Abstract
Cutaneous squamous cell carcinoma (cSCC) differs from SCC of other organs in its strong association with chronic sun exposure. However, the specific driver mutations in cSCC remain unknown. Fusion genes in established cSCC cell lines (A431 and DJM-1) were predicted by transcriptome sequence, and validated by Sanger sequence, fluorescence in situ hybridization and G-banding. By transcriptome sequencing, we identified fusion gene EGFR-PPARGC1A in A431, which were expressed in 31 of 102 cSCCs. The lesions harboring the fusion gene tended to be located in sun-exposed areas. In vivo cutaneous implantation of EGFR-PPARGC1A-expressing NIH3T3 induced tumors resembling human cSCC, indicating its potent tumorigenicity. NIH3T3 transfected with EGFR-PPARGC1A as well as A431 showed increased cell proliferation activity. With regard to underlying mechanism, EGFR-PPARGC1A protein causes constitutive tyrosine phosphorylation, and induces the phosphorylation of wild-type full-length epidermal growth factor receptor (EGFR) by dimerization. Conversely, the RNAi-mediated attenuation of EGFR or CRISPR/Cas9-mediated knockdown of the fusion gene in A431 led to a decrease in the cell number, and may have therapeutic value. Our findings advance the knowledge concerning genetic causes of cSCC and the function of EGFR, with potential implications for new diagnostic and therapeutic approaches.
Introduction
Cutaneous squamous cell carcinoma (cSCC), which originates from the keratinocytes of the epidermis in elderly patients, is one of the most common types of skin cancer. Although SCC can be seen in many other internal organs, including the esophagus, lungs, and urinary bladder, cSCC is characterized by its strong association with chronic and cumulative sun exposure.
Most of cSCCs are considered low risk and can be treated by local excision. However, when they progress, their potential to recur and metastasize leads to a poor prognosis and is associated with significant mortality. Although an early diagnosis is important, some atypical or well-differentiated cases are difficult to diagnose clinically or histopathologically. At present, the serum SCC antigen level is the only available tumor marker. Unfortunately, this marker only becomes elevated at the late stage1. Furthermore, although radiotherapy and chemotherapy have been utilized in the treatment of advanced cSCC, the tumor is sometimes resistant to such standard treatments. Clarifying the pathogenesis of this tumor may be helpful for the development of novel biomarkers and new therapeutic strategies.
We previously reported that various cytokines and microRNAs are involved in the pathogenesis of cSCC2–5. On the other hand, other publications have reported several genetic abnormalities, including TP53 mutations, in cSCC6. However, there may be more specific driver mutations in this tumor. To date, chromosomal translocation with corresponding fusion genes has been widely identified in various tumors, especially mesenchymal neoplasms and sarcomas. Among skin tumors, dermatofibrosarcoma protuberans is associated with t(17;22) translocation. The t(17;22) translocation causes COL1A1-PDGFB rearrangement, and COL1A1-PDGF fusion protein stimulates PDGF receptor, leading to malignant transformation7. Because COL1A1-PDGFB is highly specific to dermatofibrosarcoma protuberans, the detection is clinically helpful for the diagnosis. In addition, imatinib has been found to have anti-tumor effect through its inhibition of fusion gene-induced PDGF receptor activation. Accordingly, the identification of fusion genes in malignant tumors is important for the development of novel treatments. However, no specific fusion genes have been identified in cSCC.
In the present study, we describe fusion gene EGFR-PPARGC1A, which was found in 31 of 102 cSCC patients. We analyzed the clinical significance, and proved that the fusion play causal roles in tumor development in vitro and in vivo.
Result
The identification of cSCC-specific fusion genes in A431
High-quality RNAs were isolated from NHEKs or two cSCC cell lines (A431 and DJM-1), and were analyzed by paired-end transcriptome sequencing on Illumina HiSeq2000 platform: among all of the reads, 36,796,013 (90.48%) for NHEKs, 36,748,245 (89.80%) for A431, and 39,474,844 (90.38%) for DJM-1 were mapped to the reference genome (GRCh37/hg19).
Initially, for the analysis of the differential gene expression between A431 and NHEKs, fragments per kilobase per million map reads (FPKM) values were calculated, and the statistical significance was evaluated using Cuffdiff (Supplementary Fig. S1A). As shown in Supplementary Table S1, when the cut-off value was set at 28-fold-change with false discovery rate (FDR) of <0.01, 16 or 54 genes were estimated to be substantially up- or down-regulated in A431 in comparison to NHEKs, respectively. Several of these changes in A431 that were found in the present study were compatible with previous reports: For example, MAGEA4, MMP13, and SPP1, which have been reported to be overexpressed in cSCC cells in vivo and in vitro 8–10, were also up-regulated in A431 in our transcriptome analysis. However, the cluster analysis did not indicate a specific signature of signal pathways or cellular activities (Supplementary Fig. S1B).
Thus, we next examined the presence of A431-specific point mutations in seven genes related to SCC of other organs and 36 oncogenes. In addition to several neutral polymorphisms and amino acid substitutions that represent common variations, 13 single nucleotide variations were detected in A431 when compared to NHEKs and DJM-1, as well as the reference genome in bioinformatic analysis (Supplementary Table S2). Among these, TP53 R273H point mutation had already been described in A43111.
To identify more specific gene changes, we then examined the presence of novel fusion transcripts in A431. We focused on four candidates that were only predicated in A431 and not in NHEKs or DJM-1, using both of two different softwares, deFuse and Fusion Hunter (Supplementary Table S3). Among these, 16 spanning reads and 8 spanning mate pairs indicated fusion between exon16 of epidermal growth factor receptor (EGFR) and exon2 of PPARγ coactivator 1-α (PPARGC1A), which are located at 7p12 and 4p15.1, respectively (Fig. 1A and Supplementary Fig. S2A-B). It is noteworthy that the gene fusion caused a frameshift change of PPARGC1A, and that the stop codon appears at only 23 amino acids from the breakpoint. Specific primers designed for EGFR exon16 and PPARGC1A exon2 were used for RT-PCR to validate the finding (Fig. 1B). The expected fragment was amplified by PCR using RNA from A431, but not RNA from NHEKs, DJM-1, cultured human endothelial cells, or dermal fibroblasts. Sanger sequencing of the amplified fragments confirmed the presence of gene fusion between EGFR and PPARGC1A (Fig. 1A). The above-mentioned gene expression analysis indicated that EGFR level in A431 was not substantially up-regulated in comparison to NHEKs (2.95-fold change, q-value = 0.99).
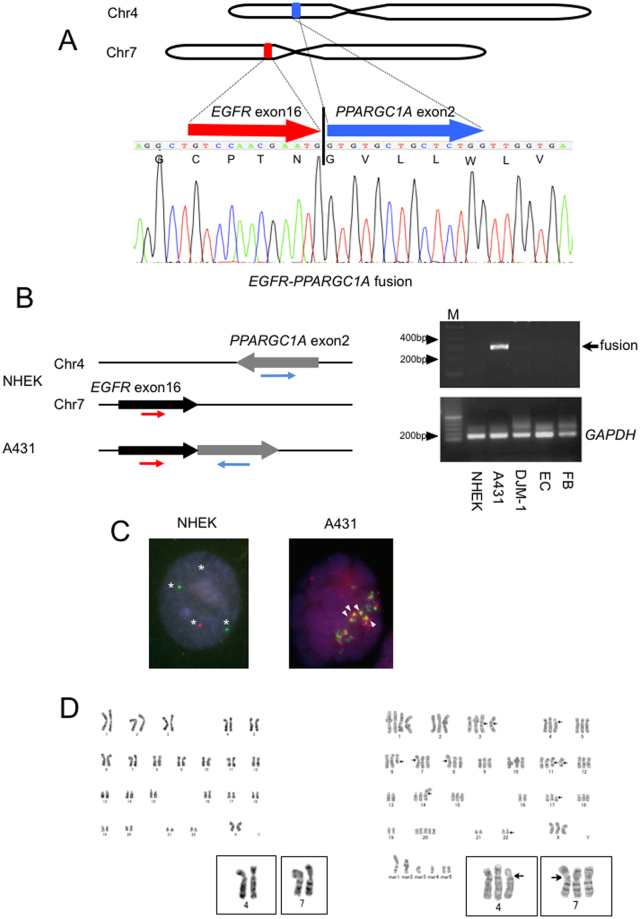
Figure 1.
The identification of a novel EGFR-PPARGC1A fusion in A431. (A) (Upper panel) A schematic diagram showing gene structures of EGFR (red) and PPARGC1A (blue) in relation to the formation of the EGFR-PPARGC1A fusion transcript. (Lower panel) The partial sequence of the EGFR-PPARGC1A fusion transcript at the break point, along with the predicted amino acid sequence. The transcript is a fusion of EGFR exon 16 to PPARGC1A exon 2. (B) (Left panel) The RT-PCR-based detection of the EGFR-PPARGC1A fusion. The presence of the EGFR-PPARGC1A fusion transcript was verified by RT-PCR using the EGFR forward (red) and PPARGC1A reverse primers (blue). The fusion gene-specific primers were designed to not amplify the wild-type full-length EGFR or PPARGC1A. (Right panel) The PCR products by the fusion gene-specific primers or GAPDH primers and ladder marker were run out on agarose gels containing ethidium bromide. Lane M, 100-bp marker; lane 5, cultured human dermal microvascular endothelial cells (EC); lane 6, normal human dermal fibroblasts (FB). The arrow indicates the PCR product of the EGFR-PPARGC1A-specific primers (287-bp). The GAPDH levels (118-bp) were shown as the control. (C) A FISH analysis with human EGFR- (green) and PPARGC1A-specific (red) probes. NHEKs (left) had two green or red signals (indicated by asterisks), while increased green/red signals were detected in A431 (right). The white arrowheads indicate the fusion signals (yellow). (D) The result of a chromosome analysis. (Left) The karyotypes of NHEKs, which were diploid: 46, XX. (Right) A431 had a complex abnormal karyotype and was aneuploid. The karyotype was 72-80,XXX,+1,+3,i(3)(q10)×2,add(4)(p11),add(6)(q13), add(7)(p13),add(8)(p11.2),+11,add(11)(q13)×2,-13,i(14)(q10),-16,add(17)(p11.2),-19,+20,+20,-21,-22,add(22)(q13),+mar1,+mar2,+mar3,+mar4,+mar5×2 [cp20]. The arrows indicate the putative break points. The insets depict chromosomes 4 and 7.
EGFR-PPARGC1A was not found in eight normal skin samples or in other tumor tissue specimens, including seven melanoma and five basal cell carcinoma (BCC) by RT-PCR (Supplementary Table S3). Thus, the fusion may be specific to cSCC. Furthermore, the other three candidate fusion genes predicted in A431 by transcriptome analysis (Supplementary Table S3) or PPARGC1A-EGFR fusion were not detected in the cell line by RT-PCR validation.
To characterize the rearrangements at the chromosomal level, two-color fluorescence in situ hybridization (FISH) analysis was performed (Fig. 1C). Two separate green (EGFR) and red (PPARGC1A) signals could be detected in NHEKs, while increased single signals were detected in A431, indicating their aneuploidy. The presence of EGFR-PPARGC1A fusion was confirmed by several yellow signals that were found in all of 100 interphase or metaphase cells determined.
G-banding demonstrated that there was aneuploidy in many chromosomes, including chromosome4 and 7, of A431 (Fig. 1D). However, for example, gene expression analysis showed that only 3 of 1,436 chromosome7 genes were substantially up-regulated, while five genes were down-regulated in A431 cell. Thus, there seems to be no correlation between aneuploid chromosome7 and the expression levels of chromosome7 genes (e.g. due to DNA methylation).
The clinical significance of EGFR-PPARGC1A
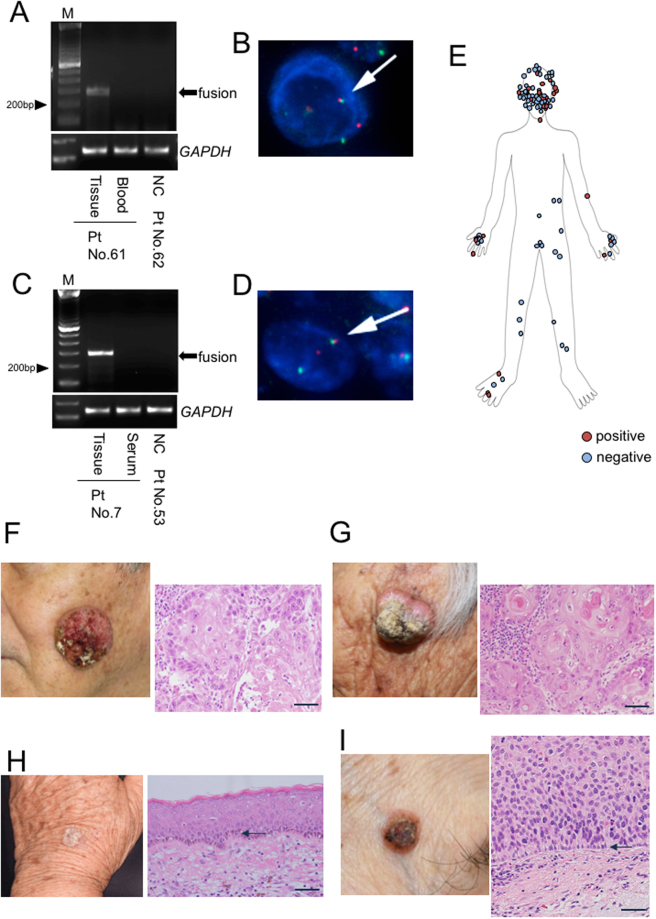
To determine whether the EGFR-PPARGC1A fusion was recurrent, we evaluated the frequency using RNAs obtained from an increased number of paraffin sections of human tumors using fusion-specific RT-PCR. We screened 102 cSCC specimens and 106 other tumors (BCC, n = 38; melanoma, n = 20; Paget disease, n = 20; dermatofibrosarcoma protuberans, n = 14; and angiosarcoma, n = 14), and detected the fusion gene specifically in 31 cSCC without junctional variability (Table 1). Each of these detections was confirmed by Sanger sequencing and FISH: the sensitivity and specificity of the fusion for cSCC was 30.4% and 100%, respectively (Supplementary Table S4). For example, the amplification by fusion gene-specific primer pair was observed in a cSCC tumor RNA sample from patient No. 61 (Fig. 2A), but not in the peripheral blood cell RNA of the same individual. Thus, the rearrangement was somatically acquired. FISH analysis indicated the single allele localization of chromosomes 4 and 7 as well as their aneuploidy in the tissue DNA specimen of the patient (Fig. 2B). On the other hand, the fusion gene was present in the tumor tissue of patient No.7, but not in the patient’s serum (Fig. 2C), suggesting that the detection of EGFR-PPARGC1A was not useful as a serum tumor marker. The gene fusion in the tissue sample was also confirmed by FISH analysis (Fig. 2D). As the clinical significance of the fusion gene, we found that the lesions positive for EGFR-PPARGC1A were located in sun-exposed areas substantially more frequently than those negative for the fusion (42.9% vs 0%, P < 0.05, Fig. 2E). Furthermore, three cSCCs of the genital region were negative for the fusion. The other features of the patients with (Fig. 2F) and without the fusion gene (Fig. 2G) were basically similar: both patient groups clinically presented with nodules, tumors, or ulcers. A known genesis, such as burns or traumas, was not correlated with fusion positivity (Table 1). Histopathologically, both poorly- and well-differentiated lesions consisted of pleomorphic and hyperchromatic cells were seen in both of patients with and without fusion gene (Fig. 2F,G). Nuclear atypia and mitotic figures were also found. Both groups included two in situ forms of cSCC, actinic keratosis and Bowen’s disease (Fig. 2H,I).
Table 1.
The clinical and pathological data for the cSCC patients with and without the EGFR-PPARGC1A fusion gene.
| No. | Age | Sex | Diagnosis | Duration | Region | Size | Appearance | SCC antigen | Genesis | T | N | M | Stage | EGFR-PPARGC1A | FGFR3-TACC3 | AML4-ALK | NSD3-NUT | GOLM1-MAK10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 | M | AK | 36 | cheek | 1 cm | erythema | is | 0 | 0 | 0 | + | − | − | − | − | ||
| 2 | 88 | M | Bowen | 0 | cheek | 1 cm | tumor | 1.6 | is | 0 | 0 | 0 | + | − | − | − | − | |
| 3 | 93 | F | AK | 12 | cheek | 1 cm | tumor | 0.9 | is | 0 | 0 | 0 | − | − | − | − | − | |
| 4 | 73 | M | AK | foot | 2 cm | tumor | 0.6 | is | 0 | 0 | 0 | − | − | − | − | − | ||
| 5 | 68 | F | AK | 48 | hand | 3.3 × 2.2 cm | tumor | 0.5 | is | 0 | 0 | 0 | + | − | − | − | − | |
| 6 | 81 | M | Bowen | cheek | 1.2 × 1.0 cm | erythema | is | 0 | 0 | 0 | − | − | − | − | − | |||
| 7 | 90 | F | AK | 1 | head | 1.2 cm | tumor | 1.1 | is | 0 | 0 | 0 | + | − | − | − | − | |
| 8 | 68 | M | SCC | 24 | hand | 0.5 cm | ulcer | 1.2 | burn | 1 | 0 | 0 | I | + | − | − | − | − |
| 9 | 87 | F | SCC | finger | 1 cm | tumor | 1 | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 10 | 91 | F | SCC | cheek | 1 cm | tumor | 1 | 0 | 0 | I | + | − | − | − | − | |||
| 11 | 95 | F | SCC | cheek | 2 cm | tumor | 0.9 | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 12 | 94 | F | SCC | 60 | cheek | tumor | 1 | 0 | 0 | I | − | − | − | − | − | |||
| 13 | 86 | F | SCC | 12 | cheek | 1.2 × 1.5 cm | tumor | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 14 | 82 | F | SCC | 4 | lip | 1 cm | tumor | 1.1 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 15 | 64 | M | SCC | cheek | 1.2 × 0.8 cm | tumor | 0.6 | burn | 1 | 0 | 0 | I | − | − | − | − | − | |
| 16 | 54 | F | SCC | 1 | eye lid | 2 cm | tumor | 0.9 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 17 | 82 | M | SCC | 5 | foot | 1.1 × 1.0 cm | tumor | 1.3 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 18 | 81 | M | SCC | 7 | finger | 2 × 1.5 cm | tumor | 4.2 | trauma | 1 | 0 | 0 | I | + | − | − | − | − |
| 19 | 67 | M | SCC | 36 | inguinal | 2 cm | tumor | 1 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 20 | 77 | M | SCC | 3 | hand | 1.0 × 1.2 cm | tumor | 1.1 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 21 | 86 | M | SCC | 4 | nose | 0.5 cm | tumor | 2.4 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 22 | 89 | M | SCC | 24 | lip | 1 cm | tumor | 2.2 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 23 | 77 | M | SCC | hand | 2 cm | tumor | 1.4 | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 24 | 66 | M | SCC | 12 | ear | 0.7 cm | erosion | 1.1 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 25 | 72 | M | SCC | 6 | ear | 1 cm | tumor | 1 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 26 | 63 | M | SCC | 1 | nose | 0.5 cm | tumor | 1.7 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 27 | 72 | M | SCC | 0 | cheek | 0.8 cm | tumor | 1.4 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 28 | 73 | F | SCC | 3 | head | 2 cm | tumor | 0.9 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 29 | 91 | F | SCC | 36 | cheek | 1 cm | tumor | 0.8 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 30 | 81 | F | SCC | 3 | cheek | 0.3 cm | tumor | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 31 | 83 | M | SCC | 12 | cheek | 1.1×0.8 cm | tumor | 1.7 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 32 | 74 | M | SCC | 3 | hand | 1 cm | tumor | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 33 | 87 | F | SCC | 6 | cheek | 1.2×0.8 cm | tumor | 1.2 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 34 | 82 | M | SCC | 6 | head | 1 cm | tumor | 0.9 | 1 | 0 | 0 | I | + | + | − | − | − | |
| 35 | 72 | M | SCC | 12 | foot | 0.5 cm | ulcer | 0.8 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 36 | 76 | M | SCC | 6 | hand | 0.7 cm | tumor | 2.9 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 37 | 85 | F | SCC | 600 | cheek | 2 cm | tumor | 1.5 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 38 | 71 | M | SCC | 96 | hand | 0.8 cm | tumor | 1.5 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 39 | 94 | F | SCC | 3 | cheek | 2 cm | tumor | 1.2 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 40 | 86 | M | SCC | 2 | hand | 1 cm | tumor | 2.5 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 41 | 80 | F | SCC | hand | 1 cm | tumor | 2.5 | 1 | 0 | 0 | I | + | − | − | − | − | ||
| 42 | 90 | M | SCC | 3 | cheek | 1 cm | tumor | 2.2 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 43 | 77 | F | SCC | 0 | cheek | 0.3 cm | tumor | 0.5 | 1 | 0 | 0 | I | − | + | − | − | − | |
| 44 | 95 | F | SCC | 3 | ear | 1 cm | tumor | 3.1 | 1 | 0 | 0 | I | + | − | − | − | − | |
| 45 | 76 | M | SCC | 4 | cheek | 1.3 × 1.0 cm | tumor | 1.2 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 46 | 91 | F | SCC | hand | 1.5 cm | tumor | 1.9 | 1 | 0 | 0 | I | − | − | − | − | − | ||
| 47 | 84 | M | SCC | 12 | ear | 0.8 × 0.7 cm | tumor | 1 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 48 | 72 | M | SCC | 6 | cheek | 1.2 × 1.3 cm | tumor | 1.1 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 49 | 78 | M | SCC | 2 | cheek | 1.1 cm | tumor | 1.5 | 1 | 0 | 0 | I | − | − | − | − | − | |
| 50 | 90 | M | SCC | 12 | head | 2 cm | erosion | 1.7 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 51 | 85 | M | SCC | head | 4.5 × 4.6 cm | tumor | 0.7 | 2 | 0 | 0 | II | + | − | − | − | − | ||
| 52 | 91 | F | SCC | 2 | inguinal | 3.5 × 4 cm | tumor | 5.8 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 53 | 66 | M | SCC | 0 | foot | 5.5 × 2 cm | ulcer | 1.5 | burn | 2 | 0 | 0 | II | − | − | − | − | − |
| 54 | 59 | M | SCC | foot | 2 cm | tumor | 0.9 | 2 | 0 | 0 | II | + | − | − | − | − | ||
| 55 | 76 | F | SCC | 48 | lip | 3 cm | tumor | 3.2 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 56 | 76 | M | SCC | 48 | head | 2.1 × 1.3 cm | tumor | 0.6 | burn | 2 | 0 | 0 | II | + | − | − | − | − |
| 57 | 78 | M | SCC | 4 | elbow | 10 cm | tumor | 1.4 | trauma | 2 | 0 | 0 | II | + | − | − | − | − |
| 58 | 85 | M | SCC | 1 | finger | 2.5 × 2.0 cm | erosion | 4.3 | 2 | 0 | 0 | II | + | − | − | − | − | |
| 59 | 91 | F | SCC | nose | 2.1 × 1.6 cm | tumor | 1.4 | 2 | 0 | 0 | II | − | − | − | − | − | ||
| 60 | 81 | M | SCC | 3 | ear | 2 × 2 cm | tumor | 2.3 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 61 | 75 | F | SCC | 3 | lip | 2 cm | tumor | 0.8 | 2 | 0 | 0 | II | + | − | − | − | − | |
| 62 | 72 | M | SCC | 1 | foot | 4 cm | ulcer | 3 | trauma | 2 | 0 | 0 | II | − | − | − | − | − |
| 63 | 53 | M | SCC | 6 | lip | 2 cm | tumor | 0.8 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 64 | 76 | M | SCC | hand | 2 cm | tumor | 1.2 | 2 | 0 | 0 | II | − | − | − | − | − | ||
| 65 | 71 | F | SCC | 120 | inguinal | 2 cm | ulcer | 1.7 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 66 | 85 | M | SCC | 1 | neck | 2 cm | tumor | 2 | 0 | 0 | II | + | − | − | − | − | ||
| 67 | 90 | M | SCC | 120 | abdomen | 3.5 × 2.5 cm | tumor | 0.8 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 68 | 59 | F | SCC | 48 | abdomen | 8 cm | tumor | 11.5 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 69 | 69 | M | SCC | 24 | knee | 8 cm | scar | 3.1 | burn | 2 | 0 | 0 | II | − | − | − | − | − |
| 70 | 61 | M | SCC | 4 | joe | 2 cm | tumor | 1.5 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 71 | 79 | M | SCC | 48 | head | 2 cm | tumor | 2.4 | burn | 2 | 0 | 0 | II | − | − | − | − | − |
| 72 | 87 | M | SCC | 24 | hand | 2 cm | tumor | 1.3 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 73 | 84 | F | SCC | 6 | foot | 2 cm | tumor | 0.5 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 74 | 89 | F | SCC | 24 | finger | 2 × 1.5 cm | tumor | 0.8 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 75 | 91 | F | SCC | 2 | foot | 2 × 1.5 cm | tumor | 1.3 | 2 | 0 | 0 | II | + | − | − | − | − | |
| 76 | 70 | M | SCC | 60 | head | 3 cm | tumor | 1.4 | burn | 2 | 0 | 0 | II | − | − | − | − | − |
| 77 | 85 | F | SCC | 4 | head | 2.5 cm | tumor | 1.2 | 2 | 0 | 0 | II | + | − | − | − | − | |
| 78 | 94 | M | SCC | 12 | foot | 7.5 cm | erosion | 1.9 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 79 | 56 | M | SCC | 0.5 | nose | 2.5 cm | tumor | 1.4 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 80 | 83 | M | SCC | 12 | ear | 2.9 × 2.0 cm | tumor | 0.7 | 2 | 0 | 0 | II | − | − | − | − | − | |
| 81 | 88 | F | SCC | 24 | head | 2.5 × 1.7 cm | tumor | 1.1 | burn | 2 | 0 | 0 | II | − | − | − | − | − |
| 82 | 78 | F | SCC | cheek | 2.5 cm | tumor | 0.8 | 2 | 0 | 0 | II | − | − | − | − | − | ||
| 83 | 69 | M | SCC | 60 | back | 2 cm | tumor | 9.4 | 2 | 1 | 0 | III | − | − | − | − | − | |
| 84 | 75 | M | SCC | 3 | cheek | 4 cm | tumor | 2 | 2 | 1 | 0 | III | + | − | − | − | − | |
| 85 | 91 | F | SCC | 12 | head | 2.7 cm | tumor | 1.8 | 2 | 1 | 0 | III | − | − | − | − | − | |
| 86 | 94 | F | SCC | 6 | head | 4 × 3 cm | tumor | 2.9 | 3 | 0 | 0 | III | − | − | − | − | − | |
| 87 | 76 | M | SCC | 3 | lip | 4.5 × 2 cm | tumor | 1.4 | 2 | 1 | 0 | III | + | − | − | − | − | |
| 88 | 44 | M | SCC | 3 | foot | tumor | 2 | burn | 4 | 1 | 0 | III | − | − | − | − | − | |
| 89 | 73 | F | SCC | 3 | head | 0.8 × 0.8 cm | ulcer | 1.8 | 1 | 1 | 0 | III | − | − | − | − | − | |
| 90 | 83 | M | SCC | 1 | cheek | 2.2 cm | tumor | 2.4 | x | 1 | 0 | III | + | − | − | − | − | |
| 91 | 82 | F | SCC | 1 | cheek | 3 × 2 cm | tumor | 1.5 | 3 | 0 | 0 | III | + | − | − | − | − | |
| 92 | 63 | F | SCC | 0 | head | 0.8 | 3 | 0 | 0 | III | − | − | − | − | − | |||
| 93 | 71 | F | SCC | 0 | cheek | 1.1 | x | 1 | 0 | III | − | − | − | − | − | |||
| 94 | 84 | M | SCC | 36 | ear | ulcer | 1.4 | 4 | 0 | 0 | III | − | − | − | − | − | ||
| 95 | 54 | F | SCC | 3 | head | 8 × 7 cm | tumor | 3.9 | burn | 4 | 0 | 0 | IV | − | − | − | − | − |
| 96 | 68 | M | SCC | 6 | foot | 21.1 × 7.8 cm | tumor | 1.3 | x | 2 | 0 | IV | − | − | − | − | − | |
| 97 | 83 | M | SCC | 1 | foot | 0.5 × 0.3 cm | tumor | 0.5 | 1 | 1 | 1 | IV | + | − | − | − | − | |
| 98 | 80 | M | SCC | 0 | cheek | 3.7 | 1 | 2 | 0 | IV | − | − | − | − | − | |||
| 99 | 81 | M | SCC | 48 | foot | tumor | x | 1 | 1 | IV | − | − | − | − | − | |||
| 100 | 95 | F | SCC | 8 | cheek | 2 cm | ulcer | 3 | x | x | X | − | − | − | − | − | ||
| 101 | 71 | M | SCC | nose | ulcer | X | − | − | − | − | − | |||||||
| 102 | 94 | F | SCC | 12 | cheek | 2 cm | tumor | 1.2 | 2 | X | − | − | − | − | − |
AK, actinic keratosis; Bowen, Bowen’s disease; SCC, squamous cell carcinoma.
Figure 2.
The clinical and histopathological features of cSCC patients with or without EGFR-PPARGC1A. (A) The RT-PCR-based detection of EGFR-PPARGC1A-specific 287-bp fragment in the RNA of cSCC tumor tissue, but not in the blood RNA from the same individual (patient No. 61). M, 100-bp ladder; NC, negative control (patient No.62). The arrow indicates the PCR product of the EGFR-PPARGC1A-specific primers. The GAPDH levels were shown as the control. (B) A FISH analysis of the cSCC tissue sample of patient No.61 with human EGFR- (green) and PPARGC1A-specific (red) probes. The white arrowhead indicates the fusion signal. (C) The RT-PCR-based detection of EGFR-PPARGC1A-specific 287-bp fragment in the RNA of cSCC tumor tissue, but not in the serum RNA from the same individual (patient No. 7). M, 100-bp ladder; NC, negative control (patient No.53). The arrow indicates the PCR product of the EGFR-PPARGC1A-specific primers. The GAPDH levels were shown as the control. (D) A FISH analysis of the cSCC tissue sample of patient No.7 with human EGFR- (green) and PPARGC1A-specific (red) probes. The white arrowhead indicates the fusion signal. (E) The tumor distribution of 102 cSCC in our study. The red dots indicate the tumors that were positive for EGFR-PPARGC1A; the blue dots indicate those that were negative for the fusion. (F,G) Representative clinical pictures and histopathological findings of a patient with (F, patient No.84) and without EGFR-PPARGC1A (G, patient No.30). (Left panel) The clinical presentation of the tumors. (Right panel) The results of hematoxylin and eosin staining showing masses of large pleomorphic and hyperchromatic tumor cells with variable keratinization. Nuclear atypia and mitotic figures were also seen. Bar = 50μm. H, (I) Representative clinical pictures and histopathological findings of a patient with EGFR-PPARGC1A–positive actinic keratosis (H, No.1) and a patient with EGFR-PPARGC1A–positive Bowen’s disease (I, No.2). (Left panel) The clinical presentation of the tumors. (Right panel) The results of hematoxylin and eosin staining showing pleomorphic and hyperchromatic tumor cells in the basal layer (F, actinic keratosis) or in stratum spinosum of the epidermis (G, Bowen’s disease). The arrow indicates the basal layer. Bar = 50μm.
Several studies have reported the detection of fusion genes in SCC of other organs (AML4-ALK in lung SCC, FGFR3-TACC3 in head and neck SCC, NSD3-NUT in NUT midline carcinoma, and GOLM1-MAK10 in esophageal SCC). We determined their frequency in our cSCC patients. Two cases were positive for FGFR3-TACC3 fusion gene (the patients were also positive for EGFR-PPARGC1A or ADCK4-NUMBL), but no other fusion genes were detected (Table 1).
The functional analysis of EGFR-PPARGC1A
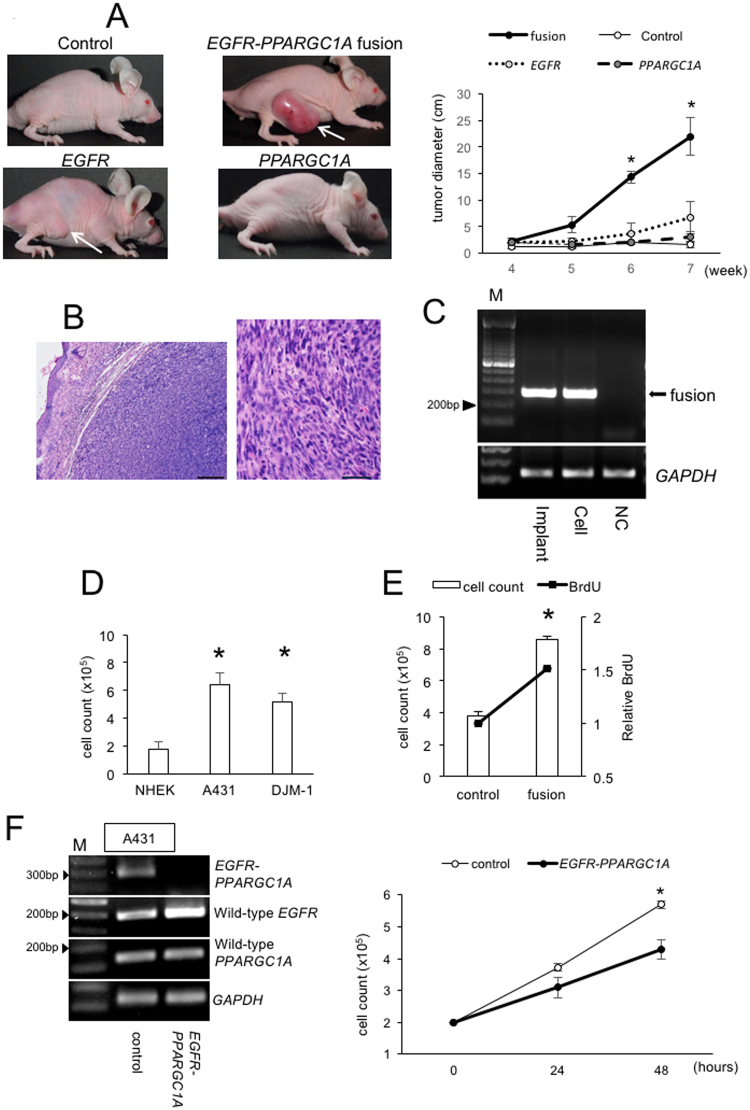
To clarify the function of EGFR-PPARGC1A protein (Supplementary Fig. S3A), the fusion gene was amplified by using PCR and cloned into lentiviral vector. We first tried to overexpress the fusion protein in NHEKs by lentiviral transfection; however, the cells were very fragile and stopped proliferating after transfection. Instead, we performed a tumorigenicity assay using NIH3T3, according to the methods of a previous study12. NIH3T3 that were stably transfected with the control vector, EGFR-PPARGC1A, full-length wild-type EGFR, or full-length PPARGC1A showed no apparent differences in cell shape (Supplemental Fig. S3B). Following subcutaneous implantation, tumorigenesis was observed in 11 of 14 samples of NIH3T3 transfected with the fusion gene, while the 12 samples transfected with the control vector did not cause tumorigenesis (Fig. 3A). The histopathological findings of the tumor tissue showed a mass of atypical, pleomorphic and hyperchromatic cells with mitosis (Fig. 3B), which were similar to the histopathological features of cSCC (Fig. 2F). We confirmed that the tumor tissues, as well as the cells cultured from them, were positive for EGFR-PPARGC1A by RT-PCR with fusion-specific primers (Fig. 3C). Tumor formation was also observed in 5 of the 7 mice with cells that overexpressed full-length EGFR gene, although their tumor size was smaller than fusion gene-overexpressing cells (Fig. 3A). Any of the 7 mice with cells overexpressing PPARCG1A did not show tumorigenesis. Taken together, these results suggest that EGFR-PPARGC1A drives tumorigenesis, probably through EGFR signaling.
Figure 3.
The forced overexpression of EGFR-PPARGC1A in NIH3T3. (A) NIH3T3 cells transfected with the control vector, EGFR-PPARGC1A fusion gene, full-length EGFR gene, or full-length PPARGC1A gene were injected into mouse skin (1 × 105 cells). (Left) After five weeks, tumor formation (white arrow) was observed in 11 of 14 mice that had been injected with cells overexpressing the fusion gene and 5 of 7 mice injected with cells overexpressing full-length EGFR. Tumor formation was not detected in the 7 mice injected with PPARCG1A-overexpressing cells or 12 mice with control vector. (Right) Tumor diameter was determined at the indicated times. Data are mean values and SD. *P < 0.05 (n = 3). (B) Hematoxylin and eosin staining of the implants of NIH3T3 cells that had been transfected with EGFR-PPARGC1A. The tumor cells were pleomorphic and hyperchromatic. Mitotic figures were also seen. (Left) Low magnification, bar = 200 μm, (Right) High magnification, bar = 50 μm. (C) The RT-PCR-based detection of EGFR-PPARGC1A-specific 287-bp fragments in RNA from implanted NIH3T3 cells that had been transfected with the fusion gene (Implant) and cultured cells from the implants (Cell). The arrow indicates the PCR products of the fusion gene-specific primers. M, 100-bp ladder; NC, PCR-negative control using NIH3T3 transfected with the control vector. (D) NHEKs, A431, and DJM-1 (1 × 105 cells) were plated and grown in six-well plates. After 72 hours, the cells were detached from the wells by trypsin treatment and counted. *P < 0.05 in comparison to NHEKs (n = 3). (E) For the proliferation analysis, NIH3T3 cells that were stably transfected with a lentiviral control or EGFR-PPARGC1A fusion gene were counted as described in the Fig. 4D. The cells were also labeled with BrdU and analyzed by ELISA. The white bars indicate the cell numbers, and the black line represents the relative absorbance as determined by BrdU ELISA. *P < 0.05 (n = 3). (F) The expression of EGFR-PPARGC1A was knocked down in A431 using CRISPR/Cas9 system targeting their junctions. (Left) To show the transfection efficiency, the PCR products obtained using the primer pairs for EGFR-PPARGC1A, wild-type EGFR, or wild-type PPARGC1A were run out on agarose gels containing ethidium bromide. The GAPDH levels were shown as the control. M, 100-bp ladder. (Right) Cell number was determined at the indicated times. Data are mean values and SD. *P < 0.05 (n = 3).
We attempted to address the mechanism by which EGFR-PPARGC1A causes tumorigenesis. The cell numbers of A431 and DJM-1 were substantially increased in comparison to NHEKs (Fig. 3D). Considering the reported potential role of mitogenesis in the development of cSCC13, we focused on the possibility that the proliferation of A431 was activated by EGFR-PPARGC1A. The stable overexpression of the fusion gene in NIH3T3 led to a substantial increase in the cell numbers and the incorporation of BrdU (Fig. 3E). On the other hand, EGFR siRNA inhibited the expression of the fusion gene (Supplementary Fig. S4A), as well as the full-length wild-type EGFR in A431, and substantially reduced the cell number in comparison to control siRNA (Supplementary Fig. S4A), indicating therapeutic value of the siRNA. For the more specific inhibition of fusion gene, EGFR-PPARGC1A was knocked down using CRISPR/Cas9 system targeting their junctions (Fig. 3F). The expression levels of full-length EGFR or PPARGC1A were not affected. The inhibition of EGFR-PPARGC1A substantially reduced cell number of A431, demonstrating that EGFR-PPARGC1A is required for the growth of A431.
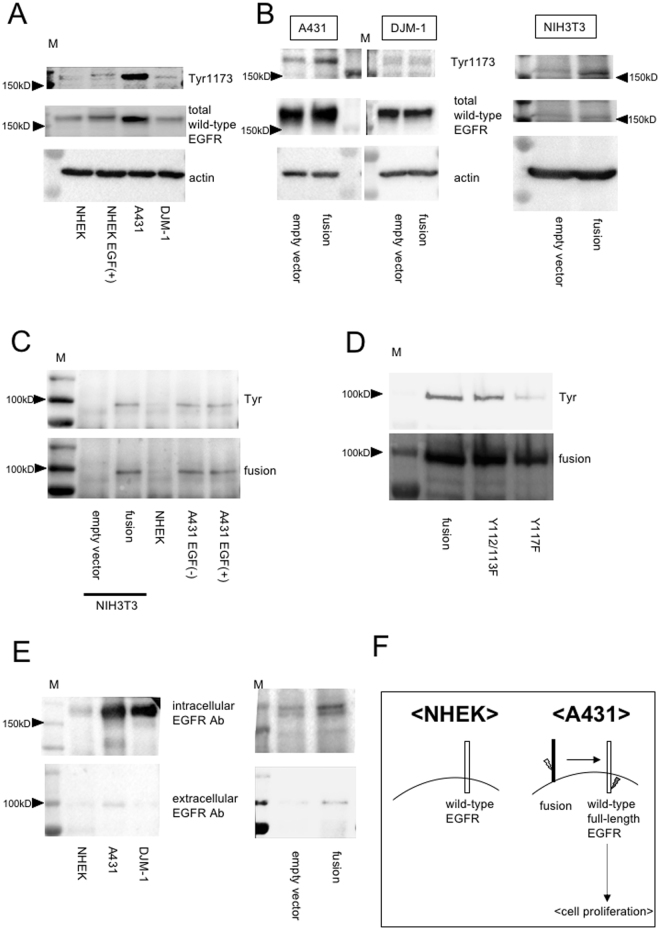
Next, to investigate the mechanism underlying the activation of A431 proliferation by EGFR-PPARGC1A, we focused on EGFR signaling, because tyrosine phosphorylation of the wild-type full-length EGFR has been well-implicated in the keratinocyte proliferation14. In many of the tyrosine phosphorylation residues of wild-type EGFR, Y1173 (the major phosphorylation site) was hardly phosphorylated in NHEKs, but its phosphorylation was induced by ectopic EGF stimulation (Fig. 4A). On the other hand, as described previously15, unstimulated A431 or DJM-1 showed strong or weaker constitutive Y1173 phosphorylation, respectively, which is consistent with their cellular proliferation activity (Fig. 3D). The Y1173 phosphorylation levels in A431 were further enhanced by the viral overexpression of EGFR-PPARGC1A fusion gene (Fig. 4B), but not in DJM-1. Furthermore, NIH3T3 is reported to express a small amount of wild-type EGFR16, and its phosphorylation level was also induced by the fusion gene (Fig. 4B). Accordingly, it is possible that the constitutive tyrosine phosphorylation of wild-type full-length EGFR in A431 may be induced by the presence of fusion gene.
Figure 4.
The role of the EGFR-PPARGC1A fusion gene in the constitutive phosphorylation of wild-type EGFR in A431. (A) Immunoblots of cell lysates with antibodies to phospho-EGFR Tyr1173 and total wild-type full-length EGFR. NHEKs were treated with or without EGF (100ng/μl) for 15 minutes before lysis. Actin was shown as the loading control. M, molecular marker. Cropped images were displayed and original blots are shown in the Supplementary Fig. S5. (B) Lysates were obtained from A431, DJM-1 (Left) or NIH3T3 (Right) stably transfected with empty vector or the EGFR-PPARGC1A fusion gene. Immunoblotting was performed using antibodies against phospho-EGFR Tyr1173, total wild-type EGFR, and actin. M, molecular marker. Cropped images were displayed and original blots are shown in the Supplementary Fig. S6. (C) Cell lysates were immunoprecipitated with antibody to the extracellular domain of EGFR, followed by immunoblotting with anti-phosphotyrosine (4G10) antibody (Tyr). The same membrane was then stripped and reprobed with anti-EGFR extracellular domain antibody to determine the abundance of total EGFR-PPARGC1A fusion protein. A431 were treated with or without EGF for 3 hours before lysis. M, molecular marker. Cropped images were displayed and original blots are shown in the Supplementary Fig. S7. (D) The EGFR-PPARGC1A fusion gene or the fusion genes with the indicated mutations were transfected into NIH3T3 cells using lentiviral constructs. Cell lysates were immunoprecipitated with antibody to the extracellular domain of EGFR, followed by immunoblotting with anti-phosphotyrosine (4G10) antibody (Tyr). The same membrane was stripped and reprobed with anti-EGFR extracellular domain antibody. Cropped images were displayed and original blots are shown in the Supplementary Fig. S8. (E) Immunoblots of lysates from NHEKs, A431, or DJM-1 (left) and NIH3T3 stably transfected with empty vector or the EGFR-PPARGC1A fusion gene (right), following immunoprecipitation with antibody (Ab) to the intracellular domain of wild-type EGFR. The blots were incubated with antibody to the extracellular domain of EGFR to detect the EGFR-PPARGC1A fusion protein. M, molecular maker. Cropped images were displayed and original blots are shown in the Supplementary Fig. S9. (F) Our hypothetical model of the role of the EGFR-PPARCG1A fusion gene in A431. The EGFR-PPARGC1A protein shows constitutive tyrosine phosphorylation, but cannot transduce cellular signals due to the lack of an intracellular domain. Instead, the fusion protein phosphorylates the wild-type full-length EGFR through their interaction, which stimulates cellular proliferation without ectopic stimulation.
NUP214-ABL1 fusion causes its own phosphorylation, which contributes to the pathogenesis of T-cell acute lymphoblastic leukemia17. Thus, we determined the phosphorylation levels of EGFR-PPARGC1A protein (Supplementary Fig. S3A). Immunoprecipitation with the antibody for the extracellular domain of wild-type EGFR revealed that the fusion protein (approximately 100kDa) could only be detected in NIH3T3 overexpressing the fusion gene and A431, and showed tyrosine phosphorylation (Fig. 4C). Ectopic EGF stimulation did not affect the phosphorylation of the fusion. The antibody for EGFR extracellular domain did not react with wild-type full-length EGFR, while the detection of the fusion protein was confirmed by immunoprecipitation with antibody against His-tag attached to the fusion protein (Supplementary Fig. S4B). A Y117F substitution mildly reduced the tyrosine phosphorylation levels of the fusion protein (Fig. 4D), but Y112/113 substitution did not. Thus, the phosphorylation at least partly involves Y117. On the other hand, although EGFR-PPARGC1A also has several serine/threonine residues, serine/threonine phosphorylation was not detected.
Given that wild-type EGFR phosphorylation is correlated with their dimerization14, we next hypothesized that the EGFR-PPARGC1A fusion protein (approximately 100kDa) can form a dimer with wild-type full-length EGFR (approximately 170kDa), and that they would interact with each other. Immune complexes generated with antibody to EGFR intracellular domain from unstimulated A431 lysates contained fusion protein (Fig. 4E), which was detected using EGFR extracellular domain antibody: because the fusion protein lacks an intracellular domain according to NCBI database (Supplementary Fig. S3A), this result suggested an interaction between wild-type EGFR and the fusion protein. Similarly, the interaction was found in NIH3T3 by overexpressing fusion gene (Fig. 4E).
Taken together, our hypothetical model regarding the role of the fusion gene in A431 is shown in Fig. 4F. The EGFR-PPARGC1A protein shows constitutive tyrosine phosphorylation, but cannot transduce signals inside the cells due to the lack of an intracellular domain. Instead, the fusion protein phosphorylates wild-type full-length EGFR by dimerization, which stimulates cellular proliferation via the activation of wild-type EGFR.
On the other hand, Lin et al. noted that A431 is sensitive to gefitinib, an EGFR tyrosine kinase inhibitor, but that other SCC cell lines are not18. Consistently, we also found that the cell number of A431 was reduced by gefitinib, but that DJM-1 was not sensitive to gefitinib treatment (Supplementary Fig. S4C). Gefitinib inhibited the phosphorylation levels of wild-type full-length EGFR as described previously19, but not the fusion protein phosphorylation (Supplementary Fig. S4D). Accordingly, the effect of gefitinib seems to occur via wild-type EGFR.
Discussion
Recently, we first reported the presence of novel NUP160-SLC43A3 fusion gene in angiosarcoma9. In addition, we previously found a case of dermatofibrosarcoma protuberans that was positive for novel COL1A1 exon 14/PDGFB fusion20.
In this study, we performed global gene expression analysis and global mutation detection using A431 and DJM-1. Because these are the most commonly-used SCC cell lines, these analyses will be useful for future studies. Transcriptome sequencing is powerful tool for fusion gene detection as well as gene expression analysis and discovery of point mutations. Fusion genes are often found in mesenchymal tumors or sarcomas, and in-frame fusion genes are the most common. However, EGFR-PPARGC1A fusion gene in A431 is characterized by its presence in epithelial carcinoma and the frameshift of its 3′ fusion partner. A similar well-characterized fusion gene in epithelial tumor, MYB-NFIB fusion, was reported in adenoid cystic carcinoma21. Furthermore, several studies have reported out-of-frame fusion genes in malignancies, including acute myeloid leukemia22.
Wild-type full-length EGFR is a transmembrane glycoprotein that acts as a receptor for EGF. Through ligand binding, the EGFR dimerized and phosphorylated, which led to cellular proliferation. EGFR signaling is thought to play a central role in the pathogenesis of SCC in various organs including cSCC, and its constitutive phosphorylation has been reported in A43123. It is noteworthy that a mutated EGFR lacking an extracellular domain, so-called ΔEGFR, was found to contribute to the pathogenesis of glioblastoma24. When it is considered that EGFR-PPARGC1A causes frameshift, although PPARGC1A acts as a transcriptional coactivator that regulates genes involved in energy metabolism, it is not likely to express the function in the fusion protein. Instead, EGFR-PPARGC1A may function via phosphorylation, probably through the truncation of EGFR intracellular domain and conformation change or through an interaction with wild-type endogenous EGFR, which are novel insights of EGFR function.
EGFR-PPARGC1A was found in keratinocytes, but also induced tumor formation in mouse fibroblasts NIH3T3, suggesting its potent tumorigenicity. The increased phosphorylation and dimer formation of wild-type EGFR was also seen in NIH3T3 with the overexpression of the fusion gene, which supports our hypothesis. EGFR-PPARGC1A-negative DJM-1 also showed mild wild-type EGFR phosphorylation, but it was not further induced by EGFR-PPARGC1A overexpression. Thus, the mild phosphorylation may be mediated by different mechanism in DJM-1.
Other fusion genes that have been reported in SCCs of other organs were rare in cSCC. This may be because cSCC differs from other SCCs in terms of the strong correlation with chronic sun exposure, and EGFR-PPARGC1A may be the fusion gene that is associated with chronic sun exposure. This hypothesis is supported by the detection of EGFR-PPARGC1 in cSCCs of sun-exposed areas. Furthermore, genital SCC, which is often correlated with papilloma virus infection, was negative for the fusion. Increasing attention is being paid to the role of ultraviolet irradiation in skin carcinogenesis, and the fusion gene may be the link between sun-exposure and carcinogenesis.
The diagnosis of atypical, early-stage, or well-differentiated cSCC is sometimes challenging because of the lack of specific cell markers. The detection of EGFR-PPARGC1A by RT-PCR or variant assays may be useful for the diagnosis of such challenging cases or the evaluation of surgical margins, because the fusion can be detected at the in situ stage (actinic keratosis or Bowen’s disease). Furthermore, EGFR siRNA, EGFR-PPARGC1A CRISPER/Cas9, or gefitinib substantially decreased the cell number of A431, indicating that full-length wild-type EGFR and/or the EGFR-PPARGC1A fusion may represent potential therapeutic targets. Lung SCC with or without EGFR mutations has been treated using various EGFR-targeted therapies, such as gefitinib, erlotinib, or afatinib, because EGFR signaling is the key pathway as described above. In addition, cetuximab is reported to have therapeutic effects against cSCC, although its mechanisms are still unknown25. Our results may become the basis for the use of EGFR-targeted drugs for cSCCs. The efficacy of EGFR-targeted drugs may vary due to the phosphorylation state of wild–type EGFR of each cSCC lesion, and may be predicted by the detection of the fusion gene. Such personalized medicine would be helpful for avoiding side effects in elderly patients.
As the limitation of this study, there is no junction variability in EGFR-PPARGC1A. This is different from other well-established fusions such as BCR-ABL in chronic myelogenous leukemia, TMPRSS2-ERG in prostate cancer, or EWS-FLI1 in Ewing’s sarcoma with junctional variability. However, FISH, functional experiments, and tumorigenicity assay indicated EGFR-PPARGC1A is the causative fusion gene. Further studies will be needed to determine the mechanism through which EGFR-PPARGC1A in cSCC regulate tumor formation, which may lead to a better understanding of the pathogenesis of cSCC, new diagnostic methods, and new targeted cancer therapies.
Materials and Methods
This study was approved by the medical ethics research committee of Kumamoto University and written informed consent was obtained from each patient. This informed consent include a statement about publication of images in an online open-access publication. We confirmed that all methods were carried out in accordance with the relevant guidelines and regulations.
Cell culture
A431 (an established human cSCC cell line) and NIH3T3 were obtained from American Type Culture Collection (ATCC, Manassas, VA)26. DJM-1 (another cSCC cell line) was purchased from RIKEN BRC (Tsukuba, Japan). Normal human epidermal keratinocytes (NHEKs) were from Lonza (Walkersville, MD).
RNA isolation
The RNA was extracted from paraffin-embedded sections using RNeasy FFPE kit (Qiagen, Valencia, CA). For the transcriptome sequencing, high quality RNAs were obtained from cultured cells using RNeasy mini kit (Qiagen)27.
Library preparation and sequencing
The transcriptome analysis was performed using Illumina TruSeq RNA Sample Preparation Kit (San Diego, CA) according to the protocol provided by Riken Genesis (Kanagawa, Japan)27. In brief, poly-A-containing mRNA was purified from high quality total RNA using poly-T oligo-conjugated magnetic beads. The mRNA was fragmented by divalent cations, and the cleaved RNA fragments were transcribed into first-strand cDNA using random primers and SuperScriptII Reverse Transcriptase (Invitrogen, Carlsbad, CA), followed by second strand cDNA synthesis using DNA polymerase I and RNase H. The cDNA fragments were then subjected to an end repair process, A-base addition, and the ligation of adapters, to convert them into a library of template molecules suitable for the subsequent cluster generation. The product was purified and enriched by PCR to create final cDNA library. The library was sequenced in a paired-end 100-bp configuration on Illumina HiSeq2000 platform. Adapter sequences and low-quality sequences were eliminated using Cutadapt (v1.0) software28. After performing quality control, the poly-A/T sequences were also eliminated using PRINSEQ (v0.16)29.
Transcriptome analysis
The cleaned and trimmed reads were aligned to the reference human sequence (GRCh37/hg19) using TopHat (v1.4.0), a fast splice junction mapping software that uses Bowtie alignment to align reads30–34. The mapped reads were assembled using Cufflinks (v2.0.0) software, and the transcripts across all samples were merged using Cuffmerge program. The reference GTF annotation file used in Cufflinks was downloaded from iGenomes database.
The differential expression between samples was analyzed using Cuffdiff software by calculating the fragments per kilobase per million map reads (FPKM) and by testing the statistical significance of the differences. A bioinformatic analysis was performed to detect mutations using Samtools (v1.0). Potential gene fusion transcripts were identified and filtered using deFuse (v0.61) and Fusion Hunter (v1.4).
RT-PCR
RT-PCR was performed for 40 cycles (denaturation for 15 seconds at 94 °C, annealing for 30 seconds at 55 °C, and extension for 30 seconds at 68 °C) using total RNAs, primers, and SuperScript One-Step RT-PCR System with Platinum Taq Polymerase (Invitrogen). The sequences of the EGFR-PPARGC1A-specific primers were: (forward) ATCCAGTGTGCCCACTACATTG and (reverse) GCTGTCTGTATCCAAGTCGTTC. GAPDH primers were (forward) GCACCGTCAAGGCTGAGAAC and (reverse) TGGTGAAGACGCCAGTGGA. The PCR products were run on agarose gels containing ethidium bromide. The detection of other fusion genes was performed as previously described35–37.
Fluorescence in situ hybridization (FISH)
A FISH analysis of cultured cells was performed according to the protocol provided by Nihon Gene Research Laboratories (NGRL, Miyagi, Japan)27. The cells were fixed with freshly made Carnoy’s solution. The fixed cells were dropped onto glass slides and air-dried. The probes were designed by Empire Genomics (Buffalo, NY): the probe for EGFR was labeled with fluorescein, whereas that for PPARGC1A was labeled with ROX. The slides were treated with probes, and were denatured by heating, followed by incubation at 37 °C overnight for hybridization.
Dual-color FISH analysis of formalin-fixed, paraffin-embedded tissue sections was performed by Chromosome Science Lab (Sapporo, Japan)38. Sections were deparaffinized, washed, digested in pepsin solution and dehydrated. Bacterial artificial chromosome clones RP11-752O3 and RP11-118N21 were labeled with SpectrumRed-dUTP and were used as probes for PPARGC1A. RP11-81B20, RP11-815K24 and RP11-1055P9 were labeled with SpectrumGreen-dUTP and used as probes for EGFR. The probes were applied to the sections and were simultaneously denatured. Hybridization was carried out at 37 °C overnight.
The nuclei were stained with DAPI, and fluorescence was detected by fluorescence microscopy.
G-banding
A karyotype analysis using cultured cells was carried out by G-banding according to the protocol of NGRL27. In brief, cultured cells on the slides were treated with 0.2% trypsin in Hanks' balanced salt solution. The slides were then washed with phosphate-buffered saline, and stained with Giemsa solution.
Virus generation and transfection
pCMV-VSV-G-RSV-Rev, CSII-EF-RfA, and pHIVgp, which are essential for lentiviral gene expression, were kindly donated by Dr. Hiroyuki Miyoshi (RIKEN, Wako, Japan)39,40. cDNA fragments of full-length EGFR, PPARGC1A, or EGFR-PPARGC1A fusion gene were amplified by PCR and cloned into CSII-EF-RfA. Substitution mutations were generated using Quick Change lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) and were confirmed by sequencing. Lentiviral vector-mediated gene transfer was performed as described previously41.
Tumorigenicity assay
NIH3T3 cells were implanted into six-week-old female athymic nu/nu mice (BALB/cAJcl-nu/nu, CLEA, Tokyo, Japan) by skin injection using a 25-gauge needle12. The implants were removed at eight weeks after xenografting, fixed in 10% buffered formalin, embedded in paraffin, and sliced into sections. Protocols of animal experiments were approved by the Committee on the Animal Research at Kumamoto University.
Cell count and BrdU ELISA
Cultured cells were detached from the wells by trypsin treatment and counted using TC20 Automated Cell Counter (Bio-Rad Laboratories, Hercules, CA)42. The proliferation activity of cells was confirmed using Cell proliferation ELISA BrdU kit (Roche, Basel, Switzerland), as described previously43,44.
siRNA transfection
siRNA against EGFR was purchased from Qiagen. For reverse transfection, siRNAs mixed with Lipofectamine RNAiMAX (Invitrogen) was added when the cells were plated: the plates were then incubated at 37°C in 5% CO2.
CRISPR/Cas9
Custom CRISPR RNAs targeting the junctions of EGFR-PPARGC1A (CAACGAAUGGUGUGCUGCUCGUUUUAGAGCUAUGCUGUUUUG) and trans-activating RNAs (5′-AACAGCAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUUUU-3’) were co-transfected into cultured cells with DharmaFECT Duo Transfection Reagent (Dharmacon)45.
Western blotting
Protein was extracted from harvested cells with RIPA buffer (Nacalai tesque, Kyoto, Japan). Immunoblotting was performed with antibodies for phosphotyrosine, phosphoserine/threonine (Abcam, Cambridge, United Kingdom), EGFR, phosphoEGFR, or β-actin (Santa Cruz Biotechnology, Santa Cruz, CA)46.
Immunoprecipitation
Cells were lysed in RIPA buffer with Phosphatase Inhibitor Cocktail (Nacalai tesque). The lysates were precleared with protein A/G-agarose (Santa Cruz Biotechnology) and control IgG at 4 °C for 30 minutes.
The lysates were then incubated with anti-EGFR antibodies (Santa Cruz Biotechnology) at 4 °C, followed by incubation with protein A/G-agarose. The immunoprecipitated proteins were washed with RIPA buffer, and were subjected to electrophoresis.
Statistical analysis
Bar graphs were created using the means+standard deviation (SD) from at least three experiments. Mann-Whitney U-test was used to compare median values, while Fisher’s exact probability test was used to compare frequencies. P values of <0.05 were considered to indicate statistical significance.
The P values in the differential expression analysis of the transcriptome sequencing were calculated using the Cuffdiff software47. Q values, estimated false discove ry rates using Benjamini-Hochberg procedure, were also calculated.
Electronic supplementary material
Acknowledgements
We would like to thank Dr. H. Miyoshi for kindly providing the lentiviral vector. We also thank Ms. C. Shiotsu for her valuable technical assistance.
Author Contributions
S.E. and M.J. contributed drafting of manuscript. S.E., M.J., and H.I. contributed critical revision. S.E., M.A., S.O. Y.T., A.H., M.I. contributed acquisition of data. S.E., M.J., N.S, J.A., M.H., and T.I. contributed analysis and interpretation of data. S.E, M.J., I.K., S.M., S.F., and H.I. contributed study conception and design.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12836-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yagi H, Danno K, Maruguchi Y, Yamamoto M, Imamura S. Significance of squamous cell carcinoma (SCC)-related antigens in cutaneous SCC. A preliminary report. Arch Dermatol. 1987;123:902–906. doi: 10.1001/archderm.1987.01660310070017. [DOI] [PubMed] [Google Scholar]
- 2.Yamane K, et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J Mol Med (Berl) 2013;91:69–81. doi: 10.1007/s00109-012-0935-7. [DOI] [PubMed] [Google Scholar]
- 3.Yamada S, et al. Cytokine expression profiles in the sera of cutaneous squamous cell carcinoma patients. Drug Discov Ther. 2016;10:172–176. doi: 10.5582/ddt.2016.01032. [DOI] [PubMed] [Google Scholar]
- 4.Aoi J, et al. Angiopoietin-like protein 2 is an important facilitator of inflammatory carcinogenesis and metastasis. Cancer Res. 2011;71:7502–7512. doi: 10.1158/0008-5472.CAN-11-1758. [DOI] [PubMed] [Google Scholar]
- 5.Aoi J, et al. Angiopoietin-like protein 2 accelerates carcinogenesis by activating chronic inflammation and oxidative stress. Mol Cancer Res. 2014;12:239–249. doi: 10.1158/1541-7786.MCR-13-0336. [DOI] [PubMed] [Google Scholar]
- 6.Kubo Y, Urano Y, Hida Y, Arase S. Lack of somatic mutation in the PTEN gene in squamous cell carcinomas of human skin. J Dermatol Sci. 1999;19:199–201. doi: 10.1016/S0923-1811(98)00058-9. [DOI] [PubMed] [Google Scholar]
- 7.Simon MP, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- 8.Muehleisen B, Schaerer L, Dummer R, Burg G, Hofbauer GF. Cancer/testis antigen MAGE-A4 expression pattern differs in epithelial skin tumors of organ-transplant recipients and immunocompetent patients. J Cutan Pathol. 2007;34:1–6. doi: 10.1111/j.1600-0560.2006.00576.x. [DOI] [PubMed] [Google Scholar]
- 9.Xu N, et al. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem. 2012;287:29899–29908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooley TP, Reddy SP, Wilborn TW, Davis RL. Biomarkers of human cutaneous squamous cell carcinoma from tissues and cell lines identified by DNA microarrays and qRT-PCR. Biochem Biophys Res Commun. 2003;306:1026–1036. doi: 10.1016/S0006-291X(03)01099-4. [DOI] [PubMed] [Google Scholar]
- 11.Kwok TT, Mok CH, Menton-Brennan L. Up-regulation of a mutant form of p53 by doxorubicin in human squamous carcinoma cells. Cancer Res. 1994;54:2834–2836. [PubMed] [Google Scholar]
- 12.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 13.Mansoor A, McKee PH, Simpson JA, McGuire B, Hobbs C. Prognostic significance of Ki-67 and p53 immunoreactivity in cutaneous squamous cell carcinomas. Am J Dermatopathol. 1996;18:351–357. doi: 10.1097/00000372-199608000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–8. doi: 10.1016/S0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 15.Cichocki M, Szaefer H, Krajka-Kuźniak V, Baer-Dubowska W. The effect of resveratrol and its methylthio-derivatives on EGFR and Stat3 activation in human HaCaT and A431 cells. Mol Cell Biochem. 2014;396:221–228. doi: 10.1007/s11010-014-2157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishayee A, Beguinot L, Bishayee S. Phosphorylation of tyrosine 992, 1068, and 1086 is required for conformational change of the human epidermal growth factor receptor c-terminal tail. Mol Biol Cell. 1999;10:525–536. doi: 10.1091/mbc.10.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graux C, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 18.Lin HQ, Katsifis A, Meriaty H. Tumour response to gefitinib is associated with EGF- and gefitinib- but not radiation-modulated EGFR expression. Anticancer Res. 2010;30:4899–4905. [PubMed] [Google Scholar]
- 19.Lu Y, Liang K, Li X, Fan Z. Responses of cancer cells with wild-type or tyrosine kinase domain-mutated epidermal growth factor receptor (EGFR) to EGFR-targeted therapy are linked to downregulation of hypoxia-inducible factor-1alpha. Mol Cancer. 2007;6:63. doi: 10.1186/1476-4598-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muchemwa FC, et al. A novel COL1A1 exon 14/PDGFB fusion gene in dermatofibrosarcoma protuberans. Eur J Dermatol. 2010;20:390–391. doi: 10.1684/ejd.2010.0906. [DOI] [PubMed] [Google Scholar]
- 21.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsey H, Zhang DE, Richkind K, Burcoglu-O'Ral A, Hromas R. Fusion of AML1/Runx1 to copine VIII, a novel member of the copine family, in an aggressive acute myelogenous leukemia with t(12;21) translocation. Leukemia. 2003;17:1665–1666. doi: 10.1038/sj.leu.2403048. [DOI] [PubMed] [Google Scholar]
- 23.Jelovac DB, et al. The amplification of c-erb-B2 in cancer-free surgical margins is a predictor of poor outcome in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2016;45:700–705. doi: 10.1016/j.ijom.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Piao Y, et al. Oncolytic adenovirus retargeted to Delta-EGFR induces selective antiglioma activity. Cancer Gene Ther. 2009;16:256–265. doi: 10.1038/cgt.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stratigos A, et al. Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus-based interdisciplinary guideline. Eur J Cancer. 2015;51:1989–2007. doi: 10.1016/j.ejca.2015.06.110. [DOI] [PubMed] [Google Scholar]
- 26.Sharma A, et al. Secretome Signature Identifies ADAM17 as Novel Target for Radiosensitization of Non-Small Cell Lung Cancer. Clin Cancer Res. 2016;22:4428–4439. doi: 10.1158/1078-0432.CCR-15-2449. [DOI] [PubMed] [Google Scholar]
- 27.Shimozono N, et al. NUP160-SLC43A3 is a novel recurrent fusion oncogene in angiosarcoma. Cancer Res. 2015;75:4458–4465. doi: 10.1158/0008-5472.CAN-15-0418. [DOI] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjounal. 2009;17:10–12. [Google Scholar]
- 29.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou JB, Zhang T, Wang BF, Gao HZ, Xu X. Identification of a novel gene fusion RNF213‑SLC26A11 in chronic myeloid leukemia by RNA-Seq. Mol Med Rep. 2013;7:591–597. doi: 10.3892/mmr.2012.1183. [DOI] [PubMed] [Google Scholar]
- 33.Jiang YZ, Li QH, Zhao JQ, Lv JJ. Identification of a novel fusion gene (HLA-E and HLA-B) by RNA-seq analysis in esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2014;15:2309–2312. doi: 10.7314/APJCP.2014.15.5.2309. [DOI] [PubMed] [Google Scholar]
- 34.Sherborne AL, et al. Somatic and Germline TP53 Alterations in Second Malignant Neoplasms from Pediatric Cancer Survivors. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita, M., Inoue, M., Tanizawa, O., Iwamoto, S. & Enomoto, T. Alterations of the p53 gene in human primary cervical carcinoma with and without human papillomavirus infection. Cancer Res 52, 5323-5328 (1992). [PubMed]
- 36.Antonescu CR, et al. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009;69:7175–7179. doi: 10.1158/0008-5472.CAN-09-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jinnin M, et al. Suppressed NFAT-dependent VEGFR1 expression and constitutive VEGFR2 signaling in infantile hemangioma. Nat Med. 2008;14:1236–1246. doi: 10.1038/nm.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noguchi H, et al. Fluorescence in situ hybridization analysis of extraskeletal myxoid chondrosarcomas using EWSR1 and NR4A3 probes. Hum Pathol. 2010;41:336–342. doi: 10.1016/j.humpath.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 39.Kitamura T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. doi: 10.1016/S0301-472X(03)00260-1. [DOI] [PubMed] [Google Scholar]
- 40.Miyoshi H, Blömer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tahara-Hanaoka S, Sudo K, Ema H, Miyoshi H, Nakauchi H. Lentiviral vector-mediated transduction of murine CD34(−) hematopoietic stem cells. Exp Hematol. 2002;30:11–17. doi: 10.1016/S0301-472X(01)00761-5. [DOI] [PubMed] [Google Scholar]
- 42.Jinnin M, et al. Regulation of fibrogenic/fibrolytic genes by platelet-derived growth factor C, a novel growth factor, in human dermal fibroblasts. J Cell Physiol. 2005;202:510–517. doi: 10.1002/jcp.20154. [DOI] [PubMed] [Google Scholar]
- 43.Makino T, et al. Basic fibroblast growth factor stimulates the proliferation of human dermal fibroblasts via the ERK1/2 and JNK pathways. Br J Dermatol. 2010;162:717–723. doi: 10.1111/j.1365-2133.2009.09581.x. [DOI] [PubMed] [Google Scholar]
- 44.Thangaretnam KP, Gopisetty G, Ramanathan P, Rajkumar T. A polypeptide from the junction region sequence of EWS-FLI1 inhibits Ewing's sarcoma cells, interacts with the EWS-FLI1 and partner proteins. Sci Rep. 2017;7:7172. doi: 10.1038/s41598-017-07482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, Wu JJ, Tang T, Liu KD, Dai C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep. 2017;7:7489. doi: 10.1038/s41598-017-07871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashima T, et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS One. 2010;5:e14334. doi: 10.1371/journal.pone.0014334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.