Abstract
The human blastocyst forms 5 days after one of the smallest human cells (the sperm) fertilizes one of the largest human cells (the egg). Depending on the sex-chromosome contribution from the sperm, the resulting embryo will either be female, with two X chromosomes (XX), or male, with an X and a Y chromosome (XY). In early development, one of the major differences between XX female and XY male embryos is the conserved process of X-chromosome inactivation (XCI), which compensates gene expression of the two female X chromosomes to match the dosage of the single X chromosome of males. Most of our understanding of the pre-XCI state and XCI establishment is based on mouse studies, but recent evidence from human pre-implantation embryo research suggests that many of the molecular steps defined in the mouse are not conserved in human. Here, we will discuss recent advances in understanding the control of X-chromosome dosage compensation in early human embryonic development and compare it to that of the mouse.
This article is part of the themed issue ‘X-chromosome inactivation: a tribute to Mary Lyon’.
Keywords: X-chromosome inactivation, pluripotent stem cells, X-chromosome dampening, Xist, Xact
1. The X-chromosome state of the human pre-implantation embryo
Somatic cells of an adult female human have two X chromosomes, but most genes on one of them are silenced at the level of transcription, so that the X-chromosome gene dosage in female XX cells is equal to that of male XY cells. The silenced X chromosome can be either the paternally or the maternally inherited one, making the adult female a natural mosaic. This random pattern of X-chromosome inactivation (XCI) is established in early embryogenesis. The X chromosomes inherited from the egg (maternal) and the sperm (paternal) are both active in very early female development [1,2] before each cell commits to transcriptionally silencing one X chromosome for the rest of the cell's and its progeny's life. It is not known exactly when this choice is made in human development, but based on mouse studies it is hypothesized to happen shortly after the embryo implants [3]. Surplus pre-implantation embryos from in vitro fertilization clinics donated to research have made ex vivo studies of human pre-implantation development possible. Combined with advances in single-cell transcriptome profiling, these have recently enabled a closer look at the X-chromosome biology in early human development [1,2,4–6].
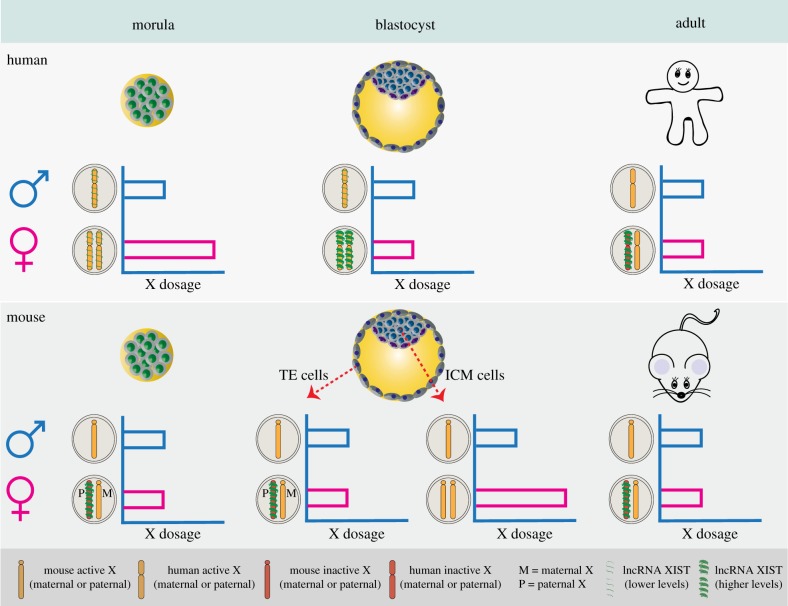
Petropoulos and colleagues studied the transcriptome of the largest number of human pre-implantation embryos reported to date, and performed sex-specific analysis of human development at days 3–7 post fertilization (E3–E7) at the single-cell level [2]. Their analysis revealed that immediately after zygotic gene activation (ZGA) at E4, female embryos had almost double expression of X-linked genes compared with males, consistent with females having two active X chromosomes (figure 1). However, with increasing developmental time from E4 to E7, this roughly 2 : 1 female : male ratio decreased, reaching nearly 1 : 1 in all cells of the embryo at E7 (figure 1), just in time for the commencement of implantation. Surprisingly, this drop in X-linked gene expression level was not due to the onset of X-chromosome-inactivation, because allelic expression analysis by single-cell RNA-sequencing revealed that both X chromosomes were active at all times [2]. Evidence for the presence of two active X chromosomes in female human pre-implantation embryos was extended further by RNA fluorescent in situ hybridization (RNA-FISH) [1,2,5,6]. Thus, Petropoulos et al. uncovered a novel mechanism of X-chromosome dosage compensation, at the mRNA level, in human pre-implantation development where female to male expression is equalized not by inactivating one of the two X chromosomes in the female, but rather by dampening the expression of both female X chromosomes (figure 1). This X-chromosome dampening (XCD), which has not been observed in mice, is reminiscent of the dosage compensation system occurring in a model organism further removed from the human on the evolutionary scale—the roundworm Caenorhabditis elegans. Both X chromosomes of XX hermaphrodite C. elegans undergo condensin-mediated three-dimensional structural remodelling, resulting in reduced transcriptional output to match X-linked gene dosage to that of the single X in XO males [7,8]. However, whether XCD in human and C. elegans are mechanistically similar remains an open question (see §2). In any case, together these findings indicate that X-chromosome dosage compensation in human is regulated by two different and sequential processes: first XCD and later XCI. Interestingly, moderate but significant expression asymmetry between the two X chromosomes was detected from E5, suggesting that X-linked gene silencing may initiate in a progressive manner at this developmental stage [5].
Figure 1.
X-chromosome dosage compensation in mouse and human. In human pre-implantation development, XIST becomes expressed from all X chromosomes upon zygotic gene activation. As pre-implantation development progresses, XIST expression from both female X chromosomes increases, but remains low in males. The former correlates with dampened gene dosage from both X chromosomes of the blastocyst, equalizing X-linked gene dosage of females to that of males. Upon implantation, all cells undergo random XCI, again resulting in dosage-compensation. In mice, XCI happens in two waves. First, Xist is induced only on the paternally inherited X chromosome (P), causing imprinted XCI in all cells of early pre-implantation embryos (morula). As the blastocyst forms, Xist expression becomes suppressed in the ICM cells (but not in the TE), and the Xi reactivates, leading to increased X-linked gene dosage in females compared to males. As the embryo implants, the maternal or the paternal X chromosome becomes randomly chosen to undergo XCI, similar to humans.
2. XIST expression correlates with X-chromosome dampening
A hallmark of the inactive X chromosome (Xi) is expression and accumulation of the cis-acting long non-coding RNA (lncRNA) XIST (X inactive specific transcript) [9–11], which, as its name suggests, was thought until recently to always correlate with the inactive status of the X chromosome. However, an unexpected finding was made in 2011, when Edith Heard's group used RNA-FISH to demonstrate that both male and female human pre-implantation embryos express the lncRNA XIST without any evidence of X-inactivation (figure 1) [1]. This was the first report of long-term expression (over several days) and accumulation of XIST RNA that does not lead to chromosome-wide silencing, and was indeed very intriguing. This finding inspired further studies of the X-chromosome state in the human pre-implantation embryo, which validated the presence of XIST-expressing active X chromosomes [2,4–6]. While XIST was expressed from both X chromosomes in the majority of cells in female blastocysts, a proportion of the cells, however, displayed mono-allelic XIST expression pattern [1,2,5]. In RNA-FISH studies, XIST was also found accumulating on the single X in male embryos, although contrasting results were obtained between studies in the proportion of XIST-expressing cells—from a majority of male cells in the blastocyst expressing XIST [1,5] to most cells being devoid of XIST expression [2]. This discrepancy is perhaps due to differences in the sensitivity of the RNA-FISH assays employed, and might be related to the fact that XIST was found at much lower amounts in male cells compared with female cells in RNA-sequencing experiments [2].
Human XIST expression initiates as early as at the 4–8-cell stage of the embryo and coincides with the onset of ZGA [2,4,5]. XIST levels increase over time up to E7, in a manner that correlates with X-linked dampening (figure 1). This correlation is also observed in naive human embryonic stem cells (hESCs), where cells with two active chromosomes and no XIST expression have overall higher X-linked gene expression compared with cells with two active Xs and XIST expression (see §6) [12]. Whether XCD in human is mediated by XIST remains an open question, but in the worm other mechanisms are involved as XIST is not conserved beyond placental mammals [13]. Should XIST mediate XCD in the female human pre-implantation embryo, one would have to assume that the lower level of XIST in male embryos is not sufficient for the induction of XCD on the male single X chromosome.
3. Differences between mouse and human XCI
In contrast to the human, mouse embryos are more easily attainable in larger numbers; hence our understanding of mouse pre- and post-implantation development, including the regulation of X-chromosome dosage, is more advanced. It is well established that female mice undergo X-chromosome dosage compensation via XCI in two waves. At the 4-cell stage mouse embryos initiate paternally imprinted XCI, which is completed by the morula stage; hence only the maternally inherited X chromosome is active in all cells (figure 1; reviewed by Takagi [14]). Imprinted XCI is maintained in the cells of the trophectoderm, which will eventually give rise to extra-embryonic tissues such as the placenta [15]. By contrast, as the embryo develops into the mid-stage blastocyst, the inactive X chromosome is reactivated in cells of the inner cell mass (ICM) that give rise to the epiblast [16–18], resulting in cells with two active X chromosomes (figure 1). These cells then undergo a second wave of XCI, which is not imprinted, but rather the maternally or the paternally inherited X chromosome is chosen at random. Both imprinted and random XCI depend on Xist, which acts in cis in both cases to silence the X chromosome from which it is expressed [19–21], and the reactivation of the imprinted Xi is accompanied by Xist silencing (figure 1) [16–18].
Early reports addressing the question of whether human early development follows what is observed in the mouse with respect to imprinted XCI have been mixed, but recent studies using more advanced techniques and larger sample sizes agree that human pre-implantation embryos lack imprinted XCI [1,2,22], and that, instead, human pre-implantation embryos reduce X-linked gene dosage by XCD on both X chromosomes [2]. Thus, in addition to XCD and XIST expression from an active X chromosome, the lack of imprinted XCI in human pre-implantation embryos is a key difference between mouse and human embryonic development. Interestingly, XIST expression and lack of imprinted XCI are also observed in rabbit pre-implantation development, despite the closer evolutionary distance between mouse and rabbit compared with rabbit and human [1].
Another distinguishing feature between mouse and human in the epigenetic regulation of the X chromosome is the presence of the long non-coding RNA Tsix in mice but not in humans. Tsix is transcribed antisense to Xist and, in imprinted XCI, is expressed from the active, maternal X chromosome in mouse pre-implantation embryos and extra-embryonic annexes, where it is required to maintain Xist repressed on this chromosome [23,24]. Similar to imprinted XCI, Tsix represses Xist expression from the active X chromosome during random XCI [25,26]. Despite the role of Tsix in both imprinted and random XCI, there is a Tsix-independent repression of Xist at play during embryo cleavage stages of mouse development, because the maternal Xist is repressed in the absence of Tsix expression [24]. Although a TSIX gene has been annotated in the human genome, a recent study shows that it is not transcribed in human pre-implantation embryos [2]. The lack of TSIX expression and function may be related to the expression of XIST from the active X chromosomes in the human pre-implantation embryo. Thus, human cells seem to have evolved a different mechanism to control the function of XIST during the initiation of random XCI and to cope with XIST expression in the pre-implantation embryo: it is the silencing ability of XIST rather than XIST expression that is prevented in these cells. This contrasts to the mouse, where Xist expression systematically leads to silencing, unless certain regions of the Xist gene are deleted [27]. A strong candidate for repressing XIST's ability to silence the X chromosomes in the pre-implantation embryo is the recently identified human- and pluripotency-specific lncRNA XACT (X active coating transcript; see §7) [28].
4. Mouse ESCs perfectly recapitulate the X-chromosome state of the mouse blastocyst
Much of our understanding of XCI comes from mouse studies mainly because mouse embryonic stem cells (mESCs), derived from the pre-implantation blastocyst, perfectly capture the X-chromosome state of in vivo development [29]. Cells of the ICM and mESCs have two active X chromosomes and, upon implantation in vivo or differentiation in vitro, Xist expression is induced from one of the two X chromosomes, chosen at random, which leads to chromosome-wide inactivation in cis (figure 1). The in vitro model system has been ideal for unravelling the molecular mechanism behind the initiation of random XCI and the transition from the XaXa (Xa for active X chromosome) to the XaXiXist+ state (Xi for inactive X chromosome). For instance, mESCs were used to perform extensive Xist RNA domain deletion studies that suggested a modular structure of Xist RNA, with different RNA domains mediating different functions [27]. More recently, mESCs were used to reveal that, at the onset of XCI, Xist spreads to regions on the X chromosome spatially closest to the Xist transcription locus, highlighting the importance of three-dimensional modelling of the X chromosome [30]. Moreover, two groups independently identified protein partners of Xist at the onset of XCI [31,32], beginning to provide a detailed mechanistic understanding of how Xist function is mediated [31–34].
The mouse model has also contributed immensely to our understanding of pluripotency—the ability to differentiate into all three germ layers. Pluripotent cell identity is not fixed but rather represents a spectrum of states, perhaps because pluripotency in vivo spans multiple days of development instead of a fixed singular time point [35]. This became obvious when pluripotent stem cells (PSCs) with characteristics rather distinct from those of mESCs were isolated from the mouse post-implantation epiblast (EpiSCs for epiblast stem cells) [36,37]. Although both are pluripotent, mESCs capture the naive pluripotent state of the pre-implantation blastocyst and EpiSCs the developmentally more advanced primed pluripotent state of the post-implantation embryo [35].
5. Limitations of conventional human ESCs in modelling the pre-implantation X-chromosome state and initiation of XCI
Unlike mESCs, conventional hESCs, which are derived in the presence of basic fibroblast growth factor, do not recapitulate the X-chromosome state of the naive pluripotent cells in the human blastocyst. When comparing to what we know from mouse studies, conventional hESCs resemble mouse EpiSCs instead of naive mESCs, although, like mESCs and unlike mouse EpiSCs, they are derived from the pre-implantation and not the post-implantation blastocyst (see [38] for a detailed review). This resemblance extends to cell morphology, signalling pathway dependence with global transcriptional signature, and the post-XCI state [38]. Hence, similar to mouse EpiSCs, conventional hESCs are in primed pluripotency [35].
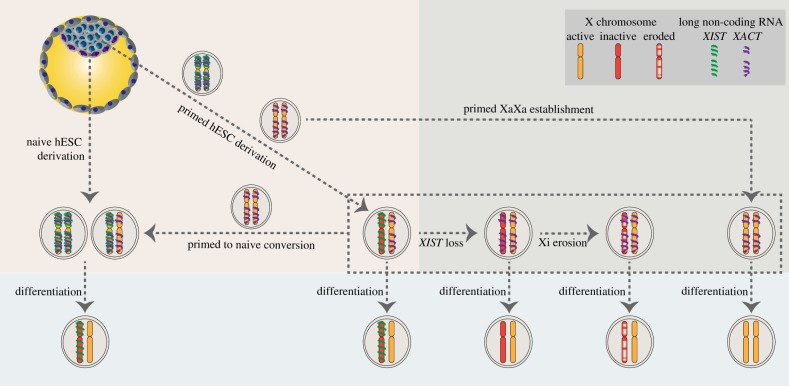
The X-inactivation status of hESCs has been very controversial, likely due to the epigenetic instability of the inactive X chromosome in primed hESCs. Two X-chromosome patterns can be observed at early passage, when hESCs are derived from the pre-implantation embryo in conventional conditions: one with two active X chromosomes (XaXa) and no XIST expression, and one with one active and one inactive X chromosome from which XIST is expressed (XaXiXIST+), the latter being more frequent (figure 2) [6,39–43]. The XIST-negative XaXa state was initially reported to be the pristine state, due to its resemblance to the mouse situation [40,41]. However, in our study, induction of differentiation of XaXa hESCs is accompanied neither by XIST induction nor by XCI [6] (figure 2). Because of this, we classified this XIST-negative XaXa state as an abnormal state, probably due to the permanent silencing of the XIST gene during the derivation of primed hESCs [6]. Previous studies contradicting this conclusion and reporting de novo XCI from such cells [40,41] may be explained by the heterogeneity of most hESC lines, with both XaXa and XaXiXIST+ cells present in the same culture before induction of differentiation. Following cells through the derivation process from human blastocysts by the analysis of a few time points suggested that the transition from the pre-implantation embryo state with two active, XIST-expressing X chromosomes to a post-XCI state involves transient silencing of XIST on both X chromosomes and its subsequent reactivation from one X only, to induce XCI [6] (figure 2). In this model, it may be possible that effective upregulation of XIST is only possible in a brief developmental window and, in cases when this window is missed in vitro, both XIST alleles become permanently silenced, leading to the stabilization of the XaXa state without XIST expression.
Figure 2.
X-chromosome states of human pluripotent stem cells. The X chromosome state of conventional (primed) hESCs differs from the ICM of human pre-implantation blastocysts from which they are derived. Primed hESCs are in a post-XCI state with an XIST-coated Xi. Over time in culture, the Xi loses expression of XIST and partially reactivates, undergoing XCI erosion. Primed hESCs with two active X chromosomes can also be derived from ICM outgrowths (far right), potentially capturing an intermediate state of the X chromosome in the transition to XCI. Differentiation does not change the X-chromosome state of any of these primed hESCs. When hESCs are derived from the blastocyst under naïve culture conditions, or when primed hESCs, regardless of their X state, are converted to naïve pluripotency, the X-chromosome state resembles that of the blastocyst, with two active X chromosomes and XIST expression (on one or both X chromosomes). Like normal development, differentiation of naïve hESCs induces XCI. Similar to primed hESC derivation, an XIST-negative state with two active X chromosomes is an intermediate in the primed to naïve hESC conversion, suggesting stepwise reversal of events. The lncRNA XACT is co-expressed with XIST in naïve pluripotency and might be responsible for inhibiting XIST-mediated silencing. XIST and XACT occupy non-overlapping territories on the active X chromosome (green and purple) in naïve hESCs. XACT is also expressed in primed hESCs both from active and eroding/eroded X chromosomes, and it might be driving erosion by interfering with XIST expression or accumulation. XACT is not expressed in differentiated cells.
The other, more common XaXiXIST+ state in early passage hESC lines appears to resemble the post-XCI state of somatic cells, as shown, for example, by the occurrence of methylation of CpG islands on the Xi [6,39,44]. However, it changes in culture over time: in nearly all cases, XIST expression on the Xi is gradually lost in these cells, and the inactive X is partially reactivated, resulting in double dosage of a subset of X-linked genes (figure 2) [6,39,44–47]. This erosion of XCI is accompanied by the loss of DNA methylation specifically in the CpG islands of affected genes, and its extent varies in different cell lines, ranging from only a handful of genes to almost the entire inactive X chromosome [6,44]. The determinants of XCI erosion are currently poorly understood, but certain regions on the Xi are more likely to erode than others [44]. Interestingly, chromatin signatures, such as H3K27me3 and H3K9me3 modifications, are good predictors of erosion, with genes enriched for H3K27me3 and relatively depleted for H3K9me3 on the Xi having an increased likelihood of reactivation upon XCI erosion [47]. A defining feature of XCI erosion is that it cannot be undone, even during differentiation [6,44,46] (figure 2). In other words, the aberrant X-chromosome state of these cells is locked in place so that, upon differentiation, the reactivated parts of the inactive X chromosome cannot be re-silenced, resulting in differentiated cells with a double dose of the X-linked genes that fall in eroded regions. This has not only been problematic for basic researchers who wish to study the onset of XCI in the human system, but also influences studies of X-linked diseases and use of female induced pluripotent stem cells (iPSCs) for disease modelling (see below) [46]. Furthermore, XCI erosion may affect cell replacement and regenerative therapies, because inappropriate dosage compensation of X-linked genes is a hallmark of female-specific cancers [48].
The X-chromosome state of human iPSCs, and whether reprogramming of somatic cells to pluripotency is accompanied by Xi-reactivation, has been heavily debated in the literature. Data from us and others argue that human iPSCs are XaXi with XIST at early passage, but over time in culture XIST expression is lost and the Xi is partially reactivated due to XCI erosion, similar to XIST-expressing XaXi hESCs [6,44,45,49,50]. Thus, despite various reports of complete Xi-reactivation in human iPSCs [51–54], our data suggest that the XaXa state is not achieved in human iPSC cultures but is unique to hESCs, consistent with the idea that it is due to the expansion of this transient state unique to the transition from the blastocyst to primed pluripotency [6].
6. Naive human PSCs capture features of the X chromosome of the blastocyst
Mouse PSCs can transition from one pluripotent state to the other in vitro. For instance, over-expression of specific transcription factors, such as Klf4 [55] or deriving stem cells from post-implantation epiblasts in leukaemia inhibitory factor (LIF) and fetal calf serum [56] achieves primed to naive conversion. The ability to convert mouse cells in vitro from one pluripotent state to the other inspired researchers to screen for naive culture conditions appropriate for hESCs, with the idea that establishment of the primed pluripotent state was due to culture conditions and not intrinsic to the pre-implantation human blastocysts from which these cell lines are derived. Different approaches were used in the search for media formulations supporting naive pluripotency, with most of them using small molecule inhibitors, building upon naive condition of the mouse. Hanna and co-workers [57] demonstrated that the serum-free naive culture formulation for mESCs on its own—inhibition of both glycogen synthase kinase 3 beta and extracellular signal-regulated kinase 1/2 in combination with LIF (2i/LIF)—was not enough to support human naive PSCs, and constant expression of the pluripotency transcription factors OCT4, SOX2 and KLF4 was required in combination with 2i/LIF to support naive-like human PSCs (hPSCs). They screened for small molecule inhibitors of additional pathways that could stabilize the naive-like state in the absence of exogenous OCT4, SOX2 and KLF4 expression and formulated the first naive hPSC condition termed NHSM (naive human stem cell medium) [57]. This was followed by the development of several other formulations based on different combinations of small molecule inhibitors and cytokines [58–60]. Each newly devised culture condition resulted in cells with transcriptional profiles different from the human primed PSCs and similar, to various degrees, to the naive PSCs of the human pre-implantation blastocyst, likely reflecting the stabilization of various pluripotency states by each method. To address this systematically, Huang et al. [61] used an unbiased approach of comparing the transcription signature of each of these naive in vitro states to that of early human pre-implantation development, including oocyte, 1-, 2-, 4-, 8-cell stage embryos, morula and the blastocyst. In this analysis, two of the naive conditions—devised by Takashima et al. [58] and Theunissen et al. [59]—had the most significant gene expression overlap with the human blastocyst. Moreover, we demonstrated that the X-chromosome state of hESCs in these two culture conditions resembles that of the blastocyst, where XIST is expressed and accumulates on active X chromosomes [5,12]. Furthermore, the naive condition devised by Theunissen et al. allowed direct derivation of naive hESC lines from pre-implantation blastocysts [59], and the stabilization of the blastocyst X-chromosome state in culture [12]. Hence, we conclude that the X-chromosome state—mainly expression of XIST from active X chromosomes—is a reliable way of testing for true naivety of hPSCs that should be employed in assessing new naive formulations in the future. Importantly, the ability to capture the naive status of XIST expression in hPSCs provides a unique system to investigate the inability of XIST to silence the X chromosome.
When primed hPSCs harbouring one active and one inactive X chromosome (with or without XIST expression from the Xi) are converted to naive pluripotency, the inactive X reactivates first, giving rise to XaXa cells, and only after several passages does XIST become expressed from either one or both X chromosomes, although the mono-allelic XIST pattern is dominant [12]. Interestingly, XaXa XIST-positive naive hESCs exhibited overall dampened X-linked gene expression levels compared with those not expressing XIST [12]. Thus, the correlation of XCD and XIST observed in human pre-implantation embryos appears to be recapitulated in vitro in the transition from primed to naive hESCs. These observations suggest that naive hESCs will also serve as model system for further exploring the novel X-linked gene dosage compensation mechanism of XCD.
In addition to serving as an in vitro model of the pre-implantation human embryo, naive culture conditions also provide a means of overcoming the XCI anomalies observed in primed PSCs (discussed in §5). When primed hESCs with either a slight or very high degree of XCI erosion, or even those that are trapped in the XIST-negative XaXa state, are adapted to the naive culture condition described by Theunissen et al. [59], and then subjected to differentiation, regardless of the starting primed XCI state, all of them result in cells with the proper somatic-like X-chromosome state: with an Xa and an XIST-expressing Xi [12]. These findings demonstrate that XCI erosion in primed hPSCs is truly just an anomaly caused by imperfect culture conditions and can be reversed given the right media formulation. Moreover, the ability to induce de novo XCI upon differentiation of naive hESCs (figure 2) [12] now opens opportunities of studying this epigenetic process in the human system for the first time.
7. The novel lncRNA XACT and its potential role in regulating human-specific aspects of X-chromosome dosage compensation
The puzzling differences in the way dosage compensation is established in the human compared with the mouse raise the intriguing hypothesis that some regulators of the process may differ between species. Tsix, the Xist antisense transcript identified in the mouse and described in §3, is one such example, having an important contribution to the regulation of murine XCI and no functional orthologue in the human. More recently, through RNA-sequencing analysis, we identified a novel X-linked lncRNA—XACT—which shares with XIST the capacity to accumulate on the chromosome from which it is expressed [28]. The appearance of XACT seems to be a recent event on the evolutionary scale, which took place in the higher primate branch, suggesting that it might fulfil primate (or human)-specific function [28].
Insights into such function came from the analysis of hPSCs with various X-chromosome states. In fact, expression of XACT is restricted to pluripotent cells: XACT gets silenced when the cells are induced to differentiate and reactivates upon induction of pluripotency (figure 2) [28]. In primed XaXiXIST+ cells, XACT is expressed from the active X only, while in XIST-negative XaXi cells, XACT is accumulating on both X chromosomes (figure 2). While this shift in XACT expression profile could simply reflect the partial reactivation of the Xi that characterizes XCI erosion, capturing the transition between the two states suggested an alternative scenario. Indeed, re-expression of XACT from the Xi undergoing erosion occurs before loss of XIST expression and prior to extended X-chromosome reactivation [47]. XACT reactivation from the Xi is thus not a mere consequence of erosion but is instead one of the earliest markers of this phenomenon. Pushing the reasoning further, XACT could causally participate in the erosion, by interfering with XIST expression or accumulation. In agreement with this hypothesis, when XACT was artificially inserted onto one X chromosome in female mESCs, XCI was biased towards the untargeted X chromosome. In other words, forced expression of XACT from one X reduced the likelihood of Xist accumulating on the very same chromosome, at least in a heterologous system [5].
What about XACT in the human embryo? Combining analysis of multiple datasets of single-cell RNA-sequencing and RNA-FISH confirmed that XACT is not an artefact of hPSC in culture, and that it is expressed in pre-implantation embryos [5]. Its expression is in fact strongly correlated to that of XIST in the early developmental stages (up to early E5), where it accumulates, together with XIST, on every X chromosome in both male and female embryos (figure 2). This pattern of active X chromosomes simultaneously decorated by XIST and XACT is recapitulated to some extent in naive hPSCs derived either in 5iLAF or in t2iL + Gö conditions [5,12,58,59], further reinforcing the idea that these naive conditions indeed bookmark the in vivo situation. Intriguingly, in both cases XIST RNA was found more dispersed in the nucleus compared with cells in which XIST coats the Xi [5,12]. This altered distribution of XIST might be linked to its inability to properly silence X chromosome at these stages. As it correlates with the simultaneous presence of XACT, it is also tempting to speculate that XACT might impair proper XIST accumulation in human cells, as it does in the heterologous mouse system described earlier.
8. Other potential mechanisms preventing XIST-mediated silencing
XACT is one strong candidate for preventing XIST from silencing the X chromosome during human pre-implantation development, but additional, non-mutually exclusive scenarios can be envisioned based on recent advances in studying the mechanism of action of mouse Xist. For instance, Patil et al. [34] demonstrated that a reversible RNA modification of adenosine residues—N6-methyladenosine (m6A)—is enriched on Xist and required for its silencing ability. Differences in this or perhaps even other RNA modifications or downstream readers of such modifications in early pre-implantation versus later post-implantation stages of human development might contribute to the functional differences of XIST.
RNA antisense purification followed by next-generation sequencing has allowed mapping of chromatin contacts made by mouse Xist at the onset of XCI, and combined with chromosome conformation studies, uncovered that Xist first contacts distal regions on the X chromosome that are spatially close to the Xist transcription locus [30]. Hence one can postulate that the three-dimensional structure of the X chromosome is important when considering how Xist can spread along the X chromatin. Therefore, another speculation is that, due to different three-dimensional folding of the X chromosome in the pre-implantation embryo and/or expression of XACT, the chromatin structures might be unfavourable for XIST spreading and thus silencing of the X chromosome in naive pluripotency.
Several independent groups recently confirmed known and identified novel proteins that bind to mouse Xist RNA at the onset of XCI initiation or on the already established Xi [31–33,62–65]. Functional experiments have demonstrated that some of these Xist binding proteins are absolutely required for Xist-mediated silencing of the X chromosome. Hence it is plausible that one or more of key XIST interacting proteins required for its silencing ability are simply not expressed in the naive context, or that XIST is somehow unable to bind to such key protein factors, due to alternative splicing, the presence of competing proteins/RNAs or to chemical modifications.
9. Conclusion
The emerging studies of XCI in the human revealed a quite surprising flexibility in the way dosage compensation is established in various mammalian species [1,2,5,12]. Not only does XCI differ in kinetics and parental origin between human and mouse, but the strategies per se by which X-chromosome dosage imbalance is compensated for follow different routes, even if only transiently. XCD is reminiscent of the worm dosage compensation system, but the underlying mechanisms in human are still largely mysterious. We have seen that there are good reasons to believe that XIST could also contribute to this process. In this context, XACT could act as a switch for XIST function, from dampening X-chromosome expression (when XACT is present) to fully silencing it (in the absence of XACT). Our understanding of human dosage compensation has for long been impaired by the paucity of relevant biological material. Recent developments in the field of human naive pluripotency will help in uncovering molecular mechanisms, although it should be kept in mind that the field in still in its infancy. In this context, and as mentioned in §§6 and 7, we believe that rigorous assessment of the X-chromosome status through monitoring XIST and XACT expression will be instrumental in assessing true naivety of hPSCs and identifying novel conditions to robustly trigger and, importantly, maintain naive pluripotency.
Acknowledgements
We apologize to researchers who were not directly cited due to limited space.
Authors' contributions
A.S., K.P. and C.R. conceptualized, drafted, edited and proofread the manuscript.
Competing interests
We have no competing interests.
Funding
A.S. is supported by the Ruth L. Kirschstein NRSA F31 Fellowship (GM115122) and the Fowler Fellowship. K.P. is funded by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, the UCLA David Geffen School of Medicine, the Jonsson Comprehensive Cancer Center, the NIH (R01 GM115233), and is a Faculty Scholar of the Howard Hughes Medical Institute. Research in the Rougeulle lab is funded by the Agence Nationale pour la Recherche (ANR) and Ligue contre le Cancer.
References
- 1.Okamoto I, et al. 2011. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472, 370–374. ( 10.1038/nature09872) [DOI] [PubMed] [Google Scholar]
- 2.Petropoulos S, et al. 2016. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026. ( 10.1016/j.cell.2016.03.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monk M, Harper MI. 1979. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281, 311–313. ( 10.1038/281311a0) [DOI] [PubMed] [Google Scholar]
- 4.Briggs SF, Dominguez AA, Chavez SL, Reijo Pera RA. 2015. Single-cell XIST expression in human preimplantation embryos and newly reprogrammed female induced pluripotent stem cells. Stem Cells 33, 1771–1781. ( 10.1002/stem.1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallot C, et al. 2017. XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell 20, 102–111. ( 10.1016/j.stem.2016.10.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel S, et al. 2017. Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep. 18, 54–67. ( 10.1016/j.celrep.2016.11.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer BJ, Casson LP. 1986. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 47, 871–881. ( 10.1016/0092-8674(86)90802-0) [DOI] [PubMed] [Google Scholar]
- 8.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244. ( 10.1038/nature14450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. ( 10.1038/349038a0) [DOI] [PubMed] [Google Scholar]
- 10.Brockdorff N, et al. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329–331. ( 10.1038/351329a0) [DOI] [PubMed] [Google Scholar]
- 11.Borsani G, et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. ( 10.1038/351325a0) [DOI] [PubMed] [Google Scholar]
- 12.Sahakyan A, et al. 2017. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20, 87–101. ( 10.1016/j.stem.2016.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653–1655. ( 10.1126/science.1126316) [DOI] [PubMed] [Google Scholar]
- 14.Takagi N. 2003. Imprinted X-chromosome inactivation: enlightenment from embryos in vivo. Semin. Cell Dev. Biol. 14, 319–329. ( 10.1016/j.semcdb.2003.09.027) [DOI] [PubMed] [Google Scholar]
- 15.Takagi N, Sasaki M. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642. ( 10.1038/256640a0) [DOI] [PubMed] [Google Scholar]
- 16.Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669. ( 10.1126/science.1092674) [DOI] [PubMed] [Google Scholar]
- 17.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, 644–649. ( 10.1126/science.1092727) [DOI] [PubMed] [Google Scholar]
- 18.Williams LH, Kalantry S, Starmer J, Magnuson T. 2011. Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development 138, 2049–2057. ( 10.1242/dev.061176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. ( 10.1038/379131a0) [DOI] [PubMed] [Google Scholar]
- 20.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166. ( 10.1101/gad.11.2.156) [DOI] [PubMed] [Google Scholar]
- 21.Borensztein M, et al. 2017. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat. Struct. Mol. Biol. 24, 226–233. ( 10.1038/nsmb.3365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira de Mello JC, Araújo ÉSS, Stabellini R, Fraga AM, Souza JES, Sumita DR, Camargo AA, Pereira LV. 2010. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE 5, e10947 ( 10.1371/journal.pone.0010947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JT, Davidow LS, Warshawsky D. 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21, 400–404. ( 10.1038/7734) [DOI] [PubMed] [Google Scholar]
- 24.Sado T, Wang Z, Sasaki H, Li E. 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Dev. Camb. Engl. 128, 1275–1286. [DOI] [PubMed] [Google Scholar]
- 25.Lee JT, Lu N. 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99, 47–57. ( 10.1016/S0092-8674(00)80061-6) [DOI] [PubMed] [Google Scholar]
- 26.Gayen S, Maclary E, Buttigieg E, Hinten M, Kalantry S. 2015. A primary role for the Tsix lncRNA in maintaining random X-chromosome inactivation. Cell Rep. 11, 1251–1265. ( 10.1016/j.celrep.2015.04.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. ( 10.1038/ng820) [DOI] [PubMed] [Google Scholar]
- 28.Vallot C, Huret C, Lesecque Y, Resch A, Oudrhiri N, Bennaceur-Griscelli A, Duret L, Rougeulle C. 2013. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat. Genet. 45, 239–241. ( 10.1038/ng.2530) [DOI] [PubMed] [Google Scholar]
- 29.Monk M. 1981. A stem-line model for cellular and chromosomal differentiation in early mouse-development. Differ. Res. Biol. Divers. 19, 71–76. ( 10.1111/j.1432-0436.1981.tb01131.x) [DOI] [PubMed] [Google Scholar]
- 30.Engreitz JM, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 ( 10.1126/science.1237973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHugh CA, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. ( 10.1038/nature14443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. ( 10.1016/j.cell.2015.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C-K, et al. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. ( 10.1126/science.aae0047) [DOI] [PubMed] [Google Scholar]
- 34.Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. ( 10.1038/nature19342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols J, Smith A. 2009. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. ( 10.1016/j.stem.2009.05.015) [DOI] [PubMed] [Google Scholar]
- 36.Brons IGM, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195. ( 10.1038/nature05950) [DOI] [PubMed] [Google Scholar]
- 37.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. ( 10.1038/nature05972) [DOI] [PubMed] [Google Scholar]
- 38.Davidson KC, Mason EA, Pera MF. 2015. The pluripotent state in mouse and human. Development 142, 3090–3099. ( 10.1242/dev.116061) [DOI] [PubMed] [Google Scholar]
- 39.Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. 2008. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl Acad. Sci. USA 105, 4709–4714. ( 10.1073/pnas.0712018105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva SS, Rowntree RK, Mekhoubad S, Lee JT. 2008. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 4820–4825. ( 10.1073/pnas.0712136105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lengner CJ, et al. 2010. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141, 872–883. ( 10.1016/j.cell.2010.04.010) [DOI] [PubMed] [Google Scholar]
- 42.O'Leary T, et al. 2012. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat. Biotechnol. 30, 278–282. ( 10.1038/nbt.2135) [DOI] [PubMed] [Google Scholar]
- 43.de Oliveira Georges JA, et al. 2014. Aberrant patterns of X chromosome inactivation in a new line of human embryonic stem cells established in physiological oxygen concentrations. Stem Cell Rev. Rep. 10, 472–479. ( 10.1007/s12015-014-9505-4) [DOI] [PubMed] [Google Scholar]
- 44.Nazor KL, et al. 2012. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10, 620–634. ( 10.1016/j.stem.2012.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchieu J, et al. 2010. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7, 329–342. ( 10.1016/j.stem.2010.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. 2012. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 10, 595–609. ( 10.1016/j.stem.2012.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallot C, et al. 2015. Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell 16, 533–546. ( 10.1016/j.stem.2015.03.016) [DOI] [PubMed] [Google Scholar]
- 48.Dunford A, et al. 2016. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 49, 10–16. ( 10.1038/ng.3726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheung AYL, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. 2011. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Human Mol. Genet. 20, 2103–2115. ( 10.1093/hmg/ddr093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pomp O, Dreesen O, Leong DFM, Meller-Pomp O, Tan TT, Zhou F, Colman A. 2011. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell 9, 156–165. ( 10.1016/j.stem.2011.06.004) [DOI] [PubMed] [Google Scholar]
- 51.Anguera MC, et al. 2012. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11, 75–90. ( 10.1016/j.stem.2012.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomoda K, et al. 2012. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell 11, 91–99. ( 10.1016/j.stem.2012.05.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim K-Y, Hysolli E, Tanaka Y, Wang B, Jung Y-W, Pan X, Weissman SM, Park I-H. 2014. X Chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Rep. 2, 896–909. ( 10.1016/j.stemcr.2014.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barakat TS, et al. 2015. Stable X chromosome reactivation in female human induced pluripotent stem cells. Stem Cell Rep. 4, 199–208. ( 10.1016/j.stemcr.2014.12.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069. ( 10.1242/dev.030957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. 2009. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461, 1292–1295. ( 10.1038/nature08534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gafni O, et al. 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. ( 10.1038/nature12745) [DOI] [PubMed] [Google Scholar]
- 58.Takashima Y, et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269. ( 10.1016/j.cell.2014.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Theunissen TW, et al. 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471–487. ( 10.1016/j.stem.2014.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ware CB, et al. 2014. Derivation of naive human embryonic stem cells. Proc. Natl Acad. Sci. USA 111, 4484–4489. ( 10.1073/pnas.1319738111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang K, Maruyama T, Fan G. 2014. The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell 15, 410–415. ( 10.1016/j.stem.2014.09.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19, 469–476. ( 10.1016/j.devcel.2010.08.006) [DOI] [PubMed] [Google Scholar]
- 63.Minajigi A, et al. 2015. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 ( 10.1126/science.aab2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moindrot B, et al. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572. ( 10.1016/j.celrep.2015.06.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561. ( 10.1016/j.celrep.2015.06.067) [DOI] [PMC free article] [PubMed] [Google Scholar]