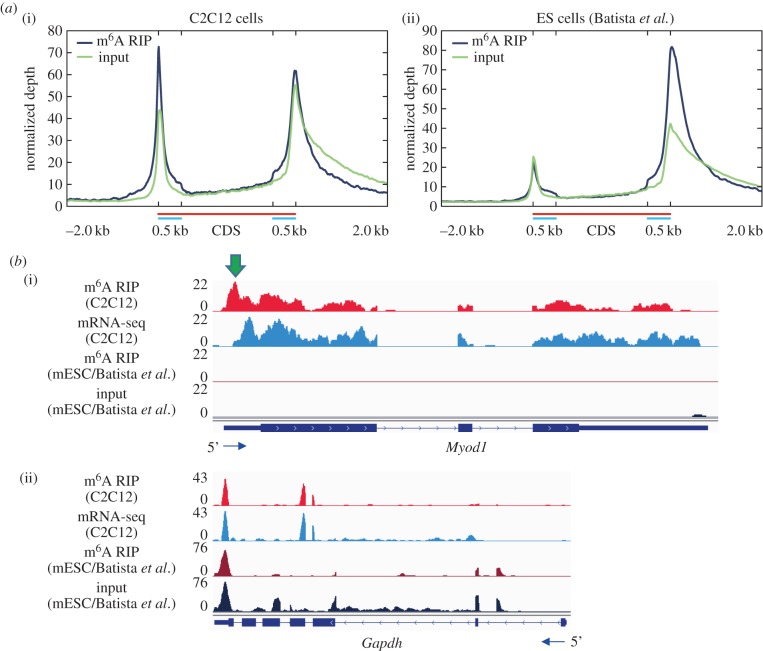
To further investigate the m6A modification of Myod1 mRNA, m6A-sequencing (m6A-seq) was conducted by pulling down transcripts with an antibody that specifically recognizes m6A modifications [31]. Published studies have reported that m6A modifications are distributed in 3′ UTRs, stop codons and internal long exons, but rarely in 5′ UTRs [19,31,37,38] (figure 4a(ii)), while we observed enriched m6A signals in C2C12 cells both near the start codon and stop codon (figure 4a(i)). Integrative Genomics Viewer plots showed that while m6A modifications on the majority of RNAs such as Gapdh and Srf were enriched around 3′ UTRs, stop codons and long exons (figure 4b; electronic supplementary material, figure S4), m6A modifications of Myod1 were notably enriched around the 5′ UTR (figure 4b(i), green arrow).
Figure 4.
m6A modifications on MyoD mRNA are primarily found in the 5′ UTR. (a) Distribution of m6A signals across all mRNAs in (i) C2C12 cells and (ii) mouse ES cells. The lengths of all coding sequences (CDS) were scaled to 2 kb for visualization (indicated by red bars), except for 500 bp at both ends (blue bars). (b) Integrative Genomics Viewer tracks displaying reads coverage of (i) Myod1 and (ii) Gapdh in m6A-seq and mRNA-seq data for C2C12 cells and mouse embryonic stem cells (mESC) (SRR1207291 (input), SRR1207292 (m6A RIP), Batista et al. [19]).
We also found that C2C12 cells and ES cells had many analogous loci for m6A signal enrichment (figure 4b(ii); electronic supplementary material, figure S4, upper panel). However, several skeletal muscle-specific genes such as Myod1 and Myog were enriched in m6A signals only in C2C12 cells, whereas m6A signal enrichment in pluripotent genes such as Nanog was observed only in ES cells (figure 4b(i); electronic supplementary material, figure S4, middle and lower panels), reflecting cell type-specific regulations.
2.6. Myod1 5′ UTR is required for the maintenance of processed Myod1 mRNA during cell proliferation
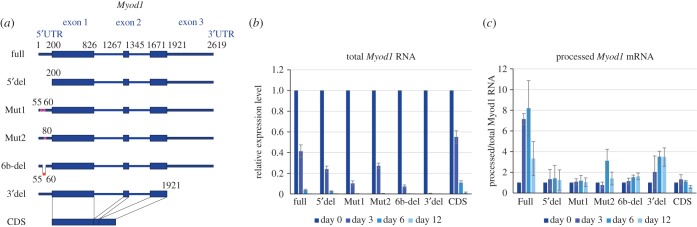
To investigate the function of m6A modification on MyoD mRNA, especially around the 5′ UTR, we constructed vectors harbouring Myod1 with various mutations and deletions in the 5′ UTR. ‘RRAC’ has been shown to be a common sequence of m6A sites, with ‘GGAC’ the most frequent of these [18,24,31,37,38]. m6A-seq revealed the presence of some ‘GGAC’ motifs within m6A modifications on Myod1 5′ UTR. Therefore, deleted or mutated versions of three ‘GGAC’ motifs in the Myod1 5′ UTR were generated, as well as an entire 5′ UTR deletion mutant, and an entire 3′ UTR deletion mutant (figure 5a). These mutants were transiently transfected into NIH3T3 cells, and qRT-PCR detected total (both processed and unprocessed) and processed MyoD RNA using primer pairs within exon 1 of Myod1 and between exon 1 and exon 2, respectively.
Figure 5.
Conservation of the Myod1 5′ UTR stabilizes processed Myod1 mRNA. (a) Schematic of Myod1 mutants. ‘Full’ consists of full-length Myod1 DNA. ‘5'del’ and ‘3'del’ are mutants with deleted 5′ UTRs and 3′ UTRs, respectively. In ‘Mut1,’ both 55A and 60A were replaced with C. In ‘Mut2,’ 80A was replaced with C. ‘6b-del’ carries a 55A to 60A deletion. ‘CDS’ contains only Myod1 CDS (i.e. the 5′ UTR, 3′ UTR, and all introns were deleted). (b) qRT-PCR analysis of total Myod1 RNA with seven Myod1 mutants. NIH3T3 cells were transfected with vectors shown in (a), and total Myod1 RNA levels were analysed at days 0, 3, 6 and 12 using a primer set within exon 1. Data are represented as the mean ± s.d. (n = 3). (c) Accumulation levels of processed Myod1 mRNA. Processed Myod1 mRNA levels were analysed using a primer pair between exon 1 and exon 2. Processed Myod1 mRNA was normalized to total Myod1 RNA. Data are represented as the mean ± s.d. (n = 3).
The ectopic introduction of each vector could be monitored by total MyoD RNA levels, all of which led to a similar pattern of decline (figure 5b). To quantify the accumulation of processed Myod1 mRNA, these levels were normalized against those of total Myod1 RNA (figure 5c). When transfecting vectors containing full-length Myod1 and 3′ UTR deletion mutants, which retained the complete 5′ UTR, we observed an accumulation of processed Myod1 mRNA until day 6 (figure 5c, Full and 3′ del). By contrast, all 5′ UTR deletion mutants showed no significant accumulation of processed mRNA upon transfection (figure 5c, 5′del, Mut1, Mut2 and 6b-del). Collectively, these data suggest that MyoD RNA is actively stabilized by its 5′ UTR through the efficient processing of Myod1 mRNA during cell proliferation.
3. Discussion
In this study, we showed that MyoD mRNA expression reduces upon cell cycle arrest. We also found that Mettl3 regulates the maintenance of MyoD mRNA levels through m6A modification of the 5′ UTR during cell proliferation. Taken together, these results suggest that the myogenic potential is maintained throughout the cell cycle, at least in part by the Mettl3-mediated stabilization of processed MyoD mRNA during proliferative phases.
m6A-seq data showed that MyoD mRNA contains m6A modifications, mainly in its 5′ UTR, while previous studies reported that m6A is more commonly seen in 3′ UTRs, stop codons and internal long exons (figure 4a(ii)) [19,31,37–39]. The function of m6A is largely dependent on modifications around the 3′ UTR; for example, YTHDF2 typically targets the stop codon region (42%), the coding region (36%) and the 3′ UTR (14%), while the proportion of all YTHDF2-binding sites in the 5′ UTR is only 2% [18]. However, a recent study found that a 5′ UTR m6A promoted cap-independent translation [23]. These lines of evidence suggest that the functions of m6A modification may differ between 5′ UTRs and 3′ UTRs.
Because Mettl3 knockdown in this study caused the immature processing of MyoD RNA, we evaluated the involvement of YTHDC1, which is a nuclear m6A reader that regulates mRNA splicing by recruiting splicing factors [32,33]. Our results showed, however, that knockdown of YTHDC1 did not change MyoD RNA level significantly (electronic supplementary material, figure S3b). Previous studies reported that target RNAs for YTHDF2 are destabilized by m6A modification and stabilized upon Mettl3 knockdown [18,19]. Our results, however, showed the opposite effects, and thus suggest that MyoD RNAs are unlikely to be a direct target of YTHDF2. This raises the possibility that YTHDF2 functions to indirectly stabilize MyoD mRNA through its many (greater than 3000) targets [18]. Future work should attempt to uncover the mechanisms involving these factors.
Although HuR and Mettl3 mRNA expression was reduced by thymidine-mediated cell cycle arrest, this decrease was non-significant following treatment with RO-3306. Cell cycle arrest was planned for the standard duration of 48 h. While a single treatment of thymidine was adequate to achieve cell cycle arrest because its effects increase gradually for at least 48 h, RO-3306 acts rapidly within 8 h, but its effects are diminished by 12 h such that additional treatments were required at intervals of 8–12 h. Between additional doses, reactivated cells may have affected mRNA levels, which may explain the more prominent effects of thymidine compared with RO-3306. To directly examine the involvement of HuR and Mettl3 in the regulation of MyoD mRNA expression, we conducted siRNA-mediated knockdown experiments and found that knockdown of Mettl3, but not of HuR, significantly affected MyoD mRNA expression. Previous studies reported that HuR stabilizes MyoD mRNA by binding to its AU-rich elements during the early stages of differentiation [25]. The discrepancy between these results and our own remains to be resolved. Because our investigation was conducted during the growth state, HuR may have had little effect on MyoD mRNA expression before the onset of differentiation. Interestingly, it was previously reported that HuR is significantly associated with m6A bait [31], but the exact relationship remains to be elucidated.
In summary, MyoD mRNA levels appear to be maintained by Mettl3-mediated m6A modifications, suggesting that Mettl3 is a critical regulator for skeletal muscle differentiation. These findings help understand the mechanisms underlying the maintenance of myogenic potential in proliferative skeletal muscle progenitors. Moreover, the methods developed in this study may be beneficial in evaluating different cell types from various species.
4. Material and methods
4.1. Cell culture and drug treatment
The mouse myoblast cell line C2C12 was maintained in a growth state in Dulbecco's modified Eagle's medium (DMEM) (Gibco by Thermo Fisher Scientific) containing 1 g l−1 glucose, 1% l-glutamine and 1% penicillin/streptomycin supplemented with 20% fetal bovine serum (FBS). NIH3T3 cells were maintained in DMEM containing 4.5 g l−1 glucose, 1% l-glutamine and 1% penicillin/streptomycin supplemented with 20% FBS. Cells were grown in a 5% CO2 cell culture incubator at 37°C. Differentiation stimuli were induced by exchanging media with 2% HS instead of 20% FBS.
C2C12 myoblasts were arrested in S phase by thymidine (Sigma-Aldrich, T1895) and in G2 phase by Cdk1 inhibitor (RO-3306, Sigma-Aldrich, SML0569) for 48 h. A single treatment of thymidine was applied, while additional RO-3306 was added at intervals of 8–12 h.
4.2. Immunocytochemistry
Cells were cultured on µ-Plate 24 Well (ibidi), washed twice with phosphate-buffered saline (PBS), fixed with 1% paraformaldehyde in PBS, permeabilized with 0.5% Triton X-100 in PBS and washed twice with PBS. A 15 min incubation with Blocking One (Nacalai Tesque Inc.) was followed by a 2 h incubation with mouse anti-myosin heavy chain (MF20, eBioscience, 1 : 200) diluted with 10% Blocking One in PBS at room temperature. The µ-Plate 24 Well was then washed three times with PBS and incubated for 30 min at room temperature with CF568-labelled goat anti-mouse antibody (1 : 1000; Biotium Inc.) and Bisbenzimide H33342 Fluorochrome Trihydrochloride (Hoechst) (1 : 2000; Nacalai Tesque Inc.) diluted with 10% Blocking One in PBS. The µ-Plate 24 Well was again washed three times in PBS and mounted in ibidi Mounting Medium (ibidi). Images were visualized using a fluorescence microscope (BZ-9000; Keyence). Co-localization was evaluated using BZ-II Analyzer software (Keyence).
4.3. Fluorescence activated cell sorting
C2C12 cells in a growth state were isolated by trypsin treatment and centrifuged for 5 min at 190g. The cell pellet was resuspended in 1 ml of PBS supplemented with Hoechst 33342 stain (Nacalai Tesque Inc.) and incubated for 15 min. This suspension was analysed by a cell sorter (SONY, SH800).
4.4. Quantitative RT-PCR
Total RNA was extracted from cells using Sepasol-RNA I Super G (Nacalai Tesque Inc.) and ethanol precipitation. Total RNA (1 µg) was used for reverse transcription with the PrimeScript RT reagent kit (Takara Bio Inc., RR047A). qRT-PCR was performed using Thunderbird SYBR qPCR Mix (Toyobo Co., Ltd.) with the PikoReal 96 Real-Time PCR System (Thermo Fisher Scientific) as previously described [40]. Primers are listed in the electronic supplementary material, table S1. qRT-PCR data were normalized to Gapdh or Eef1a1 expression levels and presented as the mean ± s.d. of three independent experiments. For qRT-PCR of processed MyoD mRNA in NIH3T3 cells with transfected MyoD mutants, differences in transfection efficiencies were normalized by using total MyoD RNA in each sample as an internal control. Total MyoD RNA levels were quantified by performing qRT-PCR using primer pairs within exon 1 of Myod1, while processed MyoD mRNA was quantified using primer pairs between exon 1 and exon 2.
4.5. Western blotting
Cells were harvested and disrupted in a Sample Buffer Solution with 2-ME (Nacalai Tesque Inc.). Samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis using a 5–20% Extra PAGE One Precast Gel (Nacalai Tesque Inc.) and electrotransferred to a polyvinylidene fluoride membrane with the Trans-Blot Turbo Transfer System (Bio-Rad Laboratories, Hercules, CA; 2.5 A, 25 V, 7 min). The membrane was blocked for 1 h in 5% (w/v) skimmed milk in Tris-buffered saline containing 0.05% (v/v) Tween 20, then incubated with primary antibodies in Hikari Solution A (Nacalai Tesque Inc.), followed by incubation with secondary antibodies and detection using Chemi-Lumi One Ultra (Nacalai Tesque Inc.).
The following primary antibodies were used for western blotting: rat anti-MyoD (5F11, Millipore 1 : 500), anti-Mettl3 (15073-1-AP, Proteintech, 1 : 1000), anti-Hsp90 (H-114, Santa Cruz Biotechnology, 1 : 1000) and mouse anti-HuR (3A2, Santa Cruz Biotechnology, 1 : 200). Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit and anti-mouse IgG antibodies (GE Healthcare, 1 : 5000).
4.6. siRNA-mediated knockdown of Mettl3, Elavl1 and m6A readers
Transient knockdown of target genes by siRNA was performed with Lipofectamine RNAi MAX (Invitrogen) and opti-MEM (Gibco by Thermo Fisher Scientific). The following siRNAs were used: siMettl3 (Dharmacon, ON-TARGETplus SMARTpool #L-049446), siElavl1 (Dharmacon, ON-TARGETplus SMARTpool #L-053812), siYthdf2 (Dharmacon, ON-TARGETplus SMARTpool #L-058271), siYthdc1 (Dharmacon, ON-TARGETplus SMARTpool #L-167076), siHnrnpa2b1 (Dharmacon, ON-TARGETplus SMARTpool #L-040194) and siHnrnpc (Dharmacon, ON-TARGETplus SMARTpool #L-044147).
4.7. Transfection
Seven MyoD mutants were generated as DNA inserts. Each insert DNA was amplified by PCR using KOD FX neo DNA polymerase (Toyobo Co., Ltd.) and ligated with the pCAGGS vector. A list of primers for cloning is shown in the electronic supplementary material, table S1. Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The sequences of all mutants were confirmed by Sanger sequencing.
4.8. mRNA-seq
Total RNA was extracted from cells using Sepasol-RNA I Super G (Nacalai Tesque Inc.) and ethanol precipitation. Purified RNA samples underwent library construction using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, NEB #7420S). Library preparation and sequence analysis followed a previously described protocol [41].
Sequenced reads were mapped onto the mouse genome (mm9) using TopHat (version 2.0.8). Gene expression levels (fragments per kilobase of exon per million mapped sequence reads) were estimated using the Cuffdiff program in Cufflinks (version 2.0.1) using mapped reads and the software's default parameters [42].
4.9. m6A immunoprecipitation
m6A immunoprecipitation was performed according to a previously described protocol [31,43]. Total RNA was extracted from cells with Sepasol-RNA I Super G (Nacalai Tesque) and ethanol precipitation, and 348 µg was used for independent experiments. RNA was fragmented by mixing with 10 × fragmentation buffer (1 M ZnCl2, 1 M Tris–HCl pH 7.0 and RNase-free water) at 94°C for 5 min using a preheated thermal cycler. The fragmentation mix was ethanol precipitated by adding 1/10 volumes of 3 M sodium acetate, pH 5.2, glycogen (100 µg ml−1 final), and 2.5 volumes of 100% ethanol, mixing well, then incubating at –80°C overnight. Fragmented RNA was mixed with an anti-m6A antibody (Synaptic Systems, 202003, 12.5 µg) and 5 × immunoprecipitation buffer (0.05 M Tris–HCl, pH 7.4, 0.75 M NaCl, 0.5% Igepal CA-630, and RNase-free water). Ribonucleoside-vanadyl complexes (Sigma-Aldrich, R3380, 2 mM final) and RNasin Plus RNase inhibitor (Promega, N2611, 200–400 U, final) were mixed with immunoprecipitation reagents to prevent RNA degradation. Immunoprecipitation mixtures were incubated for 2 h at 4°C on a rotating wheel to allow the formation of antibody-RNA complexes. These complexes were mixed with Dynabeads Protein A (Novex by Life Technologies) and incubated for 2 h at 4°C on a rotating wheel. Immunoprecipitated RNA was eluted by competition with N6-methyladenosine, 5′-monophosphate sodium salt (m6A, Sigma-Aldrich, M2780, 6.7 mM final).
4.10. m6A-seq data analysis
Purified RNA fragments from m6A RNA immunoprecipitation underwent library construction using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, NEB #7420S). The m6A-seq library was sequenced on an Illumina HiSeq 1500 sequencing system. Sequence reads were aligned to the reference mouse genome (GRCm38) using HISAT2 software (version 2.0.4) [44]. The software deepTools2 (version 2.3.5) [45] was used to create the coverage tracks (BigWig file) of m6A-modified/input RNA on the mouse genome with the options: bamCoverage --binSize 1 --normalizeTo1x2267226534 (the effective mouse genome size for 50 bp reads)’, and the m6A signal profiles on all coding sequences (taken from UCSC refSeq genes) with the options: computeMatrix -m 2000 -b 2000 -a 2000 --unscaled5prime 500 --unscaled3prime 500 --skipZeros --missingDataAsZero.
4.11. Statistical analysis
Statistical significance in qRT-PCR data was evaluated using the two-sided Welch's t-test.
Supplementary Material
Acknowledgements
We thank H. Kimura, J. Odawara, M. Hayashi, Y. Semba and T. Iwasaki for supplying materials and advice; M. Kato, N. Urasaki, Y. Nakajo, S. Hirata and K. Fukuyama for technical support; and the ‘Advanced Computational Scientific Program’ of the Research Institute for Information Technology, Kyushu University and the National Institute of Genetics (NIG) for providing high-performance computing resources.
Data accessibility
mRNA-seq and m6A-seq data were deposited into the DNA Data Bank of Japan database (accession no.: DRA005057). The other datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
K.K. and A.H. performed the experiments. J.N. and K.M. performed statistical analysis. K.K., T.K., H.S., E.O., Y.M. and Y.O. designed the experiments and data analysis. K.K., T.K. and Y.O. wrote the manuscript.
Competing interests
We have no competing interests.
Funding
This work was supported by JSPS KAKENHI grant nos. JP25116010, JP26290064, JP15K18457, JP16H01219, JP16K18479, JP16H01577, JP16H01550, JP17H03608 and JP17K19356, and the Core Research for Evolutional Science and Technology (CREST).
References
- 1.Niwa H, Burdon T, Chambers I, Smith A. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060. (doi:10.1101/gad.12.13.2048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. 2008. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227. (doi:10.1038/nature06863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. 2011. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875–889. (doi:10.1016/j.cell.2011.05.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Sanjuan I, Brivanlou AH. 2002. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3, 271–280. (doi:10.1038/nrn786) [DOI] [PubMed] [Google Scholar]
- 5.Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. 2009. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS ONE 4, e6286 (doi:101371/journal.pone.0006286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood WM, Etemad S, Yamamoto M, Goldhamer DJ. 2013. MyoD-expressing progenitors are essential for skeletal myogenesis and satellite cell development. Dev. Biol. 384, 114–127. (doi:10.1016/j.ydbio.2013.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. 2016. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev. Cell 36, 375–385. (doi:10.1016/j.devcel.2016.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada A, et al. 2012. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 31, 2994–3007. (doi:10.1038/emboj.2012.136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zammit PS, Partridge TA, Yablonka-Reuveni Z. 2006. The skeletal muscle satellite cell: the stem cell that came in from the cold. J. Histochem. Cytochem. 54, 1177–1191. (doi:101369/jhc.6R6995.2006) [DOI] [PubMed] [Google Scholar]
- 10.Janssen I, Heymsfield SB, Wang ZM, Ross R. 2000. Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. J. Appl. Physiol. 89, 81–88. [DOI] [PubMed] [Google Scholar]
- 11.Yaffe D, Saxel O. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270, 725–727. (doi:10.1038/270725a0) [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Liu W, Nie Y, Qaher M, Horton HE, Yue F, Asakura A, Kuang S. 2017. Loss of MyoD Promotes Fate Transdifferentiation of myoblasts into brown adipocytes. EBioMedicine 16, 212–223. (doi:10.1016/j.ebiom.2017.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. (doi:10.1016/0092-8674(87)90585-X) [DOI] [PubMed] [Google Scholar]
- 14.Qin RF, Mao TQ, Gu XM, Hu KJ, Liu YP, Chen JW, Nie X. 2007. Regulation of skeletal muscle differentiation in fibroblasts by exogenous MyoD gene in vitro and in vivo. Mol. Cell. Biochem. 302, 233–239. (doi:10.1007/s11010-007-9446-1) [DOI] [PubMed] [Google Scholar]
- 15.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465–477. (doi:10.1016/S1097-2765(04)00260-6) [DOI] [PubMed] [Google Scholar]
- 16.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25, 3997–4009. (doi:101128/MCB.25.10.3997-4009.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36, 738–743. (doi:10.1038/ng1378) [DOI] [PubMed] [Google Scholar]
- 18.Wang X, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. (doi:10.1038/nature12730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batista PJ, et al. 2014. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. (doi:10.1016/j.stem.2014.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. 1997. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. 2014. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198. (doi:10.1038/ncb2902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fustin JM, et al. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. (doi:10.1016/j.cell.2013.10.026) [DOI] [PubMed] [Google Scholar]
- 23.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010. (doi:10.1016/j.cell.2015.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. 2015. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. (doi:10.1016/j.cell.2015.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. 2003. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cel. Biol. 23, 4991–5004. (doi:10.1128/MCB.23.14.4991-5004.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. 2010. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS ONE 5, e11201 (doi:101371/journal.pone.0011201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. 2003. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J. Biol. Chem. 278, 47 119–47 128. (doi:10.1074/jbc.M308889200) [DOI] [PubMed] [Google Scholar]
- 28.Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. 1998. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 142, 1447–1459. (doi:10.1083/jcb.142.6.1447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18, 662–674. (doi:10.1016/j.devcel.2010.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cammas A, et al. 2014. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 5, 4190 (doi:10.1038/ncomms5190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominissini D, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. (doi:10.1038/nature11112) [DOI] [PubMed] [Google Scholar]
- 32.Roundtree IA, He C. 2016. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Trends Genet. 32, 320–321. (doi:10.1016/j.tig.2016.03.006) [DOI] [PubMed] [Google Scholar]
- 33.Xiao W, et al. 2016. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519. (doi:10.1016/j.molcel.2016.01.012) [DOI] [PubMed] [Google Scholar]
- 34.Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. 2015. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308. (doi:10.1016/j.cell.2015.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. 2015. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. (doi:10.1038/nature14234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T. 2016. N(6)-Methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J. Mol. Biol. 428, 822–833. (doi:10.1016/j.jmb.2015.08.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S, Choe J, Du P, Triboulet R, Gregory RI. 2016. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. (doi:10.1016/j.molcel.2016.03.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke S, et al. 2015. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053. (doi:101101/gad.269415.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. (doi:10.1016/j.cell.2012.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada A, Maehara K, Sato Y, Konno D, Tachibana T, Kimura H, Ohkawa Y. 2015. Incorporation of histone H3.1 suppresses the lineage potential of skeletal muscle. Nucleic Acids Res. 43, 775–786. (doi:10.1093/nar/gku1346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odawara J, Harada A, Yoshimi T, Maehara K, Tachibana T, Okada S, Akashi K, Ohkawa Y. 2011. The classification of mRNA expression levels by the phosphorylation state of RNAPII CTD based on a combined genome-wide approach. BMC Genomics 12, 516 (doi:101186/1471-2164-12-516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. (doi:10.1038/nprot.2012.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. 2015. Transcriptome-Wide Mapping of N(6)-Methyladenosine by m(6)A-Seq. Methods Enzymol. 560, 131–147. (doi:10.1016/bs.mie.2015.03.001) [DOI] [PubMed] [Google Scholar]
- 44.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. (doi:10.1038/nmeth.3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. (doi:10.1093/nar/gkw257) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
mRNA-seq and m6A-seq data were deposited into the DNA Data Bank of Japan database (accession no.: DRA005057). The other datasets supporting this article have been uploaded as part of the electronic supplementary material.