Abstract
Gram-negative bacteria such as Escherichia coli are protected by a complex cell envelope. The development of novel therapeutics against these bacteria necessitates a molecular level understanding of the structure-dynamics-function relationships of the various components of the cell envelope. We use atomistic MD simulations to reveal the details of covalent and noncovalent protein interactions that link the outer membrane to the aqueous periplasmic region. We show that the Braun’s lipoprotein tilts and bends, and thereby lifts the cell wall closer to the outer membrane. Both monomers and dimers of the outer membrane porin OmpA can interact with peptidoglycan in the presence of Braun’s lipoprotein, but in the absence of the latter, only dimers of OmpA show a propensity to form contacts with peptidoglycan. Our study provides a glimpse of how the molecular components of the bacterial cell envelope interact with each other to mediate cell wall attachment in E. coli.
Introduction
The cell envelope of Escherichia coli is composed of two membranes separated by a region known as the periplasm or the periplasmic space (1). The outer membrane (OM) is composed of lipopolysaccharide (LPS) molecules in the outer leaflet and a mixture of phospholipids, both zwitterionic and anionic, in the inner leaflet (2). Proteins that are integral to this membrane are almost invariably β-barrels in architecture (3). The periplasm contains the sugar-peptide polymer, peptidoglycan (PGN), as well as many different periplasmic proteins. The PGN network is attached to the OM and the integral membrane via both covalent and noncovalent interactions (4, 5, 6).
The only known protein that provides a covalent link to PGN is Braun’s lipoprotein (BLP, also known as “Lpp” and “murein lipoprotein”), which is one of the most abundant proteins in E. coli (7, 8). BLP is anchored in the OM via a lipidated N-terminus, whereas the C-terminus is covalently attached to the peptide chain of PGN. BLP exists in PGN-bound and PGN-unbound states, with the former representing approximately one-third of the population (9, 10, 11, 12). Crystallographic data revealed that the E. coli BLP forms a stable homotrimer with a tight coiled coil motif held together by an alanine zipper unit (13). Recently, electron microscopy and electron cryomicroscopy studies showed that the length of BLP has a direct influence on the distance between the peptidoglycan layer and the outer membrane of E. coli (14). However, how the BLP trimer is positioned with respect to the OM and the PGN network, remains unknown at the individual molecule level.
In addition to the covalent linkage provided by BLP, PGN is also attached noncovalently to several OM and integral membrane proteins such as OmpA-like domains (15), PGN-associated lipoproteins (16), and flagella motor proteins (17). The E. coli outer membrane porin OmpA is a multidomain protein whose N-terminal domain (NTD) is made of a β-barrel and C-terminal domain (CTD) is a globular periplasmic unit that binds to PGN, connected by an unstructured 20-residue linker region (18). The NTD has been subject to numerous functional and structural studies (19, 20, 21, 22, 23), whereas the structure of the CTD has recently emerged from a NMR study (24). Experimental evidence suggests that the full-length OmpA can form a homodimer (25, 26), the model of which has been proposed and validated by mass spectrometry and MD simulations (27, 28, 29). The mechanism of PGN attachment to OmpA CTD has been elucidated by crystal structures of a homolog from Acinetobacter baumannii bound to a short PGN peptide (30).
Although BLP and OmpA, and their interactions with PGN, have been extensively studied individually and it is likely that both proteins form simultaneous interactions with the PGN network in vivo, very little is known about any interaction between the two proteins as the spatial arrangement of both proteins within the cell envelope is still largely unexplored at a resolution of individual molecules. Based on the x-ray structures, each helix of the BLP trimer is ∼90 Å in length (13). If this represents the separation of the OM and the PGN layer, the OmpA linker would have to be fully extended to allow for CTD interaction with PGN. Such a conformation, however, is likely to be entropically unfavorable. Further studies including both proteins in the same environment are therefore crucial to understand the balance between covalent and noncovalent bonding between the OM and the PGN cell wall.
To this end, here we built atomistic models representing a portion of the E. coli cell envelope, namely the OM and the periplasm containing PGN, BLP, and OmpA. Unlike our previous study, the PGN sheet was positioned ∼90 Å from the lower leaflet of the OM, unbound to the OmpA CTD, allowing us to examine how the latter can interact with PGN in the presence of BLP. This distance was specifically chosen, as it is the length of fully extended, unkinked BLP. Our simulations show that BLP lifts the PGN layer upwards by tilting and bending its helices. This in turn reduces the gap between the OM and the cell wall, thereby facilitating the initial contact between the OmpA CTD and PGN, especially in its monomeric form. OmpA dimers on the other hand are able to interact with the cell wall even in the absence of BLP by extending their linker domain. We also identify interactions between BLP and OmpA as well as showing the interaction of the latter with the cell wall.
Methods
The models
The full-length OmpA monomer and dimer models were obtained from Carol Robinson (27); their structural stability in a model OM has been verified in our previous work (28, 29). The OM model was asymmetric: the upper leaflet was made entirely of full-rough Ra LPS lipids of the R1 core type (31, 32), whereas the lower leaflet comprised a mixture of phospholipids (i.e., 90% 1-palmitoyl 2-cis-vaccenic phosphatidylethanolamine, 5% 1-palmitoyl 2-cis-vaccenic phosphatidylglycerol, and 5% 1-palmitoyl 2-cis-vaccenic 3-palmitoyl 4-cis-vaccenic diphosphatidylglycerol, otherwise known as cardiolipin) (2, 33, 34, 35). This OM model has been validated in our previous simulations studies (28, 29, 36). The OmpA structure was inserted into the OM model using g_membed (37), following the procedure previously described (29).
A PGN network consisting of three strands of 10 repeating NAG-NAM-peptide units was constructed and positioned ∼90 Å from the surface of the lower leaflet of the OM. The BLP homotrimer was built based on the structure from Shu et al. (13) (PDB: 1EQ7) with the last residues on both the N- and C termini manually added back using PyMOL (38). The N-terminus was in turn attached to the tripalmitoyl-S-glyceryl-cysteine residues to incorporate the BLP to the inner leaflet of the OM. The parameters for tripalmitoyl-S-glyceryl-cysteine were constructed from the standard GROMOS 54A7 force field (39) with the GROMOS 53A6OXY (40) ether parameters used for the linkage region. PGN was then covalently linked to the Lysine on one of the C termini of the BLP trimer via its m-DAP residue. The linkage was constructed using the standard GROMOS 54A7 parameters.
Atomistic MD simulations
All simulations were performed using the GROMACS 5 code (41), the GROMOS 54A7 force field (39) with the SPC water model (42). Each simulation was run for 100 ns, and at least one independent repeat of each simulation was performed, giving at least 200 ns for each system simulated. Temperatures of 310 and 323 K were maintained using the velocity rescale thermostat (43) using a time constant of 1 ps. The pressure was maintained semiisotropically at 1 atm using the Parrinello-Rahman barostat (44) with a time constant of 1 ps. All bonds were constrained using the LINCS algorithm (45) to allow for an integration time step of 2 fs. Long-range electrostatics were described using the particle mesh Ewald method (46). The short-range electrostatic cutoff used was 1.2 nm and the short-range van der Waals cutoff was also 1.2 nm.
Short equilibration simulations were performed for each system in both the NVT and the NPT ensembles. The NVT equilibration was first run for 500 ps, followed by the NPT equilibration for another 1 ns, after which the pressure of the systems reached a plateau. No positional restraints were imposed on the proteins during these simulations. These equilibration simulations utilized the same thermostat and barostat as mentioned above.
Results
Simulation systems
Four atomistic simulation systems were constructed as described in Fig. 1 and Table 1, containing either the full-length OmpA monomer or dimer (27), in the presence or absence of BLP (13). One system of only the BLP trimer in the absence of OmpA was also built as a control. The OmpA NTD was inserted into a biologically relevant model of the OM (2, 33, 34, 35) described in the Methods. The binding of PGN to OmpA CTD occurs in a noncovalent fashion mediated by two residues, D241 and R256, in E. coli OmpA, as indicated by recent crystallographic data (30) and a simulation study (29). These residues, however, are located deep within the OmpA CTD, suggesting that it is likely that other residues on the surface of the protein might be involved in initial binding. We therefore started all our simulations with the CTD of OmpA positioned around 30 Å above a mesh of PGN network to observe the initial binding event. In the presence of BLP, the C terminus of one of the BLP trimer was covalently linked to a PGN peptide chain. For systems with both OmpA and BLP, they were separated by ∼30 Å at the beginning of the simulations. Two independent simulations of each system, each for 100 ns, were performed at 310 and 323 K; the two temperatures were used as a means to enhance sampling with independent simulations. One simulation for the systems containing OmpA dimer (Systems 3 and 4) at 310 K was extended to 500 ns (Fig. S1), although we did not observe any significant changes after the first 100 ns. Therefore, for simplicity, all analyses were performed using the 100-ns simulations at the two temperatures mentioned in Table 1.
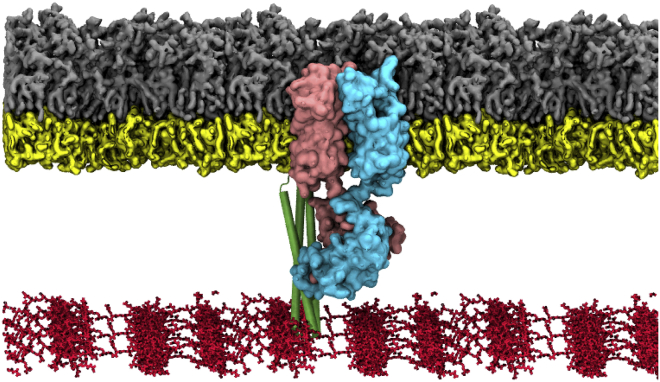
Figure 1.
Simulation setup. Given here is a snapshot of a simulation system with the full-length OmpA dimer (cyan and pink), BLP trimer (green), and PGN network (red). OmpA and BLP are embedded within an asymmetric bilayer containing Ra LPS in the upper leaflet (gray), and a mixture of phospholipids in the lower leaflet (yellow). To see this figure in color, go online.
Table 1.
Summary of Simulations Performed
| System | OmpA | BLP | Temperature (K) | Duration (ns) | OmpA Contact with PGN |
|---|---|---|---|---|---|
| Control | no | yes | 310 | 2 × 100 | — |
| Control | no | yes | 323 | 2 × 100 | — |
| 1 | monomer | yes | 310 | 2 × 100 | yes |
| 1 | monomer | yes | 323 | 2 × 100 | yes |
| 2 | monomer | no | 310 | 2 × 100 | no |
| 2 | monomer | no | 323 | 2 × 100 | no |
| 3 | dimer | yes | 310 | 2 × 100 | yes |
| 3 | dimer | yes | 323 | 2 × 100 | yes |
| 4 | dimer | no | 310 | 2 × 100 | yes |
| 4 | dimer | no | 323 | 2 × 100 | yes |
BLP tilts and kinks with respect to the membrane
At the beginning of the simulations, BLP was positioned at a right angle with respect to the plane of the membrane. Intriguingly, at the end of all simulations the BLP helices were observed to tilt, the degree of which was dependent on the presence of OmpA (Fig. 2 A). We measured the distribution of BLP tilt angle, and found that in the absence of OmpA, BLP stabilized at ∼80°, whereas in the presence of OmpA monomer, the BLP tilted slightly more at ∼75°. Adding OmpA dimer to the system resulted in BLP tilted even more at ∼65°, suggesting that the BLP helices can flexibly adjust their orientation with respect to the plane of the membrane to adapt to other nearby proteins.
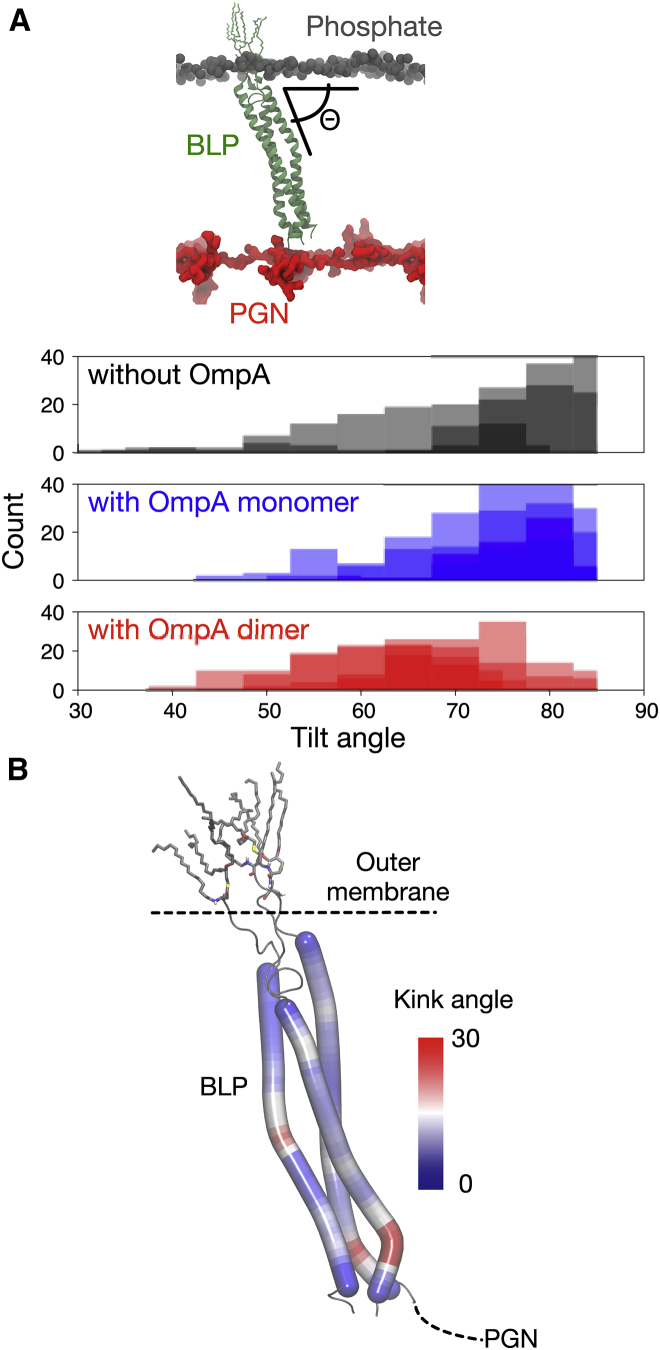
Figure 2.
BLP tilting and bending. (A) Shown here is the distribution of BLP tilt angle throughout the 100-ns simulations for systems without OmpA (black), with OmpA monomer (blue), and with OmpA dimer (red). This is measured as the acute angle between the centers of geometry of the phosphorus atoms in the inner leaflet of the OM, the N-terminal residues of the BLP, and its C-terminal residues (illustrated at the top of the graph). Independent repeats were plotted separately, and a bin size of 5° was used. (B) BLP helix kink angles were calculated along each of the three helices using VMD Bendix plugin (54). The figure shows the final snapshot of BLP from one of the BLP-only simulations, colored based on the degree of helix kink. To see this figure in color, go online.
Inspecting the shape of the BLP trimer, we also found that each helix kinked with respect to its helical axis (Fig. 2 B). These helix kinks were most prominent on the C-terminal end of the BLP helices, which bent to ∼30°. Although most helix kinks can be attributed to the presence of certain residues like proline and glycine (47, 48, 49), we could not find these residues on BLP. The position of the kink on each of the three helices also differed slightly, with helix 1 and 3 showing most bending around residue Y55, which was not the case with helix 2 (Fig. S2). Despite these different bending properties, BLP remains stable as a trimer throughout the entire simulations. Taken together, these BLP helix kinks and tilts resulted in a lift of the PGN network closer to the OM, effectively closing the gap between PGN and OmpA CTD.
BLP facilitates interactions of OmpA monomer with PGN
Our previous simulations showed that the OmpA CTD in its monomeric state has a high propensity to interact with the OM (29). If such interaction occurs in vivo with PGN bound to the CTD, it would cause a severe distortion to the PGN network. Missing from our previous model, however, is BLP, which can potentially maintain the PGN within a certain distance from the OM and therefore avoid such distortions. To understand how OmpA monomer behaves in the presence of BLP, we started our simulations with the OmpA CTD unbound from the PGN in the presence of one copy of the BLP trimer.
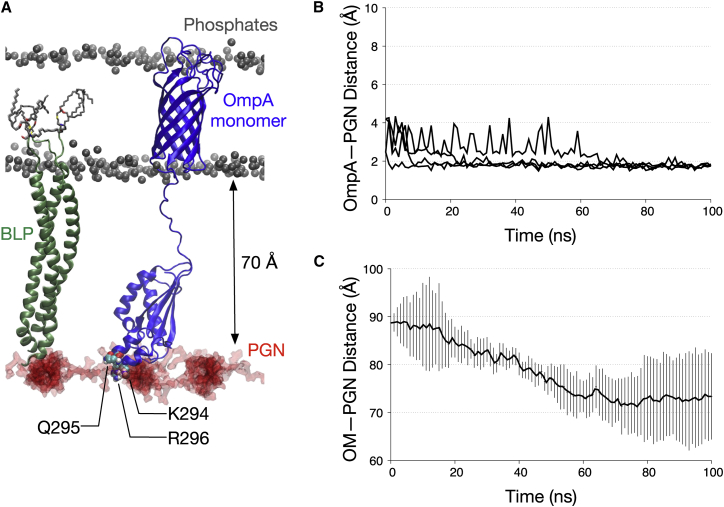
We found the distance between the OmpA CTD and the surface of the PGN network was reduced during equilibration simulations, and in all four subsequent independent production runs, the CTD contacted the PGN surface (Fig. 3, A and B). The BLP tilting and bending shifted the PGN layer toward the OmpA CTD, and concomitantly resulted in their interactions. The gap between the PGN layer and the OM was reduced to ∼70 Å at the end of the simulations (Fig. 3 C). This upward movement of the PGN network therefore eased its interactions with the OmpA monomer. Initial examination of the contact interface pointed toward a group of basic and polar residues, specifically K294, Q295, and R296, on the OmpA CTD that interacted with the negatively charged glycan moieties. This implies that the initial binding of OmpA to PGN is mediated by a long-range electrostatic force.
Figure 3.
OmpA monomer interactions with PGN. (A) A snapshot depicts the end of one of the simulations of the OmpA monomer (blue) in the presence of BLP (green), highlighting the interactions of the CTD with the PGN network (red). Given here are residues involved in PGN contacts illustrated in van der Waals representation. (B) Shown here is the minimum distance between OmpA CTD and the PGN network for all four simulations of OmpA monomer with BLP. (C) Given here is the distance between the PGN network and the OM measured along the z axis between the centers of geometry of the PGN sugar strands and the phosphorus atoms on the lower leaflet of the OM. This is averaged over all four independent simulations and the error bars indicate SDs. To see this figure in color, go online.
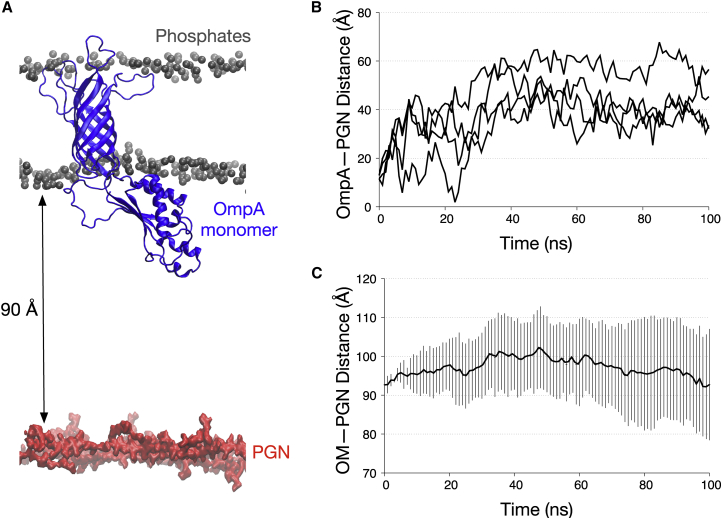
We then repeated these analyses with a similar system without the BLP trimer. Interestingly, we found in all simulations the linker region connecting the OmpA NTD and CTD contracted relative to the original starting conformation, resulting in a binding to the lower leaflet of the OM instead of PGN (Fig. 4, A and B). In contrast to the simulations with BLP, the distance along the z axis between the PGN network and the OM remained at ∼90 Å (Fig. 4 C). This observation agrees with Samsudin et al. (29, 50), who demonstrated a similar linker contraction and membrane binding event of the OmpA monomer.
Figure 4.
OmpA monomer interactions with the OM. (A) A snapshot depicts the end of one of the simulations of the OmpA monomer (blue) without BLP, highlighting the interactions of the CTD and the membrane (gray spheres represents the phosphorus atoms). (B) Minimum distance is given between OmpA CTD and the PGN network for all four simulations of OmpA monomer without BLP. (C) Distance between the PGN network and the OM is measured as described in Fig. 3, averaged over all four independent simulations. Error bars indicate SDs. To see this figure in color, go online.
The simulations of Samsudin et al. (29, 50) also revealed that the binding of OmpA CTD to the OM is mediated by residues 270–300, which intriguingly include the three key residues for initial interactions with PGN in the presence of the BLP. As this region is highly positively charged, we would expect it to form interactions with a negatively charged surface. This could either be the PGN layer underneath or the phosphate groups of the OM. Our simulations suggest that the latter is a more likely option in the absence of the BLP, perhaps due to the large energetic cost for the linker to extend and usher the CTD toward the underlying PGN network. The presence of BLP, however, lowers this energetic cost by reducing the distance between the PGN network and the CTD, making their interactions more likely to occur. Our data therefore illustrate how BLP facilitates initial binding of the OmpA monomer to the cell wall.
The OmpA dimer readily binds to PGN
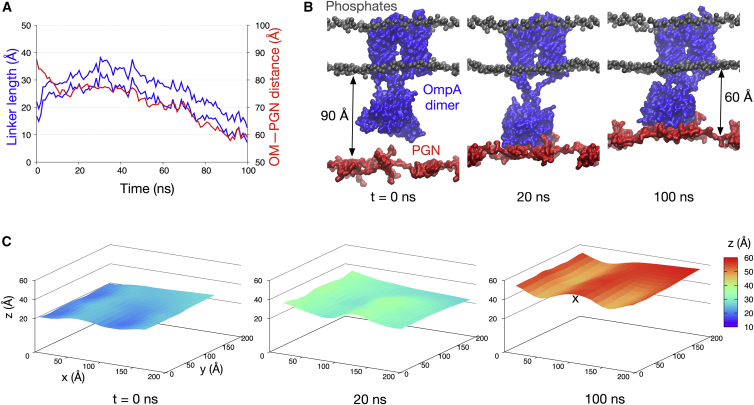
Whereas earlier experimental and computational work often considered OmpA as a monomer (19, 20, 21), evidence from several recent studies suggests that full-length OmpA can form a homodimer (25, 26, 27). We therefore built a similar simulation system using a model of OmpA dimer proposed by Marcoux et al. (27). In contrast to the monomer simulations, we found that the OmpA dimer was able to bind the PGN network with or without BLP. Again, the unstructured linker connecting the NTD and the CTD played an essential role in initiating PGN binding. In simulations without BLP, the linker first extended by ∼15 Å compared to its original length to reach the underlying PGN sheet (Fig. 5 A). Once binding occurred the linker then contracted, concomitantly shifting the PGN layer upwards by ∼30 Å (Fig. 5 B). Although the entire PGN layer was lifted, the area around the CTD showed a more pronounced upward shift, resulting in a small undulatory pattern on the PGN surface (Fig. 5 C).
Figure 5.
OmpA dimer interactions with PGN. (A) Shown here is the length of the unstructured linker connecting OmpA NTD and CTD (blue), plotted against the distance between the PGN network and the OM (red) for one of the simulations of OmpA dimer without BLP. The former is measured as previously described (28), whereas the latter is measured as in Fig. 3. (B) Given here are snapshots of this simulation at three different time points, highlighting the interactions between OmpA CTD (blue) and the PGN network (red). (C) The z coordinates of the PGN network are projected into a surface representation at these three time points to illustrate undulations observed during the simulations. The “x” indicates a local buckling effect induced by interactions with OmpA CTD. To see this figure in color, go online.
We have previously shown that the linker regions of the OmpA dimer in the absence of BLP are quite flexible, with the ability to extend and contract without disrupting the secondary structure of either the N- or C-terminal domains (28, 29). In these simulations, comparing the systems with and without BLP, we found an intriguing difference. The presence of BLP reduced the amount of extension required by the linker to initiate interactions between OmpA CTD and PGN. In simulations with the BLP, the linker extended by only ∼5 Å, compared to 15 Å in the OmpA-only simulation (Fig. S3). This is concordant with the ability of the BLP to lift the PGN networks upwards and reduce the distance between the latter and the OmpA CTD.
Next, we attempted to systematically elucidate the key residues involved in initial binding of OmpA and PGN by performing a contact analysis, combining data from simulations of both monomer and dimer (Fig. S4). Similar to the monomer simulations, a stretch of basic and polar residues, namely N203, K294, Q295, and R296, at the bottom of the CTD showed the highest degree of contacts. The latter three residues are part of a large insert only found in OmpA from certain bacteria, which comprises the least stable region of the protein as shown by a NMR study (24) and previous computational simulations (29). The lysine and arginine residues are conserved in homologs from Salmonella enterica and Neisseria meningitides and, based on their crystal structures, are positioned similarly compared to the ones in E. coli (Fig. S5) (51), suggesting a potentially conserved PGN binding mechanism in these three species.
Decomposing the nonbonded energies of the OmpA and PGN interaction into their Coulombic and Lennard-Jones components revealed that the former contributes 10 times more than the latter (Fig. S6), further corroborating the role of electrostatic interactions for initial binding of OmpA to PGN. Mutations of the key residues (N203, K294, Q295, and R296) to alanine indeed altered the way OmpA interacted with PGN (Fig. S7). The timescale of interaction was longer with the mutant compared to wild-type (50 ns instead of 10 ns). Also, instead of forming a stable binding interface involving both subunits of the CTD, only one of them contacted the underlying PGN network, suggesting that these polar and basic residues play a key role in the initial binding process. That the OmpA dimer is able to bind PGN without BLP implies that the electrostatic force from these basic residues, which is stronger than the monomer due to the dimerization, is able to attract the oppositely charged PGN network over the timescale of these simulations.
OmpA CTD binds to the BLP
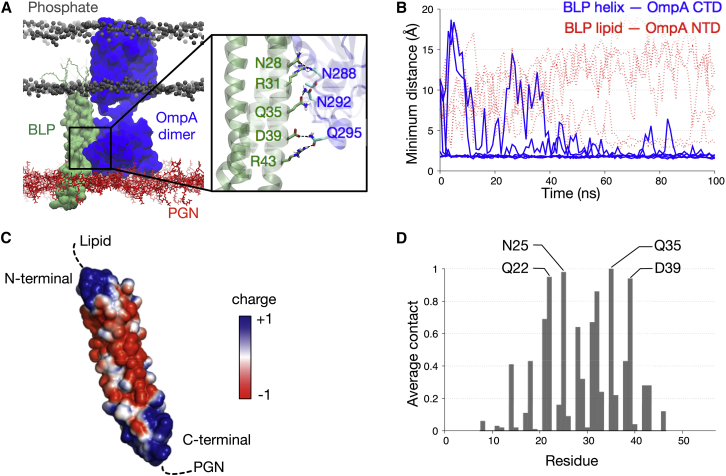
The periplasm is a crowded environment with myriad proteins surrounding the PGN cell wall. In addition to OmpA interacting with PGN, it is also likely that the ubiquitous BLP molecules make contact with OmpA. We therefore examined our simulation trajectories for such interactions and found that in three out of four dimer simulations and in one out of four monomer simulations, BLP interacted with OmpA. In all of these simulations, the BLP helices played a key role in contacting the OmpA CTD whereas the BLP lipid tails remained further away from the OmpA NTD (Fig. 6, A and B). This form of interactions involving only the helical part of the BLP and not the lipidated region was possible due to the tilted configuration adopted by the BLP. The higher frequency of interactions with the BLP of the OmpA dimer compared to the monomer is likely caused by the larger size of the former, which increased the possibility of OmpA to be within close proximity of the BLP.
Figure 6.
OmpA interactions with BLP. (A) A snapshot depicts the end of one of the simulations of OmpA dimer (blue) with BLP (green), highlighting their interactions. Enlarged image shows residues involved in these interactions from both proteins. (B) Minimum distance between the BLP helices and the OmpA CTD for all simulations of OmpA dimer is shown in blue, whereas the minimum distance between the BLP lipid tails and the OmpA NTD is shown in red dashed lines. (C) Electrostatic profile of the BLP is calculated using APBS (55) in PyMOL (18). (D) Shown here is contact analysis performed for each residue of the BLP averaged over all simulations where OmpA-BLP interactions were observed. A score of 1 indicates contacts throughout the entire 100-ns simulation. A distance cutoff of 4 Å was used for this analysis. To see this figure in color, go online.
Delineating the key residues for OmpA-BLP interactions was more challenging, due to the different ways the OmpA CTD contacts the BLP helices. In most simulations, however, BLP formed multiple salt bridges with residues on OmpA. We therefore mapped the electrostatic profile of the BLP helices and found that the surface was indeed highly charged, whereby a group of basic residues clustered toward both the N- and C termini of the helices and the center of the helices was populated by acidic residues (Fig. 6 C). This suggests that most residues on the surface of the BLP helices are able to form electrostatic interactions with OmpA CTD. To corroborate this, we performed a contact analysis and found that there was no single prominent residue responsible for this interaction, but instead most of the polar and charged residues in the middle of the helices showed a high degree of contact with OmpA (Fig. 6 D). Our results therefore suggest that BLP and OmpA CTD are able to form nonspecific electrostatic interactions in the periplasm.
Discussion
We have constructed an atomistic model of the E. coli OM bound to a network of PGN molecules via both noncovalent interactions with the outer membrane porin OmpA, and covalent linkage with the BLP. Our simulations uncover important insights into the initial binding of OmpA with the PGN cell wall and the role of BLP in facilitating these interactions. OmpA has been shown to bind PGN in a labile manner (29), and therefore it is likely that the CTD is in equilibrium between PGN-bound and PGN-unbound states. We demonstrated that from the unbound state, BLP helps the binding of OmpA monomer to the PGN network by lifting the latter closer to the former. Without BLP, the CTD of the OmpA monomer binds to the lower leaflet of the OM instead on the timescale of the simulations presented here. OmpA homodimer, on the other hand, can readily contact the PGN network by extending the linker between the NTD and the CTD, even in the absence of BLP. For both the monomer and dimer, binding is mediated by electrostatic interactions via several basic and polar residues, which are conserved in two other OmpA homologs from S. enterica and N. meningitides (51). Intriguingly these residues, and the mobile insert in which they are found, are not conserved in species like A. baumannii (30), and are also absent in other OmpA-like domains such as the E. coli PGN-associated lipoproteins (52, 53) and MotB (17). This suggests that whereas PGN interactions in the binding pocket of OmpA-like domains are conserved across species (29, 30), the initial interactions are likely to differ owing to the different residues found on the surface of these proteins. We acknowledge that in our simulations, PGN did not make any significant contact with the two key residues in the binding pocket (30), D241 and R256, most likely due to the relatively short timescale of the simulations. We note here that much longer simulations or enhanced sampling methods are needed to allow the peptide chain on PGN to enter the binding pocket and form stable interactions with these residues.
To date, OmpA is the only integral membrane eight-stranded β-barrel protein that coexists as both monomers and dimers (26). Although a model of the full-length homodimer has been proposed by mass spectrometry (27), and has been shown to interact with PGN from simulation studies (29) the physiological role of dimerization is still to be confirmed. That the dimeric interface was localized within the CTD (26, 27) suggests a functional importance of dimerization to the role of the CTD and its interactions with the cell wall. A homolog of OmpA CTD from N. meningitides, RmpM, may also exist as a dimer, as indicated by both the crystal structures and solution experiments, which the authors suggest would promote more efficient binding to PGN (51). Indeed, our OmpA dimer simulations revealed that the CTD was able to form a stable interaction with the underlying PGN layer even in the absence of the BLP. We conjecture that this is caused by the dimerization increasing the negatively charged surface area at the bottom of the CTD, and thereby strengthening the electrostatic attraction toward the PGN network. In its monomeric form, the OmpA CTD has only half as many basic residues in this region. This weaker electrostatic attraction is inadequate to surpass the energetic penalty of extending the linker connecting the NTD and the CTD, which therefore leads to contraction of the linker and subsequently interaction of the CTD and the OM. Taken together, our simulations suggest that OmpA dimerization increases the possibility of initial contact with the PGN cell wall, and therefore directly contributes toward maintaining the integrity of the cell envelope.
The BLP is one of the most abundant proteins in Gram-negative bacteria, whereby ∼7.2 × 105 molecules are found within each cell (10). Similarly, OmpA is one of the most ubiquitous outer membrane porins in E. coli (18). As both proteins play a critical structural role in preserving the robustness of the PGN cell wall, it is highly likely that BLP and OmpA function cooperatively. We show in this study that indeed BLP is required for OmpA to interact with the PGN cell wall in its monomeric state. In regions without BLP, OmpA forms homodimers to maintain this interaction.
We rationalize the differences between the behavior of OmpA monomer and dimers in the absence of BLP as follows: there is a fine balance between the energy required to extend the OmpA linker regions and the favorable electrostatic interactions formed between the C-terminal domain and PGN. When only the monomer is present, the linker will not extend; instead, the C-terminal domain forms electrostatic interactions with the lower leaflet of the outer membrane. In the case of the dimer, the combined C-terminal domains of the two monomers now provide a larger area for electrostatic interaction with PGN, and this gain in electrostatic interactions is sufficient to overcome the energy barrier required to extend the linkers. This is further augmented by the steric hindrance imposed by the dimerization interface toward interaction with the lower leaflet of the outer membrane (50).
Crucially, our results uncover some important insights into the interplay between the molecular components of the Gram-negative bacterial cell envelope, toward a better structure-function understanding of the barrier protecting the bacteria from antibiotics.
Author Contributions
S.K. and F.S. designed the research and wrote the paper. F.S. and A.B. performed the research. T.J.P. developed the parameters for BLP attachment to the Om and helped to write portions of the paper.
Acknowledgments
We acknowledge use of the Iridis 3 and 4 High Performance Computing Facilities at Southampton.
F.S. was supported by UK Biotechnology and Biological Sciences Research Council grant No. BB/M029573/1. This work was supported by NAMRIP, University of Southampton.
Editor: D. Peter Tieleman.
Footnotes
Seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30869-X.
Supporting Material
References
- 1.Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lugtenberg E.J., Peters R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim. Biophys. Acta. 1976;441:38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 3.Tsirigos K.D., Bagos P.G., Hamodrakas S.J. OMPdb: a database of β-barrel outer membrane proteins from Gram-negative bacteria. Nucleic Acids Res. 2011;39:D324–D331. doi: 10.1093/nar/gkq863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vollmer W., Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Vollmer W., Blanot D., de Pedro M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 6.Vollmer W., Höltje J.V., Ho J. The architecture of the murein (peptidoglycan) in Gram-negative bacteria: vertical scaffold or horizontal layer(s)? J. Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta. 1975;415:335–377. doi: 10.1016/0304-4157(75)90013-1. [DOI] [PubMed] [Google Scholar]
- 8.Braun V., Wolff H. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur. J. Biochem. 1970;14:387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 9.Bosch V., Braun V. Distribution of murein-lipoprotein between the cytoplasmic and outer membrane of Escherichia coli. FEBS Lett. 1973;34:307–310. doi: 10.1016/0014-5793(73)80818-x. [DOI] [PubMed] [Google Scholar]
- 10.Braun V., Sieglin U. The covalent murein-lipoprotein structure of the Escherichia coli cell wall: the attachment site of the lipoprotein on the murein. Eur. J. Biochem. 1970;13:336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 11.Inouye M., Shaw J., Shen C. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J. Biol. Chem. 1972;247:8154–8159. [PubMed] [Google Scholar]
- 12.Lee N., Inouye M. Outer membrane proteins of Escherichia coli: biosynthesis and assembly. FEBS Lett. 1974;39:167–170. doi: 10.1016/0014-5793(74)80043-8. [DOI] [PubMed] [Google Scholar]
- 13.Shu W., Liu J., Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 Å resolution. J. Mol. Biol. 2000;299:1101–1112. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- 14.Cohen E.J., Ferreira J.L., Hughes K.T. Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science. 2017;356:197–200. doi: 10.1126/science.aam6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koebnik R. Proposal for a peptidoglycan-associating α-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 16.Parsons L.M., Lin F., Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- 17.Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. USA. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith S.G.J., Mahon V., Fagan R.P. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2007;273:1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- 19.Arora A., Rinehart D., Tamm L.K. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 2000;275:1594–1600. doi: 10.1074/jbc.275.3.1594. [DOI] [PubMed] [Google Scholar]
- 20.Arora A., Abildgaard F., Tamm L.K. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat. Struct. Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 21.Bond P.J., Faraldo-Gómez J.D., Sansom M.S.P. OmpA: a pore or not a pore? Simulation and modeling studies. Biophys. J. 2002;83:763–775. doi: 10.1016/S0006-3495(02)75207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalid S., Bond P.J., Sansom M.S. OmpA: gating and dynamics via molecular dynamics simulations. Biochim. Biophys. Acta. 2008;1778:1871–1880. doi: 10.1016/j.bbamem.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 23.Pautsch A., Schulz G.E. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 24.Ishida H., Garcia-Herrero A., Vogel H.J. The periplasmic domain of Escherichia coli outer membrane protein A can undergo a localized temperature dependent structural transition. Biochim. Biophys. Acta. 2014;1838:3014–3024. doi: 10.1016/j.bbamem.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Stenberg F., Chovanec P., Daley D.O. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 2005;280:34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C., Yang L., Bruce J.E. Cross-linking measurements of in vivo protein complex topologies. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006841. M110.006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcoux J., Politis A., Robinson C.V. Mass spectrometry defines the C-terminal dimerization domain and enables modeling of the structure of full-length OmpA. Structure. 2014;22:781–790. doi: 10.1016/j.str.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Suarez M.L., Samsudin F., Khalid S. Full-length OmpA: structure, function, and membrane interactions predicted by molecular dynamics simulations. Biophys. J. 2016;111:1692–1702. doi: 10.1016/j.bpj.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samsudin F., Ortiz-Suarez M.L., Khalid S. OmpA: a flexible clamp for bacterial cell wall attachment. Structure. 2016;24:2227–2235. doi: 10.1016/j.str.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Park J.S., Lee W.C., Kim H.Y. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the Gram-negative bacterial outer membrane. FASEB J. 2012;26:219–228. doi: 10.1096/fj.11-188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appelmelk B.J., An Y.Q., de Graaf J. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology. 1994;140:1119–1124. doi: 10.1099/13500872-140-5-1119. [DOI] [PubMed] [Google Scholar]
- 32.Vinogradov E.V., van der Drift K., Holst O. The structures of the carbohydrate backbones of the lipopolysaccharides from Escherichia coli rough mutants F470 (R1 core type) and F576 (R2 core type) Eur. J. Biochem. 1999;261:629–639. doi: 10.1046/j.1432-1327.1999.00280.x. [DOI] [PubMed] [Google Scholar]
- 33.Aibara S., Kato M., Kito M. Changes in positional distribution of fatty acids in the phospholipids of Escherichia coli after shift-down in temperature. Biochim. Biophys. Acta. 1972;270:301–306. [PubMed] [Google Scholar]
- 34.Kito M., Ishinaga M., Sawada S. Metabolism of the phosphatidylglycerol molecular species in Escherichia coli. Eur. J. Biochem. 1975;54:55–63. doi: 10.1111/j.1432-1033.1975.tb04113.x. [DOI] [PubMed] [Google Scholar]
- 35.Yokota K., Kanamoto R., Kito M. Composition of cardiolipin molecular species in Escherichia coli. J. Bacteriol. 1980;141:1047–1051. doi: 10.1128/jb.141.3.1047-1051.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piggot T.J., Holdbrook D.A., Khalid S. Electroporation of the E. coli and S. aureus membranes: molecular dynamics simulations of complex bacterial membranes. J. Phys. Chem. 2011;115:13381–13388. doi: 10.1021/jp207013v. [DOI] [PubMed] [Google Scholar]
- 37.Wolf M.G., Hoefling M., Groenhof G. g_membed: efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- 38.DeLano, W. 2002. The PyMOL Molecular Graphics System. http://www.pymol.org.
- 39.Schmid N., Eichenberger A.P., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 40.Horta B.A.C., Fuchs P.F., Hünenberger P.H. New interaction parameters for oxygen compounds in the GROMOS force field: improved pure-liquid and solvation properties for alcohols, ethers, aldehydes, ketones, carboxylic acids, and esters. J. Chem. Theory Comput. 2011;7:1016–1031. doi: 10.1021/ct1006407. [DOI] [PubMed] [Google Scholar]
- 41.Abraham M.J., Teemu M., Erik L. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 42.Berendsen H.J.C., Postma J.P.M., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel; Boston, MA: 1981. pp. 331–342. [Google Scholar]
- 43.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 44.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 45.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 46.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 47.Barlow D.J., Thornton J.M. Helix geometry in proteins. J. Mol. Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 48.Deville J., Rey J., Chabbert M. Comprehensive analysis of the helix-X-helix motif in soluble proteins. Proteins. 2008;72:115–135. doi: 10.1002/prot.21879. [DOI] [PubMed] [Google Scholar]
- 49.Langelaan D.N., Wieczorek M., Rainey J.K. Improved helix and kink characterization in membrane proteins allows evaluation of kink sequence predictors. J. Chem. Inf. Model. 2010;50:2213–2220. doi: 10.1021/ci100324n. [DOI] [PubMed] [Google Scholar]
- 50.Samsudin F., Parker J.L., Fowler P.W. Accurate prediction of ligand affinities for a peptide transporter. Cell Chem. Biol. 2015;23:299–309. doi: 10.1016/j.chembiol.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grizot S., Buchanan S.K. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 2004;51:1027–1037. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 52.Abergel C., Walburger A., Lazdunski C. Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 2001;57:317–319. doi: 10.1107/s0907444900019739. [DOI] [PubMed] [Google Scholar]
- 53.Gourlay L.J., Peri C., Bolognesi M. Exploiting the Burkholderia pseudomallei acute phase antigen BPSL2765 for structure-based epitope discovery/design in structural vaccinology. Chem. Biol. 2013;20:1147–1156. doi: 10.1016/j.chembiol.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Dahl A.C.E., Chavent M., Sansom M.S.P. Bendix: intuitive helix geometry analysis and abstraction. Bioinformatics. 2012;28:2193–2194. doi: 10.1093/bioinformatics/bts357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.