Abstract
Resistance to medical triazoles in Aspergillus fumigatus is an emerging problem for patients at risk of aspergillus diseases. There are currently two presumed routes for medical triazole-resistance selection: (i) through selection pressure of medical triazoles when treating patients and (ii) through selection pressure from non-medical sterol-biosynthesis-inhibiting (SI) triazole fungicides which are used in the environment. Previous studies have suggested that SI fungicides can induce cross-resistance to medical triazoles. Therefore, to assess the potential of selection of resistance to medical triazoles in the environment, we assessed cross-resistance to three medical triazoles in lineages of A. fumigatus from previous work where we applied an experimental evolution approach with one of five different SI fungicides to select for resistance. In our evolved lines we found widespread cross-resistance indicating that resistance to medical triazoles rapidly arises through selection pressure of SI fungicides. All evolved lineages showed similar evolutionary dynamics to SI fungicides and medical triazoles, which suggests that the mutations inducing resistance to both SI fungicides and medical triazoles are likely to be the same. Whole-genome sequencing revealed that a variety of mutations were putatively involved in the resistance mechanism, some of which are in known target genes.
Keywords: Aspergillus fumigatus, experimental evolution, sterol-biosynthesis-inhibiting (SI) fungicides and medical triazoles, triazole resistance, evolutionary trajectory
1. Introduction
Triazole resistance of the saprophytic mould Aspergillus fumigatus is an emerging health problem. This fungus is an important cause of invasive fungal infections in immunocompromised patients, and has spread globally in recent decades [1–8]. Triazoles are part of the class of antifungal drugs used to treat human infections, of which itraconazole (ITR), posaconazole (POS) and voriconazole (VOR) are the most widely used [9]. These triazoles inhibit the enzyme sterol 14α-demethylase, which is encoded by the cyp51A gene, thereby blocking its function in the ergosterol biosynthesis pathway. This results in ergosterol depletion and accumulation of toxic sterols [10]. The most common resistance mechanism involves alterations in the cyp51A gene [4,11–15]. In the clinic, widespread resistance has emerged against ITR, and more recently to VOR, and numerous resistant strains have been isolated from patients.
Interestingly, highly resistant A. fumigatus is also found in soil, flowerbeds and other non-medical environments [16]. In fact, several sterol-synthesis-inhibiting (SI) fungicides are widely used in the environment to control various fungal plant pathogens, such as Botrytis and Fusarium, and as preservatives of materials such as wooden fences. These SI fungicides are chemically similar to medical triazoles, target various parts of the same metabolic pathway for ergosterol biosynthesis and have different active doses. Triazole derivatives are unique in that they are the only class of antifungals that are used both in the environment and in medicine [5,17,18]. The volume of SI fungicides sold almost doubled between 1995 and 2007 to 130 000 kg per year in the Netherlands alone, against a yearly use of around 400 kg of medical triazoles [19]. Resistant A. fumigatus isolates from patients and the environment share the same mutations in the cyp51A gene cluster that underpin the resistance mechanism [20]. This suggests that there are potentially two routes of selection for resistance in A. fumigatus strains found in patients [21]. The first route is resistance arising in patients with chronic aspergillus disease, such as in those with chronic obstructive pulmonary disease, who are under prolonged treatment with medical triazoles and have cavitary aspergillus disease [22]. The second route to resistance is through exposure of A. fumigatus to SI fungicides used in the environment, which could give rise to cross-resistance to medical triazoles. On the basis of the shared mechanism for resistance, several studies have argued that SI fungicides used in the environment can select for cross-resistance to medical triazoles [5,18,23,24]. For instance, Snelders et al. [5] discussed the highly resistant strain (isolated from a medical environment) carrying the mutations TR34/L98H that showed resistance both to SI fungicides and to medical triazoles, which target the same active sites the CYP51 enzyme enzymes of the ergosterol pathway of A. fumigatus.
However, to date there is no direct evidence that exposure of A. fumigatus to SI fungicides can lead to cross-resistance to medical triazoles. In the present study, we assessed the levels of cross-resistance to three medical triazoles of A. fumigatus strains that had evolved in the presence of one of five SI fungicides over seven weeks [5,25]. The first medical triazoles were introduced in 1997 and already by 1998 resistance to these new drugs was observed in clinical isolates. Interestingly, these resistant clinical isolates were also resistant to five SI fungicides with similar chemical structure in use for environmental applications [5]. Furthermore, triazole-resistant strains have been isolated from patients who had never been exposed to triazoles. From this, it has been suggested that exposure of A. fumigatus to SI fungicides had selected for cross-resistance to triazoles. In this study, we set out to answer the question whether exposure to SI fungicides can select for cross-resistance. To do this, we tested strains from previous work [25] that had evolved resistance in the presence of SI fungicides for cross-resistance to three medical triazoles and linked this to population dynamics and to observed mutations at the genome level. This information is essential for understanding the potential routes of development of resistance to medical triazoles and the associated mechanisms of genetic changes under the selection pressure of SI fungicides used for non-medical applications.
2. Methods summary
Below is a summary of the main aspects of our methods. Full details of the materials and methods are provided as part of the electronic supplementary material.
(a). Fungal isolates, medical triazoles and SI fungicides
All evolved fungal cultures used in this study were from a previous study that focused on the effect of variation in fungal reproduction on adaptation: in that study, the development of resistance to SI fungicides [25]. The five SI fungicides used for the selection were bromuconazole, tebuconazole, epoxiconazole, difenoconazole and propiconazole at a concentration of 1 mg l−1. In the present study we used the medical triazoles itraconazole (ITR), posaconazole (POS) and voriconazole (VOR) in various concentrations to determine minimal inhibitory concentration (MIC) values for every SI fungicide evolved lineage. A Kruskal–Wallis test was performed to test differences in resistance level (MIC) between the lineages evolved in the presence of different SI fungicides after seven weeks as groups and a second set of tests was performed to test for all pairwise differences for MIC values of all three medical azoles. The pairwise correlations of mean MIC values on the three medical azoles of the five SI fungicide and control lineages that had evolved in the absence of any fungicides were tested using Spearman's rank correlation test (α = 0.05/3 (for three pairwise correlations)).
(b). Susceptibility testing of evolved cultures to the medical triazoles
We assayed the level of resistance of all lineages that had evolved in the presence (or absence) of one of five different SI fungicides over seven weeks against the three medical triazoles, following the EUCAST reference method [26]. Briefly, resistance level was defined as the MIC that results in 100% growth inhibition of fungi. The relative MIC was defined as the individuals' MIC divided by the MIC of the triazole sensitive ancestor.
(c). Population dynamics in the evolutionary lineages over time
We selected three lineages from the difenoconazole evolution treatment that showed the highest resistance increase (lineages D1, D3 and D6) for a more in-depth study of the evolutionary dynamics focusing on morphological changes of the fungal colonies. Firstly, we propagated these lineages for two additional rounds of selection to verify whether the relative abundance of the different types would further change. Secondly, about 100 spores from samples from different time points of each D1, D3 and D6 lineage were spread onto MEA (malt extract agar) plates with 1 mg l−1 of difenoconazole and representative morphotypes were isolated.
(d). Whole-genome sequencing
We performed whole-genome sequencing of each morphological type found in lineages D1, D3 and D6 (described above). In addition, we sequenced three lineages that had evolved in the absence of SI fungicides (C2, C4 and C5). For sequencing, we picked three colonies of the same morphotype and performed a combined DNA extraction protocol as described previously [14]. Illumina HiSeq2500 125 bp paired-end reads were generated by BGI in China (electronic supplementary material, table S4a). After quality assessment [27] and subsequent quality trimming (trim-fastq.pl, -q 30, -m 70, [28]), reads were aligned to a reference genome of A. fumigatus [29] using BWA-mem (v. 0.7.10, default settings [30]). Raw BAM files were used for structural variant analysis (BreakDancer [31], SVDetect [32] cn_mops [33]) but no structural variants between ancestral and evolved lines were found, although these were found between reference genome and ancestor. Subsequently, aligned reads were filtered for quality (MAPQ > 50) and reads were only kept if both reads in pairs were aligned. Duplicates were removed and realignment was performed [34] (http://broadinstitute.github.io/picard). SNP calling was performed using SAMtools mpileup, bcftools and vcfutils.pl (v. 1.31 [35]). Resulting single-nucleotide polymorphisms (SNPs) and indels were filtered for being variable between the ancestor and any of the evolved lines (with minimum coverage 25× and minimum pairwise FST of 0.05, [36, page 308]). After this, all variants were verified in IGV [37]. For a total list of SNPs, indels and their characteristics see electronic supplementary material, table S4b.
(e). Tracing-back mutations from the different time points of the evolutionary experiment
After whole-genome sequencing analysis, 10 potential candidate genes were found from different evolved phenotypes (electronic supplementary material, table S4c). To investigate the dynamics of the mutations in the experimental evolution experiment over time, colonies of each phenotype from different time points in weeks 2, 4, and 6 in the lineages D1 and D6 were used. Dynamics and presence of three mutations were determined using PCR followed by Sanger sequencing (genes and primers for specific genes are listed in electronic supplementary material, table S5b) and, subsequently, the frequency of colonies harbouring specific fixed mutations was determined.
3. Results
(a). Susceptibility testing of evolved cultures against medical triazoles
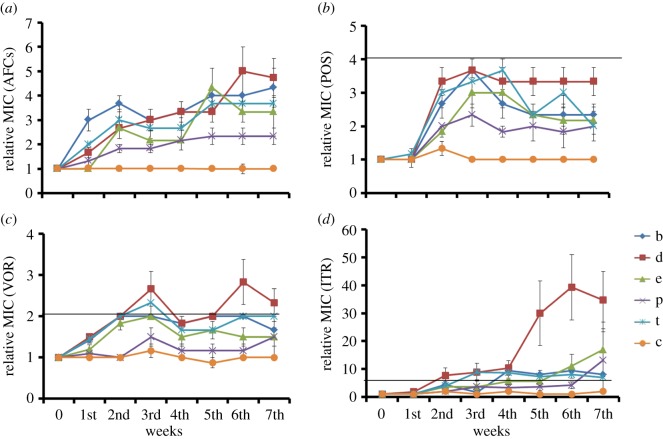
We assayed the susceptibility to three medical triazoles (ITR, VOR, POS) of replicate lineages that had evolved under laboratory conditions over seven weeks in the presence (or absence) of one of five SI fungicides. Figure 1 shows the evolutionary dynamics of cross-resistance to medical triazoles (defined as the MIC of a given evolved culture relative to the MIC of the ancestor) averaged for the six replicate lineages that had evolved under the same conditions. For completeness, figure 1a includes data of resistance to the SI fungicide used for selection, showing the development of resistance over time (taken from [25]) and electronic supplementary material, figure S1, shows the median rather than the mean MIC data. The resistance (relative MIC values) against the three medical triazoles increased by 2–3.3-fold for POS, 3–4.6-fold for VOR, and strikingly 4.4–35-fold for ITR at the end of seven weeks of experimental evolution. This cross-resistance is of the same order of magnitude or higher than the resistance increase to SI fungicides that was used as the selective agent during the seven week evolutionary experiment [25]. Since our MIC data violated the assumptions of a normal distribution required for an ANOVA, we proceeded with a non-parametric Kruskal–Wallis test to quantify differences between evolved cultures. For all three medical azoles MIC values were significantly different between the groups at week seven (ITR: χ2 = 16.865, d.f. = 5, p < 0.005; POS: χ2 = 19.915, d.f. = 5, p < 0.005; VOR: χ2 = 17.733, d.f. = 5, p < 0.005). We then tested all pairwise differences (electronic supplementary material, table S1a) which indicated differences between all SI fungicide evolved lineages combined and the no-fungicide control treatment in MIC values on ITR (see electronic supplementary material, table S1b, for all values at week 7). SI fungicides difenoconazole and tebuconazole led to increased MIC values on VOR and POS, while only bromuconazole was significantly different from control on POS (p < 0.003, see electronic supplementary material, table S1b). Lastly, the mean MIC values correlated significantly when pairwise compared between the three medical azoles, indicating that evolution on environmental azole similarly increases the degree of cross-resistance to different medical azoles.
Figure 1.
Evolution of cross-resistance. Each line shows the average of six replicate lineages that had evolved in the presence (or absence) of one of five SI fungicides over seven weekly transfers (error bars indicate the standard error of the mean, s.e.m.). Resistance is given by the relative MIC value of an evolved strain compared with the ancestor. (a) Resistance of evolved lineages against the SI fungicide used for the selection (AFC); each series of six lineages assayed for resistance against their respective SI fungicide used for selection (data from [36]); b = bromuconazole, t = tebuconazole, e = epoxiconazole, d = difenoconazole, p = propiconazole, c = no SI fungicide present. (b) Cross-resistance of evolved lineages against the medical triazole POS. (c) Cross-resistance against the medical triazole VOR. (d) Cross-resistance against the medical triazole ITR. In panels (b–d) the horizontal solid line shows the boundary above which strains are considered resistant by the clinical breakpoint [38]. (Online version in colour.)
According to the medical-resistance breakpoints of different triazoles for A. fumigatus using the proposed EUCAST susceptibility testing methodology, a strain with either a POS MIC of >0.25 mg l−1, a VOR MIC and ITR MIC of >2 mg l−1 is considered resistant to that drug [38] (solid lines in figure 1b–d). While all lineages increased their resistance to medical triazoles over seven weeks, not all were considered resistant in a clinical sense. Out of 253 cultures, we obtained a total of 97 that were clinically resistant to ITR, eight to VOR and no resistant cultures to POS. This is in line with the results described above that suggest that difenoconazole imposes the strongest induction of cross-resistance to medical triazoles (figure 1). These results further suggest that POS is the most active medical triazole with the least cross-resistance with SI fungicides, and ITR is the least effective. Notably, this matches with the time that has passed since these medical triazoles were clinically licensed: ITR in 1997, VOR in 2002 and POS in 2006 [39].
(b). Evolutionary patterns of evolved cultures on SI fungicides and correlated responses for medical triazoles
Under the selection pressure of five SI fungicides, the resistance level of each lineage to different triazoles increased over the seven-week selection period probably through the successive fixation of mutations. This was reflected in a step-wise increase in relative MIC and fitness over time, as is typically observed in laboratory experimental evolution [40,41]. We measured relative MICs against the SI fungicide used for selection and used visual inspection to estimate the time points at which an upward step in relative MIC occurred. We then used these evolutionary trajectories to ask whether a fixation of a mutation to SI fungicide led to a simultaneous increase in cross-resistance to the three medical triazoles. Linear regression analysis showed that lineages on average exhibited the same stepwise increase in relative MIC to the SI fungicide used for experimental evolution and cross-resistance to the three medical triazoles (statistical analyses using correlation coefficients between the timing of relative MIC increase of SI fungicides and the medical triazoles are shown as electronic supplementary material, table S2). While we do not have the statistical power for a full analysis involving fitting of step functions to evolutionary trajectories, the apparent parallel increases in relative MICs supported the notion that the genetic changes that underlie the increased resistance to SI fungicides also led to cross-resistance to medical triazoles.
(c). The presence and dynamics of different morphotypes within lineages that had evolved resistance to difenoconazole
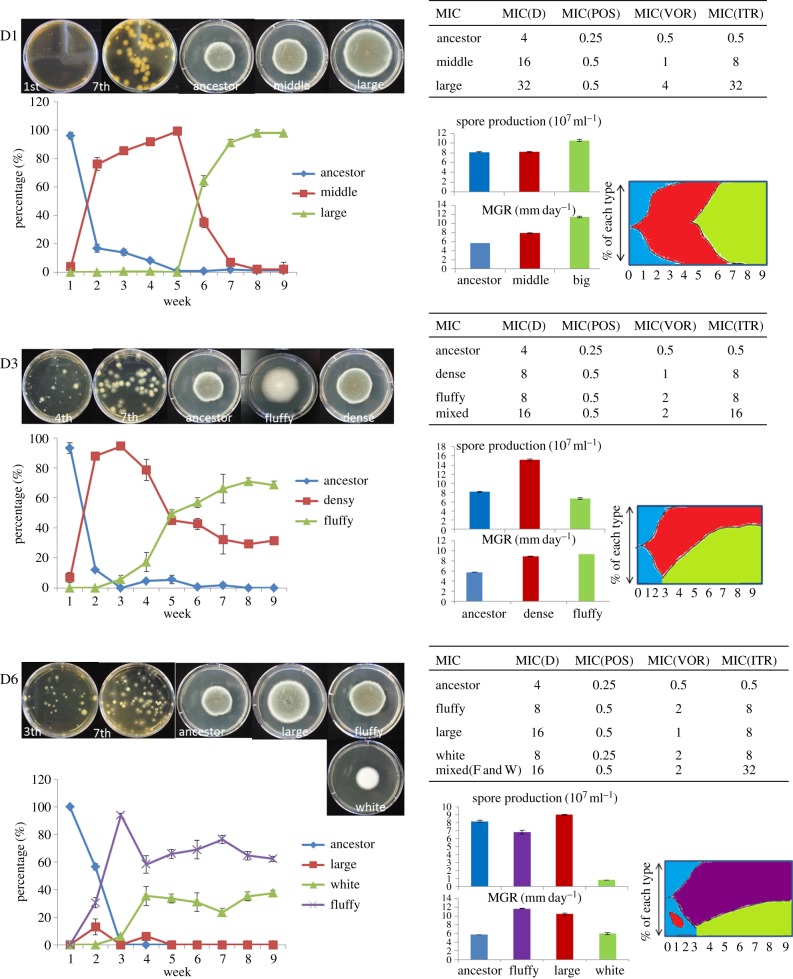
Lineages D1, D3 and D6 (three lineages that evolved in the presence of difenoconazole), which reached high (cross) resistance after seven weeks of experimental evolution, were selected for whole-genome sequencing (see next section). To decide whether to sequence the entire (mixed) population or a single clone from each population, we assayed the populations for multiple morphotypes by diluting and plating the lineage cultures from different time points. We found that the lineages consisted of a mixed population of different morphological types. This sparked a further investigation into the dynamics of the different morphotypes during the evolution experiment and the cross-resistance to medical triazoles of each morphotype in isolation as well combinations of observed morphotypes.
In lineage D1, three morphological types appeared: the ancestor, and middle and large sized colonies (figure 2). The ancestral morphotype declined from the first week and was completely replaced by the middle-sized morphotype by the fifth week that had initially emerged in the second week. This type declined sharply at the sixth week and disappeared by the seventh week, when it was completely replaced by the large-sized colony that had emerged in the fifth week. The middle-sized colony appeared to exhibit intermediate levels of mycelium growth rate (MGR) and spore production compared with the ancestor and large-sized phenotype. The shift in morphotypes suggested that the middle-sized colony carried at least one beneficial mutation compared with the ancestral type, and that the large type carried at least two mutations compared with the ancestor. This idea was supported by the outcome of a sexual cross between D1-large and the ancestor, showing segregation of at least two loci among the progeny (electronic supplementary material, figure S2). This was in line with an increased resistance to both difenoconazole and the medical triazoles (MIC susceptibility test) and increased MGR compared with the ancestral type (ANOVA: F2,6 = 1463.769, p < 0.01, post hoc LSD test, Pancestor-middle-large < 0.01, electronic supplementary material, table S3a), and increased spore production (ANOVA: F2,6 = 69.095, p < 0.01, post hoc LSD test, Pancestor-large < 0.01, Pancestor-middle = 0.891; figure 2, electronic supplementary material, table S3b).
Figure 2.
Population dynamics within evolving lineages. Pictures and data show characteristics (frequency changes, growth rates, spore production and MIC value) of different morphotypes in three difenoconazole lineages that evolved in the presence of difenoconazole. Top left shows the morphology of different types in each lineage when grown on MEA plates mixed with other types or alone. Bottom left shows the frequency dynamics of each different morphotypes over the experimental evolutionary time of nine weeks (extended two more weeks to confirm trend changes). Spore production was measured after 7 days of growth at 37°C and MGRs were measured after 4 days. Resistance level was checked following the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Error bars indicate the standard error of the mean (s.e.m.). The colour diagram is based on the frequency of each morphotype. (Online version in colour.)
In lineage D3, two morphological types (dense and fluffy) emerged during the evolutionary experiment (figure 2). The ancestral type declined sharply after the first week and was completely replaced by the dense morphotype by the third week. Between the third and fifth week the dense morphotype declined in frequency to reach 35% in the seventh week. In the third week, a fluffy morphotype emerged and reached a frequency of 70% by the seventh week. Resistance levels to difenoconazole and cross-resistance to the three medical triazoles showed that the mixed population of fluffy and dense morphotype was more resistant to ITR and difenoconazole (MIC test) than the morphotypes in isolation. The growth rates (MGR) of three types of colonies showed that the fluffy colony had a faster growth rate than the other two types (ANOVA: F2,6 = 10299, p < 0.01, post hoc LSD test, Pancestor–dense-fluffy < 0.01, electronic supplementary material, table S3a). The dense morphotype colony produced significantly more spores than the other two types (ANOVA: F2,6 = 586.204, p < 0.01, post hoc LSD test, Pancestor-dense < 0.01; Pancestor-fluffy < 0.05; figure 2, electronic supplementary material, table S3b).
Within lineage D6, we observed four morphotypes. The ancestral type sharply declined to zero, to be replaced by a fluffy morphotype by the third week. This fluffy type emerged in the second week and completely fixed by the third week, followed by a decline in the fourth week, after which it remained stable at a frequency of 60%. A large size morphotype appeared by the second week, reached a frequency of 20% before disappearing in the fifth week. The white morphotype had a sporulation delay and emerged in the third week, reaching a stable frequency of 40%. Resistance levels to the three medical triazoles were highest for the mixture of fluffy and white types, which showed a higher fitness at high concentrations of ITR and difenoconazole (MIC test). The MGR of the four types of colonies showed that the fluffy type had a faster growth rate than the other three types (ANOVA: F3,8 = 1771.202, p < 0.01, post hoc LSD test, Pancestor-large-fluffy < 0.01; Pancestor-white = 0.103, electronic supplementary material, table S3a). During culture, this white morphotype did not sporulate until the fifth day and ultimately produced 10 times fewer spores than the ancestral morphotype (ANOVA: F3,8 = 771.498, p < 0.01, post hoc LSD test, Pancestor-fluffy-white < 0.01; Pancestor-fluffy < 0.05, electronic supplementary material, table S3b).
(d). Whole-genome sequencing of lineages evolved in the difenoconazole environment
We sequenced a total of nine genomes: the ancestor, three lineages that had evolved in the absence of triazoles for seven weeks (control lineages C2, C4 and C5) and the five morphotypes observed in the lineages D1, D3 and D6 (described above and in figure 2). Descriptive statistics of the sequencing are in electronic supplementary material, table S4a.
We found mutations in all evolved lineages, which are listed in electronic supplementary material, table S4b. These SNPs were all non-synonymous and invariable in the ancestor. As described above, in the sequenced lineages, we observed that different morphotypes had appeared during the evolutionary experiment. These lineages had acquired both shared and unique genomic changes (electronic supplementary material, table S4c). D1-large carried the G138S-CYP51A mutation, a gene known for its potential to harbour resistance mutations, and had higher fitness (in terms of MGR and spore production) compared with D1-middle. Interestingly, compared with the ancestor, in the genome of D1-large a second mutation was found in HMG-CoA reductase. Surprisingly, the PCR trace-back experiment showed the P>L substitution at amino acid 320 in HMG-CoA reductase in D1-middle (electronic supplementary material, table S5a). This suggests that the D1-middle and D1-large first acquired the HMG mutation and that the increase in the frequency of D1-large was subsequently caused by the acquirement of a second mutation in the CYP51A gene. The fact that we find evidence for two segregating loci when crossing D1-large back with the ancestor further supports this (electronic supplementary material, figure S2).
Furthermore, the P>L substitution at amino acid 320 in HMG-CoA reductase was also found to be fixed in D3-dense and C5, suggesting that this mutation may be a general adaptation to the experimental evolution conditions with an associated increase in triazole resistance. However, the C5 genotype was not similar to that of D1-middle and D3-dense as a hypothetical conserved protein contained an acquired deletion causing a frameshift and stop codon (see electronic supplementary material, table S4b). Furthermore, HMG-CoA reductase, a key enzyme in mevalonate biosynthesis, is associated with a rise in ergosterol content in A. fumigatus and Candida species [42]. In D3-fluffy we found two additional SNPs that were not fixed and not found in any other isolate. In lineage C2 we found three SNPs that were still polymorphic, of which one was also found in D3-fluffy (374 P>S in putative amidohydrolase ytcJ).
Similarly, in D6-fluffy three new unique SNPs were found (compared to their ancestor), while in D6-white we found two unique SNPs that were completely fixed. The PCR trace-back experiment confirmed that PtaB Q264STOP was related to the white phenotype, i.e. delayed sporulation, while hypothetical protein G167D was associated with the increased triazole MIC. This apparent balanced mixed population within the D3 and D6 lineages can potentially be explained by cross-feeding or complementation between these two phenotypes [43]. The function of each type in this balance needs further study. Also, to establish that the mutations we detected through sequencing are the causal mutations responsible for increased resistance, additional knock-out/in experiments and complementation tests need to be performed. Both were, however, beyond the scope of this current study.
4. Discussion
In this study, we tested the consequences of prolonged exposure of A. fumigatus to one of five non-medical SI fungicides for cross-resistance to three medical triazoles. Overall, we found widespread cross-resistance to medical triazoles in experimental populations after seven weeks of experimental evolution under pressure of environmental SI fungicides. We observed that evolutionary lineages show similar phenotypic resistance patterns to SI fungicides and the three medical triazoles, which further supports the idea that exposure to SI fungicides can confer cross-resistance to medical triazoles. When looking at specific SI fungicide treatments, only lineages that evolved in the presence of difenoconazole and tebuconazole were significantly different from the non-fungicide control when tested for resistance in the three medical azole conditions, indicating that they imposed the highest selection for cross-resistance. This was corroborated by the significant correlation of mean MIC values, indicating similar degrees of cross resistance between medical azoles. The strong selection by difenoconazole could be caused by the higher decrease in growth in terms of MGR imposed by the set concentration of 1 mg l−1 used for all SI fungicides during the selection (electronic supplementary material, figure S3). We further detected population dynamics changes in evolving lineages, with lineages diversifying into several morphological phenotypes. Whole-genome sequencing revealed the presence of a mutation known for its effect of increased triazole MIC (G138S mutation in the Cyp51 gene) as well as several other mutations.
Interestingly, when assaying resistance levels against medical triazoles at various evolutionary time points, the resistance to medical triazoles shows similar patterns of step-wise increase in MIC as to SI fungicides. These changes are mirrored by a step-wise increases in fitness, indicative of the fixation of a mutation [44]. This supports the idea that mutations conferring resistance to SI fungicides also confer cross-resistance to medical triazoles, and fits the fact that medical triazoles and SI fungicides share the same site of action (binding site) and have a similar molecule structure [5]. These observations support the notion that SI fungicides can induce cross-resistance to medical triazoles in A. fumigatus and is consistent with previous findings that mutations in the cyp51 gene (G138S) confer resistance both to SI fungicides and to medical triazoles [45].
(a). The environmental origin of resistance of A. fumigatus to medical azoles in patients is likely
There has been a long-standing debate about the interaction between environmental and clinical selection routes for triazole resistance [20,46,47]. A. fumigatus grows and sporulates frequently in natural habitats such as decaying plant material, the place that might contain (residues of) SI fungicides used to control a wide variety of fungal plant diseases or used to preserve materials. Our work supports the idea that resistance development to medical triazoles through exposure to these five SI fungicides in the environment is a plausible route. It has been suggested that the development of resistance in agricultural environments is less likely than in medical environments owing to the 10- to 100-fold lower concentrations of triazoles used in agriculture than in medical environments [48], and these concentrations probably fluctuate in the field. However, our experiments indicate that exposure to low concentrations of SI fungicides, i.e. 1 mg l−1, can result in the emergence of phenotypes with 40-fold increased MIC compared with the ancestor. We also have evidence that this increase is due to a limited number of genetic changes, similar to the case of high concentrations, and not due to many changes each with small effect. These observations suggest that exposure to a high SI fungicide concentration is neither essential nor required for resistance development. In fact, exposure to low SI fungicide concentrations might permit a greater number of fungal growth cycles, thereby increasing the opportunity for (resistance) mutations to arise. As a consequence, low level exposure to SI fungicides should not be disregarded as a risk environment for triazole resistance development in A. fumigatus. Decaying plant material of different origins, such as compost, self-heated hay and corn heaps, and litter of, for example, ferns, cotton, barley, cabbage and conifers are important habitats for A. fumigatus. These materials may contain sufficient (residual) SI fungicides to generate a selection pressure for resistance, similar to that applied by our experimental evolution conditions.
In The Netherlands over 30 SI fungicides have been authorized for agricultural use in the past 20 years, of which all our five SI fungicides showed the potential for strong selection for cross-resistance against medical triazoles [5]. Interestingly, these five SI fungicides were applied in the environment before the appearance of the first resistant clinical isolates. In our experiment, we confirmed that these SI fungicides can induce cross-resistance to medical triazoles. Thus, the use of similar molecules for medical and non-medical applications has the risk of resistance selection through one application, which then negatively affects the use of similar molecules in other applications. Cross-resistance has been found in other fields as well, for instance where the use of antibacterial agents for cattle rearing has selected for antibiotic resistance for extended-spectrum beta-lactamase (ESBL) producing Escherichia coli [49]. The resistant bacteria are transferred to humans through consumption of food containing ESBL E. coli, which may then cause infections in humans that are difficult to treat due to cross-resistance to medical antibacterial drugs [49]. Therefore, the problem of fungal resistance should be approached from a one-health perspective that advocates an interdisciplinary approach similar to research addressing antibacterial resistance.
(b). Population biology of resistance, resistance mutations and potential new resistance mechanisms
Mutation is the driving force for triazole resistance evolution. Population dynamics characterization at the level of morphological types showed that the colony size (large and small), the colony texture (dense and fluffy) as well as sporulation (early and delayed) changed over the evolutionary experiment in the different lineages, which suggested that A. fumigatus adapts to the SI fungicide environment through a variety of genetic changes. The mutated genes may relate to growth, sporulation or triazole-resistance phenotypes, and all deserve further study. Interestingly, the sporulation delay mutant in the D6 lineage had a high resistance level which could be explained by a trade-off between hyphal growth and resistance traits [50]. Further, it is interesting to note that in two lineages mixed populations of multiple morphotypes that originated from one ancestral type dominated at the end of the evolution experiment and that the mixture of types gave higher (cross-)resistance than the types on their own. This might have implications for clinical settings where multiple A. fumigatus morphotypes may be recovered from patients with pulmonary aspergillus infection [51]. In reality, these multiple morphotypes are capable of coexisting, possibly in heterokaryons, and in this way exhibit high (cross-)resistance to medical triazoles [43].
Previous work has shown that mutations in the cyp51A gene of the ergosterol synthesis pathway can confer resistance both to SI fungicides and to medical triazoles, owing to the similarities in chemical structure [5,20]. With our whole-genome sequencing, we detected mutations in the cyp51A gene, as well as mutations in other targets. It is known that other mutations than those in the cyp51A gene can confer resistance to medical triazoles [52]. In D6-white, a hypothetical protein mutation is potentially associated with triazole resistance, and needs further characterization. D3-dense and D1-middle contain a mutation in HMG-CoA reductase, a rate-controlling enzyme of the mevalonate pathway that produces ergosterol (electronic supplementary material, tables S4c and 5a).
Mutations in D3-fluffy and D6-fluffy with variable SNPs were difficult to trace back in our evolution experiment, suggesting that these mutations were not causal to the resistance phenotype. Because no other mutations were found an alternative explanation for the morphotype evolution is needed. An epigenetic effect of methylation on the fluF locus can give rise to a fluffy phenotype, at least in the related species A. nidulans [53,54]. While methylation is only present at low levels in aspergilli, the fluF locus is an notable exception. Methylation is semi-stably inherited and persists during meiosis, segregating 1 : 1 in crosses. Interestingly, this may point to a potential mechanism of epigenetic resistance development. In addition, in D6-fluffy, three mitochondrial mutations were found: MnSOD is a mitochondrial superoxide dismutase, Mdm31 is an inner membrane protein of the mitochondria while Tom70 is located in the outer membrane. These mitochondrial mutations strongly suggest an association between mitochondrial dysfunction and triazole resistance. However, the mechanism by which the mutations found in these two phenotypes confer resistance needs to be further studied. Nevertheless, our experimental evolution study combined with whole-genome sequencing of various phenotypes helped to identify potential hot spots for triazole resistance mutations.
5. Conclusion
Our results clearly demonstrate that exposure to SI fungicides for non-medical use sparks cross-resistance to medical triazoles. As we observed differences in selection pressure for the various SI fungicide compounds, the implications of both the exposure level and the molecule structure should be taken into account in future studies. Knowing the exact effect of adaptive (i.e. azole resistance) mutations in A. fumigatus will be an essential avenue of future research, which will further unravel the evolutionary dynamics of A. fumigatus azole resistance both in-patient and in the environment. Crucially, understanding the key factors that facilitate resistance selection in A. fumigatus is essential to designing strategies that prevent or overcome this clear and present danger.
Supplementary Material
Acknowledgements
We thank Bertha Koopmanschap and Marijke Slakhorst for technical assistance, and Claudio Valero-Jimenez and Martin Kapun for providing bioinformatics scripts.
Data accessibility
Experimental data are available via Dryad. doi:10.5061/dryad.rj6gq. Raw sequence data are available via NCBI, Bio-ProjectID: PRJNA401150. http://www.ncbi.nlm.nih.gov/bioproject/401150.
Authors' contributions
B.J.Z., P.E.V., J.Z., A.J.M.D. and S.E.S. initiated the project; J.Z., A.J.M.D. and S.E.S. designed the methodology; J.Z. carried out the evolutionary experiments and formal analyses. J.v.d.H. analysed the whole-genome sequences. W.J.G.M. joined the discussion and gave advice on whole-genome sequencing. J.Z. and S.E.S. prepared the draft of the manuscript. All authors contributed to the commenting and editing of subsequent versions of the manuscript.
Competing interests
We have no competing interests.
Funding
We gratefully acknowledge funding from the China Scholarship Council for J.Z. and funding from a Marie Curie Fellowship to S.E.S.
References
- 1.Latgé J-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12, 310–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhodes JC. 2006. Aspergillus fumigatus: growth and virulence. Med. Mycol. 44, 77–81. ( 10.1080/13693780600779419) [DOI] [PubMed] [Google Scholar]
- 3.Lockhart SR, Frade JP, Etienne KA, Pfaller MA, Diekema DJ, Balajee SA. 2011. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrob. Agents Chemother. 55, 4465–4468. ( 10.1128/AAC.00185-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Linden JW, et al. 2011. Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg. Infect. Dis. 17, 1846–1854. ( 10.3201/eid1710.110226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snelders E, Camps SM, Karawajczyk A, Schaftenaar G, Kema GH, van der Lee HA, Klaassen CH, Melchers WJ, Verweij PE. 2012. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS ONE 7, e31801 ( 10.1371/journal.pone.0031801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermeulen E, Maertens J, Schoemans H, Lagrou K. 2012. Azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation emerging in Belgium, July 2012. Euro Surveill. 17, pii20326. [PubMed] [Google Scholar]
- 7.Lavergne R.-A., Morio F, Favennec L, Dominique S, Meis JF, Gargala G, Verweij PE, Le Pape P. 2015. First description of azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation in France. Antimicrob. Agents Chemother. 59, 4331–4335. ( 10.1128/AAC.00127-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiederhold NP, et al. 2015. First detection of TR34/L98H and TR46/Y121F/T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J. Clin. Microbiol. 54, 168–171. ( 10.1128/JCM.02478-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albarrag AM, Anderson MJ, Howard SJ, Robson GD, Warn PA, Sanglard D, Denning DW. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55, 5113–5121. ( 10.1128/AAC.00517-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph-Horne T, Hollomon DW. 1997. Molecular mechanisms of azole resistance in fungi. FEMS Microbiol. Lett. 149, 141–149. ( 10.1111/j.1574-6968.1997.tb10321.x) [DOI] [PubMed] [Google Scholar]
- 11.Howard SJ, Webster I, Moore CB, Gardiner RE, Park S, Perlin DS, Denning DW. 2006. Multi-azole resistance in Aspergillus fumigatus. Int. J. Antimicrob. Agents 28, 450–453. ( 10.1016/j.ijantimicag.2006.08.017) [DOI] [PubMed] [Google Scholar]
- 12.Howard SJ, et al. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15, 1068–1076. ( 10.3201/eid1507.090043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodriguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51, 1897–1904. ( 10.1128/AAC.01092-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJ. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob. Agents Chemother. 54, 2425–2430. ( 10.1128/AAC.01599-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelaez T, Gijon P, Bunsow E, Bouza E, Sanchez-Yebra W, Valerio M, Gama B, Cuenca-Estrella M, Mellado E. 2012. Resistance to voriconazole due to a G448S substitution in Aspergillus fumigatus in a patient with cerebral aspergillosis. J. Clin. Microbiol. 50, 2531–2534. ( 10.1128/JCM.00329-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snelders E, Huis Int Veld RA, Rijs AJ, Kema GH, Melchers WJ, Verweij PE. 2009. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 75, 4053–4057. ( 10.1128/AEM.00231-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verweij PE, Snelders E, Kema GH, Mellado E, Melchers WJ. 2009. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect. Dis. 9, 789–795. ( 10.1016/S1473-3099(09)70265-8) [DOI] [PubMed] [Google Scholar]
- 18.Stensvold CR, Jørgensen LN, Arendrup MC. 2012. Azole-resistant invasive aspergillosis: relationship to agriculture. Curr. Fungal Infect. Rep. 6, 178–191. ( 10.1007/s12281-012-0097-7) [DOI] [Google Scholar]
- 19.Verweij PE, van de Sande-Bruisma N, Kema GH, Melchers WJ. 2012. Azole resistance in Aspergillus fumigatus in the Netherlands—increase due to environmental fungicides? Ned. Tijdschr. Geneeskd. 156, A4458. [PubMed] [Google Scholar]
- 20.Gisi U. 2013. Assessment of selection and resistance risk for demethylation inhibitor fungicides in Aspergillus fumigatus in agriculture and medicine: a critical review. Pest Manag. Sci. 70, 352–364. ( 10.1002/ps.3664) [DOI] [PubMed] [Google Scholar]
- 21.Verweij PE, Kema GH, Zwaan B, Melchers WJ. 2013. Triazole fungicides and the selection of resistance to medical triazoles in the opportunistic mould Aspergillus fumigatus. Pest Manag. Sci. 69, 165–170. ( 10.1002/ps.3390) [DOI] [PubMed] [Google Scholar]
- 22.Camps SM, van der Linden JW, Li Y, Kuijper EJ, van Dissel JT, Verweij PE, Melchers WJ. 2012. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob. Agents Chemother. 56, 10–16. ( 10.1128/AAC.05088-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowyer P, Denning DW. 2014. Environmental fungicides and triazole resistance in Aspergillus. Pest Manag. Sci. 70, 173–178. ( 10.1002/ps.3567) [DOI] [PubMed] [Google Scholar]
- 24.Faria-Ramos I, Farinha S, Neves-Maia J, Tavares PR, Miranda IM, Estevinho LM, Pina-Vaz C, Rodrigues AG. 2014. Development of cross-resistance by Aspergillus fumigatus to clinical azoles following exposure to prochloraz, an agricultural azole. BMC Microbiol. 14, 155 ( 10.1186/1471-2180-14-155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Debets AJ, Verweij PE, Melchers WJ, Zwaan BJ, Schoustra SE. 2015. Asexual sporulation facilitates adaptation: the emergence of azole resistance in Aspergillus fumigatus. Evolution 69, 2573–2586. ( 10.1111/evo.12763) [DOI] [PubMed] [Google Scholar]
- 26.Kahlmeter G, et al. 2006. European Committee on Antimicrobial Susceptibility Testing (EUCAST) technical notes on antimicrobial susceptibility testing. Clin. Microbiol. Infect. 12, 501–503. ( 10.1111/j.1469-0691.2006.01454.x) [DOI] [PubMed] [Google Scholar]
- 27.Andrews S.2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc .
- 28.Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A, Kosiol C, Schlötterer C. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS ONE 6, e15925 ( 10.1371/journal.pone.0015925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438, 1151–1156. ( 10.1038/nature04332) [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen K, et al. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6, 677–681. ( 10.1038/nmeth.1363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeitouni B, Boeva V, Janoueix-Lerosey I, Loeillet S, Legoix-Né P, Nicolas A, Delattre O, Barillot E. 2010. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 26, 1895–1896. ( 10.1093/bioinformatics/btq293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klambauer G, Schwarzbauer K, Mayr A, Clevert D-A, Mitterecker A, Bodenhofer U, Hochreiter S. 2012. cn. MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 40, e69 ( 10.1093/nar/gks003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. ( 10.1093/bioinformatics/btr509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charlesworth B, Charlesworth D. 2010. Elements of evolutionary genetics. Greenwood Village, Colorado, USA: Roberts Publishers. [Google Scholar]
- 37.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. ( 10.1038/nbt.1754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verweij PE, Howard SJ, Melchers WJ.G, Denning DW. 2009. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist Updat. 12, 141–147. ( 10.1016/j.drup.2009.09.002) [DOI] [PubMed] [Google Scholar]
- 39.Siopi M, Mavridou E, Mouton JW, Verweij PE, Zerva L, Meletiadis J. 2014. Susceptibility breakpoints and target values for therapeutic drug monitoring of voriconazole and Aspergillus fumigatus in an in vitro pharmacokinetic/pharmacodynamic model. J. Antimicrob. Chemother. 69, 1611–1619. ( 10.1093/jac/dku023) [DOI] [PubMed] [Google Scholar]
- 40.Lenski RE, Travisano M. 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl Acad. Sci. USA 91, 6808–6814. ( 10.1086/285289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoustra SE, Bataillon T, Gifford DR, Kassen R. 2009. The properties of adaptive walks in evolving populations of fungus. PLoS Biol. 7, e1000250 ( 10.1371/journal.pbio.1000250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macreadie IG, Johnson G, Schlosser T, Macreadie PI. 2006. Growth inhibition of Candida species and Aspergillus fumigatus by statins. FEMS Microbiol. Lett. 262, 9–13. ( 10.1111/j.1574-6968.2006.00370.x) [DOI] [PubMed] [Google Scholar]
- 43.Ribeck N, Lenski RE. 2015. Modeling and quantifying frequency-dependent fitness in microbial populations with cross-feeding interactions. Evolution 69, 1313–1320. ( 10.1111/evo.12645) [DOI] [PubMed] [Google Scholar]
- 44.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341. ( 10.1086/285289) [DOI] [Google Scholar]
- 45.Chassot CHU, Sierotzki H, Gisi U. 2008. Sensitivity of cyp51 genotypes to DMI fungicides in Mycosphaerella graminicola. In 15th Int. Reinhardsbrunn Symp., Friedrichroda (eds Dehne HW, Gisi U, Kuck KH, Russell PE, Lyr H), pp. 129–136. Braunschweig. [Google Scholar]
- 46.Hof H. 2008. Is there a serious risk of resistance development to azoles among fungi due to the widespread use and long-term application of azole antifungals in medicine? Drug Resist. Updat. 11, 25–31. ( 10.1016/j.drup.2008.01.001) [DOI] [PubMed] [Google Scholar]
- 47.Lago M, Aguiar A, Natário A, Fernandes C, Faria M, Pinto E. 2014. Does fungicide application in vineyards induce resistance to medical azoles in Aspergillus species? Environ. Monit. Assess. 186, 5581 ( 10.1007/s10661-014-3804-8) [DOI] [PubMed] [Google Scholar]
- 48.Kuck KH, Stenzel K, Vors J-P. 2012. Sterol biosynthesis inhibitors. In Modern crop protection compounds, vol. 2. Fungicides (eds Kramer W, Jeschke P, Witschel M, 2nd rev.), pp. 761–805. Weinheim, Germany: Wiley-VCH Verlag. [Google Scholar]
- 49.Ewers C, Bethe A, Semmler T, Guenther S, Wieler L. 2012. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. ( 10.1111/j.1469-0691.2012.03850.x) [DOI] [PubMed] [Google Scholar]
- 50.Mille-Lindblom C, Fischer H, Tranvik L. 2006. Antagonism between bacteria and fungi: substrate competition and a possible tradeoff between fungal growth and tolerance towards bacteria. Oikos 113, 233–242. ( 10.1111/j.2006.0030-1299.14337.x) [DOI] [Google Scholar]
- 51.Verweij PE, Zhang J, Debets AJ, Meis JF, van de Veerdonk FL, Schoustra SE, Zwaan BJ, Melchers WJ. 2016. In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: a dilemma for clinical management. Lancet Infect. Dis. 16, e251–e260. ( 10.1016/S1473-3099(16)30138-4) [DOI] [PubMed] [Google Scholar]
- 52.Camps SM, Dutilh BE, Arendrup MC, Rijs AJ, Snelders E, Huynen MA, Verweij PE, Melchers WJ. 2012. Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS ONE 7, e50034 ( 10.1371/journal.pone.0050034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamame M, Antequera F, Villanueva J, Santos T. 1983. High-frequency conversion to a ‘fluffy’ developmental phenotype in Aspergillus spp. by 5-azacytidine treatment: evidence for involvement of a single nuclear gene. Mol. Cell. Biol. 3, 2287–2297. ( 10.1128/MCB.3.12.2287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arratia-Quijada J, Sánchez O, Scazzocchio C, Aguirre J. 2012. FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 11, 1132–1142. ( 10.1128/EC.00101-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Experimental data are available via Dryad. doi:10.5061/dryad.rj6gq. Raw sequence data are available via NCBI, Bio-ProjectID: PRJNA401150. http://www.ncbi.nlm.nih.gov/bioproject/401150.