Abstract
The Xist gene produces a long noncoding RNA that initiates chromosome-wide gene repression on the inactive X chromosome in female mammals. Recent progress has advanced the understanding of Xist function at the molecular level. This review provides an overview of insights from genetic approaches and puts the new data in the context of an emerging mechanistic model as well as the existing literature. Some consideration is given on how independent biochemical studies on X inactivation help to advance on the wider question of chromatin regulation in the mammalian dosage compensation system.
This article is part of the themed issue ‘X-chromosome inactivation: a tribute to Mary Lyon’.
Keywords: noncoding RNA, gene regulation, chromatin, histone modification, X chromosome inactivation, Xist
1. Introduction
Noncoding RNAs appear as regulators of cell signalling and gene expression in many biological contexts. Lack of sequence bias and conservation associated with open reading frames makes functional RNAs more difficult to identify. Proving that an RNA rather than a protein is the functional product of a gene appears to be a formidable task. Xist is among the first regulatory RNAs discovered and implicated in formation of the inactive X chromosome (Xi). This review focuses on the analysis of Xist function and its molecular mechanism as an important model for functional RNAs.
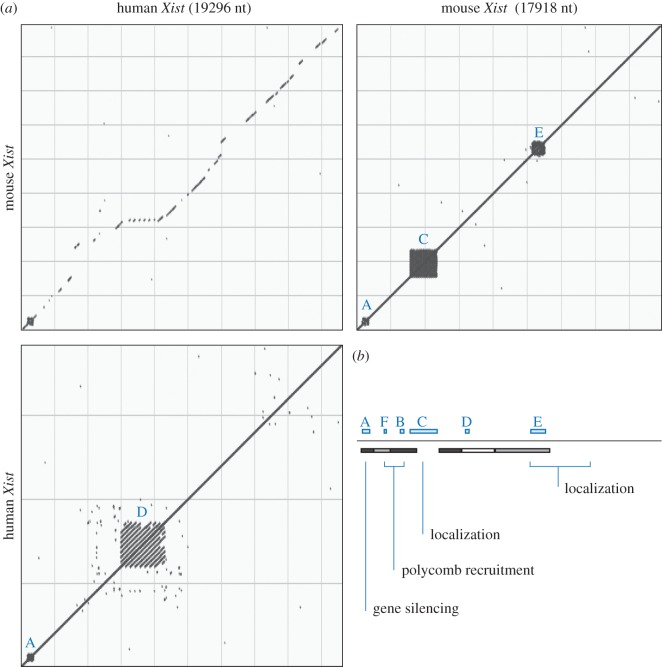
The discovery of Xist follows the investigation of the genetic basis of X chromosome inactivation. One of the two X chromosomes in female mammals is inactivated in a random manner [1]. X chromosome inactivation (XCI) serves as a dosage compensation mechanism and adjusts for the different number of X chromosomes between the sexes. The X inactivation centre (Xic) has been genetically defined as a locus that facilitates inactivation of one of the two X chromosomes in female cells [2]. The Xic locus also contains the Xist gene [2–4]. Xist produces an RNA, which is localized to the nucleus, shows no association with ribosomes, and paints the inactive X chromosome [5–7]. Together these observations make a convincing case for the functional product of the Xist gene being an RNA and not a protein. Xist is conserved in the large majority of placental mammals and contains a number of repeated sequence elements [7–10] (figure 1a). Its requirement for female development has been shown in mice [11]. Thereby Xist is required for inactivation of the X chromosome and for choosing the chromosome to inactivate [12]. An X chromosome bearing a mutation of Xist remains active and does not express the mutant Xist [13].
Figure 1.
Xist RNA sequence conservation and sequence repeat elements. (a) Dot plot analysis of mouse and human Xist RNA shows overall conservation (upper left panel) and repeated sequence elements (right and lower panels). The analysis was performed using NCBI nucleotide Blast with default search parameters for detecting somewhat similar sequences (blastn, v. 2.5.0). Accession codes for human and mouse sequences are NR_001564.2 and NR_001463.3, respectively. (b) Schematic overview of mouse Xist RNA. Sequence repeats A to F are indicated above the black line. Estimated contribution of regions to localizing and stabilizing Xist RNA from a study of the mouse Xist cDNA [10] are shown below the line (greyscale, darker indicated greater function in focal Xist cluster formation). Below assigned functions to indicated Xist sequences in independent studies (see text). The scheme is drawn to scale and aligned with mouse Xist in panel (a). (Online version in colour.)
Analysis of Xic sequences by transgenic experiments in mouse embryonic stem (ES) cells has further provided evidence that functions of the Xic can be grafted to other chromosomal contexts. Using yeast artificial chromosomes carrying approximately half a megabase of Xic sequences, inactivation of genes on autosomes has been achieved [14,15]. Also, smaller transgenes derived from Xic sequences show that elements for chromosomal inactivation reside in a confined genomic interval [16]. Results obtained with a cosmid derived transgene spanning the Xist gene indicate that some function might reside within the Xist locus itself [17]. These studies show that aspects of Xic activity can be reconstituted by transgenes on autosomes and provide a starting point for investigating the underlying mechanisms.
2. Functional analysis of Xist RNA sequences
The role of Xist in chromosome-wide gene silencing is difficult to investigate as it is under the control of complex regulation. Deletions within the Xist gene often prevent expression such that the function of the mutant Xist RNA cannot be studied [12,18]. To circumvent this issue, heterologous promoters have been used for driving Xist expression from transgenes. Using the tetracycline inducible expression system [19], it could be shown that expression of mouse Xist from a cDNA transgene is sufficient to elicit signs of inactivation in male mouse ES cells [20]. In this system, chromosomal hallmarks of X inactivation as well as evidence for gene repression could be demonstrated. Following the idea of the inducible transgenic system, a series of Xist deletions have been characterized by insertion of several Xist cDNA transgenes into the Hprt locus on the X chromosome of male ES cells [10]. The ability of mutant Xist RNAs to localize and to elicit X chromosome silencing could be analysed (summary in figure 1b). The latter was measured indirectly through the induction of cell death by the loss of expression from the single X chromosome in male ES cells. This investigation led to the identification of repeat A as an important element within Xist for gene repression. Repeat A also appears to achieve silencing in human cells whereby cooperation between monomeric repeat A motifs is observed [21]. Xist repeats F and B have been implicated in the recruitment of Polycomb complexes [22] (figure 1b). Use of peptide linked nucleic acid (PNA) and locked nucleic acid (LNA) interference techniques have also provided evidence that interference with repeat C can lead to delocalization of Xist RNA from the chromosome [23,24]. Albeit results with both PNA and LNA reagents are in agreement with each other, the precise mode of action of the synthetic oligonucleotides remains unclear and does not seem to resemble a simple deletion of repeat C sequences [10]. In addition, there is evidence that repeat C might act as a binding site for YY1 [25]. Deletion of sequences within the Xist gene locus has shown a contribution of Xist exon 7 to stable localization of Xist RNA to the X chromosome [26]. Furthermore, Xist repeat D has been suggested to have some function in XIST expression and gene silencing in transformed 293FT human embryonic kidney cells [27]. Sequence elements throughout Xist that are not restricted to the identified repeats A to F act in a parallel or redundant manner for localizing Xist RNA [10].
The above cited studies have also facilitated to separate different functions of Xist through deleting specific sequences. Notably, expression of Xist lacking repeat A has been shown to lead to the recruitment of chromatin modifications without affecting gene expression [28–30]. Based on this observation a model has been put forward that Xist might establish a chromatin compartment that resembles the facultative heterochromatin of the Xi. At the initiation of X inactivation this compartment would initially not affect gene expression. In a subsequent and separate event that is dependent on Xist repeat A sequences gene repression would be achieved and finally repressed genes would associate with the repressive chromatin compartment [28]. The idea of such a repressive compartment is consistent with several observations including the location of X-linked genes outside the Xist covered nuclear territory at early time points in X inactivation. Furthermore, partial histone H4 acetylation pattern can be visualized by microscopy of the X chromosome that expresses Xist RNA lacking repeat A. The latter might account for gene activity on the X chromosome that else recapitulates the heterochromatin composition of the Xi [30]. However, these models are based on results that were largely obtained in ES cells. Caution is therefore needed with generalizing these results as the situation in ES cells and the embryo might differ. Furthermore, it should be considered that induced Xist expression from a strong artificial promoter might not reflect physiological expression levels of Xist. Future studies in mouse embryos are therefore desirable for addressing these limitations and investigating Xist RNA sequences in different lineages and stem cell systems.
3. Evidence for developmental regulation of the gene silencing pathways of Xist
To evaluate the influence of the type or differentiation state of a cell on Xist function inducible Xist expression in differentiating ES cells was investigated initially. Gene repression was observed if induction of Xist had occurred within the first 2 days of differentiation [20]. In fully differentiated cells Xist appeared to have lost its ability to initiate repression. This result is further consistent with an analysis in embryonic fibroblasts suggesting Xist function is restricted in cell differentiation [20]. Using a mouse line that carried an inducible promoter in the endogenous Xist gene a more extensive analysis of Xist function in development was performed based on the idea that chromosome-wide repression of the single X chromosome in male mice by forced Xist expression translates into cell death and an anatomically scorable phenotype [31]. From this study it appeared that Xist caused widespread cell loss in embryos when induced early. Xist induction at E9.5 was still capable to initiate X-inactivation at least to some extent, when normally XCI is thought to be initiated at E5.5 in the female embryo. It is not clear if this phenotype was entirely caused by defects in the embryonic lineages or also defects in the extraembryonic tissues contributed. Progressively later induction of Xist in development resulted in less and less severe malformations. Induction at or after day 12.5 gave life born mice that appeared anatomically normal but died shortly after birth. A likely cause of death is a lack of blood cells as it could be shown that haematopoietic progenitors in all blood lineages are susceptible to Xist-mediated gene repression even in adult mice [31]. The haematopoietic context for repression for Xist could independently be observed in a thymic lymphoma mouse model [32]. Xist induction in tumours or tumour cells resulted in rapid cell death and tumour regression. The observation that gene repression by Xist was regulated in haematopoietic differentiation prompted an attempt to identify genes that are differentially expressed between Xist responsive and resistant tumours. The latter could be obtained by culture of tumour cells over many cell doublings. SATB1 could be identified as a factor that correlated with Xist function in gene repression. SATB1 further correlated with the thymic progenitor population that was susceptible to gene repression by Xist [32] suggesting a potential silencing factor of Xist. However, later work showed that Satb1 appears not to be critical for X inactivation in the embryo [33]. Whereas Satb1 can reprogram cells to reactivate the context for Xist-mediated gene repression, it appears to be required only in blood cells suggesting other factors might perform similar functions in the embryo [33]. Notably, XIST expression can initiate gene repression also in the human tumour derived HT1080 fibrosarcoma cell line [34] and the immortalized human embryonic kidney 293 cell line [35]. These observations could indicate that transformed and embryonic cells might have access to pathways that Xist uses for initiating gene repression. The cell type specificity of Xist function further suggests that the underlying pathways might be under developmental control and therefore could be isolated and manipulated for studying associated molecular components.
4. Identification of factors in X inactivation through genetic screening
Genetic screening can be an efficient tool to identify factors that interact with Xist in gene silencing. The development of methods for conducting genome-wide screens in mammals and the availability of a variety of model systems provide a basis for experimental strategies to unravel the regulatory mechanisms of Xist-mediated silencing.
Structural maintenance of chromosomes hinge domain containing 1 (Smchd1) was the first factor that was identified in a genetic screen and could be linked to X chromosome inactivation. In this pioneering work an autosomal transgene reporter was used for identifying epigenetic modifiers of gene expression in mice [36]. Blewitt et al. used mice that carry a variegating green fluorescence protein (GFP) transgene as a model system, whereby 55% of the erythrocyte population shows metastable GFP expression. Mutations were generated by using N-ethyl-N-nitrosourea (ENU) as the mutagenic agent. Mutant offspring were selected based on inherited epigenetic alterations of GFP transgene activity that could be measured by flow cytometry of blood samples. One of these mutations, called Momme D1 for Modifier of Murine Metastable Epiallele D1, showed increased GFP expression and also revealed a homozygous female-specific lethal phenotype at embryonic day 11. The Momme D1 mutation mapped to the Smchd1 gene in chromosome 17 [37]. The implication of Smchd1 in X inactivation was confirmed in mice carrying an X-linked EGFP transgene and a maternal mutant Xist allele. The latter modification prevents the randomness of XCI and forces the silencing of the paternal X chromosome harbouring the GFP marker gene. Upon genetic truncation of Smchd1, EGFP expression was detected not only in embryonic but also in extra-embryonic tissues at early developmental stages. In addition CpG islands that are normally subject to DNA methylation on the Xi were found to be hypomethylated in Smchd1 mutant embryos. Enrichment of SMCHD1 on the Xi was detected by immunofluorescence analysis suggesting a direct role of Smchd1 in regulating DNA methylation and gene expression on the Xi [37]. Whether SMCHD1 is a component of the DNA methylation system or is more generally involved in the chromatin configuration of the Xi remains to be established in future studies. However, this discovery highlights an impressive advance that has been facilitated by genetic screening in mice.
Recent studies have aimed to extend screening for silencing factors into different cell systems. This approach is motivated in part by the availability of selection strategies and also by consideration of the high resource and time requirements for screens in animals. A suitable model system for studying X inactivation can be found in ES cells that represent the developmental context where Xist can initiate silencing. Moindrot et al. introduced an inducible Xist transgene and a GFP reporter into chromosome 17 of a male ES cell line [38]. Addition of doxycycline to the culture results in Xist transgene expression that causes repression of genes including the GFP reporter on the transgenic autosome. Viral shRNA libraries were used for depletion of genes through RNA interference. After Xist induction the cells showing the highest GFP expression were isolated and the enriched shRNAs identified. The rationale behind this screen is that whereas GFP is repressed by Xist expression in the parental cell line, GFP expression might persist if a silencing factor for Xist is depleted. This strategy led to the identification of several candidates including the RNA binding protein RBM15, a subunit of an RNA methyltransferase complex WTAP, and a mammalian homologue of Split Ends SPEN. In addition, several genes were identified that might be related to the GFP reporter system used or related to technical aspects of the cell line. Spen, Wtap and Rbm15 were validated in differentiating female ES cells since the screen had been performed in an autosomal context.
Development of haploid ES cells provided additional opportunities for genetic exploration of X inactivation [39]. Monfort et al. employed mouse haploid ES cells carrying an inducible promoter in the endogenous Xist locus for conducting a screen for silencing factors [40]. Mutagenesis in these haploid cells through retroviral gene-trap vectors facilitated the generation of large mutant pools that were selected through phenotypic interrogation following a simple rationale. Induction of Xist expression from the single X chromosome results in cell death in wild-type haploid cells, whereas cells carrying mutations in silencing factors cannot initiate X inactivation and survive. Amplification of the flanking genomic regions of viral integrations from cell pools and sequence analysis allowed the identification of genes that are overrepresented in the surviving population. Overall this is an efficient approach but requires the use of large numbers of haploid ES cells as well as high titre retroviruses to obtain robust statistics. In addition, repeated screening in independent pools can provide further support for potential candidate factors. Identification of expected technical hits including the Xist gene and the expression cassette for the tetracycline regulated transactivator provide evidence for the feasibility and efficiency of the screening set-up. A ranking of several factors was obtained among which Spen was identified in a high ranked position [40]. Subsequently, the function of Spen was investigated through nuclease mediated deletion in ES cells. Spen was required for repression of X-linked genes by Xist and its mutation reduced the recruitment of chromatin modifications. In contrast, Xist localization was apparently not dependent on Spen.
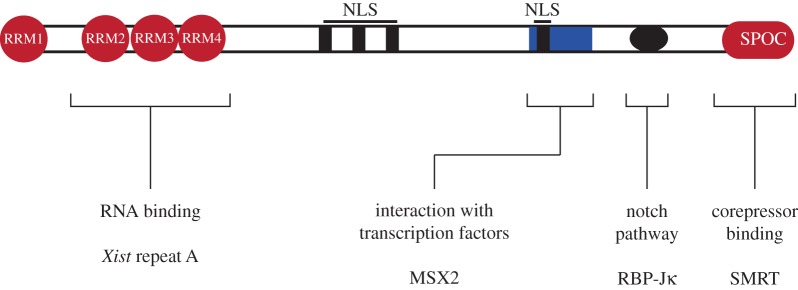
Interestingly, the candidate lists obtained from the screens by Moindrot et al. and Monfort et al. share few genes. This observation likely reflects the differences in screening system as well as mutagenic strategy. It should also be considered that both screens are unlikely saturated to a genome-wide level and might have different false positive and false negative rates. Notably, Spen (also named Mint, Sharp, or Rbm15c) has been identified in both screens suggesting a very robust genetic silencing factor. SPEN is an RNA binding protein and has been implicated in gene repression before. SPEN is a nuclear protein that contains four N-terminal RNA recognition domains (RRM) and a conserved Spen paralog and orthologue (SPOC) C-terminal domain that interacts with co-repressor complexes and histone deacetylases [41] (figure 2). This interaction has also been suggested to contribute to the function of Spen in X inactivation [42]. Spen has further been implicated in negative regulation of the Notch and nuclear receptor signalling pathways [43,44]. Furthermore, RNA-protein cross-linking strategies suggest a direct interaction of SPEN with Xist [42,45–47]. Consistent with expectations for a silencing factor SPEN can interact with repeat A sequences of Xist [40,45,48]. A mutation of Spen is embryonic lethal at embryonic day 12.5 in mice [43]. Therefore, loss of Spen appears to be less severe than a paternally inherited Xist mutation [11]. Possible explanations might be maternal contribution of SPEN protein or Spen independent X inactivation in extraembryonic trophectoderm lineages that could mask an early phenotype. Alternatively, partially compensating pathways might exist in the embryo that would not be prominent in ES cells.
Figure 2.
Domains and known interactions of SPEN protein. The scheme represents the 400 kDa (3643 amino acids) mouse SPEN protein and approximate position of computationally identified domains (NLS; nuclear localization signal). Known interactions of SPEN are indicated. The RNA binding domains and corepressor interacting domains have been implicated in Xist function, whereas the relevance of the other SPEN sequences to X inactivation remains to be investigated. (Online version in colour.)
5. Factors and pathways contributing to silencing the Xi
Presently, it appears that a key pathway that might be engaged by repeat A of Xist and involves Spen is essential for gene repression. However, Spen and repeat A sequences appear not required for Xist localization and have little effect on establishing the chromatin modifications on Xi. Additional factors have been identified that are involved in these aspects of Xist function. Immunofluorescence studies in female somatic cells permitted the identification of a specific histone composition of the Xi that is characterized by H3K9 methylation and hypoacetylation, H3K4 hypomethylation [49], a lack of histone H4 acetylation [50], increased mono-methylation of histone H4 lysine 20 [29], and enrichment of the histone variant mH2A [51]. Although the chromatin configuration contributes to the Xi, loss of function analysis indicates that not all factors might have a crucial function. mH2A is largely dispensable for X inactivation [52] and there are two related genes in mice. Combined mutations in mH2A.1 and mH2A.2 have little effect on viability and fertility of mice [51] suggesting redundant chromatin regulation.
A large number of proteins of the Polycomb group are recruited to the X chromosome upon Xist expression and have a prominent effect on histone modifications of the Xi. The initial observations implicating the PcG gene Eed in X inactivation were a female-specific phenotype in the extraembryonic lineages [53]. Eed is a component of Polycomb repressive complex 2 (PRC2) that has histone methylase activity. Subsequently, enrichment of EED and tri-methylation of histone H3 lysine 27 (H3K27me3) could be demonstrated on the Xi of several cell types and in mice [54,55]. The Xist-mediated recruitment of PRC2 to the X-chromosome was recently shown to involve the JARID2 protein, which also might mediate crosstalk between PRC1 and PRC2 [22,56]. PRC1 complexes mediate mono-ubiquitination of histone H2A lysine 119 (H2AK119ub) on the Xi [57]. Absent, small or homeotic discs 2 like (ASH2 L) is classified as a Trithorax group protein but apparently also is enriched on the Xi possibly through association with PcG proteins [30]. These observations suggest that a large set of chromatin associated proteins contribute to a unique chromatin configuration of the Xi. RNA interference mediated depletion of Ash2l did not abrogate silencing of an autosomal marker gene by transgenic Xist suggesting that Ash2l is not essential for gene repression in XCI [30]. Similarly, mutation of the PcG genes Eed or Ring1b did not abrogate Xist-mediated silencing despite disruption of PRC1 and PRC2 function [58,59]. Loss of PRC2 function was further shown not to impair X inactivation in embryonic lineages in mice [60]. Taken together the findings suggest that chromatin modifying complexes are relevant for X inactivation but are likely acting in a redundant manner or possess modulatory function. It needs to be considered that most of the experiments have been performed in cell systems that might just not be sensitive enough to detect subtle functions. However, one thought provoking result stands. Expression of a mutant Xist that lacks repeat A still appears to be able to recruit chromatin modifiers to establish a heterochromatin configuration similar to that of the Xi. Therefore, it is possible that the chromatin configuration per se is not sufficient for gene repression and X inactivation might require additional contributions such as the three-dimensional arrangement of gene loci.
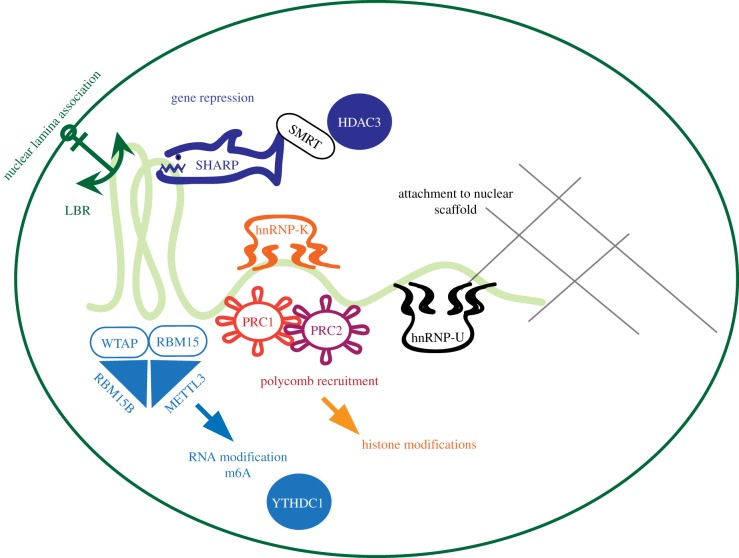
Nuclear attachment factors are enriched in the Xi territory and contribute to localization of Xist transcripts over the entire X chromosome and chromatin organization. Scaffold attachment factor A/heterogeneous nuclear ribonucleoprotein U (SAF-A/HNRNP U) has been identified in association with the nuclear scaffold of the Xi [61]. Mutation of Saf-A in mice result in early embryonic lethality at day 11.5 [62]. Depletion of Saf-A by RNA interference in mouse neuroblastoma cells led to the loss of Xist accumulation on the Xi and a diffuse distribution throughout the nucleus [63]. Similarly Xist became diffusely localized in female fibroblasts and ES cells after SAF-A depletion. These results were independently confirmed in a male ES cell line with an inducible promoter inserted into the endogenous Xist gene [42] suggesting SAF-A as a crucial factor for Xist localization. Recent data indicate that in different cell systems SAF-A related proteins might also provide compensatory function [64]. Independent biochemical studies have confirmed SAF-A as an interactor of Xist [42,45,46]. Conversely, Xist appears also required for SAF-A enrichment on the Xi [30]. Genetic deletion of Xist in female fibroblasts derived from mouse embryos homozygous for a conditional Xist allele resulted in a loss of SAF-A enrichment. The same study also found that SAF-A enrichment could be induced independent of repeat A of Xist and therefore gene repression [30]. Taken together these results demonstrate separable genetic requirements for localization of Xist and gene repression (figure 3).
Figure 3.
Separable genetic requirements and biochemical pathways for functions of Xist RNA. Graphic overview of factors and pathways discussed in the text. The green curved line in the centre represents Xist RNA. Separable pathways contributing to Xist function are grouped by colour.
HNRNP K, another member of the HNRNP family, has been identified in a comprehensive investigation of Xist RNA binding proteins by mass spectrometry [45]. Depletion of HNRNPK in male ES cells measurably impaired the ability of an inducible Xist transgene on chromosome 11 to repress the imprinted Grb10 gene on the transgenic autosome. Unlike depletion of SAF-A, depletion of HNRNP K did not affect the localization of Xist RNA. Notably, accumulation of H2AK119ub and H3K27me3 was strongly reduced specifically on the Xi upon HNRNAP K depletion suggesting a critical role for hnrnpk for establishing histone modifications in X inactivation (figure 3).
Another attachment factor with a role in XCI is the Lamin B Receptor (LBR) a component of the nuclear lamina that recruits Xist to the nuclear periphery. LBR was identified in an RNA antisense purification and mass spectrometry (RAP-MS) approach for Xist binding proteins [42]. A defect in silencing was readily observed in differentiating female ESCs where LBR had been disrupted [48]. An arginine-serine motif was postulated as a novel RNA binding domain for the interaction between Xist and LBR. A truncated LBR protein lacking this RS-motif did not rescue the depletion of LBR and Xist function. In addition, the interaction site of LBR could be mapped to the 5′-region of Xist. Deletion of this interaction site resulted in a silencing defect. Notably, expression of a version of Xist containing a BoxB motif, could be shown to cause gene silencing only if the mutated LBR protein lacking the RS-motif was fused to the heterologous λN RNA binding domain, which recognizes the BoxB sequence. This series of experimentation establishes LBR as a direct binder of Xist through a novel RNA binding domain. Its function is likely related through positioning of the X chromosome towards the nuclear lamina. Interestingly, the association with the nuclear lamina through LBR and with the nuclear scaffold through SAF-A are both required for X inactivation and, thus, suggest parallel but not redundant mechanisms of attachment (figure 3).
Identification of WTAP and its proposed interactors RBM15 and RBM15B by genetic screening as well as by biochemical purification hinted at a potential involvement of RNA N6-adenosine methylation (m6A) in Xist function [38,42,45]. RBM15 and RBM15B form a complex with WTAP and METTL3, which has catalytic RNA adenine methylase activity [65]. It could be shown that Xist RNA is methylated on adenosines and that methylation depends on METTL3 and WTAP. RBM15 and RBM15B appear to be redundant for Xist adenosine methylation. In addition the known methyl-adenine binding protein YTHDC1 is recruited to Xist and required for silencing [65]. Silencing could be rescued in the absence of Xist adenosine methylation by tethering YTHDC1 to Xist using the heterologous λN/BoxB system suggesting a role for adenosine methylation in the gene repression pathway [65] (figure 3). How the interaction with YTHDC1 results in transcriptional repression is not fully understood.
Other studies have investigated epigenetic mechanisms for the maintenance of gene silencing on the Xi in differentiated cells, when Xist is not critically required, through RNAi screens [66,67]. A number of factors with broad overlap of cellular mechanisms including DNA replication have been implicated in the maintenance of XCI. However, their role at early stages of XCI and importance in the XCI mechanism remains to be clarified in further studies.
6. Concluding remarks and future outlook
Recent studies have brought substantial advances in the understanding of the molecular basis of Xist function. Progress follows and builds on earlier results in cell biology and biochemistry that yield a much improved picture of the mammalian dosage compensation system. Whereas this is undoubtedly a gratifying time for the field it should also be a good time to remember that most of the results have been obtained in artificial systems including reporter systems in mouse ES cells. Indeed, inducible Xist expression appears not a mere convenience but the only technically feasible approach to conduct some of the analysis. This observation notwithstanding future work will need to carefully examine the relevance of the identified molecular components for X chromosome inactivation in the organism. Mice will be an obvious choice as mutations of many genes are available or can be generated with reasonable effort. Some of identified Xist silencing factors do show expected embryonic lethality including Spen at day 12.5 [43], Rbm15 at day 9.5 [68], Wtap at day 6.5 [69], and Saf-A at 11.5 [62]. These findings are consistent with a role of the relevant factors in X inactivation. However, observation of a female specific earlier lethality is the only outcome that could provide additional support for a function in XCI in the embryo. Naturally, such studies might only be conclusive for genes that play a critical role in XCI, and are redundant in other mechanisms. Death of male embryos is likely caused by functions of the genes in pathways other than X inactivation. Today few of the genes have been specifically investigated for sex specific early embryonic requirements.
A major question that needs to be resolved is if the insights obtained into X inactivation are also relevant to gene regulation in other biological systems and maybe can help to understand questions in development or human disease. Much of progress in this area will depend on precise definition of molecular mechanisms in X inactivation. The identified silencing factors provide starting points for leveraging recent screening methodology and interactome analysis techniques. Future screening in sensitized backgrounds might be useful to extend pathways where redundancy might pose a problem for identifying genetic components. Recent work has shown that screening for synthetic phenotypes can be performed in mammalian cells [70]. Other noncoding RNAs with function in chromatin or chromosome regulation have been identified including Rsx in marsupials [71], XACT in humans [72], and roX RNAs in flies [73,74]. Noncoding RNAs seem to act through different mechanisms and could point to more widespread involvement of RNA in chromatin domain regulation. It will be exciting to follow the exploration of these gene regulatory RNAs in future.
Acknowledgements
We are indebted to G. Di Minin and A. Postlmayr for stimulating discussions and critical questions during writing and apologize to the authors whose work we did not cite due to space constraints aiming to keep the text concise.
Authors' contributions
A.M. and A.W. designed the structure of the review and wrote initial drafts of the individual sections. Both authors corrected and polished the final version of the text and figures.
Competing interests
We have no competing interests.
Funding
This work was supported by Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (31003A_152814).
References
- 1.Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. ( 10.1038/190372a0) [DOI] [PubMed] [Google Scholar]
- 2.Brown CJ, et al. 1991. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349, 82–84. ( 10.1038/349082a0) [DOI] [PubMed] [Google Scholar]
- 3.Borsani G, et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. ( 10.1038/351325a0) [DOI] [PubMed] [Google Scholar]
- 4.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. ( 10.1038/349038a0) [DOI] [PubMed] [Google Scholar]
- 5.Clemson CM, McNeil JA, Willard HF, Lawrence JB. 1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275. ( 10.1083/jcb.132.3.259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526. ( 10.1016/0092-8674(92)90519-I) [DOI] [PubMed] [Google Scholar]
- 7.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. ( 10.1016/0092-8674(92)90520-M) [DOI] [PubMed] [Google Scholar]
- 8.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653–1655. ( 10.1126/science.1126316) [DOI] [PubMed] [Google Scholar]
- 9.Brockdorff N, et al. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329–331. ( 10.1038/351329a0) [DOI] [PubMed] [Google Scholar]
- 10.Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. ( 10.1038/ng820) [DOI] [PubMed] [Google Scholar]
- 11.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166. ( 10.1101/gad.11.2.156) [DOI] [PubMed] [Google Scholar]
- 12.Marahrens Y, Loring J, Jaenisch R. 1998. Role of the Xist gene in X chromosome choosing. Cell 92, 657–664. ( 10.1016/S0092-8674(00)81133-2) [DOI] [PubMed] [Google Scholar]
- 13.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. ( 10.1038/379131a0) [DOI] [PubMed] [Google Scholar]
- 14.Lee JT, Jaenisch R. 1997. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386, 275–279. ( 10.1038/386275a0) [DOI] [PubMed] [Google Scholar]
- 15.Lee JT, Strauss WM, Dausman JA, Jaenisch R. 1996. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86, 83–94. ( 10.1016/S0092-8674(00)80079-3) [DOI] [PubMed] [Google Scholar]
- 16.Lee JT, Lu N, Han Y. 1999. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc. Natl Acad. Sci. USA 96, 3836–3841. ( 10.1073/pnas.96.7.3836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzing LB, Romer JT, Horn JM, Ashworth A. 1997. Xist has properties of the X-chromosome inactivation centre. Nature 386, 272–275. ( 10.1038/386272a0) [DOI] [PubMed] [Google Scholar]
- 18.Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. 2009. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136, 139–146. ( 10.1242/dev.026427) [DOI] [PubMed] [Google Scholar]
- 19.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769. ( 10.1126/science.7792603) [DOI] [PubMed] [Google Scholar]
- 20.Wutz A, Jaenisch R. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5, 695–705. ( 10.1016/S1097-2765(00)80248-8) [DOI] [PubMed] [Google Scholar]
- 21.Minks J, Baldry SE, Yang C, Cotton AM, Brown CJ.. 2013. XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics Chromatin 6, 23 ( 10.1186/1756-8935-6-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Rocha ST, et al. 2014. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316. ( 10.1016/j.molcel.2014.01.002) [DOI] [PubMed] [Google Scholar]
- 23.Sarma K, Levasseur P, Aristarkhov A, Lee JT. 2010. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl Acad. Sci. USA 107, 22 196–22 201. ( 10.1073/pnas.1009785107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beletskii A, Hong YK, Pehrson J, Egholm M, Strauss WM. 2001. PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc. Natl Acad. Sci. USA 98, 9215–9220. ( 10.1073/pnas.161173098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon Y, Lee JT. 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133. ( 10.1016/j.cell.2011.06.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada N, Hasegawa Y, Yue M, Hamada T, Nakagawa S, Ogawa Y.. 2015. Xist Exon 7 contributes to the stable localization of Xist RNA on the inactive X-chromosome. PLoS Genet. 11, e1005430 ( 10.1371/journal.pgen.1005430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lv Q, Yuan L, Song Y, Sui T, Li Z, Lai L. 2016. D-repeat in the XIST gene is required for X chromosome inactivation. RNA Biol. 13, 172–176. ( 10.1080/15476286.2015.1137420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237. ( 10.1101/gad.380906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A.. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2, E171 ( 10.1371/journal.pbio.0020171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pullirsch D, Hartel R, Kishimoto H, Leeb M, Steiner G, Wutz A. 2010. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development 137, 935–943. ( 10.1242/dev.035956) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. 2006. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol. Cell. Biol. 26, 7167–7177. ( 10.1128/MCB.00810-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agrelo R, et al. 2009. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev. Cell 16, 507–516. ( 10.1016/j.devcel.2009.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nechanitzky R, Davila A, Savarese F, Fietze S, Grosschedl R. 2012. Satb1 and Satb2 are dispensable for X chromosome inactivation in mice. Dev. Cell 23, 866–871. ( 10.1016/j.devcel.2012.09.018) [DOI] [PubMed] [Google Scholar]
- 34.Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. 2002. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl Acad. Sci. USA 99, 8677–8682. ( 10.1073/pnas.132468999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ. 2007. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc. Natl Acad. Sci. USA 104, 10 104–10 109. ( 10.1073/pnas.0610946104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blewitt ME, Vickaryous NK, Hemley SJ, Ashe A, Bruxner TJ, Preis JI, Arkell R, Whitelaw E. 2005. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl Acad. Sci. USA 102, 7629–7634. ( 10.1073/pnas.0409375102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blewitt ME, et al. 2008. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40, 663–669. ( 10.1038/ng.142) [DOI] [PubMed] [Google Scholar]
- 38.Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572. ( 10.1016/j.celrep.2015.06.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leeb M, Wutz A. 2011. Derivation of haploid embryonic stem cells from mouse embryos. Nature 479, 131–134. ( 10.1038/nature10448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561. ( 10.1016/j.celrep.2015.06.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guenther MG, Barak O, Lazar MA. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21, 6091–6101. ( 10.1128/MCB.21.18.6091-6101.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHugh CA, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. ( 10.1038/nature14443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroda K, et al. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18, 301–312. ( 10.1016/S1074-7613(03)00029-3) [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15, 1140–1151. ( 10.1101/gad.871201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. ( 10.1016/j.cell.2015.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minajigi, et al. 2015. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 ( 10.1126/science.aab2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Z, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279. ( 10.1016/j.cell.2016.04.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen CK, et al. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. ( 10.1126/science.aae0047) [DOI] [PubMed] [Google Scholar]
- 49.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107, 727–738. ( 10.1016/S0092-8674(01)00598-0) [DOI] [PubMed] [Google Scholar]
- 50.Jeppesen P, Turner BM. 1993. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74, 281–289. ( 10.1016/0092-8674(93)90419-Q) [DOI] [PubMed] [Google Scholar]
- 51.Pehrson JR, Changolkar LN, Costanzi C, Leu NA. 2014. Mice without macroH2A histone variants. Mol. Cell. Biol. 34, 4523–4533. ( 10.1128/MCB.00794-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanasijevic B, Rasmussen TP.. 2011. X chromosome inactivation and differentiation occur readily in ES cells doubly-deficient for macroH2A1 and macroH2A2. PLoS ONE 6, e21512 ( 10.1371/journal.pone.0021512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28, 371–375. ( 10.1038/ng574) [DOI] [PubMed] [Google Scholar]
- 54.Silva J, et al. 2003. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 Polycomb group complexes. Dev. Cell 4, 481–495. ( 10.1016/S1534-5807(03)00068-6) [DOI] [PubMed] [Google Scholar]
- 55.Plath K, et al. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135. ( 10.1126/science.1084274) [DOI] [PubMed] [Google Scholar]
- 56.Cooper S, et al. 2016. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 ( 10.1038/ncomms13661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. 2004. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J. Cell Biol. 167, 1025–1035. ( 10.1083/jcb.200409026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leeb M, Wutz A. 2007. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 178, 219–229. ( 10.1083/jcb.200612127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122. ( 10.1038/sj.emboj.7601187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalantry S, Magnuson T.. 2006. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2, e66 ( 10.1371/journal.pgen.0020066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helbig R, Fackelmayer FO. 2003. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma 112, 173–182. ( 10.1007/s00412-003-0258-0) [DOI] [PubMed] [Google Scholar]
- 62.Roshon MJ, Ruley HE. 2005. Hypomorphic mutation in hnRNP U results in post-implantation lethality. Transgenic Res. 14, 179–192. ( 10.1007/s11248-004-8147-8) [DOI] [PubMed] [Google Scholar]
- 63.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19, 469–476. ( 10.1016/j.devcel.2010.08.006) [DOI] [PubMed] [Google Scholar]
- 64.Sakaguchi T, Hasegawa Y, Brockdorff N, Tsutsui K, Tsutsui KM, Sado T, Nakagawa S. 2016. Control of chromosomal localization of Xist by hnRNP U family molecules. Dev. Cell 39, 11–12. ( 10.1016/j.devcel.2016.09.022) [DOI] [PubMed] [Google Scholar]
- 65.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. ( 10.1038/nature19342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhatnagar S, Zhu X, Ou J, Lin L, Chamberlain L, Zhu LJ, Wajapeyee N, Green MR. 2014. Genetic and pharmacological reactivation of the mammalian inactive X chromosome. Proc. Natl Acad. Sci. USA 111, 12 591–12 598. ( 10.1073/pnas.1413620111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan KM, Zhang H, Malureanu L, van Deursen J, Zhang Z. 2011. Diverse factors are involved in maintaining X chromosome inactivation. Proc. Natl Acad. Sci. USA 108, 16 699–16 704. ( 10.1073/pnas.1107616108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raffel GD, Mercher T, Shigematsu H, Williams IR, Cullen DE, Akashi K, Bernard OA, Gilliland DG. 2007. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc. Natl Acad. Sci. USA 104, 6001–6006. ( 10.1073/pnas.0609041104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horiuchi K, et al. 2006. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl Acad. Sci. USA 103, 17 278–17 283. ( 10.1073/pnas.0608357103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blomen VA, et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. ( 10.1126/science.aac7557) [DOI] [PubMed] [Google Scholar]
- 71.Grant J, et al. 2012. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254–258. ( 10.1038/nature11171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vallot C, Huret C, Lesecque Y, Resch A, Oudrhiri N, Bennaceur-Griscelli A, Duret L, Rougeulle C. 2013. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat. Genet. 45, 239–241. ( 10.1038/ng.2530) [DOI] [PubMed] [Google Scholar]
- 73.Amrein H, Axel R. 1997. Genes expressed in neurons of adult male Drosophila. Cell 88, 459–469. ( 10.1016/S0092-8674(00)81886-3) [DOI] [PubMed] [Google Scholar]
- 74.Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88, 445–457. ( 10.1016/S0092-8674(00)81885-1) [DOI] [PubMed] [Google Scholar]