Abstract
Family members show behavioural strategies predicted to maximize individual fitness. These behaviours depend directly on genes expressed in focal individuals but also indirectly on genes expressed in other family members. However, how sibling and parental behavioural strategies are modified by genes expressed in family members, and to what degree, remains unclear. To answer this question, we have used a split litter design in an experimental population of genetically variable mouse families, and identified loci that indirectly affected sibling and maternal behaviour simultaneously. These loci map to genomic regions that also show a direct effect on offspring behaviour. Directly and indirectly affected traits were significantly correlated at the phenotypic level, illustrating how indirect effects are caused. Genetic variants in offspring that influence solicitation also impacted their siblings' and maternal behaviour. However, in contrast to predictions from sibling competition, unrelated litter mates benefited from increased solicitation. Overall, such indirect genetic effects explained a large proportion of variation seen in behaviours, with candidate genes involved in metabolism to neuronal development. These results reveal that we need to view behavioural strategies as the result of conjoint selection on genetic variation in all interacting family members.
Keywords: parent–offspring conflict, indirect genetic effects, systems genetics, family conflicts
1. Introduction
Although social behaviours are expressed by individuals, their fitness effects depend on the behaviour shown by social partners. Understanding the evolution of social behaviour is a major challenge in biology because trait variation is not just influenced by genes expressed in focal individuals but also by genes expressed in social partners [1]. While theoretical work has shown that this dual genetic control of social traits fundamentally alters predictions about trait evolution [2], we still know little about the actual genes underlying social traits and the importance of their indirect effects [3,4].
For mammals, the most important social interactions occur during early development between parent and offspring, and among siblings [5,6]. However, family members are in conflict over resource share and level of parental investment with offspring favouring greater parental investment than is optimal for the parent, and individual siblings claiming more than their fair share of parental provisioning. Behavioural strategies affecting solicitation, provisioning, and resource share are therefore selected to maximize different fitness optima for different family members [7,8], however, fitness pay-offs are dependent on behaviours shown by all members. Thus, variation in genes expressed in individuals showing a particular behaviour and those expressed in family members will influence the response of that particular behaviour to selection and its evolution.
What remains unclear is the degree to which a particular behavioural strategy (such as the level of offspring solicitation) is influenced by genes expressed in a focal individual (a direct effect) or genes expressed in other family members (an indirect effect). Further, we do not know how genes expressed in one family member affect the behaviour of other members.
A key problem in answering these fundamental questions is to separate direct from indirect effects on behaviours during social interactions because in a genetically variable population all genotypes are likely to exert both direct and indirect effects. Our previous work [9] has shown that offspring solicitation and maternal behaviour are significantly correlated at the phenotypic level but could not demonstrate how sibling and maternal behaviour is influenced simultaneously by genes showing a direct and indirect effect. We hypothesized that indirect effects can explain a large proportion of variation in behavioural traits in all family members and designed an experiment that enabled us to determine the effects of genetic variation in offspring on traits in unrelated litter mates and unrelated foster mothers. We investigated behavioural interactions in an experimental population of genetically defined mice, in which adoptive families consist of half genetically variable (using the recombinant inbred strain BXD) and half genetically uniform offspring (using the inbred strain C57BL/6 J or B6; figure 1). We recorded in this population, from birth until weaning, maternal provisioning and activity, as well as offspring solicitation, sucking (from maternal teats), and activity, independently in both half litters following [10]. Next, we conducted a quantitative trait locus (QTL) interval mapping analysis that enabled us to locate where in the genome genetic variation causes indirect effects and investigate in more detail potential candidate genes.
Figure 1.
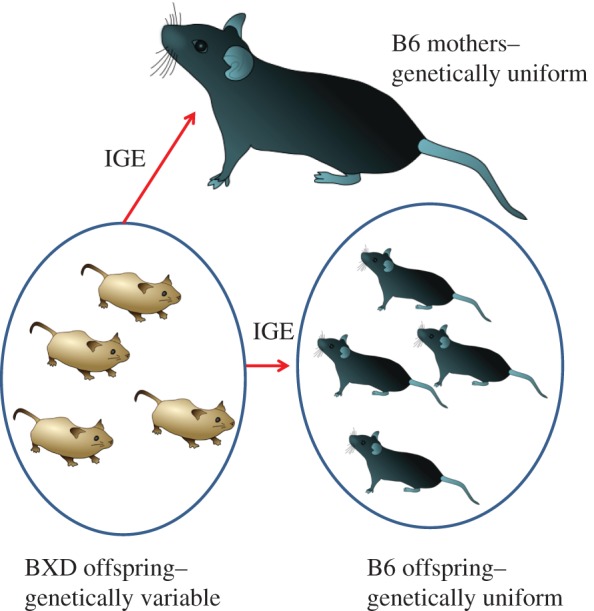
Experimental half-litter cross-fostering design. B6 mothers adopt half litters of different lines of the BXD population and half litters of B6 offspring. Genetic variation among BXD genotypes causes indirect genetic effects (IGE) in both mothers and siblings.
2. Material and methods
We investigated behavioural interactions among siblings of different genotype and their adoptive mothers on 3 days during lactation, and used single nucleotide polymorphism (SNP) interval mapping to map trait variation in genetically uniform individuals (B6 mothers and B6 offspring) as a function of genetic variation in their (BXD) siblings to investigate indirect genetic effects (IGE). By contrast, direct genetic effects were investigated by mapping behavioural variation among BXD individuals to their own genotype. The QTL analysis was followed by a systems genetics analysis to identify functional candidate genes and biological pathways. We then analysed the phenotypic correlation between the behavioural traits affected by direct and indirect genetic variation. The genetic variants are identified as social interaction loci together with the chromosome number, e.g. SocInt1 would denote a locus on chromosome 1.
(a). Experimental animals
Our project used mice of the BXD recombinant inbred population, which consists of experimentally tractable and genetically defined mouse lines capturing a large amount of naturally occurring genetic variation, which underlies variation at the phenotypic level (e.g. [11]). BXD is the largest genetic model system in mammals and consists of over 140 experimentally tractable mouse lines and has the largest phenome of any mammalian model system (5 388 traits, March 2017; greater than 1 k papers since 2003, http://www.genenetwork.org). BXD is derived from two divergent mouse strains (C57BL/6 J and DBA/2 J, hence BXD), in which different recombination patterns have been inbred, each with a fixed recombination pattern of exactly two possible alleles. BXD incorporates 4–5 million segregating single nucleotide polymorphisms, 500 k insertions and deletions, and 55 k copy number variants [12,13].
In this experiment, we used 32 lines (from BXD lines 1, 11, 24, 38, 43–45, 48a, 49, 51, 55, 56, 61, 62, 64, 65a, 65b, 65, 68–71, 73a, 73b, 73–75, 84, 87, 90, 100, and 102), which were selected to exclude very poor breeding lines. For each line, three within-line replicates were set up, although breeding success reduced this in some lines. C57BL/6 J (B6) inbred mice were used as the genetically uniform strain such that all mothers and half of each litter had the same genotype in all cases (figure 1). BXD mice were obtained from Prof. Robert W. Williams at the University of Tennessee Health Science Centre, Memphis, TN, USA, and C57BL/6 J mice were obtained from Charles River, UK. All procedures were approved by the University of Manchester Ethics Committee.
(b). Husbandry and mating protocol
Mice were maintained under standard laboratory conditions in the same room, exclusively used for the experiment, in individually ventilated cages (IVC Tecniplast Green line), maintained at 20(±2)°C with a relative humidity of 55% (±10%). Because we investigated behavioural patterns in a nocturnal species, we used a reverse dark : light cycle with red light between 10.00 and 22.00 h. Food and water was provided ad libitum. Cages were cleaned once a week but never within the first 6 days after birth to minimize disturbance. The parental mice were all sexually mature and females were nulliparous. Groups of up to five sibs were housed together in single-sex cages until mating, which occurred between six and 10 weeks of age, when females were greater than or equal to 18 g. Prior to mating bedding from the prospective mate's cage was added to the female cages to encourage synchronized oestrus [14], and individual males were moved to new cages to allow them to scent mark. Two days later two sisters were added to the male's cage. Once visibly pregnant (weight gain ≥8 g or distended abdomen), females were separated into an individual cage. This ensured that neither father nor aunt had a social interaction with the offspring.
(c). Experimental design
Females in individual cages were checked daily for new-born litters. Litters were weighed and cross-fostered, such that each B6 mother had a litter composed of half B6 and half BXD offspring (figure 1). On a few occasions (less than 10% of litters) no corresponding litters were available for cross-fostering, in which case the procedure was delayed by a day. If no corresponding litter was available on the following day, the individuals were removed from the study. Cross-fostering after birth meant that all litters were genetically, and as far as possible, environmentally, identical with the exception of the genotype of the BXD pups.
For 6 days following cross-fostering, the litter was left undisturbed, apart from visual checks from outside the cage twice daily, to minimize disturbance.
On postpartum days 6, 10, and 14, observations were conducted following [15]. Litters and mothers were weighed and then separated for 4 h. Mothers were placed in a new cage with the food and water from the original cage while the litter was left in the original cage and placed on a heat mat to keep the pups warm during the separation. This standardizes as much as possible the motivation for maternal care when they are reunited with their pups but also reduces variation in offspring motivation due to differences in care received prior to observations. To distinguish between half litters, we used colour differences where coat colours are different between the respective BXD line and B6. Where this was not the case, we used small fur clippings for one genotype but varied this randomly between B6 and BXD. Maternal and pup behaviour was recorded during the 15 min after the pups were reunited. Behaviours were separated into states (long-lasting, commonly occurring activities) and events (short, less common). States were recorded by scan sampling every 20 s, and events were recorded whenever they occurred. Offspring behaviour was recorded as the number of pups (of an individual genotype) engaged in the behaviour at any given time, and an average for the litter was used for statistical analysis. Offspring solicitation behaviour is defined as pups attempting to suck and following the mother, while sucking refers to the actual feeding behaviour while being attached to teats. Activity refers to active behaviour other than solicitation such as moving around. For all offspring measures, individual pups were not distinguished. For mothers we focused on provisioning behaviour, which is suckling, and other activity, such as digging and moving around.
(d). Quantitative trait locus mapping and candidate analysis
To account for differences between litters not due to genotype differences, residuals were calculated from a general linear model (GLM) with the following covariates: maternal bodyweight, average bodyweight of the B6 offspring (weight of the B6 litter divided by the B6 litter size), B6 litter size, average bodyweight of BXD offspring (weight of the BXD litter divided by the BXD litter size), BXD litter size, and batch. Non-significant (p > 0.05) terms were removed sequentially and in a stepwise manner until only significant covariates remained in the model following [16]. All GLMs were carried out using SPSS (v. 21, IBM Corporation, Armonk, NY, USA).
For QTL analyses, the average trait value per line was calculated and residuals from the GLMs were mapped using interval mapping as implemented in GeneNetwork (GN). Interval mapping relies on 3 795 informative SNP markers across all chromosomes, except Y. The BXD strains were genotyped using the MUGA array in 2011, along with genotypes generated earlier using Affymetrix and Illumina platforms [17], and mm9 was used. Loci are identified in GN by the computation of a likelihood ratio statistic score and significance was determined using 5 000 permutations of the phenotype data. Confidence intervals were given by a LOD drop of 1.5 from the peak marker location [18]. To investigate how indirect effects arise from direct effects, we scanned the genome for co-location of indirect effect QTL and direct effect QTL, where both the direct effect locus and indirect effect locus have to be at the same genomic location (i.e. within the same region as given by the confidence intervals; table 1). Since we analysed four traits at three different time points (overall 24 tests, with just over 1 locus expected to be a false positive) during lactation we have used the false discovery rate (FDR) criterion following [19] that applies a correction for multiple testing based on the number of rejected null hypotheses. For our study, all loci had to be significant at the genome-wide level for either a direct or indirect effect during the genome scan and pass the threshold following [19]. While it is important to protect against many false discoveries, both at the genome-wide level when scanning for one trait and when considering multiple scans, we need, at the same time, assurance we are not ignoring truly interesting effects (e.g. [20]). Thus, where a locus was identified at the genome-wide level and it passed the Benjamini and Hochberg threshold, we then investigated indirect effects in a protected test at that locus [21,22]. Here, following Benjamini & Yekutieli [23], we applied an FDR of 0.10 to identify other indirect effects at a given locus. To further validate loci, we investigated the phenotypic correlations between the directly and indirectly effect loci (see table S1, electronic supplementary material).
Table 1.
Direct and indirect genetic effects during family interactions. After the locus is identified, the phenotypes affected by the locus are listed with direct effects first, followed by the QTL position in Mb, the 1.5 LOD confidence interval, the genome-wide peak marker likelihood ratio statistic (LRS) and log of the odds (LOD) score and associated p-value, the number of genes within the interval, the allele that increases the trait value, and finally the coefficient of determination as obtained from a regression on BXD genotype. All loci have exceeded significance thresholds at the genome-wide level and corrected for multiple traits as outlined in the Material and methods except the indirect effect on B6 maternal activity at SocInt15, which narrowly missed the multiple trait threshold but was retained due to the high phenotypic correlation between the direct and indirect effect (electronic supplementary material, table S2).
| locus | phenotype | QTL position (Mb) | confidence interval (Mb) | max LRS | max LOD | max p | no. genes | allele increasing trait value | coefficient of determination (R2) |
|---|---|---|---|---|---|---|---|---|---|
| SocInt2 | BXD sibling solicitation d14 | 75.857–76.309 | 73.31–77.355 | 20.016 | 4.342 | 0.045 | 49 | D2 | 0.590 |
| B6 maternal suckling d14 | 74.928–75.515 | 74.928–75.515 | 6.426 | 1.394 | 0.992 | 49 | D2 | 0.601 | |
| SocInt4 | BXD sibling sucking d6 | 11.148–11.507 | 10.826–13.031 | 19.672 | 4.277 | 0.044 | 21 | B6 | 0.603 |
| B6 sibling activity d6 | 11.148–11.507 | 10.826–13.031 | 20.285 | 4.4 | 0.030 | 21 | B6 | 0.603 | |
| B6 sibling sucking d6 | 11.148–11.507 | 10.826–13.031 | 20.652 | 4.48 | 0.032 | 21 | B6 | 0.601 | |
| B6 maternal suckling d6 | 11.148–11.507 | 10.826–13.031 | 19.517 | 4.234 | 0.045 | 21 | B6 | 0.611 | |
| SocInt10 | BXD sibling sucking d14 | 101.578–102.633 | 99.07–103.025 | 23.537 | 5.117 | 0.035 | 17 | D2 | 0.597 |
| B6 sibling sucking d14 | 101.578–102.633 | 99.07–103.025 | 23.847 | 5.173 | 0.029 | 17 | D2 | 0.590 | |
| B6 maternal suckling d14 | 101.028 | 97.112–103.025 | 19.857 | 4.307 | 0.085 | 27 | D2 | 0.601 | |
| SocInt15 | BXD sibling sucking d14 | 3.619–4.349 | 3.229–6.298 | 14.671 | 3.189 | 0.223 | 19 | D2 | 0.597 |
| B6 maternal suckling d14 | 3.619–4.349 | 3.229–6.298 | 22.538 | 4.889 | 0.036 | 19 | D2 | 0.601 | |
| B6 sibling sucking d14 | 3.619–4.349 | 3.229–6.298 | 15.058 | 3.266 | 0.205 | 19 | D2 | 0.590 | |
| B6 maternal activity d14 | 3.619–4.349 | 3.229–7.273 | 12.373 | 2.684 | 0.551 | 26 | B6 | 0.670 |
To identify candidates, we firstly used the ‘Phenotypes, Alleles & Disease Models Search’ (http://www.informatics.jax.org/allele) on Mouse Genome Informatics [24] to find phenotypes associated with each of the genes within the loci we identified. Second, QTLminer [25] was used to summarize information about candidate genes, including if they have non-synonymous SNPs (nsSNPs) or insertions or deletions (indels) in the BXD lines. We note that in the latter case, potential causal variants may be omitted if they are, for example, regulatory variants in unidentified enhancers. Finally, to obtain a broad estimate of heritability we used genotype as a predictor and trait value as a dependent variable in an ANOVA.
3. Results and discussion
Four social interaction loci were identified that directly influenced offspring solicitation, sucking, and activity, and, at the same time, indirectly affected sibling and maternal behaviour (table 1); three of which during the weaning period (d14), and one during early lactation (d6). At the phenotypic level, we found significant correlations between directly and indirectly affected traits. We can thus not only show how genetic variation affects the phenotype of a focal individual directly, and indirectly those of social partners, but also how this indirect effect arises as a consequence of genes expressed in another individual at the phenotypic level.
Specifically, three loci on chromosomes 4, 10, and 15 directly affected offspring sucking behaviour either during early lactation (d6) or during the weaning period (d14). All three loci also indirectly influenced the sucking behaviour of their unrelated litter mates, as well as maternal provisioning in their unrelated mothers. At the phenotypic level, directly and indirectly influenced traits were positively correlated. Activity levels were indirectly affected by SocInt4 in siblings; and in mothers by SocInt15, which showed a negative correlation with both solicitation and sucking (electronic supplementary material, table S1). It is important to note that there is no genetic variation among either mothers or litter mates so we do not necessarily expect that the same traits in both half litters are affected by direct and indirect effects. Further, given we expect competition between siblings over resource share, it is surprising to see that unrelated litter mates actually benefited from increased sucking behaviour of their litter mates. Given limited access to maternal teats, we expected that genes increasing sucking behaviours would increase competition and thus reduce sucking in their litter mates. However, our results suggest that the indirect effect on increasing maternal provisioning benefited unrelated littermates who then also increased their sucking behaviour.
Finally, while we have standardized environmental conditions among both BXD and B6 mothers pre- and postnatally as much as possible, it is important to note that differences among B6 half litters may arise for reasons other than genetic differences among BXD genotypes. Firstly, pre-natal maternal (genetic or environmental [26,27]) effects among the B6 mothers may contribute to differences among B6 half litters. Secondly, BXD litters may vary in their experience of B6 mothers or siblings.
(a). Variation explained by direct and indirect effects
We next sought to establish how important IGE are in explaining variation in behavioural strategies during family interactions. Here, we calculated the proportion of variation explained, either for direct (i.e. variation in BXD phenotype) or for indirect effects (i.e. variation in B6 phenotype). Indeed, a large proportion of variation in behavioural traits could be explained by IGE (table 1). For maternal traits that are indirectly affected by offspring genotype, such as suckling, this ranges from 60% to 67% while for indirectly affected sibling traits, such as activity and solicitation, the range is 56–60%. By comparison, the values for direct genetic effects range from 59% to 62% (table 1). This clearly underlines the significance of indirect effects caused by genetic variation in other family members. For trait evolution, the impact of the significant contribution of indirect effects to trait variation is crucial as they can impose constraints on evolution [2].
(b). Candidate gene analysis
Our hypothesis was that variation in behavioural traits observed during family interactions are caused by genetic differences between the lines (i.e. genotypes). We used a systems genetics analysis for directly affected traits to identify a list of candidate genes within the loci identified in the mapping analysis. However, we note that as with any mapping study, our loci contain many genes (for an exact number see electronic supplementary material, table S2). While we focused on those candidates that have functional polymorphisms among the BXD genotypes, it may be possible that causal genes are located outside the interval, or affect expression of genes outside the locus.
We begin with SocInt2 (chromosome 2, 73.310–77.355 Mb). This locus directly affected offspring solicitation on d14, as well as maternal suckling behaviour indirectly. Genes within this locus are linked to early growth and body weight. Twenty of the 49 genes within this locus have nsSNPs or indels among the BXD genotypes and, among those 20, several alter behaviour and weight, such as Chn1 and Atf2 [28,29] and Nfe2l2 [29,30], which are thus the best candidates.
SocInt4 (chromosome 4, 10.826–13.031 Mb) affected offspring sucking directly, and maternal suckling, sibling activity, and sucking behaviour indirectly during early lactation (day 6). Only 13 genes within the locus have nsSNPs or indels, producing a limited list of potential candidates. A potential candidate is Dpy19l4, which contains 18 nsSNPS and three indels, and has been linked to early brain development [31,32]. Rbm35a (now known as Esrp1) is another candidate, as ablation causes cleft lip and palate [33], and therefore more minor changes may alter the ability to suck.
Next, SocInt10 (chromosome 10, 97.112–103.025 Mb) influenced pup sucking directly, and sibling sucking and maternal suckling indirectly. This region has been linked to several weight and obesity QTL. Only seven of the 27 genes within the confidence interval have nsSNPs or indels. Dusp6 is a possible candidate, as it contains three nsSNPs, and is involved in several metabolic pathways, and is important for normal development [34]. Cep290 is another potential candidate, as mutations in the gene have been linked to slow postnatal weight gain (https://www.jax.org/strain/013702), which may be due to reduced feeding. Mutations in the gene have also been linked to problems with vision, olfaction, and taste [35–37], possibly causing pups to be less responsive to behavioural signals from the mother.
Finally, SocInt15 (chromosome 15, 3.229–7.273 Mb) affected the level of offspring sucking behaviour directly, and, indirectly, maternal suckling and activity, and sibling sucking during the weaning period on d14. Within the locus are 11 genes with nsSNPs or indels, producing a small number of candidate genes. We find two out of eight possible candidates alter behaviour, Ghr and Sepp1. Ghr influences feeding behaviour and growth [38], while Sepp1 influences parental behaviour [39] and grooming behaviour [40]. Sepp1 has nsSNPs and Ghr has both nsSNPs and indels.
4. Conclusion
The key result of our study is that IGE significantly contribute to explaining differences between family members in behavioural strategies, and are as important as direct genetic effects, with fundamental consequences for trait evolution [2]. We have shown that indirect effects can have counterintuitive effects, for example, in sibling competition whereby litter mates can actually benefit from increased sucking behaviour in a focal individual. This result supports predictions from recent coadaptation models [41], which demonstrated that in family social environments coadaptation accelerates the evolution of cooperation rather than competition. Indeed, our empirical support for these theoretical predictions casts some doubt on the commonly adopted sibling conflict paradigm [7]. Clearly, the dynamics and consequences of selection pressures on behavioural strategies during family interactions are more complex than one might assume. Our study shows that genetic variants involved in different pathways ranging from morphology to behavioural development may affect our traits, and allelic variation at these loci increases or reduces the trait value.
By identifying the direct and indirect sources of genetic variation in key traits during family interactions our study demonstrates that trait variation is not just caused by genes expressed in a focal individual but that such direct effects influence variation in multiple traits in different family members indirectly. Thus, direct effect loci may cause indirect effects either pleiotropically or by being closely linked to variants causing indirect effects. Our results suggest that we need to view behavioural strategies really as the result of selection on genetic variation in all interacting partners.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Matthew Cobb, Tucker Gilman, and James McInerney for helpful comments, and Robert W. Williams, Lu Lu, and Arthur Centeno for providing the animals, assistance in the set-up of this project, and the use of GeneNetwork.
Data accessibility
Data are available on genenetwork.org using the following record ID numbers: 18781–18790.
Authors' contributions
D.G.A. analysed data and wrote manuscript. N.S. analysed data. R.H. conceived study, analysed data, and wrote manuscript. All authors gave final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
This research was funded by NERC, UK NE/I001395/1.
References
- 1.Schneider J, Atallah J, Levine JD. 2017. Social structure and indirect genetic effects: genetics of social behaviour. Biol. Rev. Camb. Phil. Soc. 92, 1027–1038. ( 10.1111/brv.12267) [DOI] [PubMed] [Google Scholar]
- 2.Moore AJ, Brodie ED III, Wolf JB. 1997. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362. ( 10.2307/2411187) [DOI] [PubMed] [Google Scholar]
- 3.Royle NJ, Russell AF, Wilson AJ. 2014. The evolution of flexible parenting. Science 345, 776–781. ( 10.1126/science.1253294) [DOI] [PubMed] [Google Scholar]
- 4.Dingemanse NJ, Araya-Ajoy YG. 2015. Interacting personalities: behavioural ecology meets quantitative genetics. Trends Ecol. Evol. 30, 88–97. ( 10.1016/j.tree.2014.12.002) [DOI] [PubMed] [Google Scholar]
- 5.Badyaev AV, Uller T. 2009. Parental effects in ecology and evolution: mechanisms, processes and implications. Phil. Trans. R. Soc. B. 364, 1169–1177. ( 10.1098/rstb.2008.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilner RM, Boncoraglio G, Henshaw JM, Jarrett BJM, De Gasperin O, Attisano A, Kokko H. 2015. Parental effects alter the adaptive value of an adult behavioural trait. Elife 4, e07340 ( 10.7554/eLife.07340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trivers RL. 1974. Parent-offspring conflict. Am. Zool. 14, 249–264. ( 10.1093/icb/14.1.249) [DOI] [Google Scholar]
- 8.Kölliker M, Boos S, Wong JWY, Röllin L, Stucki D, Raveh S, Wu M, Meunier J. 2015. Parent-offspring conflict and the genetic trade-offs shaping parental investment. Nat. Commun. 6, 6850 ( 10.1038/ncomms7850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashbrook DG, Gini B, Hager R. 2015. Genetic variation in offspring indirectly influences the quality of maternal behaviour in mice. Elife 4, e11814 ( 10.7554/eLife.11814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hager R, Johnstone RA. 2003. The genetic basis of family conflict resolution in mice. Nature 421, 533–535. ( 10.1038/nature01239) [DOI] [PubMed] [Google Scholar]
- 11.Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy M-F, Henry H, Schoonjans K, Williams RW, Auwerx J. 2012. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell 150, 1287–1299. ( 10.1016/j.cell.2012.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peirce JL, Lu L, Gu J, Silver LM, Williams RW. 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5, 7 ( 10.1186/1471-2156-5-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mozhui K, Wang X, Chen J, Mulligan MK, Li Z, Ingles J, Chen X, Lu L, Williams RW. 2011. Genetic regulation of Nrxn1 expression: an integrative cross-species analysis of schizophrenia candidate genes. Transl. Psychiatry 1, e25 ( 10.1038/tp.2011.24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hager R, Johnstone RA. 2005. Differential growth of own and alien young in mixed litters of mice: a role for genomic imprinting? Ethology 111, 705–714. ( 10.1111/j.1439-0310.2005.01097.x) [DOI] [Google Scholar]
- 15.Hager R, Johnstone RA. 2007. Maternal and offspring effects influence provisioning to mixed litters of own and alien young in mice. Anim. Behav. 74, 1039–1045. ( 10.1016/j.anbehav.2007.01.021) [DOI] [Google Scholar]
- 16.Grafen A, Hails R. 2002. Modern statistics for the life sciences, 1st edn Oxford, UK: Oxford University Press. [Google Scholar]
- 17.Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4, 2227–2237. ( 10.1371/journal.pbio.0040395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manichaikul A, Dupuis J, Sen S, Broman KW. 2006. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics 174, 481–489. ( 10.1534/genetics.106.061549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. ( 10.2307/2346101) [DOI] [Google Scholar]
- 20.Chen L, Storey JD. 2006. Relaxed significance criteria for linkage analysis. Genetics 173, 2371–2381. ( 10.1534/genetics.105.052506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, Wolf JB. 2008. Genomic imprinting effects on adult body composition in mice. Proc. Natl Acad. Sci. USA 105, 4253–4258. ( 10.1073/pnas.0706562105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hager R, Cheverud JM, Leamy LJ, Wolf JB. 2008. Sex dependent imprinting effects on complex traits in mice. BMC Evol. Biol. 8, 303 ( 10.1186/1471-2148-8-303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Yekutieli D. 2005. Quantitative trait loci analysis using the false discovery rate. Genetics 171, 783–790. ( 10.1534/genetics.104.036699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bello SM, Smith CL, Eppig JT. 2015. Allele, phenotype and disease data at Mouse Genome Informatics: improving access and analysis. Mamm. Genome 26, 285–294. ( 10.1007/s00335-015-9582-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberts R, Schughart K. 2010. QTLminer: identifying genes regulating quantitative traits. BMC Bioinformatics 11, 516 ( 10.1186/1471-2105-11-516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf JB, Cheverud JM. 2009. A framework for detecting and characterizing genetic background-dependent imprinting effects. Mamm. Genome 20, 681–698. ( 10.1007/s00335-009-9209-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pick JL, Ebneter C, Hutter P, Tschirren B. 2016. Disentangling genetic and prenatal maternal effects on offspring size and survival. Am. Nat. 188, 628–639. ( 10.1086/688918) [DOI] [PubMed] [Google Scholar]
- 28.Borgius L, Nishimaru H, Caldeira V, Kunugise Y, Löw P, Reig R, Itohara S, Iwasato T, Kiehn O. 2014. Spinal glutamatergic neurons defined by EphA4 signaling are essential components of normal locomotor circuits. J. Neurosci. 34, 3841–3853. ( 10.1523/JNEUROSCI.4992-13.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegmeyer H, et al. 2007. EphA4-dependent axon guidance is mediated by the RacGAP α2-chimaerin. Neuron 55, 756–767. ( 10.1016/j.neuron.2007.07.038) [DOI] [PubMed] [Google Scholar]
- 30.Xue P, et al. 2013. Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes 62, 845–854. ( 10.2337/db12-0584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiura N, Patel RG, Corriveau RA. 2001. n-methyl-d-aspartate receptors regulate a group of transiently expressed genes in the developing brain. J. Biol. Chem. 276, 14 257–14 263. ( 10.1074/jbc.M100011200) [DOI] [PubMed] [Google Scholar]
- 32.Watanabe K, Takebayashi H, Bepari AK, Esumi S, Yanagawa Y, Tamamaki N. 2011. Dpy19l1, a multi-transmembrane protein, regulates the radial migration of glutamatergic neurons in the developing cerebral cortex. Development 138, 4979–4990. ( 10.1242/dev.068155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bebee TW, Park JW, Sheridan KI, Warzecha CC, Cieply BW, Rohacek AM, Xing Y, Carstens RP. 2015. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife 4, e08954 ( 10.7554/eLife.08954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Scott DA, Hatch E, Tian X, Mansour SL. 2007. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development 134, 167–176. ( 10.1242/dev.02701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang B, et al. 2006. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 15, 1847–1857. ( 10.1093/hmg/ddl107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cideciyan AV, et al. 2011. Cone photoreceptors are the main targets for gene therapy of NPHP5 (IQCB1) or NPHP6 (CEP290) blindness: generation of an all-cone Nphp6 hypomorph mouse that mimics the human retinal ciliopathy. Hum. Mol. Genet. 20, 1411–1423. ( 10.1093/hmg/ddr022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEwen DP, Koenekoop RK, Khanna H, Jenkins PM, Lopez I, Swaroop A, Martens JR. 2007. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl Acad. Sci. USA 104, 15 917–15 922. ( 10.1073/pnas.0704140104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egecioglu E, et al. 2006. Growth hormone receptor deficiency results in blunted ghrelin feeding response, obesity, and hypolipidemia in mice. Am. J. Physiol. Endocrinol. Metab. 290, E317–E325. ( 10.1152/ajpendo.00181.2005) [DOI] [PubMed] [Google Scholar]
- 39.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. 2003. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 278, 13 640–13 646. ( 10.1074/jbc.M300755200) [DOI] [PubMed] [Google Scholar]
- 40.Raman AV, Pitts MW, Seyedali A, Hashimoto AC, Seale LA, Bellinger FP, Berry MJ. 2012. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav. 11, 601–613. ( 10.1111/j.1601-183X.2012.00794.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drown DM, Wade MJ. 2014. Runaway coevolution: adaptation to heritable and nonheritable environments. Evolution 68, 3039–3046. ( 10.1111/evo.12470) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on genenetwork.org using the following record ID numbers: 18781–18790.


