Abstract
Ribonucleotide incorporation is the most common error occurring during DNA replication. Cells have hence developed mechanisms to remove ribonucleotides from the genome and restore its integrity. Indeed, the persistence of ribonucleotides into DNA leads to severe consequences, such as genome instability and replication stress. Thus, it becomes important to understand the effects of ribonucleotides incorporation, starting from their impact on DNA structure and conformation. Here we present a systematic study of the effects of ribonucleotide incorporation into DNA molecules. We have developed, to our knowledge, a new method to efficiently synthesize long DNA molecules (hundreds of basepairs) containing ribonucleotides, which is based on a modified protocol for the polymerase chain reaction. By means of atomic force microscopy, we could therefore investigate the changes, upon ribonucleotide incorporation, of the structural and conformational properties of numerous DNA populations at the single-molecule level. Specifically, we characterized the scaling of the contour length with the number of basepairs and the scaling of the end-to-end distance with the curvilinear distance, the bending angle distribution, and the persistence length. Our results revealed that ribonucleotides affect DNA structure and conformation on scales that go well beyond the typical dimension of the single ribonucleotide. In particular, the presence of ribonucleotides induces a systematic shortening of the molecules, together with a decrease of the persistence length. Such structural changes are also likely to occur in vivo, where they could directly affect the downstream DNA transactions, as well as interfere with protein binding and recognition.
Introduction
Current opinion on the evolution of genetic information suggests that DNA was selected as the storage molecule because it is more stable with respect to its ancient precursor, RNA (1). The only difference between DNA and RNA is the presence of a hydroxyl group on the ribose of RNA monomers (rNMPs). Such group makes RNA unstable and less suitable to safely store genetic information (2).
Recent works reported that large amounts of ribonucleotides are misincorporated into chromosomes during DNA replication, even though DNA polymerases are extremely accurate enzymes (3, 4, 5). The frequency of incorporation in budding yeast is estimated to be ∼1 every 700 nucleotides, making ribonucleotides the most frequent noncanonical nucleotides incorporated into the genome (6).
The elevated levels of ribonucleotides incorporated may suggest that this is not a mere error of DNA polymerases, but that it may have some beneficial roles. Indeed, it was recently demonstrated that ribonucleotides help a specific DNA repair pathway in discriminating the newly synthesized strand from the template filament (7, 8). However, ribonucleotides are not permanent in DNA, because cells possess specific mechanisms to remove them from the genome (6, 9). The persistence of rNMPs is an endogenous source of genome instability and replication stress (5, 10, 11, 12).
RNase H enzymes are able to recognize and cleave embedded rNMPs (13), and are responsible for the major pathway that processes genomic rNMPs. Interestingly, defects in RNase H2 function represent the major cause of a rare genetic disorder, Aicardi-Goutieres syndrome (14).
How rNMPs embedded in chromosomal DNA may interfere with DNA-protein interactions has not been investigated yet, although it has been reported that nucleosomes assembly on DNA is reduced when even a single ribonucleotide is present (15). Understanding the structural changes imposed upon DNA molecules by the presence of ribonucleotides is essential to determine the biological impact of their persistence in the genome. It is thus crucial to explore and investigate those effects at the single-molecule level.
During the 1990s, the first ribonucleotides containing DNA (RC-DNA) molecules were crystallized. These were very short self-complementary oligonucleotides with embedded ribonucleotides. X-ray diffraction analyses suggested that the presence of ribonucleotides induce a complete change toward the A-conformation, both when they are in the middle or at the ends of the molecules (16, 17, 18). In solution studies, such as nuclear magnetic resonance, the transition observed was partial (19, 20). Molecular dynamics simulations suggested that even with 50% of ribonucleotide substitutions, B-DNA is not fully converted to A-DNA, although the ribose caused local perturbations (21). In addition, atomic force microscopy (AFM) measurements showed that ribonucleotides also perturb the backbone elasticity (22, 23, 24) with respect to DNA in the B-form, although these studies report different trends of the persistence length in relation to the presence of ribonucleotides. In conclusion, only a few single-molecule studies on the structural properties of RC-DNAs are available in the literature, and furthermore, in some cases apparently contradictory results are reported. And in most cases, short RC-DNA molecules are studied, although in biologically relevant cases (such as in vivo), the molecules are much longer.
Here we present the results of a systematic study of the structural effects of ribonucleotide incorporation into DNA carried out taking advantage of, to our knowledge, a novel protocol to synthesize several-hundred-of-basepairs-long RC-DNA molecules. RC-DNA molecules were produced via enzymatic synthesis by a mutant Taq polymerase (I614K), able to introduce ribonucleotides (rNTPs) in addition to deoxynucleotides (dNTPs). We were therefore able to produce numerous populations of RC-DNA molecules, as well as their controls without ribonucleotides, that have been studied at the single-molecule level by means of AFM. The main advantage of AFM is the possibility of studying large ensembles of molecules, by quantitatively analyzing each molecule individually, therefore obtaining robust average values of relevant structural and conformational observables (25, 26, 27, 28). In particular, we have characterized the scaling of the contour length with the number of basepairs, the scaling of the end-to-end distance with the curvilinear distance, the bending angle distribution, and the persistence length of DNA molecules, showing that the presence of ribonucleotides affects the DNA structure and conformation well beyond the scale of the single ribonucleotide, up to full molecular length. The observed changes are also likely to take place in physiological conditions such as the cell environment, and consequently influence DNA transactions occurring in vivo.
Materials and Methods
Taq polymerase production
The pTaq plasmid was site-directed mutagenized using primers TAQI614Kfor 5′-TGG CCC TGG ACT ATA GCC AGA AAG AGC TCA GGG TGC TGG CCC A-3′ and TAQI614Krev 5′-TGG GCC AGC ACC CTG AGC TCT TTC TGG CTA TAG TCC AGG GCC A-3′. BL21(DE3) Escherichia coli cells harboring pTaq or pTaqI614K were grown in selective medium and protein expression was induced by IPTG addition. Protein extraction and purification was done by GeneSpin (Milan, Italy).
RC-DNAs synthesis
Polymerase chain reaction (PCR) was performed with either wild-type (WT) or I614K Taq polymerase in the presence of dATP, dGTP, and dTTP 0.2 mM, dCTP 0.1 mM for normal DNAs, and plus rCTP 0.8 mM only for RC-DNAs. Nucleotides were purchased from GeneSpin. The 464- and 727-basepair (bp) fragments were amplified from pGEM3Zf plasmid using primer pairs 5′-TCG GGA AAC CTG TCG TGC C-3′/5′-CAG CGT GAG CTA TGA GAA AG-3′ and 5′-TCG GGA AAC CTG TCG TGC C-3′/5′-TCA GCA GAG CGC AGA TAC CA-3′, respectively. The 646-bp fragment was amplified from pNB187 plasmid using primers 5′-TAG TTG AAG CAT TAG GTC CC-3′/5′-CTT CTC AAA TAT GCT TCC CA-3′; the 960- and the 1079-bp fragments were amplified from pFL39.3 with primers 5′-AAA GAG TTA CTC AAG AAT AA-3′/5′-CAA AAC GGC ATT TAA GAA GC-3′ and 5′-GGA CGA GGC AAG CTA AAC AG-3′/5′-CAA AAC GGC ATT TAA GAA GC-3′, respectively. The complete sequences are reported in Fig. S8. PCR reactions were carried out in multiple independent samples and then pooled to increase the product yield. The samples were loaded onto 1% agarose gel and the band corresponding to the amplification product was excised and purified using silica columns (The Wizard SV Gel and PCR Clean-Up System; Promega, Fitchburg, WI) according to the manufacturer’s instructions. This last step was necessary to further clean the samples from template plasmid and primers. All the samples were finally resuspended in ultrapure Milli-Q water (Millipore, Billerica, MA).
PCR with radiolabeled nucleotides
PCR reactions were performed adding α32P-dCTP or α32P-rCTP (PerkinElmer, Waltham, MA) in addition to the nonradioactive dCTP or rCTP, respecting the final concentrations described above for RC-DNAs. Samples were drop-dialyzed on 0.025-μm membranes (Millipore) and further cleaned by ethanol precipitation. Samples were then run in a 2% agarose gel that was dried and exposed on a phosphor storage screen; images were acquired using a Phosphoimager (Typhoon FLA 7000; GE Healthcare, Buckinghamshire, UK).
Alkaline gel and Southern blot
Alkaline gel electrophoresis was performed as described in (29). Briefly, samples were incubated for 2 h at 55°C in 0.3 M NaOH and then run in alkaline gel (1% agarose in Milli-Q water with 1 mM EDTA and 250 mM NaOH) previously equilibrated in its alkaline running buffer (1 mM EDTA, 250 mM NaOH). DNA was transferred to a charged nylon membrane (GeneScreen Plus Hybridization Transfer Membrane; PerkinElmer) by Southern blotting and hybridized with the radiolabeled 464-bp fragment as a probe (prepared by a DECAprime II DNA Labeling Kit; Ambion, Austin, TX). Images were acquired using a Phosphoimager (Typhoon FLA 7000; GE Healthcare).
AFM imaging
The procedure is described in detail in (30). Samples were deposited on freshly cleaved mica of the highest quality (V1, ruby muscovite; Ted Pella, Redding, CA) in a Mg2+-containing buffer (5 mM MgCl2, 10 mM NaCl, 10 mM HEPES-Na, pH 7.5 in Milli-Q H2O). Incubation time ranged from 2 to 10 min at room temperature, then the samples were gently washed dropwise with 1–2 mL of Milli-Q water, and dried under a clean nitrogen stream. Images were captured in air, using a Multimode Nanoscope IV AFM (Bruker, Billerica, MA) working in tapping mode, equipped with rigid cantilevers (∼300 kHz resonance frequency) and single-crystal silicon tips with nominal radius of curvature below 10 nm. The scan rate was typically 1.5–2 Hz, the scan area 2 × 1 μm, with a sampling resolution of 1 and 2 nm/pixel in the fast and slow scan directions, respectively.
Analysis of AFM data
Raw images were first flattened by subtracting polynomials up to the third order, using only the flat mica surface as reference for the fit. DNA and RC-DNA molecules were semiautomatically traced using FiberApp software (31), to obtain the spatial coordinates of the backbones. Calibration of the scanner was checked by scanning a calibration grating and the determined correction factors (always <2–3%) were applied to the coordinates, when needed. The traces were analyzed using custom MATLAB (The MathWorks, Natick, MA) routines. We evaluated the following statistical quantities describing structural/mechanical and conformational properties of semirigid polymers, as described in our previous works (26, 30): the mean contour length 〈l〉, the mean squared end-to-end distance 〈R2(L)〉 of segments of the molecules with a curvilinear length (curvilinear distance) L, the distribution of bending angles θ(L) (as well as 〈cos(θ(L))〉 and 〈θ2(L)〉), and the persistence length P. If DNA molecules are in a well-defined form (either B or A), the 〈l〉 versus n of the bp curve is a straight line with a slope equal to rA or rB, the rise per residues of the A and B forms, respectively, with units nm/bp. A reduction of the contour length independent on bp simply shifts the curve vertically by a constant offset c, but does not alter the slope of the curve. According to the wormlike chain (WLC) model, in two dimensions the mean squared end-to-end distance 〈R2〉2D increases as the curvilinear distance L increases, and depends on the persistence length P of DNA as (25, 26):
| (1) |
Estimation of the fraction of incorporated rCTP. To estimate the percentage %rCTP of rCTP incorporated into RC-DNA molecules, we use Eq. 2, i.e., we multiply the fraction of sites along the DNA backbone available for rCTP incorporation (the GC content %GC) by the estimated frequency of incorporation, fincorporation (defined and calculated with Eq. 3 in the Results and Discussion):
| (2) |
Apparent B to A transition fraction. An apparent fraction of basepairs in the DNA molecules that have switched from B to A conformation can be calculated by assuming that whenever an rCTP is incorporated, the hosting basepair switches from the B to the A form. The B-to-A transition fraction represents the fraction of basepairs that undergo such transition. Following (32, 33), the total number of available basepairs before rCTP incorporation is N = (l0 − c)/rB, where l0 is the measured contour length; c is a possible systematic shortening of the molecules, as discussed before (the negative intercept of the 〈l〉 versus n of basepairs curve shown in Fig. 4); and rB = 0.34 nm/bp is the B-form helical rise. After rCTP incorporation, NA basepairs switch to the A conformation whereas NB basepairs remain in the B conformation, such that N = NB + NA. In terms of contour length, NB rB + NA rA = (l0−c)−|Δl|, where rA = 0.26 nm/bp is the A-form helical rise and Δl is the measured difference in contour length upon ribonucleotide introduction. It follows that the bases in A form are NA = |Δl|/(rB−rA), and the B to A transition fraction NA/N can be calculated as NA/N = [|Δl|/(l0−c)] × [rB/(rB−rA)].
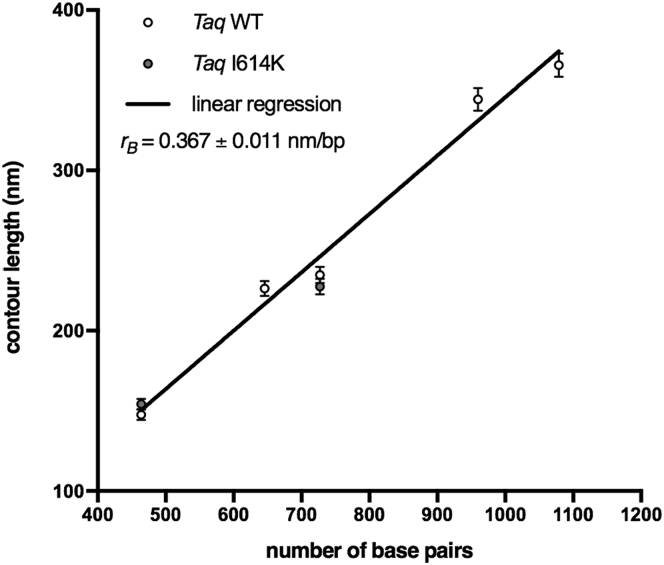
Figure 4.
DNA molecules from WT and I614K Taq pol retain canonical B conformation. Measured contour length values are plotted versus the number of basepairs of each DNA population analyzed (464, 646, 727, 960, and 1079), produced either by WT or I614K Taq polymerases. The linear fit (R2 = 0.9851) exhibits a slope (rB = 0.367 ± 0.011 nm/bp) close to the one typical of B DNA conformation. The length of the molecules produced by the mutated I614K Taq pol agrees with the length of the corresponding molecules produced by the WT Taq pol, within the error. The number of analyzed 464-, 646-, 727-, 960-, and 1079-bp WT Taq pol molecules is 146, 92, 74, 67, and 46, respectively; the number of analyzed 464- and 727-bp I614K Taq pol molecules is 176 and 47, respectively.
Statistical analysis. Length data are reported in the figures and tables as mean value ± effective error. The mean values and the SDs have been obtained by a Gaussian fit of the distributions of experimental values (see Fig. S4 for some representative distributions of contour length values). The effective errors have been calculated by summing in quadrature the SDs of the mean and a systematic error of ±1% due to the z-piezo calibration. The error associated to the persistence length, extracted by fitting Eq. 1 to the average end-to-end distance curves of the samples, has been estimated by applying the fit to a few set of data obtained by adding a Gaussian noise to the average curves, whose width was set equal to the SD of the mean associated to each experimental value (the resulting error bar is comparable to the marker size, and it is not shown in the graphs). The significance of the observed differences in the value of relevant parameters was evaluated applying a two-tailed t-test.
Results and Discussion
Synthesis of RC-DNA molecules
DNA molecules with incorporated ribonucleotides are generally synthesized chemically by a stepwise addition of nucleotides, whose limit is the chain extension step; as a result, with this methodology, only relatively short molecules are produced. Such short RC-DNAs molecules (10–30 bp) have been studied by several techniques (16, 21, 22), reproducing environments quite far from the physiological one.
We propose, to our knowledge, a new approach to synthesize RC-DNA molecules that exploits many consecutives cycles of an enzymatic reaction known as PCR. PCR is performed with the Thermus aquaticus DNA Polymerase (Taq pol), a very versatile enzyme, able to sustain multiple reaction cycles to amplify a defined DNA sequence exponentially (34). Taq pol is endowed with a high capability of discrimination between dNTPs and rNTPs; we took advantage of a known mutant version that is able to incorporate ribonucleotides more efficiently (35). We mutated the Taq pol with a single amino acid substitution at Isoleucine 614 to Lysine, making the enzyme more prone to binding and introducing rNTPs (35). The incorporation rates range from 150- to 1500-fold with respect to the WT Taq pol, depending on the rNTP species (rCTP, rATP, rGTP, or rTTP) (35). We expressed and purified both the WT and I614K Taq pols from E. coli cells, as described in Materials and Methods.
PCR allows the synthesis of a significant number of linear molecules, thanks to repetitive cycles of reaction. We set PCR conditions for the I614K Taq pol, in the presence of all four dNTPs and of rCTP, the most common ribonucleotide found in the DNA of living cells (36, 37). To verify the effective rCTP incorporation, PCRs were performed using radiolabeled α32P-rCTP, and the amplification products (464 bp) were then purified and visualized by autoradiography after agarose gel electrophoresis. The radioactive signal corresponding to a band of 464 bp indicates that the I614K Taq pol is indeed introducing α32P-rCTP, although with low efficiency compared to α32P-dCTP (Fig. 1). From the published kinetic parameters of I614K Taq pol (35) it is possible to theoretically estimate such frequency of incorporation, using
| (3) |
In our PCR conditions, the mutant Taq pol introduces one rCTP every 19 dCTP, in front of each guanosine residue (G), respecting the Watson-Crick basepairing. In fact, its misincorporation rates are small, regardless of the fact that I614K mutation confers less fidelity to the polymerase (38), as we verified by sequencing (data not shown).
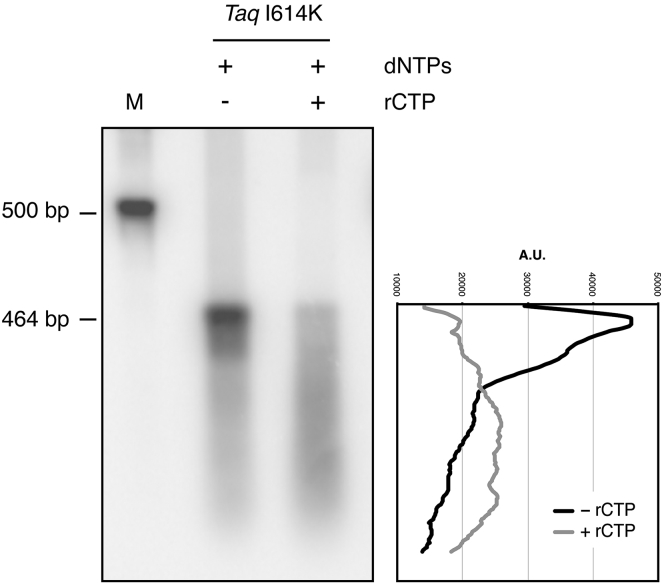
Figure 1.
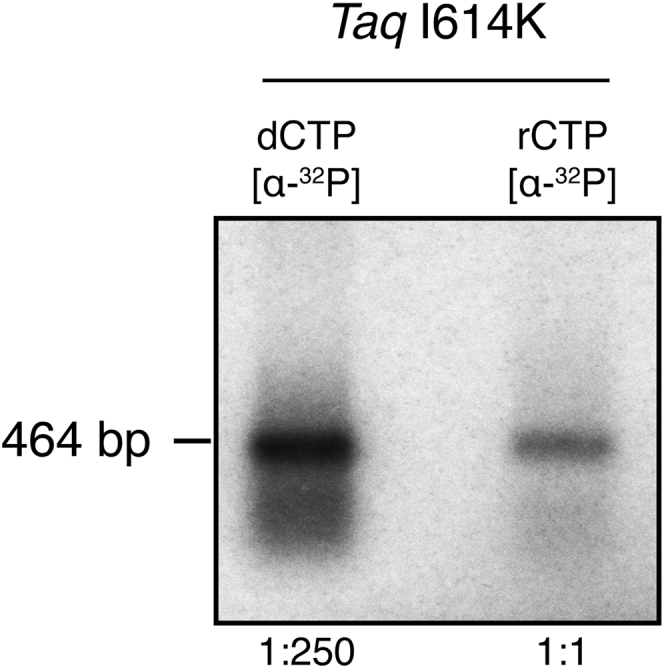
Taq polymerase I614K introduces rCTP in PCR products. DNA is amplified by I614K Taq pol mutant in the presence of radiolabeled α32P-dCTP or α32P-rCTP, and then it is separated onto an agarose gel, after purification through drop-dialysis and ethanol precipitation. The radioactive signal visible in the rCTP lane confirms that PCR products contain rNMPs.
To assess to what extent ribonucleotides are present in RC-DNA molecules, we exploited alkaline gel electrophoresis. PCR reactions were performed with I614K Taq pol in the presence or absence of rCTP and the products (see Fig. S1) were separated by gel electrophoresis in alkaline conditions; here the DNA is denatured and its backbone hydrolyzed at ribonucleotide positions, generating smaller fragments (4). Fig. 2 shows the degree of alkaline degradation of the 464-bp fragments, synthesized either in the absence or presence of rCTP. The molecules produced without rCTP migrate as a sharp band, whereas the ones containing rCTP generate a smear of smaller fragments, confirming the presence of ribonucleotides in most of them.
Figure 2.
RC-DNA molecules are sensitive to alkaline hydrolysis. DNA is amplified in the presence or absence of rCTP and run in alkaline conditions. The DNA backbone is hydrolyzed in correspondence to ribonucleotides, resulting in a population of smaller molecules. Only molecules produced in the presence of rCTP are hydrolyzed and their corresponding band is converted to a smear signal. DNA is detected by Southern blotting hybridization, using the radiolabeled 464-bp fragment as a probe. The chart on the right displays the lane profiles in the presence or absence of rCTP.
The presence of rNMPs in the template strand could interfere with the Taq pol activity in the following reaction cycles, as reported for other polymerases (39, 40). To exclude this, we tested the ability of the mutant Taq pol to bypass ribonucleotides in the template by using a primer extension assay. The I614K Taq polymerase efficiently bypasses embedded rCMP in DNA (Fig. S2); a slight pausing before the rNMP can be detected at enzyme concentrations much lower than those used in PCR reactions. Our synthesis strategy allows us, therefore, to obtain full-length amplified sequences.
AFM imaging and characterization of RC-DNAs
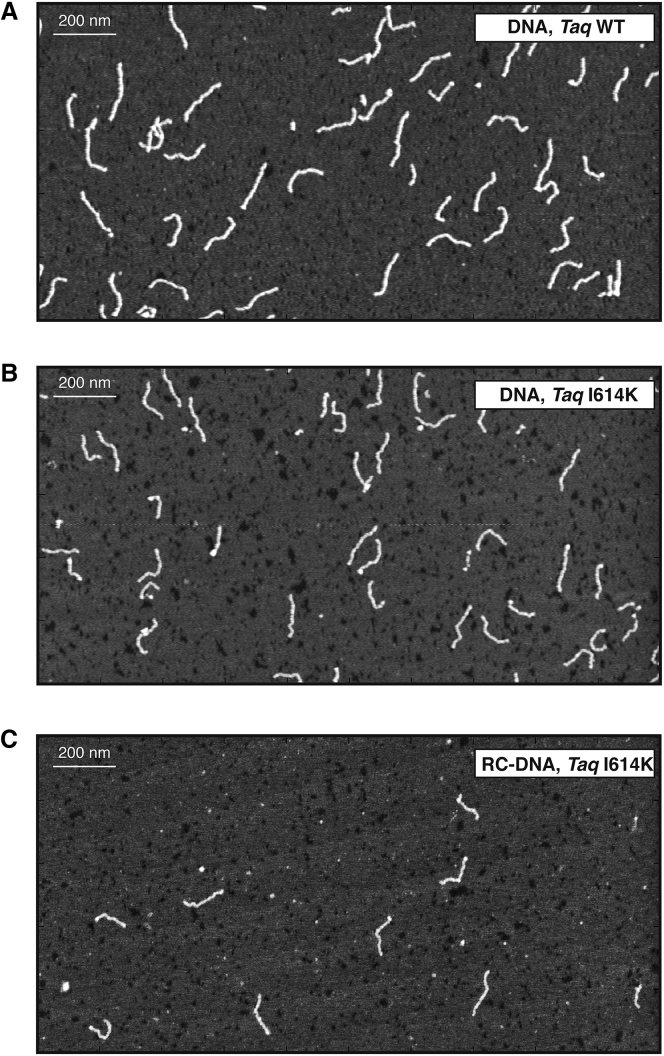
Once the RC-DNA production has been validated, we generated several DNA molecules with different lengths and features. We produced and purified DNA and RC-DNA molecules with five different lengths in basepairs (464, 646, 727, 960, and 1079), and subjected them to AFM imaging. As described in detail in the Materials and Methods, DNA molecules were deposited onto a negatively charged mica surface, where they adsorb thanks to the mediation of magnesium divalent ions, which bridge the negative charges of the DNA backbone and the mica surface. Fig. 3 shows three representative AFM images of DNA and RC-DNA molecules produced with either WT or I614K Taq polymerases. Typically, the DNA molecules are well contrasted, thanks to the Mg2+ buffer, which provides a clean way to bind the molecules to the mica surface, preserving the atomic smoothness and cleanliness of the freshly cleaved substrates. Because the efficiency of PCR decreases when the mutated Taq is used, and when ribonucleotides are added to the reaction, AFM maps typically feature a decreasing number of molecules per unit area. Some molecules from both WT and I614K Taq pol (∼20%) exhibited severe irregularities in their shape and dimensions, such as overlapping and condensed regions, protruding asperities, etc., and were not considered for the analysis. These molecules were excluded on the basis of a visual analysis as well as of the analysis of representative profiles, as shown in Fig. S3. As detailed in Materials and Methods, molecules from several AFM topographic profiles were semiautomatically traced to calculate the relevant structural and conformational quantities (contour length, rise per residue, bending angle distribution, and end-to-end distance curve).
Figure 3.
Representative AFM topographies of 464-bp molecules. Molecules are deposited on mica and imaged in air by AFM. (A) Shown here are DNA molecules synthesized with WT and (B) I614K Taq pol in the presence of dNTPs, and (C) RC-DNA molecules synthesized with the addition of rCTP by the I614K Taq polymerase. The size of the image is 2 × 1 μm2.
To validate our experimental imaging conditions, we have first characterized the conformational properties of DNA molecules produced by the WT Taq pol with dNTPs only. The scaling of the average contour length of linear DNA molecules with respect to the number of basepairs (464, 646, 727, 960, and 1079) is shown in Fig. 4.
The slope of the contour length versus number-of-basepairs curve represents the rise per residue r of the molecules. We found rB = 0.367 ± 0.011 nm/bp, which is close to the value for the B form of DNA (rB = 0.34 nm/bp), and significantly larger than the rise per residue of the A form (rA = 0.26 nm/bp). Notably, the intercept c of the fitting line in Fig. 4 is negative (c = −21.3 ± 7.2 nm), witness to a reduction of the contour length of the molecules, irrespective of the number of basepairs, of ∼60 bp in the B form. The reduction of the length of DNA molecules imaged in air is commonly observed (32, 33, 41), and the reason is still debated, although the prevalent hypothesis is that of a partial transition from B to A conformation. Such transition is often assessed based on the comparison of the measured contour length to the one expected for the B form, by considering only one molecular length (i.e., a given number of basepairs). This procedure, however, cannot capture accurately the scaling of the contour length with the number of basepairs, especially in those cases when a systematic alteration of the contour length is not attributable to a distributed, yet partial, transition. In the case of this study, we have evidence that the scaling of the DNA lengths, despite a systematic shortening, is the one typical of the B form (Fig. 4); we argue therefore that the constant shortening must be well localized within the molecule, which is at odds with the idea of a uniformly distributed shortening as expected by a uniform (yet only partial) B to A transition. Recently, Japaridze et al. (32) reported a similar evidence of molecular shortening, and by means of tip-enhanced Raman spectroscopy they were able to localize the shortened DNA tracts at the molecules’ free ends. We have carefully checked the calibration of our instrument, and we exclude this as the reason for the observed shortening (data not shown).
Reference DNA molecules synthesized by the I614K Taq pol have also been characterized (for a few selected lengths), and the measured lengths agree within error with those of the WT Taq pol, indicating that there is no alteration of DNA synthesized by the mutant Taq polymerase (Fig. 4). Our data suggest therefore that reference DNA molecules (WT and I614K Taq pol) are in the B-form, although there are shortened domains within the molecules. These samples represent the controls for the investigation of the effects of ribonucleotide incorporation. The latter have been assessed first by looking at changes of the contour length of the molecules upon ribonucleotide incorporation.
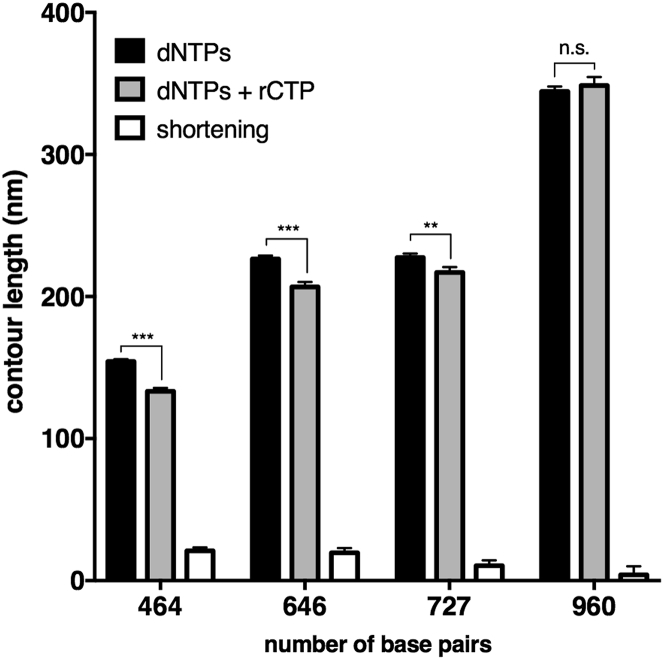
Interestingly, when comparing RC-DNA samples (464, 646, 727, and 960 bp) to their reference molecules without ribonucleotides, we observed a systematic shortening, except in the case of the 960-bp population (see Figs. 5 and S4 for the distribution of measured length values). We attribute the shortening of the contour length to the presence of rCTP. In fact, apart from the rCTP incorporation, nothing else is different from the control DNA molecules; moreover, the molecules generated by the I614K Taq pol have been shown to be equivalent in terms of length to the ones from the WT Taq (Fig. 4). We also exclude that the shortening effect is due to increased truncation of PCR products when ribonucleotides are present in the template DNA strand: the primer extension assay described above demonstrated that the I614K Taq polymerase efficiently bypasses embedded rCMP, without prematurely ending the synthesis reaction (see Fig. S2 and Supporting Materials and Methods). The measured length differences between DNA and RC-DNA molecules are all significant according to the two-tailed t-test, with the exception of the 960-bp DNA sample that is not significant. Indeed, this is the sample with the lowest expected percentage of basepairs containing an rCMP (1.8%, and reported in Table 1) that was determined by Eq. 2, which considers the GC content and the ribonucleotide incorporation frequency. As for the others, we expect a 3.1% of basepairs containing a ribonucleotide in the 464-bp, 2.3% in the 646-bp, and 3.0% in the 727-bp molecules (Table 1). These data suggest that the number of embedded ribonucleotides could be crucial to induce detectable alterations of the contour length; however, there seems to be no clear correlation of the contour length shortening with the original GC content (reported in Table 1), especially when comparing the 646- and 727-bp samples (the shortening is more pronounced in 646-bp RC-DNAs, despite the smaller GC content). In addition to the absolute amount of GC pairs, it is important to consider their spatial distribution along the molecules (see Fig. S5 for a graphical representation of the distribution of GCs). We notice that in the case of the 646-bp sequence, the majority of available positions for rCTP incorporation consist of single bases, and that G or C clusters contain a maximum of three adjacent bases (as detailed in Table 1). Although the I614K Taq polymerase could be able to insert consecutive ribonucleotides, this would be a very inefficient and unfavorable reaction (35). As a consequence, despite the higher expected quantity of rCTP in the 727 bp, this sample probably presents a lesser degree of incorporation. In its sequence, there are actually fewer single positions available, and more, and longer, GC clusters, made of two to six consecutive C or G (Table 1).
Figure 5.
The incorporation of rCTPs induces shortening of DNA molecules. Solid bars represent the mean contour length of four different sets of molecules, produced with dNTPs or with the addition of rCTP. Open bars represent the difference in contour length (shortening) observed upon ribonucleotide incorporation. In the case of 464-, 646-, and 727-bp populations, the shortening of the contour length is significant according to the two-tailed t-test (with p ≤ 0.001, 0.001, and 0.010, respectively), and this can be attributed to ribonucleotide incorporation. The 960-bp sample did not show a significant decrease in length (p = 0.50). Error bars represent the combination of the standard deviation of the mean and the calibration error, as explained in Materials and Methods. The number of analyzed 464-, 646-, 727-, and 960-bp dNTPs/dNTPs+rCTP molecules is 176/157, 92/92, 47/57, and 61/26, respectively. All plotted values are reported in Table S1.
Table 1.
Features of RC-DNA Molecules
| bp | %GC (%) | Total Number of G+C | Number of Clusters with 6–4 G or C | Number of Clusters with 2–3 G or C | Number of Single G or C | Total Number of rCTP Incorporated | %rCTP (%) |
|---|---|---|---|---|---|---|---|
| 464 | 58 | 269 | 4 | 50 | 143 | 14 | 3.1 |
| 646 | 43 | 279 | 0 | 51 | 159 | 15 | 2.3 |
| 727 | 57 | 413 | 5 | 77 | 224 | 22 | 3.0 |
| 960 | 34 | 325 | 3 | 45 | 215 | 17 | 1.8 |
GC content percentage, the GC clustering degree, and the estimated rCTP incorporation (absolute and percentage content) in the investigated DNA sequences. Clusters consist of consecutive nucleotides of the same species; here we have reported the sum of G and C clusters in one DNA strand, which is identical in the other complementary strand. Clusters are divided into two main groups: one made from 6 to 4 units, the other from 2 to 3. The number of isolated G or C present in the sequence is also reported. The total number of rCTP incorporated represents the estimated amount of rCTP incorporated into the molecule (with the percentage value) calculated using Eq. 2, as described in Materials and Methods.
Because double-stranded RNA molecules are known to be in the A form (42, 43), and RNA-containing oligonucleotides have been shown to undergo a partial B to A transition (19, 21), one can tentatively estimate an apparent fraction of basepairs that underwent a B to A transition upon rCTP incorporation, assuming that whenever an rCTP is incorporated, the hosting basepair switches from the B to the A form. The extent of such transition is calculated using the equations given in Materials and Methods. Fractions of the B-to A apparent transitions are 0.67, 0.41, 0.22, and 0.05 for 464-, 646-, 727-, and 960-bp fragments, respectively. These estimated fractions are surprisingly high, because the ribonucleotide incorporation ratio (one rCTP every 19 bp) would lead to a B to A transition fraction up to one order-of-magnitude smaller. A reasonable explanation could be that the incorporation of one rCTP triggers the transition from the B to A form not only in correspondence to that single basepair, but also along the surrounding nucleotides (a B/A junction), extending across several tens of basepairs (corresponding to a few cooperative lengths, ∼10 basepairs each (44, 45)). At present, however, we do not have clear evidence that a (partial) B to A transition is the leading mechanism behind the observed shortening of DNA molecules upon ribonucleotide incorporation.
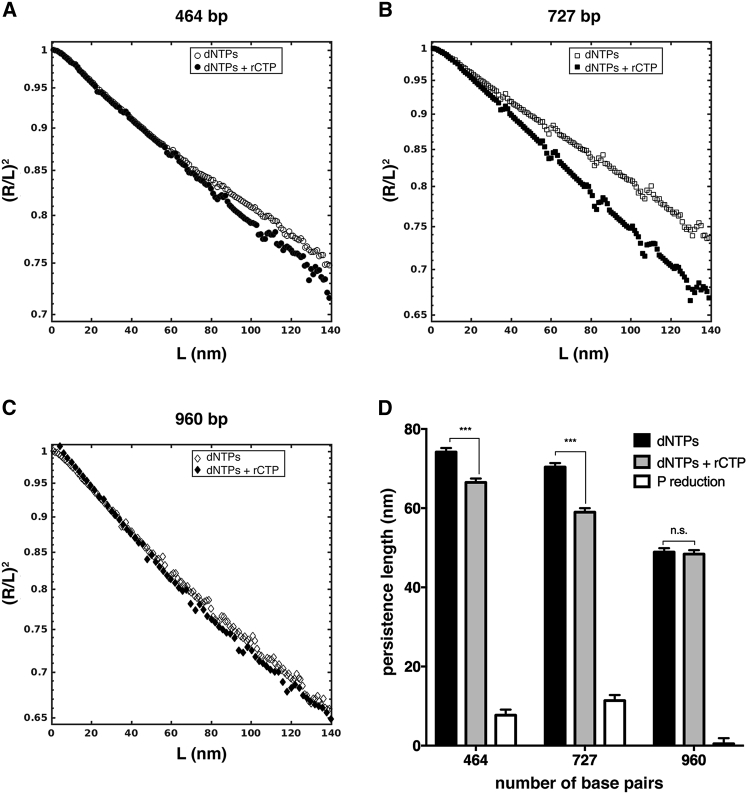
To further investigate the nature of the rCTP action on the DNA structure, we have investigated the mesoscopic conformation and elasticity of RC-DNAs. To this purpose, we have characterized the distribution of bending angles along the molecules’ backbones as a function of the curvilinear separation L, as well as the scaling of the mean squared end-to-end distance 〈R2〉2D (Eq. 1), as described in the Materials and Methods. Equation 1 is valid for molecules equilibrated on two dimensions, where P refers to the persistence length of the molecules in the three dimensions, i.e., in the bulk solution. Noticeably, the ratio (R/L)2 depends only on the ratio P/L, which also represents the asymptotic scaling of the normalized curves at large distances. The measured ratio of higher moments of the angle distribution 〈θ4〉/〈θ2〉2 is close to three (see Supporting Materials and Methods and Fig. S6), which is the theoretical WLC value for full equilibration of the molecules in two dimensions (25). The values of P for DNA and RC-DNA samples are obtained by fitting Eq. 1 to the R2 versus L curves (the representative curve for the 464-bp sample is shown in Fig. S7). Typically, the WLC model fitted with good accuracy the experimental data across the 0–120-nm distance range. Fig. 6, A–C, shows the scaling of the normalized mean squared end-to-end distance (R/L)2. The mean squared end-to-end distance R2 has been normalized by L2 to better appreciate the change in the slope (i.e., in the persistence lengths P) upon incorporation of rCTP, and the extrapolated persistence length values are plotted in Fig. 6 D and reported in Table S1. First, we noticed that for the 464- and 727-bp samples (from I614K Taq pol) the measured values of the persistence length are higher than the value of ∼50 nm, typically for DNA molecules with a ∼50% GC content (25, 26). However, this is consistent with the fact that high GC content is known to induce stiffening, with an increase of the persistence length (46). Remarkably, the incorporation of ribonucleotides into the molecules with higher GC content (464 and 727 bp) induced a significant shortening of the persistence length, according to the two-tailed t-test (Fig. 6 D; Table S1).
Figure 6.
rCTP incorporation affects end-to-end distance and persistence length of DNA molecules. (A–C) Shown here is the scaling of the normalized mean squared end-to-end distance (R/L)2 as a function of the curvilinear distance L for 464-, 727-, and 960-bp samples. Differences in the slope of the curves are appreciable for 464- and 727 bp, but not for 960 bp. In (D) are represented persistence length values of the same samples (solid bars): 464- and 727 bp showed extremely significant decrease in P upon rCTP incorporation (with p ≤ 0.001, according to the two-tailed t-test), whereas the 960-bp samples did not show any difference (p = 0.10). Open bars represent the persistence length shortening between the two compared samples. See Materials and Methods for details on error bars. The number of analyzed 464-, 727-, and 960-bp dNTPs/dNTPs+rCTP molecules is 176/157, 47/57, and 61/26, respectively. All plotted values are reported in Table S1.
The 960-bp sample is the only one not showing any appreciable reduction of the persistence length, in addition to the absence of the shortening of the contour length, as shown previously. However, as for the shortening of contour length, we could not define a clear trend of the persistence length shortening with the GC content, probably because the rCTP incorporation is not simply proportional to the available sites; the latter in turn are not uniformly distributed along the molecules, but present with different degrees of clustering and spatial distribution (Fig. S5; Table 1).
The molecules containing rCTP seem to be equilibrated to a good extent on the mica surface, and their R2 versus L curves can be fitted by the WLC model across a wide range of distances; these evidences suggest that the incorporation of rCTP exerts an influence on the DNA structure that goes well beyond the scale of the single ribonucleotide. In this study, we observe that, upon rCTP incorporation, the structure and conformation of DNA molecules change significantly and on the mesoscopic scale. In particular, the observed reduction of the persistence length P suggests that ribonucleotides induce a softening of DNA molecules (Fig. 6 D) in addition to causing a significant shortening (Fig. 5). The extension of the effects of the rCTP incorporation in dsDNA is remarkable, because one would expect an effective rCTP content of a few percent (Table 1). The action of a single ribonucleotide, namely its effect on the DNA structure, extends far beyond its linear dimension. Attributing the observed changes of conformational and elastic properties to a B to A transition—based on the fact that RNA is in the A form—is not at all straightforward, and this conclusion would not at present be fully supported by the data. On one side, DNA in the A form in solution is reported to be stiffer than in the B form (47), at odd with our observations; on the other side, DNA in the A form deposited on mica at different ethanol concentrations showed decreasing values of the persistence length at increasing B to A transition fractions, a trend similar to that observed by us, at least in the general terms. Concerning the published single-molecule studies on the bending rigidity (persistence length) of ribonucleotide-containing DNA molecules, including double-stranded RNA, either stiffer (23) or softer and slightly stiffer molecules (22) have been reported, depending on the sequence context and the positions of ribonucleotides. It emerged that besides the absolute content, which determines the extent of the incorporation, the distribution of ribonucleotide along the molecular backbone also is important in determining structural changes (22). Chiu et al. (22) concluded that the incorporation of ribonucleotides locally induces a significant (torsional) distortion of the sugar-phosphate backbone, affecting the elastic, and structural properties of the molecule as a whole. Relating such a torsional alteration to the observed mesoscopic changes of the DNA molecules conformation, namely the shortening of the molecules, and the reduction of the persistent length, is not trivial. Intuitively, a torsional alteration, especially if distributed along the molecular backbone, even if not uniformly, can lead to a shortening of the molecule. It was recently proposed that the rotational stiffening caused by the ribonucleotide-induced torsion of the sugar-phosphate backbone can hamper the rotational fluctuations, resulting in bending stiffening, rather than the opposite, at least as long as the electrostatic component of the persistence length is concerned (48). Under the hypothesis that the incorporation of ribonucleotides induces the formation of B/A junctions, which are known to be significantly bent (49), one could also consider the role of these bent domains in decreasing the apparent persistence length of DNA molecules (50). Further investigations are therefore required, by means of both experimental techniques and structural simulations, to unravel the fine mechanisms underlying the observed structural and conformational changes in DNA molecules upon ribonucleotide incorporation. In particular, the influence of the base sequence should be directly investigated and assessed, at similar GC content.
Conclusions
In this work, we present, to our knowledge, a novel approach to study the effects of ribonucleotides incorporation into DNA. Our synthesis strategy exploited the use of a mutant version of the Taq polymerase (I614K) in PCR reactions allowing us to generate populations of long (several hundreds of basepairs) RC-DNAs, which are more biologically relevant than short oligonucleotides. In particular, we have studied the ribonucleotides-induced changes in the structural and conformational properties of DNA at the single-molecule level by means of atomic force microscopy. Our systematic and statistical study highlighted the impact of ribonucleotides intrusions on the DNA double helix. We found that their presence alters DNA structural properties. All the investigated DNA molecules, with the exception of the longest molecules with the lowest GC content (960 bp), showed a significant reduction in the contour length upon rCTP introduction compared to their cognate molecules without rCTP. As observed in RNA molecules, it is the presence of the extra hydroxyl group on ribonucleotides that leads to a compaction of the DNA backbone. From the biochemical parameters of the I614K Taq polymerase (38) we estimated an incorporation frequency of 1/19 (rCTP/dCTP), representing 2–3% of the total number of basepairs. By contrast, the calculated apparent B to A transition fraction ranges between 20 and 60%. Although we do not have concluding evidence that this structural transition is the mechanism triggered by the ribonucleotide insertion, these figures suggest that the ribonucleotide effect on DNA structure extends remarkably on a scale that goes well beyond the typical dimension of a single ribonucleotide, affecting the full length of the molecule. Together with the shortening, RC-DNA molecules become more flexible, as demonstrated by the reduction of the persistence length. This is another indication that the effect of even a tiny fraction of incorporated ribonucleotides affects the DNA molecules on a global scale. A deeper understanding of the observed phenomena would require a precise quantification of the extent of rCTP incorporation, as well as of their spatial distribution along the RC-DNA molecules. Nevertheless, rCTP incorporation is nearly controlled by the number of positions available, given by the GC content.
In our system, control DNA molecules retain their native B conformation, supporting the idea that changes upon ribonucleotide introduction are likely to occur also in vivo. Along the genome of living cells there are hotspots of ribonucleotide incorporation, and the most frequently incorporated ribonucleotide is rCTP (36, 37). We speculate that the induced structural and conformational alterations can contribute to the negative outcome of ribonucleotide persistence in DNA. These alterations could be easily transferred to DNA transactions that rely on structure recognition, such as protein binding. For instance, it is reported that ribonucleotides prevent nucleosome assembly on DNA (15), probably reshaping their positioning. Therefore, our strategy to study the effect of ribonucleotide incorporation of long supported DNA fragments could contribute to a better understanding of their harmful consequences upon genome stability.
Author Contributions
M.M.-F., F.L., and A.P. conceived the study and designed the experiments. A.M. and A.P. performed the main experiments and analyzed the data. E.M. and E.C. performed the control experiment reported in Fig. S2. A.M. and A.P. wrote the manuscript. M.M.-F. and F.L. revised the manuscript. All authors participated in scientific discussions.
Acknowledgments
A.M. thanks Francesca Borghi for support in AFM imaging, and Barbara Sciandrone for support in radioactive DNA dialysis. We thank Prof. A. Aliverti and Prof. P. Plevani for critical discussions, and G. Maga for support in the control experiments.
A.P. thanks the Department of Physics of the University of Milano for financial support under the project “Piano di Sviluppo dell’Ateneo per la Ricerca 2014. Linea B: Supporto per i Giovani Ricercatori”. The M.M.-F. lab is funded by AIRC (no. 15631), MIUR, and Telethon (GGP15227). F.L. is funded by AIRC and Fondazione Cariplo (TRIDEO 2014 Id. 15724) and Fondazione CARIPLO (RIF. 2013-0798). E.M. and E.C. are supported by AIRC individual grant MFAG2016 18811.
Editor: Karin Musier-Forsyth.
Footnotes
Federico Lazzaro and Alessandro Podestà contributed equally to this article.
Supporting Materials and Methods, eight figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30811-1.
Contributor Information
Marco Muzi-Falconi, Email: marco.muzifalconi@unimi.it.
Alessandro Podestà, Email: alessandro.podesta@mi.infn.it.
Supporting Material
References
- 1.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. [Google Scholar]
- 2.Li Y., Breaker R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- 3.Hiller B., Achleitner M., Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nick McElhinny S.A., Watts B.E., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nick McElhinny S.A., Kumar D., Kunkel T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparks J.L., Chon H., Burgers P.M. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghodgaonkar M.M.M., Lazzaro F., Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lujan S.A., Williams J.S., Kunkel T.A. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N., Huang S.N., Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conover H.N., Lujan S.A., Argueso J.L. Stimulation of chromosomal rearrangements by ribonucleotides. Genetics. 2015;201:951–961. doi: 10.1534/genetics.115.181149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazzaro F., Novarina D., Muzi-Falconi M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reijns M.A.M., Rabe B., Jackson A.P. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crow Y.J., Leitch A., Jackson A.P. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 15.Hovatter K.R., Martinson H.G. Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc. Natl. Acad. Sci. USA. 1987;84:1162–1166. doi: 10.1073/pnas.84.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egli M., Usman N., Rich A. Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 17.Ban C., Ramakrishnan B., Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA-RNA chimeric decamer duplex d(CCGGC)r(G)d(CCGG) with a novel intermolecular G-C base-paired quadruplet. J. Mol. Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 18.Wahl M.C., Sundaralingam M. B-form to A-form conversion by a 3′-terminal ribose: crystal structure of the chimera d(CCACTAGTG)r(G) Nucleic Acids Res. 2000;28:4356–4363. doi: 10.1093/nar/28.21.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaishree T.N., van der Marel G.A., Wang A.H. Structural influence of RNA incorporation in DNA: quantitative nuclear magnetic resonance refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG) Biochemistry. 1993;32:4903–4911. doi: 10.1021/bi00069a027. [DOI] [PubMed] [Google Scholar]
- 20.Chou S.H., Flynn P., Reid B. High-resolution NMR studies of chimeric DNA-RNA-DNA duplexes, heteronomous basepairing, and continuous base stacking at junctions. Biochemistry. 1991;30:5248–5257. doi: 10.1021/bi00235a019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeRose E.F., Perera L., London R.E. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu H.-C., Koh K.D., Storici F. RNA intrusions change DNA elastic properties and structure. Nanoscale. 2014;6:10009–10017. doi: 10.1039/c4nr01794c. [DOI] [PubMed] [Google Scholar]
- 23.Abels J.A., Moreno-Herrero F., Dekker N.H. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys. J. 2005;88:2737–2744. doi: 10.1529/biophysj.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrero-Galán E., Fuentes-Perez M.E., Arias-Gonzalez J.R. Mechanical identities of RNA and DNA double helices unveiled at the single-molecule level. J. Am. Chem. Soc. 2013;135:122–131. doi: 10.1021/ja3054755. [DOI] [PubMed] [Google Scholar]
- 25.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 26.Podestà A., Indrieri M., Dunlap D. Positively charged surfaces increase the flexibility of DNA. Biophys. J. 2005;89:2558–2563. doi: 10.1529/biophysj.105.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lia G., Indrieri M., Dunlap D. ATP-dependent looping of DNA by ISWI. J. Biophotonics. 2008;1:280–286. doi: 10.1002/jbio.200810027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podestà A., Imperadori L., Dunlap D. Atomic force microscopy study of DNA deposited on poly L-ornithine-coated mica. J. Microsc. 2004;215:236–240. doi: 10.1111/j.0022-2720.2004.01372.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J., Russell D.W. Alkaline agarose gel electrophoresis. CSH Protoc. 2006 doi: 10.1101/pdb.prot4027. 2006 Jun 1;2006(1). pii: pdb.prot4027. http://dx.doi.org/10.1101/pdb.prot4027. [DOI] [PubMed] [Google Scholar]
- 30.Meroni A., Lazzaro F., Podestà A. Characterization of structural and configurational properties of DNA by atomic force microscopy. In: Walker J.M., editor. Methods in Molecular Biology. Humana Press; New York, NY: 2017. [DOI] [PubMed] [Google Scholar]
- 31.Usov I., Mezzenga R. FiberApp: an open-source software for tracking and analyzing polymers, filaments, biomacromolecules, and fibrous objects. Macromolecules. 2015;48:1269–1280. [Google Scholar]
- 32.Japaridze A., Vobornik D., Dietler G. Toward an effective control of DNA’s submolecular conformation on a surface. Macromolecules. 2016;49:643–652. [Google Scholar]
- 33.Rivetti C., Codeluppi S. Accurate length determination of DNA molecules visualized by atomic force microscopy: evidence for a partial B- to A-form transition on mica. Ultramicroscopy. 2001;87:55–66. doi: 10.1016/s0304-3991(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 34.Chien A., Edgar D.B., Trela J.M. Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J. Bacteriol. 1976;127:1550–1557. doi: 10.1128/jb.127.3.1550-1557.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel P.H., Loeb L.A. Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J. Biol. Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- 36.Koh K.D., Balachander S., Storici F. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat. Methods. 2015;12:251–257. doi: 10.1038/nmeth.3259. 3, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausen A.R., Lujan S.A., Kunkel T.A. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel P.H., Kawate H., Loeb L.A. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J. Biol. Chem. 2001;276:5044–5051. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
- 39.Clausen A.R., Zhang S., Kunkel T.A. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair (Amst.) 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clausen A.R., Murray M.S., Kunkel T.A. Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc. Natl. Acad. Sci. USA. 2013;110:16802–16807. doi: 10.1073/pnas.1309119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Sevilla A., Thimonier J., Barbet J. Accuracy of AFM measurements of the contour length of DNA fragments adsorbed on mica in air and in aqueous buffer. Ultramicroscopy. 2002;92:151–158. doi: 10.1016/s0304-3991(02)00128-6. [DOI] [PubMed] [Google Scholar]
- 42.Langridge R., Gomatos P.J. The structure of RNA. Science. 1963;141:1024. doi: 10.1126/science.141.3582.694. [DOI] [PubMed] [Google Scholar]
- 43.Arnott S., Hukins D.W.L., Dover S.D. Optimised parameters for RNA double-helices. Biochem. Biophys. Res. Commun. 1972;48:1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- 44.Ivanov V.I., Minchenkova L.E., Zimmer C. The detection of B-form/A-form junction in a deoxyribonucleotide duplex. Biophys. J. 1996;71:3344–3349. doi: 10.1016/S0006-3495(96)79527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov V.I., Krylov D.Y., Minchenkova L.E. B-A transition in DNA. J. Biomol. Struct. Dyn. 1983;1:453–460. doi: 10.1080/07391102.1983.10507454. [DOI] [PubMed] [Google Scholar]
- 46.Hormeño S., Ibarra B., Arias-Gonzalez J.R. Mechanical properties of high-G.C content DNA with A-type base-stacking. Biophys. J. 2011;100:1996–2005. doi: 10.1016/j.bpj.2011.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charney E., Chen H.-H., Rau D.C. The flexibility of A-form DNA. J. Biomol. Struct. Dyn. 1991;9:353–362. doi: 10.1080/07391102.1991.10507917. [DOI] [PubMed] [Google Scholar]
- 48.Dobrynin A.V. Electrostatic persistence length of semiflexible and flexible polyelectrolytes. Macromolecules. 2005;38:9304–9314. [Google Scholar]
- 49.Selsing E., Wells R.D., Arnott S. Bent DNA: visualization of a base-paired and stacked A-B conformational junction. J. Biol. Chem. 1979;254:5417–5422. [PubMed] [Google Scholar]
- 50.Rouzina I., Bloomfield V.A. DNA bending by small, mobile multivalent cations. Biophys. J. 1998;74:3152–3164. doi: 10.1016/S0006-3495(98)78021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.