Abstract
Genome dynamics are intimately linked to the regulation of gene expression, the most fundamental mechanism in biology, yet we still do not know whether the very process of transcription drives spatial organization at specific gene loci. Here, we have optimized the ANCHOR/ParB DNA-labeling system for real-time imaging of a single-copy, estrogen-inducible transgene in human cells. Motion of an ANCHOR3-tagged DNA locus was recorded in the same cell before and during the appearance of nascent MS2-labeled mRNA. We found that transcription initiation by RNA polymerase 2 resulted in confinement of the mRNA-producing gene domain within minutes. Transcription-induced confinement occurred in each single cell independently of initial, highly heterogeneous mobility. Constrained mobility was maintained even when inhibiting polymerase elongation. Chromatin motion at constant step size within a largely confined area hence leads to increased collisions that are compatible with the formation of gene-specific chromatin domains, and reflect the assembly of functional protein hubs and DNA processing during the rate-limiting steps of transcription.
Introduction
Three-dimensional (3D) organization of the genome contributes significantly to regulation of all major nuclear processes. Changes in average positions of chromosome loci in a population of cells correlate with local or global changes in DNA metabolism (1, 2, 3, 4, 5, 6, 7, 8, 9). This is notably the case for gene transcription, where active genes tend to associate with clusters of RNA polymerase II (pol2) (10). By imaging pol2, its cofactors, and mRNA, these transcription hubs have been shown to be relatively immobile (11, 12, 13, 14), but the motion of the associated DNA has not been reported. Consequently, we do not know if the observed reduced protein mobility is an intrinsic property of the transcription machinery or an indirect effect of changes in chromatin conformation. Whichever the cause, the precise kinetics of this reorganization at timescales short enough to determine chromatin physical properties have not been analyzed in mammalian cells. Indeed, real-time analysis of chromatin at short time scales relevant for the analysis of transcription activation (minutes) has been hampered by methodological limitations. Existing technologies to visualize DNA loci usually rely on highly repetitive sequences, based on the insertion of hundreds of repeats of bacterial operator sequences to which fluorescent repressor fusion proteins bind with high affinity (called FROS for fluorescent repressor operator system (8)), or using multiplexed short guide RNAs that stably recruit catalytically inactive dCas9-green fluorescent protein (GFP) fusion proteins to a large, repetitive genomic region and partially unwind the target DNA sequence (15, 16). These technologies have confirmed that transcription impacts the nuclear localization of gene domains. However, they do not allow tagging of genes within the immediate vicinity of regulatory elements for fear of disturbing their very function. Nevertheless, it was shown that, in yeast, the mobility of a gene was increased by permanently recruiting the potent activator VP16 or chromatin remodeling factors (17). This effect could stem from constitutive local decondensation of chromatin near the labeled gene. In mouse embryonic stem cells, in contrast, using dCas9-GFP targeted to MS2 sequence repeats inserted near the Nanog gene, it was reported that motion of the transcribed gene was reduced (18). In both studies gene motion was compared in different cells. To truly assess immediate changes in chromatin motion during transcription activation, DNA dynamics of a single-copy gene have to be analyzed in real-time whilst simultaneously monitoring steps of mRNA synthesis in the same cell.
To achieve this, we developed a novel ANCHOR DNA labeling system (ANCHOR3) for use in human cells. ANCHOR is based on insertion of a nonrepetitive, short (< 1 kb) DNA sequence (ANCH) to which a limited number of OR (bacterial partition protein or ParB) bind site-specifically. Oligomerization via N-terminal protein-protein interaction leads to accumulation of OR proteins, which nonspecifically and dynamically associate with adjacent DNA and form a fluorescent focus (19, 20, 21). In yeast, OR/ParB was shown to spread over DNA within chromatin up to 2 kb from the ANCH sequence (22). The ANCH site can be placed immediately next to a genomic locus of interest without disturbing DNA function in eukaryotic cells. For example, DNA resection from a HO cleavage site in yeast was normal despite the presence of an OR-bound ANCH site only 76 bp next to it, viral gene expression and subsequent infectious behavior in mammalian cells was preserved when inserting an ANCH- and OR-GFP-expressing cassette into the dense herpes virus genome (unpublished data; http://www.neovirtech.com), and transcription activation was fully functional from transgenes placed on a plasmid or within the mammalian genome when ANCH was inserted immediately next to promoters (this study and unpublished observations). Hence, stable insertion of the ANCHOR system into cells in which transcription of target genes can be activated under physiological conditions enables fluorescence imaging of a single locus in the same cell over time, without interfering with gene expression.
In this study, we demonstrate that the optimized ANCHOR3 system in combination with the MS2 system (23) is ideally suited for simultaneous visualization of DNA and mRNA at a single-gene level in living human cells at high-spatiotemporal resolution. We show that transcription initiation, not elongation, constrains local displacement of the hormone-induced Cyclin D1 gene as an immediate response to the transcription process in human cells.
Materials and Methods
Cell line
The human breast cancer cell line MCF-7 (purchased from American Type Culture Collection (ATCC)) was used to generate stable FRT/LacZeo clones. Cells were grown in Dulbecco’s modified Eagle medium (DMEM) F-12 (red DMEM/F12) completed with 10% fetal bovine serum (Gibco), 1% Sodium Pyruvate (Gibco), and 0.5% gentamicin (Gibco), or in phenol red free DMEM/F12 completed with 10% charcoal stripped serum, 1% Sodium Pyruvate (Gibco), and 0.5% gentamicin (Gibco), in a water-saturated atmosphere containing 5% CO2 at 37°C. Transfections were carried out using FuGENE HD Transfection Reagent (Promega), according to the supplier’s recommendation.
ANCHOR3 labeling system
ANCH3 corresponds to a specific chromosome partition sequence and was amplified directly by polymerase chain reaction (PCR) from the genome of an undisclosed exotic bacteria (the ANCHOR system is the property of NeoVirTech SAS; requests of use: contact@neovirtech.com). The PCR product was then cloned using BamH1/HindIII into pCDNA FRT vector digested by BglII/HindIII (Invitrogen). Insertion was verified by ApaI digestion and sequencing. OR3 corresponding to the cognate ParB protein was amplified by PCR and cloned via BglII/KpnI directly into pGFP-c1 digested by the same enzyme. Insertion was verified by digestion and sequencing. Functionality of the construct was verified by cotransfection of both vectors in HeLa cells and produced clearly identifiable fluorescent spots. To construct the ANCH3-CCND1-MS2 transgene (Fig. 1 a), the ANCH3 sequence was amplified by PCR with primers containing EcoRV restriction sites and ligated into the EcoRV-digested pCDNA5-FRT/CCND1pr-HA-CCND1/24MS2/3′UTR plasmid (kindly provided by Yaron Shav-Tal (23)).
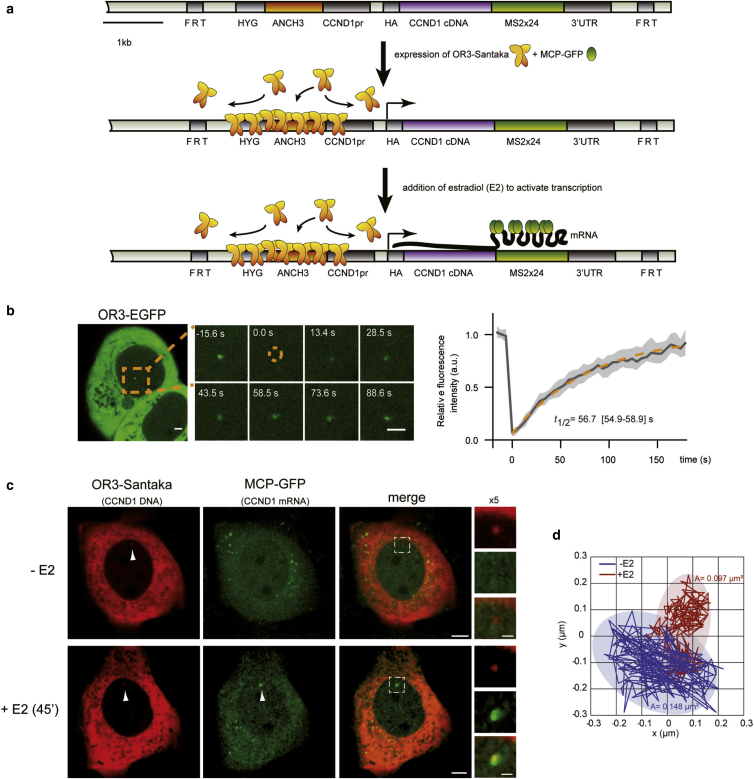
Figure 1.
Real-time visualization of a single Cyclin D1 gene locus in human cells. (a) Schematic representation of a stably inserted construct (ANCH3-CCND1-MS2) comprising the Cyclin D1 (CCND1) gene under its endogenous promoter, adjacent to a unique ANCH3 sequence, 24 × MS2 repeats within the 3′UTR, and a hygromycin selection gene (HYG). The construct is flanked by FRT sites for integration into MCF-7 FRT cells. Transient transfection with OR3 and MCP-tagged fluorescent proteins results in their accumulation at the ANCH3 and MS2 sequences (after estradiol (E2) stimulation), respectively (raw 3D images in Movie S1). (b) Fluorescent spots are easily detectable in transfected cells. A representative cell with an OR3-EGFP spot is shown. Region imaged during fluorescence recovery after photobleaching (FRAP) is indicated in orange. At time t = 0 s, a circular region enclosing the ANCHOR spot was bleached and fluorescence recovery of the spot was followed over time. Relative fluorescence intensity (RFI) was calculated as described in the Materials and Methods and Fig. S1 (right panel; solid line: mean, shadowed region: lower and upper quartile; n = 44 cells, four experiments with n ≥ 6 cells per experiment). Data were fitted to a single exponential. The 95% confidence interval is indicated in brackets. Scale bars, 2 μm. (c) Representative images of transiently transfected ANCH3-CCND1-MS2 cells expressing OR3-Santaka and MCP-EGFP (raw images in Movies S1 and S2). CCND1 DNA (red spot) colocalizes with transcribed mRNA (green spot) as MCP-EGFP associates with MS2 stem loops 45 min after adding 100 nM estradiol (E2). The same cell is shown before and after addition of E2. Scale bars, 5 and 2 μm (for cropped images). (d) Example of two-dimensional trajectories and area explored over 50 s (250 ms acquisition, 200 steps) of the OR3-Santaka-labeled CCND1 locus recorded before (−E2) and after (+E2) transcription activation. To see this figure in color, go online.
Engineering of stable FRT/LacZeo clones expressing the CCND1 transgene
MCF-7 cells were transfected using a pFRT//lacZeo kit from Invitrogen according to the supplier’s protocol. Selection of FRT clones was performed using 75 μg/mL Zeocin (InvivoGen) in red DMEM/F12 medium. Selection medium was renewed every 3 days. Three clones, showing the same growing behavior and ERα-target gene expression profiles as MCF-7 cells were kept for use: G7, A11, and D11. Clones G7, A11, and D11 were transfected with 1 μg of ANCH3-CCND1-MS2 or ANCH3 plasmids and 6 μg of plasmid pOG44 encoding Flipase (Invitrogen). Selection of positive clones was performed with 75 μg/mL Hygromycin (Invitrogen), and presence of a single fluorescent focus was verified by fluorescence microscopy 24 h after cell transfection with 500 ng OR3-Santaka. Santaka is ATUM’s synthetic nonaequorea fluorescent protein that can be amplified by PCR and cloned into any daughter expression vector of choice (https://www.atum.bio/products/expression-vectors/mammalian?hl=santaka).
ANCH3-CCND1-MS2 cells as well as control cells with only an ANCH3 insertion were maintained in red DMEM/F12 completed with 75 μg/mL Hygromycin.
Fluorescence correlation spectroscopy and fluorescence recovery after photobleaching experiments
For fluorescence correlation spectroscopy (FCS) and fluorescence recovery after photobleaching (FRAP) experiments, 2 × 104cells were seeded on 8-well Lab-Tek chambered cover glass (Nunc). To visualize spots, cells were transfected 1 h after cell seeding with a plasmid encoding OR3-EGFP using FuGENE HD transfection reagent (Promega) according to the manufacturer’s instructions for a 3:1 transfection reagent:DNA ratio. To express free mEGFP as a control for FCS, cells were transfected 1 h after cell seeding with a plasmid encoding mEGFP (kindly provided by J. Lippincott-Schwartz) using FuGene6 transfection reagent (Promega) according to the manufacturer’s instructions for a 3:1 transfection reagent:DNA ratio. FCS and FRAP experiments were performed 24–48 h after cell seeding/transfection in CO2-independent imaging medium (Gibco, custom-made) supplemented with 20% (v/v) fetal bovine serum, 1% (v/v) sodium pyruvate, and 1% (v/v) L-glutamine.
Fluorescence recovery after photobleaching
Imaging was performed on a Zeiss LSM780 ConfoCor3 confocal microscope using a 40× 1.2 NA, water Korr FCS objective. Before each FRAP time-lapse, an overview image of the measured interphase cell with a nuclear spot was taken. GFP was excited with a 488-nm laser (Ar, 25 mW, 0.4% acousto-optical tunable filter (AOTF) transmission, 2.3 μW at probe) and detected with a gallium arsenide phosphide (GaAsP) detector using a 492–552 nm detection window (Δx, Δy = 83 nm, Δz = 0.4, 60 × 60 pixels, 9 z-slices). Forty images were acquired at a 5 s time interval. After three prebleach images, a circular region of interest (ROI) (12 pixels wide) containing the ANCHOR spot was bleached with five iterations and maximal laser power. For bleaching controls, cells with spots were acquired with the same conditions as in the FRAP experiments but without bleaching the fluorescent spot. The z-position was stabilized using Zeiss Definite Focus. Due to hardware limitations, the first four images have a time interval between 3.4 and 6.6 s. These reproducible differences were taken into account in the analysis.
The image analysis was performed using FiJi (http://fiji.sc/Fiji). For each FRAP time course, the position of the OR3-EGFP ANCHOR spot was tracked in 3D using the FiJi MOSAIC plugin for 3D single-particle tracking (http://mosaic.mpi-cbg.de/?q=downloads/imageJ (24)). If a spot was not detectable after photobleaching, its position was interpolated. The mean fluorescence intensity Fs was measured in a 5 × 5 rectangular region around the tracked spot. The background fluorescence intensity Fbg was measured from a ROI in the unbleached part of the nucleus. Relative fluorescence intensity RI at time point ti (Fig. S1 d) is given by
| (1) |
whereby the data was normalized to the average of the first three prebleach images and bleach corrected using an exponential factor. The bleaching rate was estimated from the FRAP time courses without bleaching the ANCHOR3 spots. Overall bleaching was below 10% within the acquired 40 frames. The postbleach kinetics were fitted to a single exponential function as described in the legend to Fig. S1 d.
Fluorescence correlation spectroscopy and protein number estimation
Imaging and photon counts were acquired on a Zeiss LSM780 ConfoCor3 confocal microscope using a 40×, 1.2 NA, water Korr FCS objective. Cells were imaged (Δx, Δy = 83 nm, Δz = 0.4, 164 × 164 pixels, 9 z-slices) and subsequently two positions in the nucleus outside of ANCHOR3 spots were selected for FCS at the z-position of the central slice. For imaging, GFP was excited with a 488 nm laser (Ar, 25 mW, 0.4% AOTF transmission, 2.3 μW at probe) and detected with a GaAsP detector using a 492–552 nm detection window. For pre-FCS imaging, the same detector settings, pixel size, pixel dwell time, and laser settings were used as for FRAP, allowing the conversion of fluorescence intensities in the first prebleach FRAP images to concentrations and protein numbers based on an FCS calibration (Eq. 3). For FCS, GFP was excited with a 488 nm laser (Ar, 25 mW, 0.05% AOTF transmission, 0.28 μW at probe) and the photon counts were recorded for 30 s using an avalanche photodiode detector (505–590 nm detection). To estimate the effective volume, a water solution of Alexa488 (Lifetech) with a known diffusion coefficient (Dalexa = 441 μm2s−1, M. Wachsmuth, personal communication) was measured before each experiment.
The raw photon counts were processed using FluctuationAnalyzer 4G (http://www.embl.de/∼wachsmut/downloads.html). This program computes the autocorrelation function (ACF), correction factors, e.g. due to background, and fits the ACF to physical models of diffusion (see (25), for further details). The ACF was fit to
| (2) |
Equation 2 describes anomalous diffusion: N denotes the number of particles in the effective volume, is the structure factor, i.e., the ratio of axial to lateral radius of the effective volume, is the characteristic diffusion correlation time, and is the anomaly parameter. The parameter is the fraction of molecules in a nonfluorescent state and is the apparent life-time in this state. The value of was set to 100 μs and the other parameters were fit to the data.
The concentration of molecules in the effective focal volume Veff is given by , where and NA is the Avogadro’s constant. The lateral focus radius w0 is given by , where is the characteristic diffusion correlation time measured for Alexa488. The counts per molecule are given by , where is the average photon count.
For each experiment, a calibration curve (Fig. S1 c) was calculated from the mean fluorescence intensity (FI) and the concentration obtained from FCS. The mean FI was measured in a 5 × 5 px large square at the location of the FCS measurement. The data points were fit to a line that describes the relationship between fluorescence intensity FI and concentration C:
| (3) |
Based on Eq. 3, each pixel of images acquired with the same settings as the pre-FCS images can be converted to a concentration. The protein number in each voxel was obtained by multiplying the concentration C with the voxel volume Vvox and NA (see Fig. S1 e).
To estimate the protein number at the ANCHOR site, the first prebleach image of the FRAP time course was used. The FI was converted to protein number per pixel using the calibration curve in Eq. 3 and Vvox.
| (4) |
In a 14 × 14 rectangular region at the location of the ANCHOR3 site, a two-dimensional Gaussian was fit (Eq. 4), where xs and ys are the coordinates of the spot, bg is the background nuclear signal, and A is the total protein number at the ANCHOR site. For the fit, the Gauss Fit on Spot ImageJ plugin (http://imagej.nih.gov/ij/plugins/gauss-fit-spot/index.html) was used.
Fluorescence live cell imaging under stimulation
For live cell tracking during transcription activation, 100,000 cells were plated in 35 mm glass-bottom culture plates (Ibidi, Biovalley) in red DMEM/F12 medium and allowed to attach for 24 h. Red DMEM/F12 medium was changed for phenol red free DMEM/F12 medium supplemented with 10% charcoal stripped serum, without hygromycin, and cells were kept in the latter medium for 72 h. Cells were cotransfected with 500 ng OR3-Santaka and 1 μg MCP-GFP DNA vectors 24 h before observation.
To activate transcription of the ANCH3-CCND1-MS2 transgene, cells were maintained in L-15 medium (Liebovitz’s, Gibco) supplemented with 10% charcoal stripped serum, a buffer appropriate for live cell imaging.
Observations were made using a Nipkow-disk confocal system (Revolution; Andor) installed on a microscope (IX-81), featuring a confocal spinning disk unit (CSU22;Yokogawa) and a cooled electron multiplying charge-coupled device camera (DU 888; Andor). The system was controlled using the Revolution IQ software (Andor). Images were acquired using a 60× Plan Apo 1.42 oil immersion objective and a two-fold lens in the optical path. Single laser lines used for excitation were diode-pumped solid-state lasers exciting GFP fluorescence at 488 nm (50 mW; Coherent) and Santaka fluorescence at 561 nm, and a Quad band pass emission filter (Di01-T405/488/568/647-13×15×0,5; Semrock) allowed collection of the green and red fluorescence. Pixel size was 110 nm. Movies containing 200 image frames acquired with an exposure time of 250 ms were recorded. Images were processed using ICY and FIJI software (http://icy.bioimageanalysis.org/; http://fiji.sc/Fiji).
After imaging of the cells without transcription stimulation, 100 nM 17β-estradiol (E2; Sigma) was added directly under the microscope and dynamics of the OR3-Santaka spot in the same cells were recorded 45 min later. Before acquisition under E2 stimulation, presence of the MCP-GFP foci, indicating active transcription elongation, was verified in each analyzed cell (Fig. 1 c). To block transcription elongation, we added fresh L-15 medium containing 50 μM DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole; Sigma). The cells were imaged 30 min after DRB addition and disappearance of the MCP-GFP foci was verified (Fig. 3 b). To examine the impact of transcription initiation on chromatin motion in living cells, we added fresh L-15 medium containing 500 nM Triptolide (TPL, Sigma) before or after stimulation by E2. The cells were imaged under the same conditions as for DRB treatment. To study the effect of OH-Tamoxifen (OH-Tam; Sigma), cells were maintained in L-15 medium and imaged before and 45 min after 1 μM OH-Tam treatment.
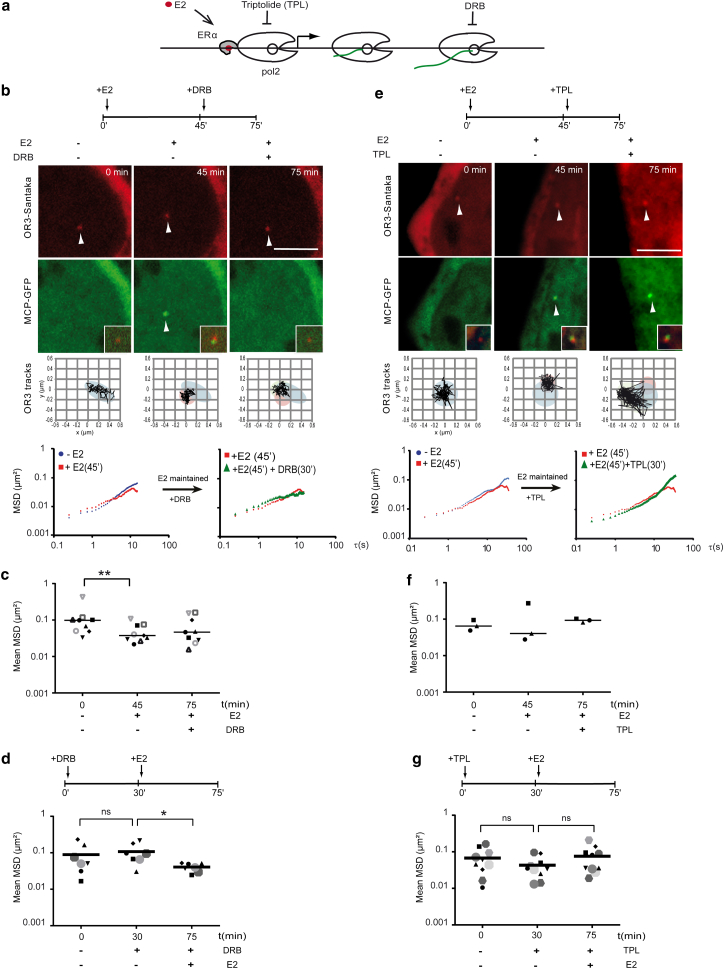
Figure 3.
Transcription initiation but not elongation confines chromatin motion. (a) Schematic representation of the steps of the E2-induced ERα-bound CCND1 gene inhibition by Triptolide (TPL) and DRB during transcription initiation or elongation, respectively. (b and e) 100 nM of E2 was used to activate transcription and 50 μM DRB to block elongation (b) or 500 nM of TPL to block initiation of transcription (e). The CCND1 locus was tracked at time points before and after treatment as specified. Representative images from live cell imaging of ANCH3-CCND1-MS2 cells (clone G7) treated first with E2 for 45 min and then with DRB or TPL for 30 min, respectively (b and e). mRNA production is activated by E2 and then blocked by inhibiting transcription initiation or elongation as evidenced by the appearance and disappearance or drifting of the MCP-EGFP signal. Scale bars, 5 μm. Successive positions of the OR3-Santaka spot are displayed in the bottom squares (122 steps, 250 ms interval). Representative single-cell MSD curves of OR3 loci detected in a cell treated with E2 and then DRB. (c) Average squared displacement between 2 and 40 s (mean MSD) of single cells (n = 9) treated successively with E2 and then DRB. (d) Mean MSD (250 ms interval, 200 steps, n = 7) of single cells pretreated with 50 μM DRB for 30 min (p-value = 0.578) and then treated with 100 nM E2 for 45 min (p-value = 0.031). (e) and (f) performed as for (b) and (c), but after timed treatment with TPL. (g) Mean MSD (250 ms interval, 200 steps, n = 10) of single cells pretreated with 500 nM TPL for 30 min (p-value = 0.117) and then treated with 100 nM E2 for 45min (p-value = 0.065). To see this figure in color, go online.
Lateral drift test
Lateral drift was analyzed by cross correlation. Cross correlation assumes that shape of the structures imaged in the sample is not expected to change significantly during the acquisition; the structure itself can be used to determine whether spatial shift between subsequent images exists. For single-cell analysis, movies were cropped and registered by the ImageJ plug-in StackReg using translation and rigid body functions for drift correction, before tracking of the DNA locus using ICY tracker (26). The translation function calculates the amount of translation (Δr) by a vectoring analysis from x = r + Δr, where x and r are the output and input coordinates. Rigid body transformation is appropriate because coordinates are x = {{cos θ, −sin θ}, {sin θ, cos θ}} ⋅ r + Δr, considering both the amount of translation (Δr) and the rotation by an angle θ(27).
Mean-square displacement
Particle tracking experiments and mean-square displacement (MSD) calculations were carried out using ICY and MatLab software (http://icy.bioimageanalysis.org/; https://fr.mathworks.com/). Tracks of 200 frames were scored. OR3-Santaka spots were detected and tracked using the Spot detector and spot tracking plug-in from ICY in single cells. MSDs were calculated in MatLab using the following equation:
| (5) |
Averaged MSD resulted from averaging the MSD at each time interval (Fig. 2 b, n = 14). Mean MSD were extracted from the average squared displacement from 2 to 40 s of the ANCH3 locus during the time of acquisition in one cell (Figs. 2 c and 3, c, d, f, and g, n = 15, n = 21, n = 9, n = 7, n = 3, and n = 10, respectively). Areas of confinement were obtained using the raw trajectories over 50 s and fitted based on an ellipse using the freely available software package GNU Octave to draw a confidence ellipse that contains 95% of all recorded positions (http://www.visiondummy.com/2014/04/draw-error-ellipse-representing-covariance-matrix/).
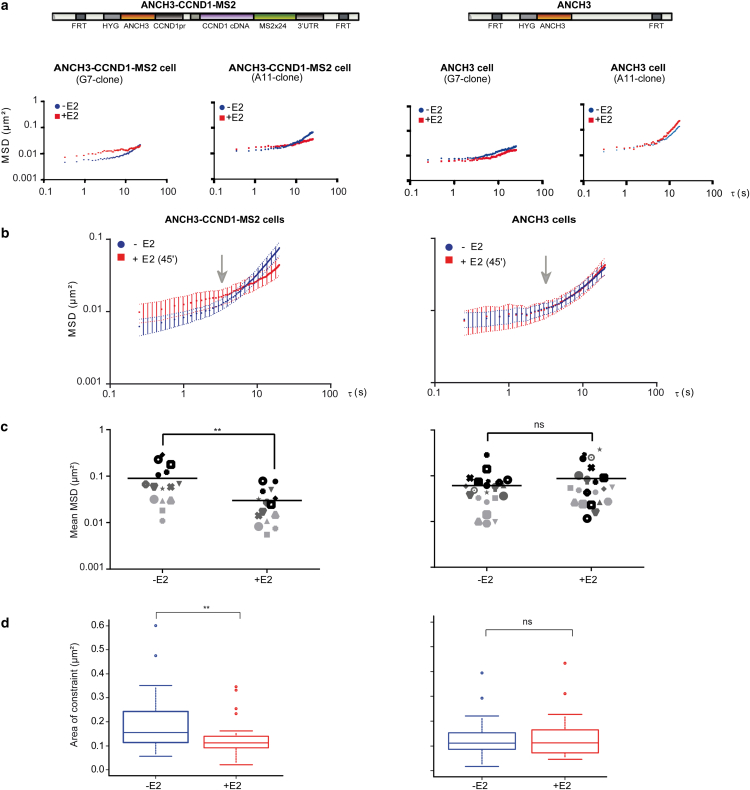
Figure 2.
Cyclin D1 motion is confined during estradiol induced transcription activation. (a) Representative single-cell MSD curves of OR3-Santaka loci in ANCH3-CCND1-MS2 and ANCH3 cells at two different chromosomal insertion sites (clones G7 and A11; distinct stable FRT insertions in MCF-7 cells) before and 45 min after E2 addition. (b) Averaged MSD curves of OR3-Santaka loci tracking in ANCH3-CCND1-MS2 (n = 14) and ANCH3 (n = 14) cells before and after E2 addition. The arrow indicates the 5 s time point separating two diffusive regimes (Fig. S4b). (c) Average squared displacement between 2 and 40 s (mean MSD) of tracking of the OR3-Santaka spot in ANCH3-CCND1-MS2 cells (left panel, n = 15, p-value = 0.006) and ANCH3 cells (right panel, n = 21, p-value = 0.3) before and 45 min after E2 addition. (d) Area of constraint of the OR3-Santaka locus in ANCH3-CCND1-MS2 cells (left panel, n = 24, p-value = 0.006) and ANCH3 cells (right panel, n = 20, p-value = 0.860) before and after E2 addition. The boxes show the median and 25–75 percentiles of the data. Asterisks indicate data points beyond the 95th percentile. Student’s t-test p-values: > 0.05 (ns), < 0.05 (∗), < 0.01(∗∗), < 0.001 (∗∗∗), and < 0.0001 (∗∗∗∗). To see this figure in color, go online.
Statistics
Results were analyzed using two different tests. A Student’s t-test with a confidence interval of 95% was used for data presented in Fig. 2, c, d, and e. A Wilcoxon signed-rank test was used for data presented in Fig. 3, c, d, f, and g.
Results
To simultaneously visualize DNA and mRNA of a single gene, we labeled a Cyclin D1 (CCND1) transgene with a new, improved ANCHOR3 system (see Materials and Methods). The ANCHOR system was derived from prokaryotic chromosome partitioning components and originally implemented in yeast (22). Specific association of a few ParB/OR protein dimers to a limited number of parS binding sites within the bacterial chromosome’s partitioning site initiates formation of a large nucleoprotein complex dependent on nonspecific, dynamic ParB/OR binding and ensuing oligomerization (19, 20, 21). The ANCHOR system thus relies simply on a short ANCH/INT sequence (< 1 kb) that can be inserted immediately adjacent, within a few basepairs, to regulatory elements.
The transgene is further composed of the endogenous CCND1 promoter, a CCND1 cDNA cassette including its 3′ enhancer region, and 24 repeats of the MS2 MCP protein-binding sequence within the CCND1 3′UTR (23) (Fig. 1 a). The construct was inserted into an FRT site within the genome of estrogen receptor alpha (ERα)-positive MCF-7 human mammary tumor cells (Fig. 1 a). In the engineered, monoclonal cells, called ANCH3-CCND1-MS2, fluorescent OR3-fusion proteins form a single focus at the ANCH3 site of the transgene that can be readily tracked in real-time (Fig. 1 b; Movies S1 and S2). To characterize the binding kinetics of OR3 proteins at and around the ANCH3 site, we used FRAP of OR3-EGFP labeled spots (Fig. 1 b). Association and dissociation of OR3-EGFP at the ANCH3-tagged site was in a dynamic steady state with a measured half-life of 57 ± 2 s (Fig. 1 b). To estimate the copy number of OR3 proteins at an ANCH3 site in steady state, we performed confocal imaging calibrated by FCS, which allowed us to convert pixel fluorescence intensity to protein concentration or number (Fig. S1, a, b, c, d, and e). The fluorescence intensity of the ANCHOR3 spot increased with OR3-EGFP abundance (Fig. S1 f). Under our OR3 expression conditions, we calculated an average of 481 ± 274 fluorescent molecules per site, corresponding to a significant amplification of the nine OR3 dimers bound specifically to the parS sites of the ANCH3 sequence (Fig. S1 f).
The MCP-EGFP signal corresponding to accumulated CCND1 transcripts is detectable near the ANCHOR3-labeled DNA site 45 min after 17β-estradiol (E2) addition to G1-synchronized cells grown in steroid-stripped media (Fig. 1 c; Movie S2), consistent with the fact that ERα target gene expression is triggered 10 min after E2 addition (28, 29). Fluorescence intensity of the OR3 spot did not vary upon transcription activation, confirming that events leading to mRNA production of the tagged locus do not interfere with OR3 accumulation. If any, displaced OR3 molecules reassociate faster than variations in intensity can be detected using our acquisition settings. Thus, the engineered cell line enabled real-time imaging of a single-copy gene during transcription activation by the endogenous ERα, under physiological conditions.
To directly test whether changes in gene expression impact local chromatin dynamics, we recorded the motion of the fluorescent ANCHOR3-Santaka-tagged gene in the same cell before and 45 min after adding 100 nM E2, whilst monitoring the appearance of MCP-EGFP-labeled mRNA signals (Fig. 1 c). Live cell tracking revealed that movement of the CCND1 gene is locally constrained upon induction of its transcription (Fig. 1 d; Table 1). We quantified the average displacement of the tagged transgene by plotting the MSD to each time interval Δt (see Materials and Methods). MSD curves calculated from time-lapse image series acquired with an interframe interval of 250 ms for a total of 50 s are shown in Fig. 2 a. We found that MSD plots of E2-activated CCND1 differed significantly from those of nonactivated cells (Fig.2 a, left panels; Figs. S2 and S3). In contrast, E2 had no effect on the behavior of a nongenic ANCH3-only construct integrated at the same genomic location (Fig. 2 a, right panels; Figs. S2 and S3), confirming that the measured decline in mobility was due to transcription of the transgene rather than to unspecific, genome-wide effects of hormone addition. We also examined the motion of the ANCH3-CCND1-MS2 or ANCH3-only constructs inserted into distinct chromosomes (G7, A11, and D11 clones; Fig. 2 a; Fig. S2). At the single-cell level, we recorded large variations in MSD of the transgene in all clones, but these variations did not correlate with any specific insertion site (Fig. S2). Coherent with the observation that the mobility of genetic loci is highly heterogeneous, modeling of nucleosome dynamics in HeLa cells suggests that subdiffusive behavior of chromatin depends on compaction levels and chromatin domain structure (30).
Table 1.
Area of Confinement Listed by Treatment
| Cell Lines | Treatment | Time (min) | Area of Confinement (μm2) |
|---|---|---|---|
| ANCH3-CCND1-MS2 cells | E2 n = 26 |
0 | 0.156 ± 0.08 |
| 45 | 0.114 ± 0.06 | ||
| 75 | 0.129 ± 0.06 | ||
| 105 | 0.101 ± 0.05 | ||
| E2 + DRB n = 8 |
0 | 0.143 ± 0.054 | |
| 45 | 0.121 ± 0.041 | ||
| 75 | 0.172 ± 0.150 | ||
| 105 | 0.173 ± 0.106 | ||
| DRB + E2 n = 7 |
0 | 0.163 ± 0.075 | |
| 30 | 0.254 ± 0.114 | ||
| 75 | 0.205 ± 0.24 | ||
| DRB n = 5 |
0 | 0.148 ± 0.040 | |
| 45 | 0.190 ± 0.064 | ||
| 75 | 0.160 ± 0.058 | ||
| 105 | 0.109 ± 0.061 | ||
| E2 + TPL n = 3 |
0 | 0.154 ± 0.057 | |
| 45 | 0.099 ± 0.045 | ||
| 75 | 0.213 ± 0.055 | ||
| 105 | 0.168 ± 0.016 | ||
| TPL + E2 n = 10 |
0 | 0.219 ± 0.189 | |
| 30 | 0.143 ± 0.083 | ||
| 75 | 0.253 ± 0.179 | ||
| no treatment n = 6 |
0 | 0.118 ± 0.035 | |
| 45 | 0.112 ± 0.066 | ||
| 75 | 0.126 ± 0.068 | ||
| 105 | 0.085 ± 0.053 | ||
| ANCH3 cells | E2 n = 20 |
0 | 0.132 ± 0.090 |
| 45 | 0.137 ± 0.096 |
Indicated drugs were added sequentially (at 45 and 75 min, or 0 and 30 min). Data are shown as average ± SD. See also Fig. 3, b, d, e, and g.
Intriguingly, motion of the ANCH3-tagged single-gene locus followed two distinct regimes with an increase in the slope of the recorded MSD at time intervals > 5 s in these human mammary tumor cells synchronized in G1 (Fig. 2 b, left panel; Fig. S2). The average MSD curves over 21 trajectories followed a nonlinear, anomalous diffusive behavior characteristic of objects moving in complex environments such as the nucleoplasm (31) (Fig. S2). Hence, we analyzed these MSDs on the basis of a generalized diffusion model obeying a power law MSD∼kτα with k as prefactor, τ the time interval, and α the anomalous diffusion exponent, a model shown to reflect chromatin motion in several cell types and under various conditions (32). Applying this model to our data, we demonstrate that at short < 5 s time intervals, diffusion of the chromatin fiber of a single-gene domain is highly anomalous (α < 0.4) and subjected to local constraints. At greater time intervals, the slope of the population-averaged MSD curve increases (α∼1), suggesting that the fiber of the nontranscribed gene locus (ANCH3-CCND1-MS2 or ANCH3 only) is rather mobile, almost freely diffusing. In contrast, the dynamic behavior of the mRNA-producing CCND1 loci differs significantly: the MSD slope remains low (α∼0.5) consistent with significant confinement of the actively transcribed locus (Fig. 2 b).
To fully exploit our ability to track a single gene, we computed an average squared displacement from 2 s to 40 s (mean MSD) before and after induction of transcription in the same cell. For both constructs, in the absence of E2, the mean MSD ranged from 0.009 to 0.290 μm2 (Fig. 2 c). This heterogeneity in the amplitude of motion is coherent with variations in the nuclear environment owing to crowding in mammalian nuclei (18, 33, 34). Independently of the initial mobility, mean MSDs recorded for each cell producing MCP-labeled mRNA were consistently reduced after addition of E2 to ANCH3-CCND1-MS2 cells (Fig.2 c, left panel; between 0.006 and 0.090 μm2; n = 15; p-value = 0.006). In contrast, variations in mean MSD of the nongenic ANCH3 locus were not significantly affected by E2 addition (Fig. 2 c, right panel; n = 21; p-value = 0.3).
To more accurately describe the behavior of the tracked chromatin locus, we determined its area of confinement and speed in addition to time-averaged MSDs, which suffer from approximating experimental errors (35). We found that spatial confinement of the transcribed locus reflected obstructed diffusion. The nuclear area explored by the tagged locus over a 50 s time interval in the absence of hormone was reduced by 27% upon addition of E2, from 0.156 ± 0.08 μm2 (no E2, no detectable MCP labeled mRNA) to 0.114 ± 0.06 μm2 (visible mRNA accumulation at the tagged locus after 45 min E2, n = 26) (Fig. 2 d; Table 1).
Confinement of the chromatin fiber could stem from steric hindrance due to protein loading or a change in the physical parameters of the fiber, or both (36). We first tested whether recruitment of large transcriptional cofactor complexes by hormone-bound ERα to the CCND1 promoter could influence chromatin mobility. Similarly to E2, OH-Tam triggers ERα binding to responsive promoters, which leads to recruitment of numerous proteins and chromatin remodeling complexes; in contrast to E2, OH-Tam-bound ERα attracts transcriptional corepressors (37, 38). After addition of 1 μM OH-Tam, motion of the ANCH3-tagged CCND1 locus was not confined. Motion was similar to the dynamic behavior of the constructs in the absence of hormone although more homogeneous, likely due to the presence of corepressor complexes (Fig. S3). We conclude that association of a multitude of transcriptional cofactors recruited by OH-Tam-bound ERα alone, in the absence of transcription, cannot explain the observed local confinement of the chromatin fiber.
We next assessed the role of pol2 activity on motion of the tagged CCND1 locus using two distinct pol2 inhibitors (Fig. 3). E2-stimulated cells were treated with an elongation inhibitor, the adenosine analog DRB. In contrast to flavopiridol, DRB action is reversible, rapid, and homogenous after 30 min (39). CCND1 mRNA signals disappeared 30 min after addition of 50 μM DRB to E2-treated ANCH3-CCND1-MS2 cells indicating that completion of elongation, i.e., transcription of the 3′UTR including MS2 repeats, was efficiently inhibited (Fig. 3 b). The appearance of the MSD plot of the CCND1 locus tracking after DRB addition to cells maintained in E2-containing medium was similar to the one in cells that had not been treated with DRB (Fig. 3 b, single-cell MSD curves), suggesting that confined motion was sustained despite blocking elongation. Mean MSD values and area of confinement for several cells analyzed 45 min after E2 stimulation and 30 min after adding DRB also did not change significantly (Fig. 3 c; Table 1). When pretreating cells for 30 min by DRB before adding hormone, we again observed a rapid decline in CCND1 motion upon mRNA production in every single cell analyzed (Fig. 3 d). It is further known that in DRB-treated cells, transcription initiation by pol2 is preserved but elongation aborts rapidly within the first transcribed exon (40). Our observations let us speculate that initiating but not elongating pol2 confines chromatin dynamics locally. To confirm this hypothesis, we analyzed the tagged gene’s motion in cells treated with 500 nM TPL, an inhibitor of TFIIH that blocks pol2 at the promoter after transcription preinitiation complex (PIC) assembly (41, 42) (Fig. 3 a). An increase in mean MSD values and in area of confinement of the ANCHOR3 spot’s track 30 min after addition of TPL to E2-stimulated mRNA-producing cells suggests that constraint of the ANCH3-tagged transgene was released by blocking initiation (Fig. 3, e and f; Table 1). Furthermore, addition of E2 to TPL pretreated cells did not alter the recorded motion of the transgene (p = 0.6; Fig. 3 g; Table 1) compared to DRB pretreated cells (Fig. 3 d). Hence, our data suggest that events linked to PIC formation induce changes in chromatin mobility, leading to increased local mobility at time scales > 5 s within a largely confined area.
Discussion
Real-time tracking of a single-copy Cyclin D1 gene in the same cell before and during hormone-induced mRNA synthesis revealed that transcription initiation rapidly confines the mRNA-producing gene and alters its diffusive behavior. Confinement was maintained even upon inhibition of pol2 elongation, but did not occur when recruitment of pol2 or transcription initiation was blocked by antiestrogens or TPL. Our results suggest that PIC formation and concomitant reorganization of the chromatin domain constrain freedom of movement of an induced gene’s promoter within minutes, compatible with the establishment of a transcriptional hub. Indeed, RNA polymerases aggregate in numerous rather immobile foci (13, 43). Several models concur in saying that a small fraction of active pol2 forms clusters with reduced mobility as transcription initiates (11, 13, 44). This clustering is dependent on the presence of initiating pol2 complexes (45). But what causes pol2 to stop moving? In principle, our observation that transcription initiation locally confines chromatin dynamics within minutes is compatible with the idea that pol2 foci assemble at active genes, and that, as the transcription initiation bubble forms, a decline in DNA freedom of movement leads to reduced pol2 mobility.
Regulation of the CCND1 locus has been shown to involve intragenic looping (46). Such conformational changes in gene domain organization, similar to those observed during glucocorticoid-stimulated transcription of MMTV tandem array gene loci in mouse adenocarcinoma cells (47), are likely to have direct consequences for chromatin dynamics. For instance, anchoring of several chromosome fibers within pol2 foci may increase the drag coefficient and hence reduce chromatin displacements. The changes in local dynamics we describe are, thus, compatible with reorganization of preexisting chromatin folding within the gene domain at the 100-kb range via long-range looping (1, 9, 48). A reduced roaming volume of the transcribed locus at constant step size increases the frequency of interaction with transcriptional cofactors and polymerases of the gene within its regulatory compartment, similar to what was recently modeled as a “nanoreactor” (49). Increased collisions are compatible with the formation of gene domain specific chromatin clustering (also called “topologically associated domains” or TADs) readily detected by crosslinking methods in mammary tumor cells (50, 51; unpublished). In turn, confined dynamics may prevent formation of unwanted long-range contacts as transcription proceeds.
At the submegabase level, TADs comprise one or a few open reading frames and their regulatory elements (gene domains) (52), particularly in human mammary tumor cells (48, 50, 53, 54) (unpublished data). If the existence of TADs is elusive in yeast, increased ligation frequencies also occur around gene bodies at the 2 kb range (55). Most of our knowledge of chromatin dynamics stems from work in budding yeast (56, 57, 58, 59). In particular, live cell chromatin motion of a series of tagged genomic yeast loci fits a Rouse model of polymer dynamics, in which the MSD increases with time with a power-law scaling and an anomaly exponent α∼0.5 (60). Assuming that within a ∼100 kb chromatin domain around any of the tagged sites, at least one gene is actively transcribed in a population of yeast nuclei, the reported dynamic behavior characterizes active chromatin. In agreement, obstructed diffusion characterizes the active CyclinD1 transgene locus in human cells here. In the absence of transcription the tagged single human transgene domain was highly dynamic, nearly freely diffusing. Similarly, our observations in yeast evidence increased motion when mutating RNA pol2 (unpublished data). Transcription-induced, pol2-dependent, intradomain contacts therefore likely result from the apparent diffusive behavior of chromatin in living cells. In mammalian cells, ATP-driven processes by chromatin remodeling factors (61) or other chromatin-associated proteins such as SAF-A (62) may contribute to the observed local motion and variation in anomaly. Anomalous diffusion was also reported for telomeres (63) and for gene arrays (64) but changes were derived from comparing motion in cells monitored under different conditions. The computed MSD curves of these loci characterized by specific structures differ from the ones we compute for a single-gene locus, which emphasizes the need for future modeling to better define physical parameters of the chromatin fiber in human cells.
The powerful real-time imaging approach of a single DNA locus undergoing functional changes presented here using the ANCHOR system is widely applicable to other loci and genomes for studying rapid biological processes with single-cell resolution, and completing the picture emerging from imaging RNA pol2 and mRNA, but also from chromosome conformation capture data.
Author Contributions
Performed Experiments, T.G., N.W., and S.K.; Analyzed Data, T.G., N.W., S.K., A.B., H.A.S., and K.B.; Analyzed and Supervised the FRAP and FCS Experiments, A.Z.P. (analyzed), J.E. (supervised); Generated Material, T.G. (cell lines), H.S. (cell lines, constructs), and F.G. (ANCHOR3 system); Wrote Article, K.B., T.G., and S.K.
Acknowledgments
We thank Y. Shav-Tal for plasmid pcDNA5/FRT/GOI-MS2. M. Dalvai is acknowledged for help during the initial phases of the study. F. Moutahir helped generate the cells. We acknowledge support from the TRI-Imaging platform Toulouse. T.G. was supported by a doctoral fellowship from the Ligue Nationale Contre le Cancer. K.B. was supported by the Human Frontier Science Program (HFSPO) RGP0044, Agence Nationale de la Recherce (ANR) ANDY, and Fondation Association pour la Recherche sur le Cancer (ARC). J.E. was supported by European Molecular Biology Laboratory (EMBL) and the EU-FP7-SystemsMicroscopy NoE (258068), and the 4DN NIH Common Fund (U01 EB021223), and N.W. was supported by the EMBL International PhD Programme (EIPP).
Editor: Tamar Schlick.
Footnotes
Thomas Germier and Silvia Kocanova contributed equally to this work.
Four figures and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30873-1.
Supporting Material
ANCH3-CCND1-MS2 cells transiently expressing OR3-Santaka were imaged in 3D (200 ms exposure, 40 frames at 200 nm intervals in z) using a Nipkow-disk confocal (ANDOR) microscope equipped with a 60x 1.4NA objective. Pixel size was 110 nm. Images were processed using ICY and FIJI software.
ANCH3CCND1-MS2 cells transiently expressing OR3-Santaka and MCP-GFP were imaged (streaming at 200 ms exposure over 200 frames) using a Nipkow-disk confocal (ANDOR) microscope equipped with a 60x 1.4NA objective. Pixel size was 110 nm. Images were processed using ICY and FIJI software.
References
- 1.Therizols P., Illingworth R.S., Bickmore W.A. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taddei A., Van Houwe G., Gasser S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 3.Kocanova S., Kerr E.A., Bystricky K. Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet. 2010;6:e1000922. doi: 10.1371/journal.pgen.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabal G.G., Genovesio A., Nehrbass U. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 5.Chambeyron S., Bickmore W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborne C.S., Chakalova L., Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 7.Schuettengruber B., Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 8.Robinett C.C., Straight A., Belmont A.S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang C.-H., Carpenter A.E., Belmont A.S. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 10.Feuerborn A., Cook P.R. Why the activity of a gene depends on its neighbors. Trends Genet. 2015;31:483–490. doi: 10.1016/j.tig.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Kimura H., Sugaya K., Cook P.R. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghamari A., van de Corput M.P., Thongjuea S. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 2013;27:767–777. doi: 10.1101/gad.216200.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cisse I.I., Izeddin I., Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 14.Darzacq X., Shav-Tal Y., Singer R.H. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B., Gilbert L.A., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma H., Naseri A., Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. USA. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann F.R., Dion V., Gasser S.M. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 2012;26:369–383. doi: 10.1101/gad.176156.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochiai H., Sugawara T., Yamamoto T. Simultaneous live imaging of the transcription and nuclear position of specific genes. Nucleic Acids Res. 2015;43:e127. doi: 10.1093/nar/gkv624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passot F.M., Calderon V., Pasta F. Centromere binding and evolution of chromosomal partition systems in the Burkholderiales. J. Bacteriol. 2012;194:3426–3436. doi: 10.1128/JB.00041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham T.G.W., Wang X., Loparo J.J. ParB spreading requires DNA bridging. Genes Dev. 2014;28:1228–1238. doi: 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez A., Cattoni D.I., Bouet J.-Y. Stochastic Self-Assembly of ParB Proteins Builds the Bacterial DNA Segregation Apparatus. Cell Syst. 2015;1:163–173. doi: 10.1016/j.cels.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Saad H., Gallardo F., Bystricky K. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 2014;10:e1004187. doi: 10.1371/journal.pgen.1004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yunger S., Rosenfeld L., Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat. Methods. 2010;7:631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- 24.Sbalzarini I.F., Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Wachsmuth M., Conrad C., Ellenberg J. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 2015;33:384–389. doi: 10.1038/nbt.3146. [DOI] [PubMed] [Google Scholar]
- 26.Chenouard N., Bloch I., Olivo-Marin J.C. Multiple hypothesis tracking for cluttered biological image sequences. IEEE Trans. Pattern Anal. Mach. Intell. 2013;35:2736–3750. doi: 10.1109/TPAMI.2013.97. [DOI] [PubMed] [Google Scholar]
- 27.Thévenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 28.Hah N., Danko C.G., Kraus W.L. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Métivier R., Penot G., Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 30.Shinkai S., Nozaki T., Togashi Y. Dynamic nucleosome movement provides structural information of topological chromatin domains in living human cells. PLoS Comput. Biol. 2016;12:e1005136. doi: 10.1371/journal.pcbi.1005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxton M.J. Single particle tracking. Fundam. Concepts Biophys. 2009;1:1–33. [Google Scholar]
- 32.Manzo C., Garcia-Parajo M.F. A review of progress in single particle tracking: from methods to biophysical insights. Rep. Prog. Phys. 2015;78:124601. doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- 33.Hancock R. Internal organisation of the nucleus: assembly of compartments by macromolecular crowding and the nuclear matrix model. Biol. Cell. 2004;96:595–601. doi: 10.1016/j.biolcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Huet S., Lavelle C., Bancaud A. Relevance and limitations of crowding, fractal, and polymer models to describe nuclear architecture. Int. Rev. Cell Mol. Biol. 2014;307:443–479. doi: 10.1016/B978-0-12-800046-5.00013-8. [DOI] [PubMed] [Google Scholar]
- 35.Kepten E., Weron A., Garini Y. Guidelines for the fitting of anomalous diffusion mean square displacement graphs from single particle tracking experiments. PLoS One. 2015;10:e0117722. doi: 10.1371/journal.pone.0117722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banks D.S., Fradin C. Anomalous diffusion of proteins due to molecular crowding. Biophys. J. 2005;89:2960–2971. doi: 10.1529/biophysj.104.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang Y., Hu X., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 38.Liu X.-F., Bagchi M.K. Recruitment of distinct chromatin-modifying complexes by tamoxifen-complexed estrogen receptor at natural target gene promoters in vivo. J. Biol. Chem. 2004;279:15050–15058. doi: 10.1074/jbc.M311932200. [DOI] [PubMed] [Google Scholar]
- 39.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gribnau J., de Boer E., Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vispé S., DeVries L., Bailly C. Triptolide is an inhibitor of RNA polymerase I and II-dependent transcription leading predominantly to down-regulation of short-lived mRNA. Mol. Cancer Ther. 2009;8:2780–2790. doi: 10.1158/1535-7163.MCT-09-0549. [DOI] [PubMed] [Google Scholar]
- 42.Jonkers I., Kwak H., Lis J.T. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darzacq X., Yao J., Singer R.H. Imaging transcription in living cells. Annu. Rev. Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stasevich T.J., Hayashi-Takanaka Y., Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell J.A., Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalvai M., Fleury L., Bystricky K. TIP48/Reptin and H2A.Z requirement for initiating chromatin remodeling in estrogen-activated transcription. PLoS Genet. 2013;9:e1003387. doi: 10.1371/journal.pgen.1003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stavreva D.A., Coulon A., Hager G.L. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res. 2015;25:845–857. doi: 10.1101/gr.184168.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mourad R., Hsu P.-Y., Li L. Estrogen induces global reorganization of chromatin structure in human breast cancer cells. PLoS One. 2014;9:e113354. doi: 10.1371/journal.pone.0113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haddad N., Jost D., Vaillant C. Perspectives: using polymer modeling to understand the formation and function of nuclear compartments. Chromosome Res. 2017;25:35–50. doi: 10.1007/s10577-016-9548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Dily F., Baù D., Beato M. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014;28:2151–2162. doi: 10.1101/gad.241422.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giorgetti L., Galupa R., Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulianov S.V., Khrameeva E.E., Razin S.V. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016;26:70–84. doi: 10.1101/gr.196006.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barutcu A.R., Lajoie B.R., Imbalzano A.N. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016;26:1188–1201. doi: 10.1101/gr.201624.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fullwood M.J., Liu M.H., Ruan Y. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsieh T.-H.S., Weiner A., Rando O.J. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bystricky K. Chromosome dynamics and folding in eukaryotes: Insights from live cell microscopy. FEBS Lett. 2015;589(20 Pt A):3014–3022. doi: 10.1016/j.febslet.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 57.Botstein D., Fink G.R. Yeast: an experimental organism for 21st Century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R., Mozziconacci J., Gadal O. Principles of chromatin organization in yeast: relevance of polymer models to describe nuclear organization and dynamics. Curr. Opin. Cell Biol. 2015;34:54–60. doi: 10.1016/j.ceb.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Taddei A., Gasser S.M. Structure and function in the budding yeast nucleus. Genetics. 2012;192:107–129. doi: 10.1534/genetics.112.140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajjoul H., Mathon J., Bancaud A. High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome. Genome Res. 2013;23:1829–1838. doi: 10.1101/gr.157008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nozawa R.S., Boteva L., Gilbert N. SAF-A regulates interphase chromosome structure through oligomerisation with chromatin- associated RNAs. Cell. 2017;169:1214–1227.e18. doi: 10.1016/j.cell.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bronshtein I., Kepten E., Garini Y. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat. Commun. 2015;6:8044. doi: 10.1038/ncomms9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Annibale P., Gratton E. Single cell visualization of transcription kinetics variance of highly mobile identical genes using 3D nanoimaging. Sci. Rep. 2015;5:9258. doi: 10.1038/srep09258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANCH3-CCND1-MS2 cells transiently expressing OR3-Santaka were imaged in 3D (200 ms exposure, 40 frames at 200 nm intervals in z) using a Nipkow-disk confocal (ANDOR) microscope equipped with a 60x 1.4NA objective. Pixel size was 110 nm. Images were processed using ICY and FIJI software.
ANCH3CCND1-MS2 cells transiently expressing OR3-Santaka and MCP-GFP were imaged (streaming at 200 ms exposure over 200 frames) using a Nipkow-disk confocal (ANDOR) microscope equipped with a 60x 1.4NA objective. Pixel size was 110 nm. Images were processed using ICY and FIJI software.