Abstract
X-chromosome inactivation (XCI) is a critical epigenetic mechanism for balancing gene dosage between XY males and XX females in eutherian mammals. A long non-coding RNA (lncRNA), XIST, and its associated proteins orchestrate this multi-step process, resulting in the inheritable silencing of one of the two X-chromosomes in females. The XIST RNA is large and complex, exemplifying the unique challenges associated with the structural and functional analysis of lncRNAs. Recent technological advances in the analysis of macromolecular structure and interactions have enabled us to systematically dissect the XIST ribonucleoprotein complex, which is larger than the ribosome, and its place of action, the inactive X-chromosome. These studies shed light on key mechanisms of XCI, such as XIST coating of the X-chromosome, recruitment of DNA, RNA and histone modification enzymes, and compaction and compartmentalization of the inactive X. Here, we summarize recent studies on XCI, highlight the critical contributions of new technologies and propose a unifying model for XIST function in XCI where modular domains serve as the structural and functional units in both lncRNA–protein complexes and DNA–protein complexes in chromatin.
This article is part of the themed issue ‘X-chromosome inactivation: a tribute to Mary Lyon’.
Keywords: XIST, X-chromosome inactivation, interactome, modularity, RNA structure domains, topological associating domains
1. Introduction
X-chromosome inactivation (XCI) comprises a series of highly organized events that result in the selective silencing of one of the two X-chromosomes in eutherian females [1–3]. The process starts with the counting of the X-chromosomes, choosing one for silencing, followed by initiation and maintenance of the inactive state. All of these steps are under the control of the X inactivation centre (Xic) and its products, especially Xist, a long non-coding RNA (lncRNA) [4,5]. Specific DNA elements within the Xic regulate counting, choice and allele-specific upregulation of the Xist transcription [1]. The Xist lncRNA is involved in multiple steps during X inactivation, including coating of the presumptive inactive X (Xi) chromosome, exclusion of RNA polymerase, removal of active histone marks and deposition of repressive histone marks, methylation of DNA, tethering of the inactive X to the nuclear periphery and packing of the chromatin. This multitude of functions requires the actions of various protein complexes because the Xist RNA itself does not possess enzymatic activities. Yet the molecular mechanisms underlying most of these functions remain elusive. A number of recent studies have employed new technologies to discover new mechanisms in XCI (see, recent reviews, [6–8]). In this review, we summarize recent progress on the identification and mechanistic studies of the functions organized by the Xi RNA and the inactive X-chromosome.
2. The XIST interactome and their functions
(a). Proteomic and genetic screens for X-chromosome inactivation factors
Xist is a 17–19 kb long RNA with complex repeat patterns that scaffold the XCI machinery (figure 1a). Previous studies have reported a number of protein complexes as Xist partners in XCI, yet some of them remain controversial. To identify the proteins that associate with Xist RNA, three laboratories developed antisense oligo-based purification of the Xist RNA [11–13]. In all three methods, the Xist RNA was isolated from cells using antisense oligos targeting the endogenous lncRNA and the associated proteins were identified by mass spectrometry. Specifically, ChRIP-MS uses formaldehyde, which cross-links both direct and indirect interactors, while RAP-MS and iDRiP use UV cross-linking, which identifies direct RNA interactors (RBP). Taken together, these three studies identified between 10 and 200 XIST-associated proteins (XIST RBP). The difference in the number of targets reflects the different cross-linking techniques, cell models and thresholds to call significant interactions. Two other laboratories have used viral-gene trap and shRNA-based genetic screens to identify factors that are required for XCI [14,15]. Monfort et al. [15] established a haploid stem cell model that expresses an inducible Xist RNA from an autosome chr11. Xist-mediated silencing of the single chr11 is lethal, which can be suppressed with disruption of critical XCI factors by viral insertion. Moindrot et al. used pooled shRNAs to silence XCI factors that are required for the suppression of a GFP gene in the vicinity of an autosomally integrated Xist gene. The proteomic screens could identify all Xist-associated proteins but not all of them are necessarily essential for XCI, while the genetic screens identify essential factors for XCI but may miss ones that are required for cell viability. Taken together, these studies validated several previously reported proteins essential for XCI and revealed new factors that have led to a deeper understanding of this complex process.
Figure 1.
Structural basis of X inactivation: XIST structure and interactions. (a) Summary of mature XIST RNA structures using the human XIST as an example. XIST exons are shown as alternating black and grey rectangles (exons 2–5 are much smaller than 1 and 6). Repeat regions shown in thick blocks were annotated based on Elisaphenko et al. [9]. Each arc represents the base pairing interaction between the two arms of a duplex. Pink rectangles mark the structure domains defined by PARIS. (b) Summary of protein complexes that interact with XIST and their functions. Some of the best-studied examples of XIST interactions are shown. Short vertical arrows indicate known interactions. Question marks indicate that the mechanisms of interactions or functions remain unknown. The WTAP–RBM15–RBM15B complex primarily binds the A-repeat, but also to other regions to a minor extent. (c) The A-repeat forms stochastic inter-repeat duplexes that bind the adapter protein SPEN. (d) Consensus inter-repeat duplex model. The highlighted sequences are the two repeats. The duplex contains eight GC base pairs at the two sides and four forced base pairs in the middle, based on SHAPE reactivity. Domain model of the XIST RNP. (e) Structure model for the interaction between A-repeat and SPEN. (f) Structure model of the XIST RNP. Question marks represent interactions that are unclear or controversial. The size of each domain is not exactly scaled to the real size. The protein complexes are placed closest to their target sites on the XIST RNA. The linker regions among the domains are flexible, so the model here only represents the topology, but not the actual rigid shape. ‘m6A mod’ represents the m6A methylase complex and associated proteins. Figure adapted from Lu et al. [10].
(b). Factors involved in Xist coating of the inactive X-chromosome
To specifically silence the X-chromosome of its origin, the Xist RNA sticks to the X-chromosome and spreads along it. Very little diffusion to the nucleoplasm occurs, ensuring that the silencing does not spill over to other chromosomes. Jeon & Lee [16] identified YY1 as a factor required for XIST localization by acting as a bridge between the Xist DNA and RNA (figure 1b). Recent studies suggest that YY1 may play a more complex role in XCI and Makhlouf et al. [17] presented evidence that YY1 activates Xist transcription by binding to the Xist promoter region. Whether or not YY1 directly binds the Xist RNA remains controversial. Although Jeon et al. detected an interaction between the two, YY1 was not identified in any of the proteomic or genetic screens in various cellular models of XCI [11–15].
Hasegawa et al. [18] identified another essential protein for Xist coating of the inactive X, HnrnpU (figure 1b). Knocking down HnrnpU in mouse cells delocalizes the Xist cloud from the Xi territory. HnrnpU is a bivalent protein that possesses both DNA- and RNA-binding activities, with the RNA binding detected by UV cross-linking experiments. Hendrickson et al. [19] used formaldehyde cross-linking to show that HnrnpU interacts with multiple broad regions on the human XIST RNA, confirming the mouse results. All three proteomics studies identified HnrnpU as a highly enriched partner of Xist, further confirming the Xist–HnrnpU interaction [11–13]. Recently, two more studies revealed redundancy in the HnrnpU-mediated Xist-coating mechanism that varies among cell lines [20,21]. An HnrnpU paralogue HnrnpUL1 compensates for HnrnpU function in some cell types when HnrnpU is knocked down. Interestingly, another HnrnpU paralogue was also purified as a component of the Xist ribonucleoprotein (RNP) [11], although knockdown studies did not support an essential role for this paralogue [20,21]. Taken together, these proteomic and functional analyses have demonstrated an essential role of the HnrnpU family of proteins in Xist coating of the Xi. Further studies will be required to dissect the relationships among the paralogues. Consistent with the analysis of Xist, Hacisuleyman et al. [22] demonstrated a role of HnrnpU in tethering Firre, another lncRNA, to the X-chromosome and other trans-chromosomal loci, suggesting a general role of HnrnpU in lncRNA localization and chromosome organization.
(c). Xist-mediated recruitment of the Polycomb complexes
The Polycomb Repressive Complexes PRC1 and PRC2 are general epigenetic regulators, catalysing the deposition of H2AK119 ubiquitination (H2AK119ub) and H3K27 trimethylation (H3K27me3), respectively [23]. PRC1 and PRC2 play important roles in XCI, but are not strictly required for transcriptional silencing [24–26]. Xist RNA is required for the recruitment of the PRC complexes to the X-chromosome [27]; however, it remains unclear how the recruitment occurs (figure 1b). Some of the controversies in PRC–Xist interactions were recently discussed in length by Brockdorff [28]. PRC1 is recruited to the inactive X by both PRC2-dependent and -independent mechanisms. Canonical PRC1 complex binds the H3K27me3 modification deposited by the PRC2 complex [29]. Alternatively, the PRC1 complex can be recruited independent of PRC2, potentially by direct interaction with Xist [24,30]. The direct interaction between PRC1 and Xist is supported by recent proteomic analysis of the Xist RNPs. Chu et al. [11] and Minajigi et al. [13] independently showed that components of the PRC1 complexes, Pcgf5, Rybp and Ring1, interact with Xist.
The recruitment mechanism for PRC2 is more complex. Several lines of evidence support an interaction between PRC2 and Xist, either directly or indirectly. For example, PRC2 can be cross-linked to Xist RNA using formaldehyde, and they form a complex using in vitro purified components. Furthermore, the A-repeat region was proposed as the PRC2-binding site [31–33]. da Rocha et al. [34] identified an RBP component of the PRC2 complex, Jarid2, as a mediator of PRC2 recruitment by Xist and showed that the recruitment depends on Xist repeats B and F. Several other studies argue against a direct role for Xist in PRC2 recruitment. Super resolution microscopy shows that the PRC2- and Xist-binding sites on Xi do not overlap, suggesting no stable interaction [35,36]. Two of the Xist proteomic screens did not yield any components of the PRC2 complex [11,12], further suggesting an indirect or transient interaction. It is also unlikely that Xist A-repeat is required for PRC2 recruitment because loss of the A-repeat region does not abolish the PRC2 enrichment on the Xi [27]. Chu et al. [11] identified HnrnpK as an abundant Xist RBP that is required for PRC1 and PRC2 recruitment to Xi; however, there is no evidence for direct interaction between HnrnpK and the PRC complexes so far and therefore it is unclear how this occurs. McHugh et al. [12] showed that the A-repeat-binding protein SPEN also promotes PRC2 recruitment to the Xi, but the mechanism is not known. Recently, Hendrickson et al. [19] analysed the RNA targets of chromatin-associated proteins using formaldehyde-assisted RNA-IP sequencing. Two of the PRC2 components, EZH2 and SUZ12, were shown to bind XIST RNA close to the E-repeat region and the end of the transcript, suggesting that there is indeed interaction, although it could be indirect and transient. The mechanisms of PRC complexes recruitment remain controversial and it would require more studies to determine the relative contribution of the multiple protein factors and of the Xist RNA regions identified so far.
(d). Tethering of the Xi to the nuclear periphery
It was known long ago that the inactive X-chromosome, also known as the Barr body, localizes to the nuclear periphery [37]. However, the mechanism was not known until recently (figure 1b). All three proteomic screens for Xist RBPs identified LBR, a nuclear membrane-associated protein, and McHugh and co-workers showed that depletion of LBR reduces XCI [11–13]. Further studies showed that the LBR protein binds Xist RNA with its arginine-serine (RS) motif and serves as a bridge between the Xist-coated X-chromosome and the nuclear lamina [38]. Interestingly, the LBR protein binds Xist RNA in three regions, the A-repeat and two regions around the E-repeat region. These multiple LBR-binding sites in Xist RNA suggest redundancy on the Xist RNA.
(e). N6-methyladenosine modification of Xist
RNA N6-methyladenosine (m6A) modification has emerged as an important regulatory mechanism for several RNA metabolism steps, including splicing, translation and stability [39–43]. m6A is widely present in the transcriptome and recent studies showed that the XIST RNA contains multiple m6A modifications (figure 1b) [44]. Consistent with this, Chu et al. [11] identified a component of the RNA methyltransferase complex, WTAP, in the Xist RNP. WTAP interacts specifically with the A-repeat region of Xist, and this interaction was observed only after differentiation, but not in undifferentiated ES cells. The developmental regulation of WTAP–Xist interaction is especially interesting, given that the stem cell differentiation process controls XCI. Further studies from the Jaffrey and Guttman labs identified RBM15 and RBM15B as critical adapters that bring the WTAP-containing methyltransferase complex to the XIST RNA and showed that the function is mediated by the m6A reader YTHDC1 [45]. The RBM15 and RBM15B proteins are paralogues of SPEN, and all of them bind primarily to the A-repeat region, although there is also weaker binding to other regions of the Xist transcript. Questions remain as to how the m6A modification regulates XCI. Potential mechanisms based on previous studies on mRNAs could involve splicing, RNA stability and other protein factors that interact with m6A readers.
(f). The A-repeat ribonucleoprotein complex
The A-repeat portion of Xist is close to the 5′-end and consists of 7.5 or 8.5 conserved repeats, each 24 nt long with sparse variations (figure 1c). The repeat units are separated by 20–50 nt variable regions that are pyrimidine-rich. The A-repeat region is one of the most conserved sequences in the entire Xist RNA and is required for the silencing activity, but not coating of the X-chromosome [46]. Previous studies have suggested PRC2 and SF2 (also known as SRSF1) as factors that bind the essential A-repeat region, but the PRC2–A-repeat interaction has been disputed (figure 1b) [28,31,32,47]. SF2 binds the A-repeat with very high specificity and is required for the efficient processing of the Xist transcript [47,48]. A number of recent studies provided definitive evidence for specific functional interaction between A-repeat and SPEN, a large RNA-binding protein. The UV cross-linking-based RAP-MS and iDRiP identified SPEN as direct XIST interactors [12,13]. Formaldehyde cross-linking-based ChRIP-MS showed that SPEN specifically binds the A-repeat region in Xist [11]. SPEN was also identified as critical for XCI using shRNA and viral-gene trap-based genetic screens [14,15]. Two following studies have used CLIP (Cross-Linking ImmunoPrecipitation) to definitively position the binding sites to the A-repeat region [10,38]. SPEN was initially identified in Drosophila as a transcriptional repressor [49–51]. Later studies showed that SPEN interacts with the SMRT–HDAC complex, which mediates histone deacetylation, RNA Pol II exclusion and transcriptional silencing [52]. Specifically, HDAC3 was shown to be the SPEN partner in XCI [12]. Taken together, these studies establish a clear link between the essential A-repeat region and the silencing activity of histone deacetylases. The identification of multiple RBPs for the A-repeat region, such as SPEN, RBM15, WTAP and SF2, suggests a multi-subunit complex, but the mechanisms of assembly and function remain to be determined.
Taken together, these studies have revealed a more comprehensive picture of the Xist RNP. Distinct RBPs and effector complexes occupy distinct sequence regions on the Xist RNA, instead of being mixed up and distributed along the RNA. This organization provides a means to structurally and functionally segregate the various functions of the Xist RNP, and also coordinate their functions during the well-organized XCI process. As new interactions are being mapped and their functions defined, we will arrive at a better understanding of this complex RNP. Yet many other questions remain, and new ones arise from these studies. What do other XIST-associated proteins do during XCI? How does the XIST RNA recruit the associated proteins? How does Xist coordinate these functions and how are they temporally organized? In the next section, we will discuss recent advances in the analysis of Xist RNA structure that have begun to address some of these questions.
3. XIST architecture and modular assembly of the ribonucleoprotein
RNA structures are increasingly being recognized as important regulators of gene expression [53,54]. Functions of lncRNAs may require sequence elements and/or structural motifs. Given the strong propensity of RNAs to form structures in cells and the overall lack of sequence conservation for lncRNAs, it is likely that the structural context plays an important role in the recruitment of RBPs and other effectors. A number of recent studies have used computational and experimental methods to determine the structure of the Xist RNA. Taken together, these studies have provided new insights into how Xist structure contributes to function and revealed a general modular organization of the Xist RNP.
(a). In vitro probing of A-repeat structure
The first structure model proposed for the Xist RNA is an intrarepeat two stem-loop model for the A-repeat region, which is essential for transcriptional repression [46]. Mutations that disrupt the proposed structure result in loss of silencing, consistent with the functional significance of the A-repeat. However, these experiments did not rule out other possible structure models, such as inter-repeat duplexes, which would also be disrupted by the mutations. Two later studies used in vitro methods to further investigate the structure of A-repeat but have obtained contradictory results [33,55]. Duszczyk et al. [55] used nuclear magnetic resonance (NMR) to investigate the structure of a single 14 nt stem-loop in the 5′-end of each A-repeat unit and showed that this stem-loop structure is remarkably stable. Contrary to the two stem-loop model proposed by Wutz et al. [46], the NMR study also suggested that the 3′-end of the repeat mediates inter-repeat duplex formation [55]. Maenner et al. [33] used a combination of single-strand and double-stand-specific enzymes and the structure selective chemicals dimethyl sulfate (DMS) and CMCT to probe the structure of purified human and mouse Xist fragments containing the A-repeat region. The sites of enzymatic and chemical reactions with the in vitro folded A-repeat RNA were determined using primer extensions. These experiments reported the base pairing status of each nucleotide and the data were then used to guide secondary structure modelling. Based on these data, Maenner et al. proposed a structure model of the A-repeat, where neighbouring repeat units form inter-repeat duplexes. Maenner et al. further used FRET (Förster resonance energy transfer) and in vitro binding experiments to validate the structure model and show that the structure binds PRC2. Both studies were conducted in vitro where the A-repeat region was isolated from its physiological context, therefore undermining the biological relevance of these conclusions.
(b). In vivo probing of the A-repeat structure
With the advent of new technologies, especially cell-permeable chemical probes and high-throughput sequencing, three more recent studies began to address the structure of the Xist RNA in living cells, revealing a more complex picture. Fang et al. [56] used DMS probing combined with high-throughput sequencing to generate a nearly complete transcript-level flexibility profile for the mouse Xist RNA and used these data to model local structures along the entire RNA. Structure models were developed for several regions, including the A-repeat region involved in silencing and the C-repeat region involved in X-chromosome coating. The A-repeat was predicted to form a combination of intrarepeat two stem-loop and inter-repeat stem-loop structures. Smola et al. [57] used selective 2′-hydroxyl acylation analysed by Primer Extension (SHAPE) chemicals 1M6, 1M7 and NMIA to probe the structure of Xist from natively purified RNA and in living cells. Similarly, the flexibility profiles were used to guide local structure modelling. Based on the SHAPE analysis, Smola et al. proposed yet another distinct model for the A-repeat region, containing inter-repeat duplexes and a pseudoknot. The lack of consensus in these studies highlighted the need for methods for direct RNA structure analysis.
(c). Direct determination of A-repeat structure by cross-linking
Conventional structure probing methods produce one-dimensional flexibility/accessibility profiles and the resulting structure models are often built with low accuracy. Several major problems plague these methods, including the ambiguity in the strength of reactivity (i.e. not a black and white difference), the size limit of the modelling and the fact that the base pairing relationships are based on prediction instead of direct experimental evidence [54]. To overcome these problems associated with chemical and enzymatic probing, Lu et al. [10] developed a new method, PARIS (Psoralen Analysis of RNA Interactions and Structures), for direct determination of base pairing helices in living cells. In addition, Lu et al. also used an improved in vivo click (ic)SHAPE chemical, NAI-N3, for structure probing [58]. The PARIS data showed that the A-repeat region forms an isolated domain with little contact with the rest of the RNA (figure 1a). Within the A-repeat domain, repeat units base pair with each other, forming a family of inter-repeat helices that are often alternative to one another (figure 1c). Furthermore, high-sensitivity icSHAPE data also supported the conserved repeat units forming duplexes while the variable spacers remain single stranded. The 12 bp stem (eight GC pairs and four internally forced pairs) in each duplex is stable (figure 1d), with lower free energy than the intrarepeat two stem-loop model originally proposed by Wutz et al. [46]. The inter-repeat nature of the new model is consistent with previous studies [33]. Interestingly, the A-repeats were originally derived from ERVB4 endogenous retroviruses [9], and the ERVB4 sequences contain two consecutive repeats that are also likely to form an inter-repeat structure.
Identification of covaried base pairs is a classical method for the detection of structures and furthermore suggests functional significance. A recent study from Rivas et al. challenged the notion of structure conservation in lncRNAs using rigorous statistical tests where phylogenetic effect on the covariation is removed [59]. Analysis of the A-repeat region did not yield statistically significant covariation support for any of the fixed A-repeat structure models previously proposed. In fact, the pattern of covariation indicates a large number of possible pairings, consistent with the stochastic formation of alternative structures detected by PARIS, which distributes and dilutes the evolutionary pressure. Taken together, the PARIS, icSHAPE and covariation analysis all support the stochastic inter-repeat duplex model for A-repeat, making it the best-understood region in the entire Xist transcript.
(d). Model of the SPEN–A-repeat complex
Recent proteomic and genetic screens have unanimously identified SPEN as an essential bridge between the A-repeat and histone deacetylases [11–15]. To further determine the mechanism of interaction between the A-repeat region and the SPEN protein, Lu et al. [10] performed in vitro individual nucleotide-resolution (i) CLIP to identify the SPEN-binding sites. Consistent with the previous mapping of SPEN to A-repeat [11], the SPEN-binding sites were clustered in the A-repeat region, in the variable spacers 2–4 nt from the 5′-end of each repeat (figure 1c). The fixed distance between the cross-linking sites and the inter-repeat duplex implies a direct contact with the duplex as well. In fact, Arieti et al. [60] solved the crystal structure of the SPEN RRM domain and showed that binding of SPEN to another known RNA target, the SRA lncRNA, requires both single- and double-stranded regions. In addition, in vitro gel shift assays suggest cooperation in the formation of the SPEN–A-repeat RNP complex [10]. Taken together, these data support a simple model for the SPEN–Xist interface, where the stochastic formation of duplexes among repeats forms a multivalent platform, where the single-stranded spacers and double-stranded repeats interact potentially with multiple SPEN proteins, which in turn recruit the histone deacetylase complex (figure 1e). Importantly, these studies revealed the assembly of a functional RNP domain, providing an example for the modularity concept. The delineation of the minimal RNP unit in the SPEN–XIST complex paves the way for solving its atomic resolution structure, which will shed light on some of the remaining questions, such as how the conserved duplex binds SPEN with high affinity despite the low specificity in reconstituted gel shift assays [10].
(e). Xist RNA architecture
PARIS also makes it possible to examine the overall architecture of the entire human XIST transcript in living cells, because it directly determines base pairing interactions and it is not constrained by the linear size of RNA. Short-range and long-range structures organize the 19 kb long XIST RNA into distinct and compact domains that span hundreds or thousands of nucleotides (figure 1a,f) [10]. In addition to the A-repeat domain, the F-repeat and downstream regions form another small domain (figure 1a). The repeats B, C and D form a giant domain that spans nearly 10 kb. E-repeat and surrounding sequences that range from the end of exon1 to the beginning of exon6 form another medium-sized domain. The remainder of exon6 forms the last domain that partially covers the E-repeat region in the beginning of exon6. These structure models and domain organization are supported by analysis of conservation in eutherian mammals using SISSIz [61], a statistical method that determines the significance of conservation (although not taking into account of the phylogenetic bias). The evolutionary conservation suggests that the domain architecture plays a role in XIST function. The elucidation of the Xist RNA architecture sets a foundation for understanding the assembly of the multi-functional Xist RNP.
(f). Mapping Xist RBPs to the Xist architecture
A few recent studies began to map the binding sites of RBPs across the transcriptome. Van Nostrand et al. [48] generated enhanced (e) CLIP data for over 100 RBPs in male and female human cell lines, and in particular, four of the RBPs were shown to bind the XIST RNA: HNRNPM, HNRNPK, PTBP1 and SRSF1. Interestingly, these proteins bind XIST in a clustered manner, rather than scattered throughout the entire transcript (figure 1a,f). For example, HNRNPK binds the BCD-repeat domain exclusively, HNRNPM binds the F-repeat domain, PTBP1 binds the E-repeat domain and SRSF1 primarily binds the A-repeat domain. The locally enriched RBP binding correlates with the domain organization of Xist RNA, although it is unclear whether the Xist modular domains contribute to the apparent RBP specificity, or whether RBP binding promotes the RNA folding.
Hendrickson et al. [19] recently used formaldehyde cross-linking to improve RNA-immunoprecipitation sequencing (fRIP-seq) and identified a number of chromatin-associated proteins as RNA-associated proteins, either directly or indirectly. This study also identified the interaction profile for a number of known human XIST-interacting proteins. For example, SUZ12 and EZH2, two core components of the PRC2 complex, bind the E-repeat domain (E-repeat and surrounding sequences) and the 3′-end of the XIST RNA, and these two clusters are physically close based on the RNA architecture. HNRNPU, the bridge between XIST RNA and the Xi chromosome, binds the BCD and exon6 domains.
Smola et al. [57] analysed a number of published CLIP datasets and observed clustered binding for several RBPs, such as CELF, PTBP1, HuR, TARDBP and RBFOX2. In particular, CELF1, PTBP1 and HuR all showed highly clustered binding to the E-repeat domain defined by PARIS. Consistent with these published studies, Chen et al. [38] recently showed similar clustered binding for SPEN (A-repeat), LBR (A-repeat + E-repeat periphery) and PTBP1 (E-repeat) on the PARIS-determined domains. Patil et al. [45] showed that RBM15 and RBM15B, two proteins in the SPEN family, also bind the A-repeat region and the E-repeat domain to a lesser extent. Taken together, these studies support a modular domain architecture that may also serve as functional units of XIST RNP.
(g). Ribonucleoprotein modularity
Modular domain organization provides a simple framework for understanding RNA functions. The physical separation of the different RNA domains and the partition of RBPs to distinct RNA domains could conceivably facilitate the regulation of the various functions coordinated by the Xist RNA. Extensive studies of the ribosomal RNAs revealed discrete domains despite the overall compact organization [62]. Cech and co-workers [63] used structure modelling and genetic manipulations to demonstrate that the large yeast telomerase RNAs form distinct functional domains each with its own conserved sequences and associated protein effectors. Importantly, the assembly of the conserved RNP in each domain is essential for function and can be relocated to different positions without affecting function. Biochemical analysis of the HOTAIR lncRNA, which is involved in the regulation of HOX gene expression, reveals two distinct regions that bind PRC2 and LSD1, two histone modification complexes, although the structural basis remains unclear [64]. Several other structural analyses of lncRNAs have also claimed the identification of domain structures; however, it is unclear whether these domains are real or artefacts of the computational modelling, given the lack of evidence for direct base pairing in the identified duplexes [65,66]. Johnson & Guigo [67] recently proposed the RIDL (Repeat Insertion Domains of LncRNAs) hypothesis to explain the potential domain organization of lncRNAs. The rapidly evolving lncRNAs could be functional by adopting modular repeat insertions that either work on the sequence level or on the structure level. Modularity is most likely more than the insertions as some of the determined domains in the XIST RNA encompass both repeats and non-repetitive sequences (figure 1a) [10]. Nevertheless, modularity is emerging as an important feature for long RNA molecules.
4. The X-chromosome architecture and insights into X-chromosome inactivation
(a). Zooming into the Barr body
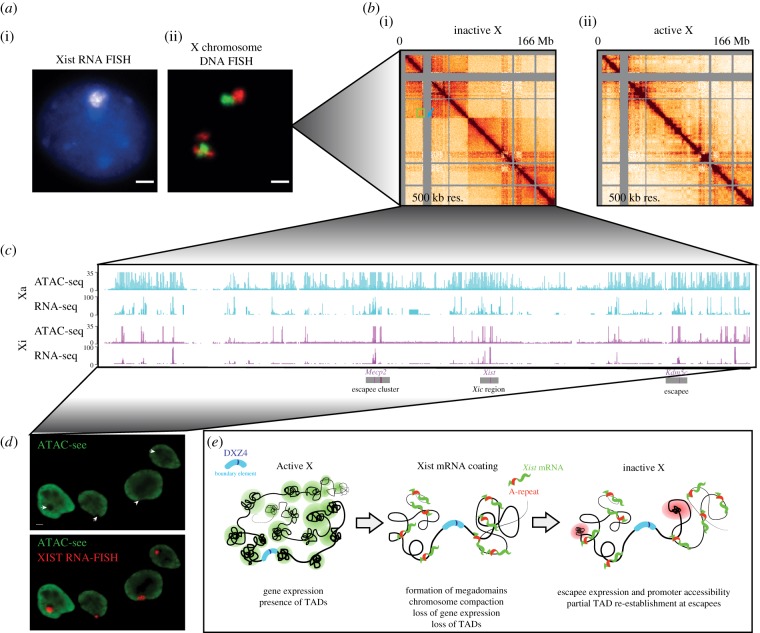
The Barr body, first observed under the microscope in 1948 [37], has been thought to be just a densely compacted, inert heterochromatic chromosome, relegated to the side of the nucleus unused (figure 2a). It has been appreciated in recent decades, however, that there is fine gene control on the inactive X-chromosome, both to maintain silencing and to promote the expression of a handful of escape genes. This gene control is mediated by long non-coding RNAs, histone modifications and DNA methylation, as well as three-dimensional chromatin structure and accessibility. Here, we review how chromatin structure changes following XCI by zooming in on the Xi at different levels of compaction.
Figure 2.
Zooming in on chromosome structure on the Xi. (a) (i) Xist RNA FISH in female mouse NPCs showing a cloud of Xist RNA coating the X-chromosome. (ii) DNA FISH on the X-chromosome showing the separation of the Xi into two domains or lobes. The Xa, shown for comparison, shows mixing of the two domains. (b) Allele-specific HiC at 500 kb resolution for the Xi (i) and Xa (ii) in NPCs. The Xi is configured in two megadomains within which TAD structure is lost. The megadomains are separated by the DXZ4 satellite element (blue arrow). A few mini-TADs are re-formed on the Xi (green box). For comparison, the Xa does not have this megadomain structure, but maintains autosome-like TAD structure. (c) Allele-specific ATAC-seq and RNA-seq in NPCs on the Xa (top) and Xi (bottom). There is a global reduction in accessibility and gene expression on the Xi compared to the Xa. Regions that retain acccessibility are located at the promoters and CTCF sites close to escape genes. A cluster of escapees near Mecp2, the Xic and the Kdm5c escape locus, are indicated below. (d) ATAC-see reveals the spatial organization of accessible chromatin in NPCs. Xist RNA FISH cloud falls in a region of depletion of accessible chromatin. (e) Model for how the megadomain structure of the Xi is formed during X inactivation. TADs on the Xa are lost and replaced by two large domains separated by the DXZ4 element. Full-length Xist containing the A-repeat region is required for this restructuring. After X inactivation, small escape TADs form. Within these TADs, escape genes make contact with one another and are regulated at highly proximal CTCF elements. Figures adapted from Giorgetti et al. [68] and Chen et al. [69].
(b). Chromatin conformation
The genome has traditionally been read and visualized in a linear fashion, but it is becoming clear that the organization of the chromatin in the nucleus and the protein-mediated contacts between distant genomic loci are necessary for gene regulation. The recent development of chromosome conformation capture (3C) technologies has led to the discovery of hierarchical chromatin looping that is essential for stable gene expression patterns. Chromosomes are organized first into active (A) and inactive (B) compartments and then into topologically associating domains (TADs) at the megabase scale [70]. Within these TADs, loops between enhancers and promoters fine-tune gene expression levels. It is known that the majority of TADs are cell-type independent but that TADs are erased and re-established during every cell cycle [71]. The direction of causality between loop or TAD formation and gene expression, however, is still unclear.
(c). Allele-specific technologies
One of the greatest challenges in studying the structure and activity of the inactive X-chromosome is distinguishing it from the active X. Sequencing analysis techniques have traditionally merged the data from the two alleles of every chromosome, assuming that the two behave identically. In the case of the X-chromosome in female cells, it has been difficult to separate sequencing reads from the inactive and active X chromsomes. A number of strategies have traditionally been used to deal with this. Male cell lines have been engineered to contain an inducible Xist transgene on the X-chromosome, which upon induction will turn on the Xist transcript and lead to chromosome silencing [46]. This approach is limited by the fact that after silencing of the X-chromosome, male cells begin to die, making it impossible to track the later stages of XCI. Female cell lines with dox-inducible Xist transgenes have also been engineered but suffer the same limitation [72]. Furthermore, in these systems, XCI proceeds much more quickly than it does in the endogenous context and thus may not follow an identical process. To study the stable Xi, it is necessary to distinguish the two X-chromosomes under physiological conditions in female differentiated cells.
To tackle this problem, we and others have developed allele-specific genomic analysis methods and applied them to highly polymorphic hybrid cell lines to get a high resolution picture of the Xi. We used allele-specific Assay for Transposase Accessible Chromatin (ATAC)-seq and HiC in hybrid undifferentiated mouse embryonic stem cells and neural progenitor cells (NPCs) to study the relationship between 3D chromatin structure, local chromatin structure and gene expression on the Xi [68].
(d). Higher-order structure of the Xi
Upon X inactivation, we and others showed that the inactive X-chromosome in the mouse undergoes massive structural rearrangement, yielding two chromosomal lobes or megadomains [13,68,73]. Within a megadomain loci interact with frequency, while there is very little interaction between chromosomal loci in opposite megadomains (figure 2b). Using DNA FISH probes spanning loci in either domain, we showed that the two megadomains are spatially separated in the nucleus forming a dumbbell-like structure (figure 2a).
Strikingly, the Xi is devoid of TADs at the megabase scale. Within each megadomain, the interactions between loci are not organized into small domains, but rather are equal across the domain. This suggests that chromatin organization is dependent on gene expression in this context and is lost upon global silencing. Furthermore, small TADs re-formed on the Xi around clusters of genes that escape XCI. These TADs do not have the same boundaries as their Xa counterparts, suggesting that their formation is driven by the Xi-specific escape gene expression pattern.
(e). Chromatin accessibility on the Xi
To understand chromatin structure at the individual locus level on the Xi, we used allele-specific ATAC-seq. ATAC reveals all active elements in the genome, including promoters, enhancers and insulators. It also provides information about transcription factor binding and nucleosome positioning [74]. On the Xi, chromatin accessibility is globally lost except at regions of escape gene expression (figure 2c). Specifically, CTCF-binding regulatory elements highly proximal to escape gene promoters escape this loss of accessibility, suggesting a local regulatory mechanism for maintaining escape gene expression within a sea of heterochromatin. This is consistent with previous findings that Xist spreads up to but not into escape gene loci and that escape genes have putative CTCF insulator elements on their flanks [75,76]. These escape elements make contact with one another on the Xi but not on the Xa, suggesting that they cooperate in this restrictive chromatin landscape to enhance expression of one another.
Chromatin accessibility can be globally visualized using a novel technique, ATAC-see, in which fluorescently tagged adapters are inserted into open chromatin regions [77]. By performing ATAC-see in conjunction with Xist RNA FISH, we can see a region depleted for open chromatin or a ‘hole’ in the ATAC-see signal where the Xi lies (figure 2d). This suggests that escaping loci may be found outside of this space, consistent with previous findings using PolII staining.
(f). The DXZ4 boundary element
The two megadomains in the mouse are separated by a hinge-like boundary which is located at the DXZ4 macrosatellite element. The human Xi is also separated at the DXZ4 homologue, despite it being in a different location on the human X [78]. This suggested that the DXZ4 element plays a conserved role in chromosomal organization. Deng and co-workers showed that this DXZ4 hinge element associated with the nucleolus, which is a common location for the Xi. This interaction was dependent on both the DXZ4 element, CTCF and the Firre RNA [22,73,79].
To interrogate the role of the DXZ4 boundary element, we deleted the boundary using allele-specific CRISPr. After boundary deletion, the two megadomains fused together, yielding a chromosome devoid of domains. Megadomain structure was also lost on the human Xi when DXZ4 was deleted [68,80]. In the mouse, the small TADs that re-formed around escape genes in the WT NPCs were lost along with escape gene expression and accessibility of escape gene promoter elements. These findings point towards a critical role for the DXZ4 element in creating the unique Xi structure in both mouse and human. Furthermore, in the mouse, the unique Xi structure plays a role in regulating escape gene expression and the contacts between escape loci.
(g). The A-repeat region modulates Xi structure
The A-repeat region of the Xist RNA is required for silencing of the X-chromosome and does this at least partially via protein binding to intricately folded RNA domains (figure 1e) [10,11,46]. In a study where the entire Xist RNA was ablated, the Xi failed to reorganize at the level of three-dimensional chromatin structure [13]. Thus, we asked whether the A-repeat region of the Xist RNA and its protein-binding factors affect both higher-order and local chromatin structure. Using a cell line in which Xist or its A-repeat deletion version is inducibly expressed, we measured three-dimensional and local chromatin structure in the early time points of X inactivation. Strikingly, in the absence of the A-repeat region, the chromatin does not begin folding into the bipartite structure characteristic of the WT Xi. Furthermore, when A-repeat is deleted, accessibility at the local level is no longer reduced. Thus, a silencing-competent, full-length Xist is required for chromatin compaction and restructuring, likely via recruitment of proteins such as SPEN that bind to the A-repeat region of the RNA [68].
The development of allele-specific HiC and ATAC-seq technologies has allowed us to zoom into a detailed picture of the chromatin of the Xi at different levels. Chromatin on the Xi is not simply compacted as was previously believed, but rather is divided into two megadomains within which certain genes are able to escape. Escape genes form a regulatory territory within which their promoter–proximal regulatory elements contact one another, co-opting these regions as Xi-specific enhancer elements. The development of these technologies has further set the stage for studying allele-specific differences in chromatin structure and activity at autosomal loci.
5. Towards a functional map for X-chromosome inactivation and beyond
The past few years have seen major progress in defining the molecular mechanisms of XCI, both on the Xist RNP side and the X-chromosome side. Further functional analysis of the proteins identified in the Xist RNP can help determine other factors that are involved in the establishment of XCI. A number of these proteins were identified in multiple proteomic and genetic screens and thus are probably bona fide Xist interactors. Some of these newly identified factors may contribute to the folding of the XIST RNA or to recruitment of the histone and DNA modification enzymes. For example, a significant number of these factors seem to be general RNA processing factors that are involved in RNA splicing and stability and have mostly been ignored [11–15]. However, it is very likely that these factors still play a specific role in XCI. For example, a recent study shows that SF2 (also known as SRSF1), a general splicing factor, couples allelic differences in Xist to splicing and thus plays a role in the random choice of which X to silence [47]. Further studies are also required to determine, on the chromatin side, how specific loci are selected for escape in the overall repressive Xi compartment. Some of the Xist interactors identified recently could play a role in this important decision.
Biological macromolecules such as DNA, RNA and proteins all form three-dimensional structures that present a new level of organization distinct from their primary sequences. Principles of folding have been studied in great details for proteins. Larger proteins are often folded into multiple domains that evolve and operate independently from one another and at the same time work together to efficiently execute their functions. Different proteins can form similar structural domains that share little sequence similarity, and this feature has been extensively used to guide structural and functional studies [81–83]. DNA molecules, although rigid at the local level, fold into robust domains in the long range in the context of chromatin [84]. The higher-order chromatin domains coordinate gene expression programmes by clustering related genes and regulatory elements. RNA molecules are more flexible than proteins, and structural analyses of RNA have mostly focused on local structures, such as simple stem loops. Segregation of domains has been suggested to underlie the multiple functions that are coordinated by lncRNAs in a number of cases. Recent studies on XIST provide the most compelling example of the principle of structural and functional modularity, which parallels that of the X-chromosome being silenced.
In this emerging picture of XCI, many questions remain unanswered. The formation of long-range structures in the Xist RNA is remarkable. These data also raise the question of how long-range structures form during transcription. Chaperone mechanisms may need to be invoked to counteract co-transcriptional folding and facilitate long-range domain formation. How does the multi-functional XIST RNP coordinate the multiple steps of XCI? The retention of multiple activities on one RNP particle indicates an active role of the Xist RNA in the process. It is likely that the different activities, such as deposition of histone and DNA modifications, occur more efficiently when coupled by one scaffold RNA. The presence of alternative structures, not only in the A-repeat region but also throughout the transcript, could result from the repetitive nature of the RNA, and/or may play a role in an allosteric regulation that couples different reactions catalysed by the XIST RNP. The technologies developed for the analysis of XCI and the principles governing this process will also be widely applicable to a wide variety of problems in gene regulation and facilitate a deeper understanding into the dynamics and functions of lncRNAs and chromatin.
Data accessibility
This article has no additional data.
Authors' contributions
All authors contributed to the drafting of the manuscript. That magnitude of the contributions is reflected in the list order of the author names.
Competing interests
We have no competing interests.
Funding
Z.L. is a Layton Family Fellow of the Damon Runyon-Sohn Foundation Pediatric Cancer Fellowship Award (DRSG-14-15) and supported by the Stanford Jump Start Award of Excellence in Postdoctoral Research. A.C.C. is supported by a T32 Aging Training Grant AG0047126. H.Y.C. is supported by NIH R01-HG004361 and P50-HG007735.
References
- 1.Avner P, Heard E. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2, 59–67. ( 10.1038/35047580) [DOI] [PubMed] [Google Scholar]
- 2.Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. ( 10.1038/190372a0) [DOI] [PubMed] [Google Scholar]
- 3.Wutz A. 2011. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12, 542–553. ( 10.1038/nrg3035) [DOI] [PubMed] [Google Scholar]
- 4.Brockdorff N, et al. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329–331. ( 10.1038/351329a0) [DOI] [PubMed] [Google Scholar]
- 5.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. ( 10.1038/349038a0) [DOI] [PubMed] [Google Scholar]
- 6.Cerase A, Pintacuda G, Tattermusch A, Avner P. 2015. Xist localization and function: new insights from multiple levels. Genome Biol. 16, 166 ( 10.1186/s13059-015-0733-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mira-Bontenbal H, Gribnau J. 2016. New Xist-interacting proteins in X-chromosome inactivation. Curr. Biol. 26, 1383 ( 10.1016/j.cub.2016.05.009) [DOI] [PubMed] [Google Scholar]
- 8.Moindrot B, Brockdorff N. 2016. RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin. Cell. Dev. Biol. 56, 58–70. ( 10.1016/j.semcdb.2016.01.029) [DOI] [PubMed] [Google Scholar]
- 9.Elisaphenko EA, et al. 2008. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS ONE 3, e2521 ( 10.1371/journal.pone.0002521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Z, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279. ( 10.1016/j.cell.2016.04.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu C, et al. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. ( 10.1016/j.cell.2015.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McHugh CA, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. ( 10.1038/nature14443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minajigi A, et al. 2015. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, pii:aab2276 ( 10.1126/science.aab2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moindrot B, et al. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572. ( 10.1016/j.celrep.2015.06.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561. ( 10.1016/j.celrep.2015.06.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon Y, Lee JT. 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133. ( 10.1016/j.cell.2011.06.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makhlouf M, Ouimette JF, Oldfield A, Navarro P, Neuillet D, Rougeulle C. 2014. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 5, 4878 ( 10.1038/ncomms5878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19, 469–476. ( 10.1016/j.devcel.2010.08.006) [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson DG, Kelley DR, Tenen D, Bernstein B, Rinn JL. 2016. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 17, 28 ( 10.1186/s13059-016-0878-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolpa HJ, Fackelmayer FO, Lawrence JB. 2016. SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell 39, 9–10. ( 10.1016/j.devcel.2016.09.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakaguchi T, Hasegawa Y, Brockdorff N, Tsutsui K, Tsutsui KM, Sado T, Nakagawa S. 2016. Control of chromosomal localization of Xist by hnRNP U family molecules. Dev. Cell 39, 11–12. ( 10.1016/j.devcel.2016.09.022) [DOI] [PubMed] [Google Scholar]
- 22.Hacisuleyman E, et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. ( 10.1038/nsmb.2764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz YB, Pirrotta V. 2013. A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 14, 853–864. ( 10.1038/nrg3603) [DOI] [PubMed] [Google Scholar]
- 24.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122. ( 10.1038/sj.emboj.7601187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva J, et al. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4, 481–495. ( 10.1016/S1534-5807(03)00068-6) [DOI] [PubMed] [Google Scholar]
- 26.de Napoles M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676. ( 10.1016/j.devcel.2004.10.005) [DOI] [PubMed] [Google Scholar]
- 27.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2, E171 ( 10.1371/journal.pbio.0020171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brockdorff N. 2013. Noncoding RNA and Polycomb recruitment. RNA 19, 429–442. ( 10.1261/rna.037598.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745. ( 10.1016/j.cell.2007.02.009) [DOI] [PubMed] [Google Scholar]
- 30.Tavares L, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678. ( 10.1016/j.cell.2011.12.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, et al. 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953. ( 10.1016/j.molcel.2010.12.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756. ( 10.1126/science.1163045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maenner S, et al. 2010. 2-D structure of the a region of Xist RNA and its implication for PRC2 association. PLoS Biol. 8, e1000276 ( 10.1371/journal.pbio.1000276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Rocha ST, et al. 2014. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316. ( 10.1016/j.molcel.2014.01.002) [DOI] [PubMed] [Google Scholar]
- 35.Cerase A, et al. 2014. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc. Natl Acad. Sci. USA 111, 2235–2240. ( 10.1073/pnas.1312951111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smeets D, et al. 2014. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenet. Chromatin 7, 8 ( 10.1186/1756-8935-7-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr ML, Bertram EG. 1949. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163, 676 ( 10.1038/163676a0) [DOI] [PubMed] [Google Scholar]
- 38.Chen CK, et al. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. ( 10.1126/science.aae0047) [DOI] [PubMed] [Google Scholar]
- 39.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR. 2015. 5' UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010. ( 10.1016/j.cell.2015.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao W, et al. 2016. Nuclear m6 a reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519. ( 10.1016/j.molcel.2016.01.012) [DOI] [PubMed] [Google Scholar]
- 41.Batista PJ, et al. 2014. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. ( 10.1016/j.stem.2014.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominissini D, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. ( 10.1038/nature11112) [DOI] [PubMed] [Google Scholar]
- 43.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. ( 10.1016/j.cell.2012.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Met. 12, 767–772. ( 10.1038/nmeth.3453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patil DP, et al. 2016. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369 ( 10.1038/nature19342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. ( 10.1038/ng820) [DOI] [PubMed] [Google Scholar]
- 47.Royce-Tolland ME, Andersen AA, Koyfman HR, Talbot DJ, Wutz A, Tonks ID, Kay GF, Panning B. 2010. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nat. Struct. Mol. Biol. 17, 948–954. ( 10.1038/nsmb.1877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Nostrand EL, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Met. 13, 508–514. ( 10.1038/nmeth.3810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dickson BJ, van der Straten A, Dominguez M, Hafen E. 1996. Mutations modulating Raf signaling in Drosophila eye development. Genetics 142, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolodziej PA, Jan LY, Jan YN. 1995. Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron 15, 273–286. ( 10.1016/0896-6273(95)90033-0) [DOI] [PubMed] [Google Scholar]
- 51.Wiellette EL, Harding KW, Mace KA, Ronshaugen MR, Wang FY, McGinnis W. 1999. Spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 126, 5373–5385. [DOI] [PubMed] [Google Scholar]
- 52.Shi Y, et al. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15, 1140–1151. ( 10.1101/gad.871201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mortimer SA, Kidwell MA, Doudna JA. 2014. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 15, 469–479. ( 10.1038/nrg3681) [DOI] [PubMed] [Google Scholar]
- 54.Lu Z, Chang HY. 2016. Decoding the RNA structurome. Curr. Opin. Struct. Biol. 36, 142–148. ( 10.1016/j.sbi.2016.01.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duszczyk MM, Wutz A, Rybin V, Sattler M. 2011. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17, 1973–1982. ( 10.1261/rna.2747411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. 2015. Probing Xist RNA structure in cells using targeted structure-seq. PLoS Genet. 11, e1005668 ( 10.1371/journal.pgen.1005668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl Acad. Sci USA 113, 10 322–10 327. ( 10.1073/pnas.1600008113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spitale RC, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. ( 10.1038/nature14263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rivas E, Clements J, Eddy SR. 2016. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Met. 14, 45–48. ( 10.1038/nmeth.4066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arieti F, Gabus C, Tambalo M, Huet T, Round A, Thore S. 2014. The crystal structure of the split end protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs. Nucleic Acids Res. 42, 6742–6752. ( 10.1093/nar/gku277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gesell T, von Haeseler A. 2006. In silico sequence evolution with site-specific interactions along phylogenetic trees. Bioinformatics 22, 716–722. ( 10.1093/bioinformatics/bti812) [DOI] [PubMed] [Google Scholar]
- 62.Wilson DN, Doudna Cate JH. 2012. The structure and function of the eukaryotic ribosome. Cold Spring Harbor Perspect. Biol. 4, a011536 ( 10.1101/cshperspect.a011536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zappulla DC, Cech TR. 2004. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA 101, 10 024–10 029. ( 10.1073/pnas.0403641101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai MC, et al. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. ( 10.1126/science.1192002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY. 2016. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 16, 3087–3096. ( 10.1016/j.celrep.2016.08.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. 2015. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361. ( 10.1016/j.molcel.2015.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson R, Guigo R. 2014. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA 20, 959–976. ( 10.1261/rna.044560.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giorgetti L, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. ( 10.1038/nature18589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen X, et al. 2016. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Met. 13, 1013–1020. ( 10.1038/nmeth.4031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lieberman-Aiden E, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. ( 10.1126/science.1181369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the mitotic chromosome. Science 342, 948–953. ( 10.1126/science.1236083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulz EG, et al. 2014. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14, 203–216. ( 10.1016/j.stem.2013.11.022) [DOI] [PubMed] [Google Scholar]
- 73.Deng X, et al. 2015. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16, 152 ( 10.1186/s13059-015-0728-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Met. 10, 1213–1218. ( 10.1038/nmeth.2688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Engreitz JM, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 ( 10.1126/science.1237973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. 2005. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell 8, 31–42. ( 10.1016/j.devcel.2004.10.018) [DOI] [PubMed] [Google Scholar]
- 77.Chen EL, et al. 2016. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 3, 1013–1020. ( 10.1038/nmeth.4031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rao SS, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. ( 10.1016/j.cell.2014.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang F, et al. 2015. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16, 52 ( 10.1186/s13059-015-0618-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darrow EM, et al. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl Acad. Sci. USA 113, E4504–E4512. ( 10.1073/pnas.1609643113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchler-Bauer A, et al. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41(Database issue), D348–D352. ( 10.1093/nar/gks1243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33(Web Server issue), W116–W120. ( 10.1093/nar/gki442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. 1998. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26, 320–322. ( 10.1093/nar/26.1.320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dixon JR, Gorkin DU, Ren B. 2016. Chromatin domains: the unit of chromosome organization. Mol. Cell 62, 668–680. ( 10.1016/j.molcel.2016.05.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.