Abstract
Squamate reptiles (lizards and snakes) are the most diverse group of terrestrial vertebrates, with more than 10 000 species. Despite considerable effort to resolve relationships among major squamates clades, some branches have remained difficult. Among the most vexing has been the placement of snakes among lizard families, with most studies yielding only weak support for the position of snakes. Furthermore, the placement of iguanian lizards has remained controversial. Here we used targeted sequence capture to obtain data from 4178 nuclear loci from ultraconserved elements from 32 squamate taxa (and five outgroups) including representatives of all major squamate groups. Using both concatenated and species-tree methods, we recover strong support for a sister relationship between iguanian and anguimorph lizards, with snakes strongly supported as the sister group of these two clades. These analyses strongly resolve the difficult placement of snakes within squamates and show overwhelming support for the contentious position of iguanians. More generally, we provide a strongly supported hypothesis of higher-level relationships in the most species-rich tetrapod clade using coalescent-based species-tree methods and approximately 100 times more loci than previous estimates.
Keywords: lizards, phylogenomics, snakes, species-tree
1. Introduction
Squamates, lizards and snakes, are a particularly important group. They include more than 10 000 extant species [1], surpassing birds as the most diverse major tetrapod clade. They are used as study systems in many areas of research, including (among many) the evolution of sex, viviparity, herbivory, body form and toxins [2]. Venomous squamates also pose a serious hazard to humans in some regions [3], whereas their venoms offer many resources for medical research [4].
There has been considerable recent research on higher-level squamate phylogeny, but some branches have remained contentious. Molecular analyses have supported a very different set of relationships relative to those suggested by morphology. Morphological analyses have placed iguanian lizards at the base of the squamate tree [5,6]. By contrast, molecular analyses have placed gekkotans and dibamids at the base, and iguanians in a more recent clade with snakes and anguimorphs [7–11]. Combined analyses of molecular and morphological data have also supported the molecular trees [12,13], and have shown that the support for the morphological tree is problematic. Nevertheless, some authors have continued to question the placement of iguanians by molecular data [6,14,15].
Further, some key aspects of higher-level squamate phylogeny have remained uncertain, even in molecular analyses. One is the placement of snakes. Many molecular analyses have placed snakes as the sister to a clade uniting iguanians and anguimorphs, but the support for the latter clade has been consistently weak [8–11]. Furthermore, previous studies generally used concatenated analyses, rather than species-tree analyses. Concatenated analyses may yield weak or even positively misleading results when there is extensive conflict among gene trees, whereas species-tree methods can yield well-supported and accurate results [16]. Such conflicts among gene trees are well documented for many major branches of squamate phylogeny [9]. Another key area of uncertainty is whether the sister group to all other squamates is dibamids [7,10,11] or a clade consisting of dibamids and gekkotans [9,12,13].
Here, we analyse higher-level squamate relationships using phylogenomic data and species-tree methods. We obtain nuclear data from 4178 loci, approximately 100 times more than previous analyses of higher-level squamate phylogeny [9,11,13]. We strongly support snakes as the sister group of a well-supported clade uniting Iguania + Anguimorpha. Thus, our analyses simultaneously resolve these two persistent controversies in squamate phylogeny. Overall, we provide a generally well-supported estimate of higher-level squamate phylogeny using phylogenomic data and species-tree methods.
2. Material and methods
We sampled 32 squamate species and five outgroup taxa, which included representatives of all major amniote clades (i.e. rhynchocephalians, crocodilians, birds, turtles and mammals). Sampling within squamates included all major groups and most families (outside of snakes, iguanians and amphisbaenians). Details of sampling are given in the electronic supplementary material, table S1.
We used anchored enrichment to sequence ultraconserved elements (UCEs, [17]) and obtained an average of 2738.7 UCEs per species. Laboratory methods (following [18,19]) are listed in the electronic supplementary material, appendix S1. Data for most outgroups (all but Sphenodon) and nine ingroup taxa were from previous studies (electronic supplementary material, table S1). We obtained new UCE data for 24 taxa. In theory, we could have included additional published data for iguanian lizards [18] and snakes [19]. However to make the analyses more computationally tractable, we included only three species from each clade, representing major lineages (i.e. scolecophidian, pythonid, and colubroid snakes; pleurodont and acrodont iguanians). We included the most complete taxon available within each major lineage.
We cleaned, assembled, identified and aligned UCE data using the software packages velvet 1.2.10 [20] and Phyluce 2.0.0 [17,21]. We included UCEs with up to 50% missing taxa, which appears to give optimal results [18]. The final dataset had 4178 UCEs (1 530 015 total aligned base pairs), with 52.4% missing data overall (including gaps). The average number of variable sites per UCE was 108.3 (range 17–455) and mean UCE length was 380.0 base pairs (range 108–878). Raw FASTQ files for new samples were uploaded to the NCBI sequence read archive (electronic supplementary material, table S1). Alignments and gene trees for all analyses are available via the Dryad Digital Repository [22].
Phylogenetic analyses were conducted using two approaches: (i) concatenated maximum likelihood, and (ii) species-tree analysis using gene trees as input (i.e. one best likelihood tree per UCE). To perform the concatenated analysis and infer gene trees, we used RAxML 8.2.9 [23] with default settings and the GTRCAT model applied to the entire alignment (partitions are not generally appropriate for UCE data, given that most loci are not protein coding [17]). The concatenated analyses used the CIPRES server [24] and branch support was assessed with bootstrapping. Species-trees analyses used the program Astral 4.10.11 [25,26], with branch support estimated using local posterior probabilities [27].
We generally included only taxa with more than 1000 UCEs. However, we included two more incomplete taxa that were potentially important for the tree's base: the dibamid Anelytropsis (90 UCEs; electronic supplementary material, table S1) and the closest squamate outgroup (Sphenodon; 596 UCEs, electronic supplementary material, table S1). Although Anelytropsis is highly incomplete, its inclusion or exclusion had no impact on the estimated topologies (see the electronic supplementary material, figures S1 and S2).
3. Results
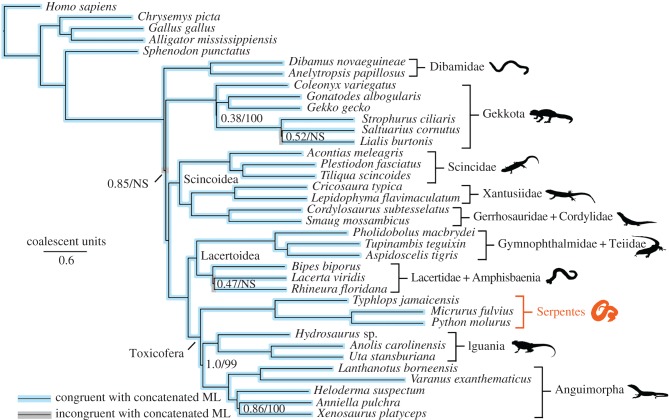
The concatenated and species-tree analyses provided strong and concordant support for most higher-level squamate relationships (figure 1; electronic supplementary material, figure S3). There was strong support for a clade uniting anguimorphs, iguanians, and snakes (Toxicofera) and strong support for a clade linking anguimorphs and iguanians. Thus, these results strongly resolve the controversial placement of snakes and iguanians with species-tree methods and a dataset of unprecedented size. There was strong, concordant support for most other relationships across the tree, such as the clade uniting scincids, xantusiids, gerrhosaurids and cordylids (Scincoidea), and the clade uniting gymnophthalmids, teiids, lacertids, and amphisbaenians (Lacertoidea). The backbone relationships among these major clades were also strongly supported.
Figure 1.
Phylogeny of squamate reptiles inferred from a species-tree analysis (Astral) of 4178 nuclear loci and 1 530 015 aligned base pairs. Tree is rooted with Homo. Light blue highlighting indicates clades congruent with the concatenated maximum-likelihood analysis (see the electronic supplementary material, figure S3). Grey highlighting indicates incongruent clades. Support values are: local posterior probabilities (species tree)/bootstrap support (concatenated). NS indicates ‘no support’ from the concatenated analysis (incongruent branch). When no numbers are shown, support values are equal to 1.0/100 (maximum support from both methods).
There were also a few remaining areas of uncertainty. Both approaches agreed that dibamids and gekkotans are at the base of the squamate tree, but differ as to whether dibamids are sister to all other squamates (Astral), or whether dibamids and gekkotans are sister taxa (concatenated). The conflicting relationships were moderately supported by each approach. There were also conflicts (and weak support from Astral) among some gekkotan families. Finally, there were conflicting results and weak support regarding monophyly of amphisbaenians relative to lacertids.
4. Discussion
In this study, we assembled a large phylogenomic dataset to address higher-level squamate phylogeny, using species-tree methods and sampling approximately 100 times more loci than previous studies. Our results strongly resolve the phylogenetic placement of snakes among lizard families, as the sister group to anguimorphs and iguanians. Concomitantly, they provide overwhelming evidence to conclusively resolve the contentious placement of iguanians. We acknowledge that some readers might consider the placement of snakes and iguanians as already known, but previous analyses had only weak support and generally used concatenated analyses [7–13]. Some authors have also argued for a snake + anguimorph clade instead [28]. More generally, we provide strong support for most higher-level relationships across squamates. We note that Squamata may be among the first major groups of vertebrates to have their higher-level relationships largely resolved by species-tree analyses of thousands of loci (along with birds [29]).
Nevertheless, we acknowledge three areas of remaining uncertainty in our tree. Two involve lower-level relationships, and can probably be resolved with increased taxon sampling (e.g. relationships among gekkotan families and among lacertids and amphisbaenians). Thus, adding more taxa within these groups should help subdivide long branches associated with these problematic lower-level relationships. The third area (gekkotans, dibamids, other squamates) is more difficult. It is unclear if adding more gekkotans or dibamids can significantly shorten the relevant branches. Resolving these relationships might require major methodological advances, or even more loci. Intriguingly, we note that the relationships for these taxa from Astral are consistent with concatenated analyses including thousands of taxa [10,11]. The results from the concatenated analysis (dibamids + gekkotans) are consistent with concatenated analyses of fewer taxa and loci [9,13].
In summary, our results here provide strong support for most higher-level squamate relationships. We resolve two contentious aspects of squamate phylogeny using approximately 100 times more loci than in previous studies, and with species-tree methods. Although some areas of uncertainty remain, squamates may now be among the most phylogenetically well-resolved of the major tetrapod clades.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the many individuals and institutions that provided tissues used here (listed in the electronic supplementary material, table S1), but especially T. Reeder.
Data accessibility
All data are accessible via Dryad (http://dx.doi.org/10.5061/dryad.66gt5) [22].
Authors' contributions
J.W.S. and J.J.W. designed the study and wrote the paper, J.W.S. collected data and performed analyses, all authors revised the paper, approved the final version to be published, and agree to be held accountable for the content therein.
Competing interests
The authors declare no competing interests.
Funding
Funding was provided by the University of Arizona.
References
- 1.Uetz P, Freed P, Hošek J (eds) 2017. The Reptile Database. See http://www.reptile-database.org (accessed 28 May 2017). [Google Scholar]
- 2.Sites JW Jr, Reeder TW, Wiens JJ. 2011. Phylogenetic insights on evolutionary novelties in lizards and snakes: sex, birth, bodies, niches, and venom. Annu. Rev. Ecol. Evol. Syst. 42, 227–244. ( 10.1146/annurev-ecolsys-102710-145051) [DOI] [Google Scholar]
- 3.Kasturiratne A, et al. 2008. Estimation of the global burden of snakebite. PLoS Med. 5, e218 ( 10.1371/journal.pmed.0050218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JW, Serrano SMT. 2007. Approaching the golden age of natural product pharmaceuticals from venom libraries: an overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr. Pharm. Des. 13, 2927–2934. ( 10.2174/138161207782023739) [DOI] [PubMed] [Google Scholar]
- 5.Conrad JL. 2008. Phylogeny and systematics of Squamata (Reptilia) based on morphology. Bull. Am. Mus. Nat. Hist. 310, 1–182. ( 10.1206/310.1) [DOI] [Google Scholar]
- 6.Gauthier JA, Kearney M, Maisano JA, Rieppel O, Behike ADB. 2012. Assembling the squamate tree of life: perspectives from the phenotype and the fossil record. Bull. Peabody Mus. Nat. Hist. 53, 3–308. ( 10.3374/014.053.0101) [DOI] [Google Scholar]
- 7.Townsend T, Larson A, Louis EJ, Macey JR. 2004. Molecular phylogenetics of Squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 53, 735–757. ( 10.1080/10635150490522340) [DOI] [PubMed] [Google Scholar]
- 8.Vidal N, Hedges SB. 2005. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C. R. Biol. 328, 1000–1008. ( 10.1016/j.crvi.2005.10.001) [DOI] [PubMed] [Google Scholar]
- 9.Wiens JJ, Hutter CR, Mulcahy DG, Noonan BP, Townsend TM, Sites JW Jr, Reeder TW. 2012. Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol. Lett. 8, 1043–1046. ( 10.1098/rsbl.2012.0703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13, 93 ( 10.1186/1471-2148-13-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Wiens JJ. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4,162 species. Mol. Phylogenet. Evol. 94, 537–547. ( 10.1016/j.ympev.2015.10.009) [DOI] [PubMed] [Google Scholar]
- 12.Wiens JJ, Kuczynski CA, Townsend T, Reeder TW, Mulcahy DG, Sites JW Jr. 2010. Combining phylogenomics and fossils in higher-level squamate reptile phylogeny: molecular data change the placement of fossil taxa. Syst. Biol. 59, 674–688. ( 10.1093/sysbio/syq048) [DOI] [PubMed] [Google Scholar]
- 13.Reeder TW, Townsend TM, Mulcahy DG, Noonan BP, Wood PL, Sites JW Jr, Wiens JJ. 2015. Integrated analyses resolve conflicts over squamate reptile phylogeny and reveal unexpected placements for fossil taxa. PLoS ONE 10, e0118199 ( 10.1371/journal.pone.0118199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Losos JB, Hillis DM, Greene HW. 2012. Who speaks with a forked tongue? Science 338, 1428–1429. ( 10.1126/science.1232455) [DOI] [PubMed] [Google Scholar]
- 15.McMahan CD, Freeborn LR, Wheeler WC, Crother BI. 2015. Forked tongues revisited: molecular apomorphies support morphological hypotheses of squamate evolution. Copeia 103, 525–529. ( 10.1643/CH-14-015) [DOI] [Google Scholar]
- 16.Edwards SV. 2009. Is a new and general theory of molecular systematics emerging? Evolution 63, 1–19. ( 10.1111/j.1558-5646.2008.00549.x) [DOI] [PubMed] [Google Scholar]
- 17.Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. 2012. Ultraconserved elements anchor thousands of genetic markers for target enrichment spanning multiple evolutionary timescales. Syst. Biol. 61, 717–726. ( 10.1093/sysbio/sys004) [DOI] [PubMed] [Google Scholar]
- 18.Streicher JW, Schulte JA II, Wiens JJ. 2016. How should genes and taxa be sampled for phylogenomic analyses with missing data? An empirical study in iguanian lizards. Syst. Biol. 65, 128–145. ( 10.1093/sysbio/syv058) [DOI] [PubMed] [Google Scholar]
- 19.Streicher JW, Wiens JJ. 2016. Phylogenomic analyses reveal novel relationships among snake families. Mol. Phylogenet. Evol. 100, 160–169. ( 10.1016/j.ympev.2016.04.015) [DOI] [PubMed] [Google Scholar]
- 20.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. ( 10.1101/gr.074492.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faircloth BC. 2016. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788. ( 10.1093/bioinformatics/btv646) [DOI] [PubMed] [Google Scholar]
- 22.Streicher JW, Wiens JJ. 2017. Data from: Phylogenomic analyses of more than 4000 nuclear loci resolve the origin of snakes among lizard families Dryad Digital Repository. ( 10.5061/dryad.66gt5) [DOI] [PMC free article] [PubMed]
- 23.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees In Proc. Gateway Comp. Environ. Workshop, 14 November 2010, New Orleans, USA, pp. 1–8. [Google Scholar]
- 25.Mirarab S, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548. ( 10.1093/bioinformatics/btu462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species-tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31, i44–i52. ( 10.1093/bioinformatics/btv234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayyari E, Mirarab S. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 33, 1654–1668. ( 10.1093/molbev/msw079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MSY. 2009. Hidden support from unpromising datasets strongly unites snakes and anguimorph lizards. J. Evol. Biol. 22, 1308–1316. ( 10.1111/j.1420-9101.2009.01751.x) [DOI] [PubMed] [Google Scholar]
- 29.Jarvis ED, et al. 2014. Whole genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1321–1331. ( 10.1126/science.1253451) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Streicher JW, Wiens JJ. 2017. Data from: Phylogenomic analyses of more than 4000 nuclear loci resolve the origin of snakes among lizard families Dryad Digital Repository. ( 10.5061/dryad.66gt5) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data are accessible via Dryad (http://dx.doi.org/10.5061/dryad.66gt5) [22].