Abstract
Grainy head (Grh) is a conserved transcription factor (TF) controlling epithelial differentiation and regeneration. To elucidate Grh functions we identified embryonic Grh targets by ChIP-seq and gene expression analysis. We show that Grh controls hundreds of target genes. Repression or activation correlates with the distance of Grh-binding sites to the transcription start sites of its targets. Analysis of 54 Grh-responsive enhancers during development and upon wounding suggests cooperation with distinct TFs in different contexts. In the airways, Grh-repressed genes encode key TFs involved in branching and cell differentiation. Reduction of the POU domain TF Ventral veins lacking (Vvl) largely ameliorates the airway morphogenesis defects of grh mutants. Vvl and Grh proteins additionally interact with each other and regulate a set of common enhancers during epithelial morphogenesis. We conclude that Grh and Vvl participate in a regulatory network controlling epithelial maturation.
KEY WORDS: Drosophila, Airway, Chip-seq, Epithelial maturation, Grainy head, Microarray
Summary: The conserved transcription factor Grh exhibits unexpected functional complexity as a regulator of epithelial morphogenesis, wound healing and immune responses.
INTRODUCTION
Genes of the Grainy head (Grh) family encode conserved transcription factors (TFs) controlling epithelial morphogenesis and wound healing. Nematodes and flies have a single grh gene, whereas mice and humans have evolved three genes encoding Grainy head-like (Grhl) factors (Paré et al., 2012). Grhl proteins can act as activators or repressors in different biological contexts. For example, Drosophila Grh activates wound repair genes such as Ddc, ple and Stit (Cad96Ca) upon injury (Mace et al., 2005; Wang et al., 2009), but it represses dpp and tll during early embryonic development (Huang et al., 1995; Liaw et al., 1995). However, the molecular mechanisms by which Grhl TFs regulate gene expression remain unclear.
Grh was originally identified in Drosophila (Bray et al., 1989; Dynlacht et al., 1989; Johnson et al., 1989; Bray and Kafatos, 1991). It is expressed in epithelial tissues such as the epidermis, the tracheal airways, the foregut and hindgut (Bray and Kafatos, 1991), but is also detected in neural stem cells of the CNS (Uv et al., 1997). grh mutants show a variety of phenotypes in epidermal barrier formation (Bray and Kafatos, 1991; Mace et al., 2005), tube size control (Hemphälä et al., 2003), neural stem cell programming (Almeida and Bray, 2005; Cenci and Gould, 2005; Maurange et al., 2008; Baumgardt, 2009; Bayraktar and Doe, 2013; Li et al., 2013) and in wing hair orientation (Lee and Adler, 2004). Grh targets in the epidermis include cell adhesion proteins and matrix components (Paré et al., 2012). Additionally, receptor kinase signaling upon injury activates Grh to facilitate wound closure and barrier restoration (Kim and McGinnis, 2011; Tsarouhas et al., 2014).
Expression of the mammalian family members Grhl1-3 (Wilanowski et al., 2002; Ting et al., 2003a) is tissue- and developmental stage-specific in epithelial organs such as the epidermis, oral epithelium, kidneys, the digestive tract and lung (Auden et al., 2006). Analysis of Grhl1 mouse mutants revealed epidermal thickening, impaired hair anchoring and desmosomal abnormalities (Wilanowski et al., 2008). Loss of Grhl2 causes early embryonic lethality and neural tube closure failure (Rifat et al., 2010), while an ENU-induced mutation in Grhl2 revealed defects in lung development (Pyrgaki et al., 2011). Grhl3 is essential for neural tube closure, epidermal barrier formation and wound healing (Ting et al., 2003a,b; Ting et al., 2005; Caddy et al., 2010). Additionally, Grhl mutant mice are extensively used to model epithelial disease, ranging from hearing loss to cancer (Gordon et al., 2014). Studies of Grhl2 downstream genes in mice and in human bronchial cells revealed its key role in epithelial morphogenesis, cell adhesion, and motility (Gao et al., 2013; Varma et al., 2014; Aue et al., 2015; Gao et al., 2015).
Our studies focus on Grh function in the Drosophila airways. The fly respiratory system (termed trachea) is an epithelial tube network that extends to internal organs to facilitate gas transport and exchange. An important step in tubular organ morphogenesis is the final acquisition of distinct and uniform branch sizes. Grh regulates tube length selectively, as indicated by the fact that grh mutants show overelongated airways without any apparent defect in tube diameter or early branch outgrowth. Instead, Grh regulates cuticle deposition and epithelial cell shape, and restricts apical cell membrane expansion during late embryogenesis (Hemphälä et al., 2003; Luschnig et al., 2006). On the other hand, Grh overexpression in all tracheal cells during development inhibits branch extension. Grh targets in airway size control have not been identified and the molecular mechanism underlying Grh control of tracheal tube size remains unknown.
In this study we identified Grh targets in Drosophila embryos by combining whole-genome ChIP-seq experiments with gene expression analysis in wild type, grh mutants and in embryos overexpressing grh in ectodermal epithelial organs. We show that in addition to genes involved in extracellular matrix assembly and junction integrity, Grh directly promotes the maturation of the epithelial innate immune responses. To identify functional Grh targets in airway morphogenesis we compared the results of a genome-wide airway-specific RNAi screen with the ChIP-seq analysis. Among hundreds of conserved Grh targets in the airways, Grh represses the expression of several key TFs promoting cell differentiation, including vvl, which encodes a POU domain TF. Reduction of vvl gene dosage in grh mutants largely ameliorates the tube overelongation defects, arguing that repression of vvl is a pivotal aspect of Grh function in airway morphogenesis.
RESULTS
Genome-wide identification of Grh targets in Drosophila embryos
To explore the regulatory roles of Grh on a genome-wide scale, we performed ChIP-seq experiments using stage 16 Drosophila embryos and either a mouse anti-Grh monoclonal antibody (Bray et al., 1989; Uv et al., 1997) or a rabbit anti-Grh polyclonal antibody (Fig. S1). The monoclonal antibody identified 1606 Grh peaks whereas the rabbit antibody revealed 11,741 binding sites compared with the control IgG sample; 1587 of these binding sites were detected with both antibodies and 92.7% (1471/1587) of these common peaks also include the predicted Grh-binding motif (Fig. 1A). Although common peaks most likely represent true Grh-binding sites, the 5599 peaks uniquely revealed by the rabbit antibody might also represent genuine Grh-binding regions because they are enriched for Grh-binding motifs.
Fig. 1.
Identification of Grh targets. (A) Venn diagram of ChIP-seq data from stage 16 Drosophila embryos showing the number of peaks obtained with rabbit (olive) or mouse (turquoise) anti-Grh antibodies, and the peaks with a predicted Grh-binding site (PWM, 80%; purple). Multiple peaks within 200 bp were collapsed. (B) Overview of Grh peak localization relative to gene region. (C) The Grh-binding motif derived from the sequences of ChIP-seq peaks from the mouse Grh antibody. (D) The distance (bp) between the Grh peaks and the transcription start sites (TSSs) of the closest genes. The red line represents genes activated by Grh, the green line genes repressed by Grh, and the black line genes that show no change in grh mutants. Thin gray lines represent Grh-binding site distances to TSSs of 100 subsets (250 genes/subset) of unchanged genes. The distance between Grh peaks and putative target genes is longer for repressed genes than for activated genes (Wilcoxon test, P=8.9×10−5). (E) Grh-repressed genes. Overlap of genes associated with Grh ChIP-seq peaks (with Grh-binding motifs) with genes showing increased expression in grhB37 mutants and decreased levels in Grh-overexpressing (69B>grh) embryos. (F) Grh-activated genes. Overlap of genes encompassing Grh ChIP-seq peaks (with Grh-binding motifs) with genes showing decreased expression in grhB37 mutants and increased levels in Grh-overexpressing (69B>grh) embryos. (E,F) All genes with expression changes compared with wild type with a threshold of P<0.01 are shown.
To validate the data, we selected Grh-bound regions from Ddc, Stit, Cht2, knk, verm, mmy, kkv, CG32699 (LPCAT) (revealed by both antibodies) and serp, ck, Hmu, Cpr11A (detected only by the rabbit antibody) for ChIP-qPCR analysis. Compared with two control regions from CG34245 and CG18559 (Cyp309a2), all of the Grh peak regions showed Grh-binding with more than 10-fold enrichment relative to control IgG (Fig. S2D), supporting that these are bone fide target sites.
Analysis of genomic distributions revealed that Grh ChIP-seq peaks are located in proximal and distal regulatory regions, introns and, to a lesser extent, in exons (Fig. 1B). Located in proximity to 3754 genes, the bound regions are predominantly found near transcription start sites (TSSs) (Fig. S2B, Table S1). An 5′-AACNGGTTT-3′ motif matching the predicted Grh-binding motif (Venkatesan et al., 2003; Gao et al., 2013; Potier et al., 2014) was enriched within the sequences of Grh mouse antibody ChIP-seq peaks (E=3.5×10−382; Fig. 1C). This argues that a significant number of the additional Grh peaks revealed only by the rabbit antiserum and containing this motif identify true Grh-binding sites in the DNA. About half of the embryonic Grh peaks with the Grh-binding motif were also detected in ChIP-seq experiments targeting a Grh-GFP fusion protein on chromatin from larval eye disks (Potier et al., 2014) (Fig. S3A, Table S2). Therefore, we focused on genes with Grh ChIP-seq peaks, which contain the consensus Grh motif. Gene ontology (GO) analysis of the Grh targets shows enrichment for chitin-binding proteins (n=55), consistent with the known function of Grh in epidermal barrier formation. The set of Grh targets is also highly enriched in metabolic enzymes (n=778), signaling proteins (n=423), cytoskeletal or cell junction components (n=176) and TFs (n=233) (Fig. S2A).
To assess the consequence of Grh-binding to chromatin, we performed RNA microarray gene expression analysis from stage 16 wild type (wt), grhB37 mutants and from embryos overexpressing UAS-grh with the epithelial 69B-Gal4 driver. The combination of the Grh ChIP-seq and gene expression data identified 240 potentially repressed and 248 activated genes (Fig. 1E,F, Tables S3 and S4). In situ hybridization confirmed that expression of several putative targets is altered in grh mutants and upon grh overexpression (Fig. S4A-P). We also tested 26 selected candidates from the ChIP-seq and microarray experiments by comparing their expression levels in wt, grhB37 and 69B>grh embryos by RT-qPCR. We found that 78% of these show changes in expression, as in the microarray analysis of grh mutants and 69B>grh embryos, compared with wt (Fig. S4Q; data not shown).
The results suggest that Grh might act both as a repressor and as an activator of genes in ectodermal epithelia, consistent with previous studies of individual Grh targets (Brown and Kassis, 2013). A search for epigenetic landmarks described by modENCODE showed differences between Grh peaks at activated and repressed targets. Grh peaks of activated genes showed a reduced overlap with binding sites for Polycomb group (PcG) proteins (P=0.010), for the GAGA factor (P<5×10−4) and for H3K27me3 modifications (P<5×10−4), compared with a background distribution of Grh peaks at genes that do not respond to grh inactivation. This reduction of repressive chromatin marks was not detected at the Grh-binding sites of repressed genes, which instead showed increased overlap for the GAGA factor (P<5×10−4) and the H3K27me3 mark (P<5×10−4). Furthermore, Grh peaks at repressed genes showed reduced overlap with insulator proteins Beaf32 (P<5×10−4) and Cp190 (P<5×10−4) and with the histone modification mark H3K4me3 (Fig. S2C). Additionally, Grh-binding sites in repressed genes are consistently located at a greater distance from the TSSs, as compared with the genes that are activated or are unaffected by Grh (Fig. 1D). These correlations suggest that Grh-activated and -repressed enhancers are organized differently in the genome, and imply different direct or indirect mechanisms for Grh-dependent transcriptional activation or repression.
Grh-responsive elements display tissue-specific expression
To determine whether regions containing Grh peaks or predicted binding sites represent genuine gene regulatory modules, we cloned 50 such regions derived from 48 targets into the pHPdest-EGFP vector and assayed for GFP expression in transgenic embryos (Fig. 2M-O). Six of these constructs contained consensus Grh-binding motifs but had only minor ChIP enrichment that did not pass the relatively strict cut-off of our initial peak selection (Table S5). We found that 44 out of 50 transgenic lines show GFP expression in embryos, indicating that the Grh ChIP-seq peaks that include binding sites identify functional regulatory elements. The six fragments with predicted binding motifs but without a ChIP peak were among those driving GFP expression in relevant tissues. Furthermore, 47 out of the 50 reporters were responsive to Grh since they were ectopically activated upon Grh overexpression in epidermal stripes driven by en-Gal4 (Fig. 2J-L). Therefore, the combination of the Grh ChIP-seq data and the presence of consensus Grh-binding motifs reliably identifies functional regulatory elements but also excludes Grh-responsive regions that can be identified by the presence of the predicted binding motifs alone. Based on the GFP expression, we divided the reporter strains into three groups: (A) an epidermal and tracheal group (Fig. 2A,D,G); (B) an epidermal group (Fig. 2B,E,H); and (C) a group expressing GFP in scattered cells or subsets of cells of internal organs (Fig. 2C,F,I).
Fig. 2.
Grh-binding sites identify Grh-responsive regulatory elements. (A,D,G) Representatives of group A reporters with GFP expression in the embryonic trachea and other ectodermal epithelial tissues. (B,E,H) Group B reporters with GFP expression in the embryonic epidermis but not in the trachea. (C,F,I) Group C reporters with GFP expression in scattered cells and in subsets of internal epithelial organs. (J-L) Overexpression of Grh by en-Gal4 activates most reporters in epidermal stripes. Single examples are shown from each of group A (J), group B (K) and group C (L). Embryos were stained with anti-GFP antibodies. Scale bars: 50 μm. (M,N,O) Representative Grh ChIP-seq peaks and regulatory regions: Cht2 (M), CG11349 (N), yellow d2 (O) loci. Red peaks denote Grh rabbit antibody peaks. Green peaks denote signals from the IgG control. Blue bars denote the presence of the Grh-binding motif. Orange blocks denote cloned fragments. The y-axes show read coverage.
Taken together, the transgenic reporter assays suggest that Grh-regulated regions drive tissue-specific gene expression in different ectodermal tissues such as epidermis, tracheal or other tissues. The tissue-specific variations in the expression of the reporters suggest that Grh co-operates with other, tissue-specific factors in regulating gene expression. The induced activation of most reporters by Grh overexpression in epidermal stripes suggests that Grh can control them directly or indirectly.
A direct role of Grh in controlling the developmental expression of PGRP-LC and innate immunity genes
Grh controls wound healing upon epidermal injury (Mace et al., 2005). A few Grh-regulated enhancers, such as Ddc-GFP, have been shown to be wound inducible. To test whether any of the 50 Grh reporters respond to injury, we wounded embryos from the transgenic strains and monitored GFP expression. Eight of the 50 tested enhancers showed GFP upregulation at the wound site. Importantly, one of these reporters contains the regulatory region of a known wound-induced gene, Duox. The remaining seven reporters include enhancers for CG33110, Dro5 (Drsl5), CG10898, CG15282, knk, Cht2 and CG33978 (Fig. 3A-J). These genes have not previously been implicated in wound healing, and it will be interesting to test whether any of them might also play a role in epithelial regeneration upon injury.
Fig. 3.
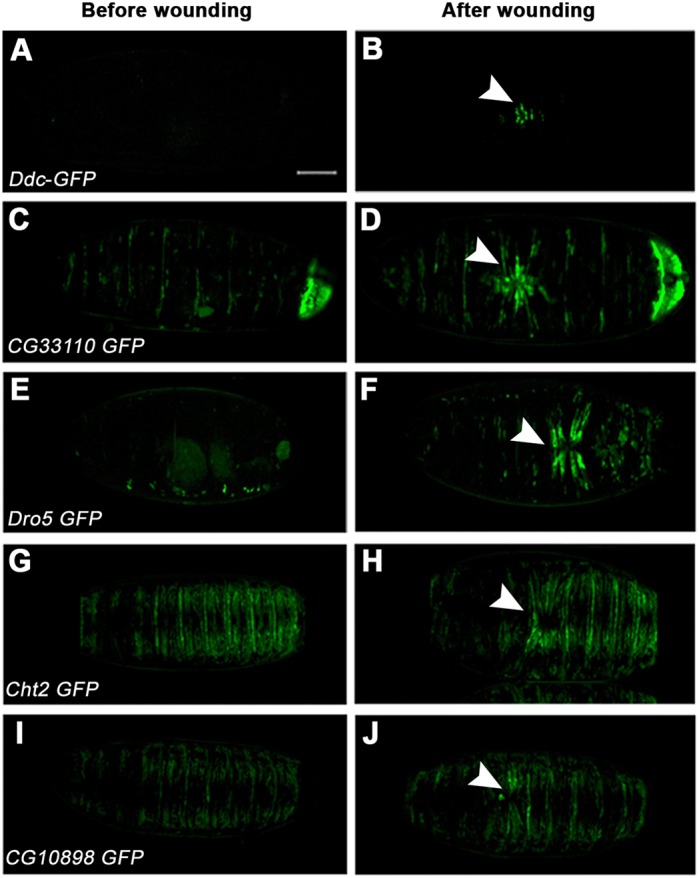
New Grh-dependent wound reporters. (A,B) GFP expression as a reporter of Ddc enhancer activity, a positive control for wounding. (C-J) CG33110-GFP (C,D), Dro5-GFP (E,F), Cht2-GFP (G,H) and CG10898-GFP (I,J) reporters before and after wounding. Arrowheads highlight the puncture area. Scale bar: 50 μm.
Previous analysis of gene expression in grh mutants and wt embryos has shown that innate immunity and stress-response gene expression are reduced upon grh inactivation (Paré et al., 2012). To investigate a potential direct role of Grh in the embryonic expression of innate immunity genes, we focused on PGRP-LC, since it is dispensable for Drosophila development (Choe et al., 2002). The only reported function of PGRP-LC is in the activation of antimicrobial peptides (AMPs) upon infection. Grh binds to the regulatory region of PGRP-LC and regulates its embryonic expression (Table S1). To test if Grh directly regulates epidermal PGRP-LC expression, we generated strains in which the GFP reporter is under the control of a 2 kb genomic region of PGRP-LC containing three Grh-binding motifs (Fig. 4A). In parallel, we mutated these motifs and generated PGRP-LCΔ-GFP flies, to test the impact of Grh-binding on GFP expression. PGRP-LC-GFP is expressed in all epidermal cells and faithfully reproduced PGRP-LC mRNA expression in embryos (Fig. 4B,C), whereas GFP expression in PGRP-LCΔ-GFP embryos was drastically reduced (Fig. 4D). Consistently, PGRP-LC-GFP expression was also dramatically reduced in grh mutants. Further, epidermal PGRP-LC mRNA was dramatically reduced in grh mutant embryos compared with wt (Fig. 4E,F). Conversely, Grh overexpression by btl-Gal4 ectopically induced PGRP-LC expression in the midline glia and the airways (Fig. 4G,H; data not shown). Therefore, the 2 kb DNA fragment in the PGRP-LC gene contains a Grh-dependent enhancer.
Fig. 4.
PGRP-LC is a direct Grh target. (A) Grh-bound regions in the PGRP-LC locus. Blue and green boxes denote PGRP-LA and PGRP-LF exons, respectively. PGRP-LC exons are shown as white (non-coding) and black (coding) boxes. PGRP-LC transcripts are indicated by lines connecting the exons. Arrows indicate transcription direction. Red arrowheads indicate predicted Grh-binding sites. The regions contained within the PGRP-LC-GFP and PGRP-LCΔ-GFP (crosses denote the three mutated Grh-binding sites) reporters are indicated beneath. (B-D) Epidermal expression of PGRP-LC-GFP in grh mutants (C) compared with wild type (B). Epidermal expression of PGRP-LCΔ-GFP is lower than that of PGRP-LC-GFP (D). (E-H) In situ hybridization for PGRP-LC mRNA in wild type (wt), btl>grh and grhB37 mutants. PGRP-LC expression is reduced in grhB37 mutants compared with wt (E,F). Grh overexpression in the trachea and midline glia (arrow) ectopically induces PGRP-LC (G,H). Scale bars: 50 μm in B-D; 100 μm in E-H.
Collectively, this analysis shows that Grh directly controls PGRP-LC epidermal expression during embryonic development. Additionally, the ChIP-seq analysis showed that Grh occupies the regulatory regions of a large battery of genes involved in the innate immune response (Table S6). Because PGRP-LC mutants do not show discernible developmental phenotypes, our analysis reveals a new direct function of Grh in ensuring the ability of developing epithelial tissues to mount effective immune responses against future infections.
Identification of functional Grh targets in airway maturation
The ChIP-seq experiments identified numerous Grh targets in the embryo but do not reveal which of these are responsible for each of the diverse defects observed in grh mutants. To identify functional Grh targets in the airways, we intersected the results of an unbiased genome-wide RNAi screen for genes involved in airway maturation and function (Hosono et al., 2015) with the genes in our ChIP-seq dataset. We found that 1152 Grh targets are required for proper gas filling of the airways (Fig. S5A, Table S7); 791 of them have homologs in vertebrates. These genes encode various enzymes, signaling molecules, TFs and cytoskeletal and adhesion proteins (Fig. S5B). Of particular interest is Mmp1, which encodes a metalloprotease. Mmp1, like Grh, is a known tracheal tube size regulator that is strongly expressed in tracheal cells (Page-McCaw et al., 2003).
Seven of the Grh tracheal targets encode proteins related to chitin biosynthesis (Gangishetti et al., 2012). Among them, mmy, knk, kkv, verm and serp and Cht2 are known tracheal tube size regulators (Devine et al., 2005; Tonning et al., 2005; Swanson and Beitel, 2006). We asked whether Grh controls tracheal tube length at least partly through regulating the expression of these genes. We first confirmed by ChIP-qPCR that Grh is recruited to the regulatory regions of these genes (Fig. S2D). Additionally, the expression of knk and Cht2 was dramatically reduced in grh mutants. Conversely, Grh overexpression by the btl-Gal4 driver induced knk and Cht2 expression in both trachea and in midline glia (Fig. S4A-H). kkv expression was also slightly reduced in grh mutants and ectopically activated in btl>grh embryos (Fig. S4I-L). Grh overexpression also induced mmy but suppressed verm and serp expression (Fig. S4M-P; data not shown). However, we did not detect consistent changes in the expression of mmy, verm or serp in grh mutants. To further confirm the transcriptional regulation of cuticle by Grh, we generated reporter strains for knk and cht2 based on the location of Grh-binding sites in their regulatory regions. Both reporters show GFP expression in a pattern mimicking the endogenous gene expression, and are both induced in epidermal stripes in en>grh embryos. Therefore, we conclude that Grh activates kkv, Cht2, knk and can promote mmy expression but suppresses verm and serp expression in the airways. These results highlight an unexpected complexity of Grh function in the regulation of genes involved in the assembly of the luminal chitinous matrix.
Taken together, Grh regulates Drosophila airway maturation through activating or repressing hundreds of targets, many of which have vertebrate homologs. We also found that Grh regulates the expression of multiple genes involved in the chitin-related tube size control pathway.
Grh controls vvl and other genes encoding TFs
In addition to the arthropod-specific, exoskeleton-related genes, the Grh targets encode a large number of other conserved proteins (Table S8). A comparison of the fly targets with the putative GRHL2 targets in human bronchial airway epithelial cells suggests that the homologs of 661 of the 3754 Drosophila Grh targets might also be regulated by GRHL2 in human airway cells (Fig. S3B, Table S9). Several of the repressed Drosophila targets encode TFs regulating the epithelial differentiation program and branch outgrowth (Matsuda et al., 2015a,b). One such example is vvl, which encodes a POU domain TF that maintains tracheal cell identity, upholds the levels of RTK signaling and promotes branching and cell differentiation (Anderson et al., 1995; de Celis et al., 1995; Llimargas and Casanova, 1997). Grh peaks are found both upstream and downstream of the vvl TSS, and vvl expression is upregulated in grh mutants and downregulated in 69B>grh. RT-qPCR (Fig. 5A), RNA in situ hybridization (Fig. 5B-D′) and ChIP-qPCR (Fig. 5E) confirmed a repressive function of Grh on the vvl locus.
Fig. 5.
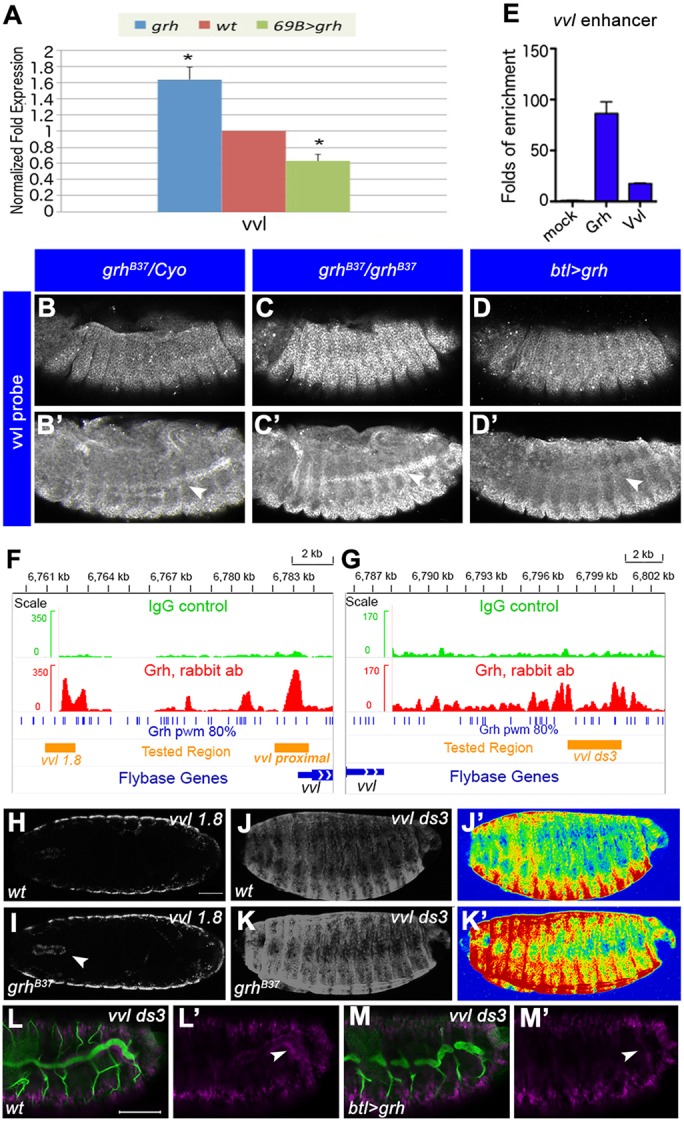
Vvl is a Grh target. (A) RT-qPCR assays for relative vvl expression in wild type, grhB37 mutants and 69B>grh embryos at stage 17. *P<0.05, Student's t-test. Error bars indicate s.e.m. (B-D′) In situ hybridization for vvl mRNA levels. (B,B′) Wild-type vvl expression. (C,C′) Increased vvl expression in grhB37 mutants. (D,D′) Decreased vvl expression in the trachea (arrowheads) of btl>grh embryos. (E) ChIP-qPCR shows Grh and Vvl enrichment in the vvl enhancer region. (F,G) vvl 1.8 (F) and vvl ds3 (G) regions. Grh rabbit antibody peaks are in red and IgG peaks in green. Blue bars denote the Grh-binding motif and orange bars the cloned enhancers. The y-axes show read coverage. (H,I) Grh represses vvl 1.8 in mouth hooks. (H) vvl 1.8 expression in wild type. (I) Higher vvl 1.8 signal in the mouth hook (arrowhead) of grhB37 mutants. (J-M′) Grh represses the vvl ds3 enhancer. (J,L) Expression of vvl ds3 reporter in wild-type embryos. (K) vvl ds3 signal increases in the epidermis in grhB37 mutants. (M) Decrease of vvl ds3 expression in btl>grh embryos. Magenta, anti-β-galactosidase; green, anti-Gasp (trachea). (J′,K′) Heat maps of signal intensities in H and I. (L′,M′) vvl ds3 expression in the trachea (arrowheads). Scale bars: 50 μm.
Several vvl enhancers have been identified by reporter strains (Fig. 5, Fig. S6A,B) (Sotillos et al., 2010). To pinpoint Grh-repressed regulatory modules we analyzed the expression of these vvl reporters in grhB37 mutants and in embryos overexpressing Grh in epidermal stripes. Two reporters, vvl 1.8 and vvl ds3 (Fig. 5F,G), showed increased expression in grhB37 mutants. vvl 1.8 is normally expressed in the trachea and epidermis at stage 17. In grhB37 embryos, vvl 1.8 was selectively upregulated in cells of the presumptive mouth hook structures. vvl ds3 was strongly expressed in the epidermis and more weakly in the trachea at stage 17. Its expression became upregulated in both tissues in grhB37 embryos. Conversely, grh overexpression by btl-Gal4 downregulated the vvl ds3 signal in the trachea (Fig. 5H-M′). The results suggest that Grh represses vvl through the vvl 1.8 fragment in the mouth hooks, likely acting together with a tissue-specific factor. Additionally, Grh represses vvl in the epidermis and trachea through the vvl ds3 enhancer, which would require cooperation with a distinct factor. Therefore, these results suggest that Grh-binding to the vvl ds3 and vvl 1.8 regulatory sequences represses the expression of vvl in distinct tissues.
Unexpectedly, besides the repressive effect of grh on the vvl 1.8 and vvl ds3 enhancers, we detected a positive effect on a new, vvl proximal reporter (Fig. 5F) and on vvl ds1.7 (Fig. S6). Both these elements could be activated in epidermal stripes in the en>grh-overexpressing embryos (Fig. S6). By contrast, grh overexpression did not influence the expression of a fifth reporter, vvl 345 (Fig. S6).
Collectively, the analysis of vvl reporters highlights the complexity of the vvl regulatory region and suggests that Grh-binding to different regulatory segments results in distinct outcomes. It also identifies two Grh-repressed regulatory modules, arguing that Grh may directly bind and, together with other factors, repress vvl expression in late embryos. Future analysis of these regulatory fragments and derivatives with mutated Grh-binding sites will address whether Grh is directly or indirectly involved in vvl repression.
Grh and Vvl co-regulate gene expression during epithelial maturation
Grh and Vvl are co-expressed during airway morphogenesis (see Fig. 7A-A″) and Grh downregulates, but does not shut off, vvl transcription. The tracheal phenotypes of vvl mutants include short branches, reflecting the early functions of Vvl in tracheal cell fate specification and branch outgrowth (Fig. S8C) (Llimargas and Casanova, 1997; Matsuda et al., 2015b). Noteworthy, 69B>vvl embryos at stage 16 show overelongated tracheal tubes, similar to grh mutants, suggesting a function of vvl in late steps of tube elongation (Fig. 6A-C). Moreover, btl>grh embryos display discontinuous trachea, mimicking the vvl mutant phenotypes (Fig. S8D). These observations suggest an antagonistic relationship between Grh and Vvl, where Vvl promotes branch outgrowth and Grh antagonizes it by downregulating vvl transcription. To test this, we generated grh mutants lacking one copy of the vvl gene, and found that this restored the overelongated tracheal tubes (Fig. 6D,E), strengthening the notion that grh antagonizes vvl to halt branch growth.
Fig. 7.
Vvl and Grh share common targets. (A-A″) Grh (green) and Vvl (red) co-expression in wild-type stage 16 embryos. (B-B″) Vvl regulates Grh-dependent enhancers. CG13377-GFP is increased in epidermal stripes in en-Gal4>vvl-overexpressing embryos. (C) ChIP-qPCR with Grh or Vvl antibodies shows enrichment of Grh or Vvl in the regulatory regions of six Grh-dependent enhancers and eight predicted common targets of Grh and Vvl. CG34245 is used as negative control. The y-axis is plotted on a logarithmic scale. (D) Co-immunoprecipitation of Grh and Vvl overexpressed in S2 cells. Grh and Vvl can form complexes. (E) RT-qPCR analysis of Grh and Vvl common targets. CG33110, CG17941 and CG17549 are activated by both Grh and Vvl. Duox is repressed by both Grh and Vvl. CG17839 and ics are repressed by Grh but activated by Vvl. Cht2, CG9363, yellow C and Gp150 are regulated by Grh but not by Vvl in stage 17 embryos. *P<0.05, **P<0.01, ***P<0.001; ns, no significant change; Student's t-test. Error bars indicate s.e.m.
Fig. 6.
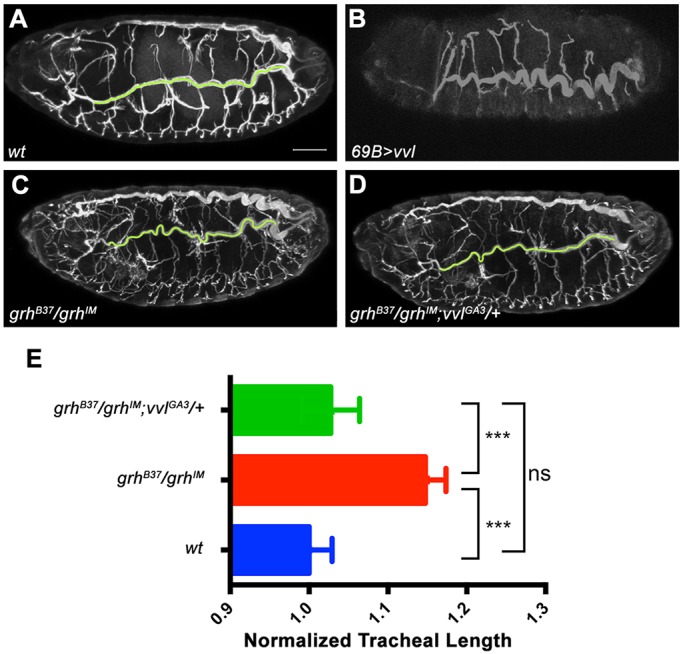
Grh and Vvl antagonize each other during tracheal morphogenesis. (A-D) Maximum projections of confocal sections of the trachea. (A) Stage 17 wild type. (B) 69B>vvl embryos show overelongated dorsal trunks. (C) grhB37/grhIM embryos with elongated airways. (D) Deletion of one copy of vvl ameliorates the tracheal phenotype in grhB37/grhIM embryos. Trachea are stained by Gasp. Green lines indicate tracings for length measurements. Scale bar: 50 μm. (E) Tracheal length quantification shows that decreased vvl expression can partially rescue grh mutant tracheal phenotype. Tracheal length was measured in confocal z-stacks, normalized to embryo length and then normalized to w1118. Error bars indicate normalized s.e.m. ***P<0.001; ns, no significant change; Student's t-test.
To further investigate the interplay between the two TFs in airway maturation, we first asked if Vvl might also control the expression of Grh targets. We found that 6 out of 11 randomly selected Grh-dependent GFP reporters were ectopically induced in epidermal stripes in en>vvl embryos (Fig. 7B-B″, Fig. S7). This suggests that vvl can regulate some Grh targets. This effect is unlikely to be due to an indirect upregulation of grh transcription by Vvl overexpression because only 6 of 11 Grh reporters responded to Vvl. Further, ChIP-qPCR with an anti-Vvl antibody and chromatin isolated from stage 17 wt embryos showed Vvl enrichment around the Grh-binding regions of these six targets (Fig. 7C).
Previous studies of Vvl function in development and upon immune responses identified Vvl-binding site sequences on its respective targets (Anderson et al., 1996; Certel et al., 1996; Junell et al., 2010). We asked whether similar motifs are present in the regulatory regions of Grh targets. We found that consensus motifs for Vvl binding frequently co-occur with Grh peaks in the genome, suggesting that the two TFs might share some of their targets (Table S1). We generated primer pairs targeting the regulatory regions of eight potential common targets and used them in ChIP-qPCR experiments with either the Grh or the Vvl antibodies. We detected more than 10-fold enrichment for both Grh and Vvl in six out of the selected eight regions, suggesting that Vvl and Grh bind to common regions in stage 16 embryos (Fig. 7C). These Grh and Vvl common targets are not only expressed in the trachea but also include genes expressed in the epidermis but not the airways, such as CG15282, CG13377 and CG33110 (Table S5).
To investigate the significance of Vvl binding at the enhancers of Grh targets, we tested the mRNA expression of potential common targets in grh and vvl mutants by RT-qPCR. CG33110, CG17941 (dachsous) and CG17549 are activated by both Grh and Vvl. Conversely, Duox is repressed both by Grh and Vvl. However, CG17839 and ics are repressed by Grh but activated by Vvl. Cht2, CG9363 (GstZ2), yellow C and Gp150 are regulated by Grh but not by Vvl in stage 17 embryos (Fig. 7E, Table S10). This suggests that Grh and Vvl share some common targets during late embryogenesis.
The gene expression analysis of target genes in vvl and grh mutants reveals a complex interaction scheme, whereby the two TFs either act independently or cooperate with each other and other TFs to induce or repress target gene expression. We therefore tested whether the two TFs can bind to each other by performing co-immunoprecipitation experiments in Drosophila S2 cells. After co-transfection, immunoprecipitation with the Grh antibody was able to precipitate HA-tagged Vvl and, conversely, the Vvl antibody could immunoprecipitate Grh (Fig. 7D). This suggests that Grh and Vvl form protein complexes to control common target gene expression.
DISCUSSION
Grh and its targets in epithelial maturation
Grh controls epithelial development and regeneration in multiple organisms. Our ChIP-seq data provide a broad view of Grh-binding to its targets in all Grh-expressing tissues. The analysis of Grh-dependent regulatory sequences indicates that the majority of the 5599 peaks that include the consensus Grh-binding sequence identify true Grh targets. Hitherto, analysis of Grh targets in development focused on proteins involved in epidermal barrier formation, adhesion molecules and junctional proteins. Our identification of functional Grh targets in the airways adds large groups of proteins involved in lipid metabolism, cell signaling and TFs. This suggests additional functions of Grh in tubulogenesis that might explain several of its additional roles. For example, the phenotype of grh mutants in the airways includes the selective expansion of the epithelial apical membranes, a phenotype that has not been detected in any of the mutants affecting junctional proteins or the formation and modification of the apical extracellular barrier (Tonning et al., 2006; Wang et al., 2009; Tiklová et al., 2010). Our definition of new Grh targets during airway maturation provides a rich resource for future studies addressing how Grh controls epithelial morphogenesis.
A prevalent group of Grh targets in the epidermis and airways includes genes involved in innate immune responses ranging from pattern recognition receptors to effectors. Interestingly, several putative GRHL2 targets in human bronchial epithelial cells, such as serpins and chitinase 3-like proteins (He et al., 2013), have been implicated in immune responses (Gao et al., 2015). Our analysis of PGRP-LC reveals a direct role of Grh in endowing epithelial cells the ability to combat infections. Although the PGRP-LC reporter expression was not inducible by wounding or bacterial injection, it remains possible that Grh also directly controls the activation of epithelial immune responses upon infection. Indeed, partial inactivation of Grh by RNAi in adult flies resulted in increased morbidity and mortality upon bacterial infection (Paré et al., 2012).
Grh as an activator or repressor of target genes
Our analysis of 47 new Grh-activated enhancers in epithelial development suggests the presence of distinct, tissue-specific Grh co-factors in the control of target genes in different epithelial cell types. The activation of some of these reporters upon injury expands the repertoire of Grh-activated enhancers and is in line with previous models proposing wound-induced interactions of Grh with other factors. These interactions could be induced by post-translational modifications of Grh or its co-factors by Rolled (Kim and McGinnis, 2011) and other kinases downstream of Stit receptor kinase signaling (Tsarouhas et al., 2014) and might facilitate the activation of transcription by Grh pre-bound to chromatin.
The ChIP-seq and gene expression analysis also reveal a potential role for Grh as a repressor. Such a repressor function of Grh is consistent with previous studies addressing Grh function on individual targets in flies and mammals (Tuckfield et al., 2002; Blastyak et al., 2006; Strubbe et al., 2011). We find a higher correlation of PcG-binding sites and repressive chromatin marks around the Grh-binding sites of repressed targets as compared with the binding sites of activated genes. The positioning of Grh-binding sites relative to the TSS of repressed versus activated or unaffected genes also differs: Grh-binding sites are usually further from the TSS in repressed target genes. This observation is supported by the analysis of vvl ds3 and vvl 1.8, the only two identified repressible enhancers, which are located more than 2 kb from the vvl TSS. The difference in the structure of the repressed and activated Grh enhancers suggests that Grh repression might require chromatin looping and involve co-repressors (Saramäki et al., 2009). Further work is needed to elucidate a potential direct function of Grh in transcriptional repression.
Grh and Vvl, transcriptional regulation and complex co-factor interactions
A characteristic group of Grh targets in the airways includes TFs involved in epithelial cell differentiation. This resembles the complex regulatory functions of Grh during neuronal specification. For instance, in neuroblasts, Grh demarcates the last time window for TF expression by repressing Castor (Baumgardt et al., 2009). In intermediate neural progenitors (INPs), Grh is detected in the ‘middle-aged’ INPs and overlaps with the expression of the TFs Dichaete and Eyeless. The three TFs cross-regulate each other (Bayraktar and Doe, 2013). Similar cross-talk between Grh and its TF targets might specify and maintain epithelial differentiation. Since reduction of vvl in grh mutants largely ameliorates the tube elongation defects, the direct or indirect repression of genes encoding TFs is likely to be a crucial function of Grh in the airways. The shared expression pattern of Vvl and Grh, their binding to a set of common enhancers and their ability to form complexes suggest that they collectively control tube growth during airway maturation. Given their co-expression in other contexts, they might also co-operate during neural cell specification and epithelial immune responses.
MATERIALS AND METHODS
ChIP-seq and ChIP-qPCR
We performed ChIP as described by Dai et al. (2013) using 13-16 h w1118 embryos. Embryos were homogenized and fixed in 1.8% formaldehyde at room temperature. After several washes, chromatin in lysis buffer was sonicated to 0.1-0.5 kb. For each immunoprecipitation, sheared chromatin was precleared with Gammabind G Sepharose (GE Healthcare) coated with BSA and incubated with pre-absorbed rabbit anti-Grh antibody (5 μg), mouse anti-Grh antibody (Bray et al., 1989; 5 μg), rabbit anti-Vvl antibody (R. Matsuda, Stockholm University, Sweden; 5 μg) or rabbit IgG (Sigma; 5 μg). Precipitated complexes were washed, eluted, and cross-links were reversed at 65°C. After proteinase K treatment, DNA was purified using QIAprep spin columns (Qiagen) and recovered in 50 µl elution buffer containing RNase A (Thermo Fisher, 5 mg/ml). DNA libraries were made using the Illumina ChIP-seq Library Kit and sequenced as individual lanes on an Illumina GAII.
Real-time PCR was performed on a Bio-Rad CFX96 using Power SYBR Green (Applied Biosystems). Primer sequences are presented in Table S11. PCR was performed in triplicate samples, and immunoprecipitated DNA was compared against standard curves obtained from serial dilutions of input DNA. Values are plotted as fold enrichment normalized to IgG control, and the standard deviation within the triplicate samples was calculated.
Quantitative RT-PCR (RT-qPCR) and microarray analysis
Stage 16 grhB37 homozygous mutant embryos and 69B>grh embryos were selected. RNA was extracted with Trizol (Thermo Fisher) and was incubated with Turbo DNase (Thermo Fisher; 2 units). Total RNA (4 µg) was reverse transcribed with Superscript III reverse transcriptase (Thermo Fisher; 200 units) using random primers. To ensure absence of genomic DNA, RT-qPCR was performed on a mock reverse-transcribed RNA sample. Primer sequences are listed in Table S11.
For microarray analysis, RNA samples from two biological replicates were labeled and hybridized to the Affymetrix GeneChIP Drosophila Genome 2.0 Array at the Genomic Core Laboratory at Memorial Sloan-Kettering.
Bioinformatics
ChIP-seq data processing was carried out according to Dai et al. (2013). Reads were mapped to the Drosophila dm3 genome using Bowtie (Langmead et al., 2009) with parameters –v 2 –m 1 (i.e. allowing two mismatches and only using uniquely mapping reads). Peaks were called using Quest (Valouev et al., 2008) with the following parameters: bandwidth, 30; region size, 300; ChIP enrichment, 5; ChIP to background enrichment, 2; and ChIP extension enrichment, 2. The set of Grh peaks was determined by combining all Grh peaks found with the two antibodies, and retaining those that covered a Grh site for further analysis. Peaks were annotated to the gene with the closest TSS using ENSEMBL transcript annotations. ChIP-seq data from Potier et al. (2014) were analyzed in the same way as for data described here.
For de novo motif finding, MEME (Bailey and Elkan, 1995) was run on the top 500 peaks, extended 100 nt in each direction from the summit of the peak, to search for 6- to 15-nt motifs using default settings. As a proxy for statistical significance of motifs, we used the E-value calculated by MEME, i.e. the number of (equally or more interesting) motifs expected by chance if the nucleotides in the input sequences were shuffled. To search for hits against the Grh site, the PWM Biostrings Bioconductor package was used with a minimum score of 80%.
Microarray data were normalized using the GCRMA Bioconductor package, and log fold change values were computed using the Bioconductor limma package. P-values were adjusted for multiple hypotheses using the false discovery rate (FDR) correction. For genes with multiple probe sets, the probe set with the lowest adjusted P-value was selected. Genes with P<0.01 after FDR adjustment were considered differentially expressed.
Enrichment of gene ontology annotations (Ashburner, 2000) and tissue-enriched expression profiles (Chintapalli et al., 2007) were computed as described (Dai et al., 2013), using Fisher's exact test with the Bonferroni correction for multiple hypothesis testing. FlyAtlas (http://flyatlas.org) gene classifications were based on tissue enrichment scores; genes with enrichment scores of at least 2 were considered enriched in a given tissue. Orthologous genes between D. melanogaster and mouse were called using the Drosophila RNAi Screening Center (DRSC) Integrative Ortholog Prediction Tool (Hu et al., 2011). Ortholog calls supported by at least three of the programs included in the meta-server were used for analysis.
Drosophila mutant strains
w1118, grhB37, grhIM and vvlGA3 are loss-of-function mutations. w1118 was used as control in all experiments. The btl-Gal4 transgene is inserted on the second chromosome and 69B-Gal4 on the third chromosome. lacZ- or GFP-marked CyO and TM6 balancers were used to identify genotypes.
Enhancer cloning and transgenic strain generation
Genomic fragments for enhancer assays were selected based on the presence of Grh ChIP-seq peaks and on the expression changes of the adjacent genes in transcriptome analysis of grh mutants or grh-overexpressing embryos. Fragments were cloned by PCR from genomic DNA of w1118 into pENTR D-TOPO (Invitrogen) and then transferred to pHPdest-EGFP vector (Boy et al., 2010). PCR primers are listed in Table S11. Constructs were integrated into the attp2 site. The resulting reporters were tested for GFP expression by live imaging and antibody staining.
The PGRP-LC GFP reporters were constructed by subcloning corresponding PCR fragments into pH-stinger (Barolo et al., 2000). The Grh-binding sites in the PGRP-LCΔ-GFP reporter were mutated using the QuikChange Site-Directed Mutagenesis Kit (Agilent) and the primers indicated in Table S11. PGRP-LC GFP transgenic lines were established by P-element-mediated transformation. The vvl ds3, vvl 1.8 and vvl ds1.7 enhancer strains were described previously (Sotillos et al., 2010).
Wounding assays
Embryos were collected during stage 15 (∼12 h after egg laying at 25°C) and pricked with a glass needle.
In situ hybridization and immunostaining
Digoxigenin-labeled RNA probes were transcribed from EST clones and hybridized to embryos as described (Lehmann and Tautz, 1994; Matsuda et al., 2015a,b). EST clones were obtained from the Drosophila Genomics Resource Center (DGRC) and are listed in Table S12.
Immunostaining was performed as described by Tsarouhas et al. (2007) with the following antibodies: rabbit anti-GFP (Invitrogen, A11122; 1:250), mouse anti-GFP20 (Sigma, G6539; 1:1000), rabbit anti-β-galactosidase (Cappel, 55976; 1:1500), guinea pig anti-Gasp (Tiklová et al., 3013; 1:2000). The polyclonal rabbit anti-Grh antiserum was generated by polyclonal genomic antibody (GAT) against the peptide QQQLISIKREPEDLRKDPKNGNIAGAATANGPGSVITQKSFDYTELCQPGTLIDANGSIPVSVNSIQQRTAVHGSQ and used at 1:1000.
Acknowledgements
We thank the fly community members who isolated, characterized or distributed valuable reagents, especially J. Castelli-Gair Hombría, Y. Engström, S. Thor, W. McGinnis, Bloomington Drosophila Stock Center (BDSC), DGRC and National Institute of Genetics (NIG) for directly sharing antibodies, strains and clones. We thank FlyBase and modENCODE for Drosophila genomic resources. We thank the Stockholm University Imaging Facility and members of the M. Mannervik, S. Åström and C.S. laboratories for support during the project, especially M. Björk for fly service.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.W., C.S.; Methodology: S.W., J.O.-W., Q.D., C.S.; Software: J.O.-W.; Validation: L.Y., S.W., Q.D., R.M., C.H.; Formal analysis: L.Y., S.W., J.O.-W., Q.D., R.M., C.H., C.S.; Investigation: L.Y., S.W., Q.D., R.M., C.H., C.S.; Resources: L.Y., S.W., J.O.-W., Q.D., R.M., C.H., S.B., E.C.L., C.S.; Data curation: L.Y., S.W., J.O.-W., Q.D., C.S.; Writing - original draft: L.Y., S.W., C.S.; Writing - review & editing: L.Y., J.O.-W., Q.D., R.M., C.H., S.B., E.C.L., C.S.; Visualization: L.Y., S.W., J.O.-W., C.H., C.S.; Supervision: S.W., E.C.L., C.S.; Project administration: C.S.; Funding acquisition: C.S.
Funding
This work was supported by grants from the Swedish Research Council (Vetenskapsrådet) (K2010-67x-21476101-3) and the Swedish Cancer Society (Cancerfonden) (CF160499) to C.S. E.C.L.'s group was supported by the National Institutes of Health (R01- NS074037, R01-NS083833) and by a Memorial Sloan-Kettering Cancer Center Core Grant [P30-CA008748]. Deposited in PMC for release after 12 months.
Data availability
ChIP-seq and microarray data have been deposited at Gene Expression Omnibus under accession numbers GSE102551 (all data), GSE102549 (expression data) and GSE102550 (ChIP data).
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.143297.supplemental
References
- Almeida M. S. and Bray S. J. (2005). Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 122, 1282-1293. 10.1016/j.mod.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Anderson M. G., Perkins G. L., Chittick P., Shrigley R. J. and Johnson W. A. (1995). drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 9, 123-137. 10.1101/gad.9.1.123 [DOI] [PubMed] [Google Scholar]
- Anderson M. G., Certel S. J., Certel K., Lee T., Montell D. J. and Johnson W. A. (1996). Function of the Drosophila POU domain transcription factor drifter as an upstream regulator of breathless receptor tyrosine kinase expression in developing trachea. Development 122, 4169-4178. [DOI] [PubMed] [Google Scholar]
- Ashburner M. (2000). A biologist's view of the Drosophila genome annotation assessment project. Genome Res. 10, 391-393. 10.1101/gr.10.4.391 [DOI] [PubMed] [Google Scholar]
- Auden A., Caddy J., Wilanowski T., Ting S. B., Cunningham J. M. and Jane S. M. (2006). Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns 6, 964-970. 10.1016/j.modgep.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Aue A., Hinze C., Walentin K., Ruffert J., Yurtdas Y., Werth M., Chen W., Rabien A., Kilic E., Schulzke J.-D. et al. (2015). A Grainyhead-Like 2/Ovo-Like 2 pathway regulates renal epithelial barrier function and lumen expansion. J. Am. Soc. Nephrol. 26, 2704-2715. 10.1681/ASN.2014080759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. and Elkan C. (1995). Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Mach. Learn. 21, 51-80. 10.1007/BF00993379 [DOI] [Google Scholar]
- Barolo S., Carver L. A. and Posakony J. W. (2000). GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726, 728, 730, 732. [DOI] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Terriente J., Díaz-Benjumea F. J. and Thor S. (2009). Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139, 969-982. 10.1016/j.cell.2009.10.032 [DOI] [PubMed] [Google Scholar]
- Bayraktar O. A. and Doe C. Q. (2013). Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498, 449-455. 10.1038/nature12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A., Mishra R. K., Karch F. and Gyurkovics H. (2006). Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol. Cell. Biol. 26, 1434-1444. 10.1128/MCB.26.4.1434-1444.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy A. L., Zhai Z. Z., Habring-Müller A., Kussler-Schneider Y., Kaspar P. and Lohmann I. (2010). Vectors for efficient and high-throughput construction of fluorescent drosophila reporters using the PhiC31 site-specific integration system. Genesis 48, 452-456. 10.1002/dvg.20637 [DOI] [PubMed] [Google Scholar]
- Bray S. J. and Kafatos F. C. (1991). Developmental function of Elf-1 - an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 5, 1672-1683. 10.1101/gad.5.9.1672 [DOI] [PubMed] [Google Scholar]
- Bray S. J., Burke B., Brown N. H. and Hirsh J. (1989). Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev. 3, 1130-1145. 10.1101/gad.3.8.1130 [DOI] [PubMed] [Google Scholar]
- Brown J. L. and Kassis J. A. (2013). Architectural and functional diversity of polycomb group response elements in Drosophila. Genetics 195, 407-419. 10.1534/genetics.113.153247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddy J., Wilanowski T., Darido C., Dworkin S., Ting S. B., Zhao Q. A., Rank G., Auden A., Srivastava S., Papenfuss T. A. et al. (2010). Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev. Cell 19, 353-353 10.1016/j.devcel.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci C. and Gould A. P. (2005). Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development 132, 3835-3845. 10.1242/dev.01932 [DOI] [PubMed] [Google Scholar]
- Certel K., Anderson M. G., Shrigley R. J. and Johnson W. A. (1996). Distinct variant DNA-binding sites determine cell-specific autoregulated expression of the Drosophila POU domain transcription factor drifter in midline glia or trachea. Mol. Cell. Biol. 16, 1813-1823. 10.1128/MCB.16.4.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J. and Dow J. A. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715-720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Choe K.-M., Werner T., Stoven S., Hultmark D. and Anderson K. V. (2002). Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296, 359-362. 10.1126/science.1070216 [DOI] [PubMed] [Google Scholar]
- Dai Q., Ren A., Westholm J. O., Serganov A. A., Patel D. J. and Lai E. C. (2013). The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 27, 602-614. 10.1101/gad.213314.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J. F., Llimargas M. and Casanova J. (1995). Ventral veinless, the gene encoding the Cf1a transcription factor, links positional information and cell differentiation during embryonic and imaginal development in Drosophila melanogaster. Development 121, 3405-3416. [DOI] [PubMed] [Google Scholar]
- Devine W. P., Lubarsky B., Shaw K., Luschnig S., Messina L. and Krasnow M. A. (2005). Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc. Natl. Acad. Sci. USA 102, 17014-17019. 10.1073/pnas.0506676102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Attardi L. D., Admon A., Freeman M. and Tjian R. (1989). Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 3, 1677-1688. 10.1101/gad.3.11.1677 [DOI] [PubMed] [Google Scholar]
- Gangishetti U., Veerkamp J., Bezdan D., Schwarz H., Lohmann I. and Moussian B. (2012). The transcription factor Grainy head and the steroid hormone ecdysone cooperate during differentiation of the skin of Drosophila melanogaster. Insect Mol. Biol. 21, 283-295. 10.1111/j.1365-2583.2012.01134.x [DOI] [PubMed] [Google Scholar]
- Gao X., Vockley C. M., Pauli F., Newberry K. M., Xue Y., Randell S. H., Reddy T. E. and Hogan B. L. M. (2013). Evidence for multiple roles for grainyheadlike 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc. Natl. Acad. Sci. USA 110, 9356-9361. 10.1073/pnas.1307589110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Bali A. S., Randell S. H. and Hogan B. L. M. (2015). GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 211, 669-682. 10.1083/jcb.201506014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon W. M., Zeller M. D., Klein R. H., Swindell W. R., Ho H., Espetia F., Gudjonsson J. E., Baldi P. F., and Andersen B. (2014). A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. J. Clin. Invest. 124, 5205-5218. 10.1172/JCI77138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. H., Lee C. G., Dela Cruz C. S., Lee C. M., Zhou Y., Ahangari F., Ma B., Herzog E. L., Rosenberg S. A., Li Y et al. et al. (2013). Chitinase 3-like 1 regulates cellular and tissue responses via IL-13 receptor alpha2. Cell Rep. 4, 830-841. 10.1016/j.celrep.2013.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphälä J., Uv A., Cantera R., Bray S. and Samakovlis C. (2003). Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development 130, 249-258. 10.1242/dev.00218 [DOI] [PubMed] [Google Scholar]
- Hosono C., Matsuda R., Adryan B. and Samakovlis C. (2015). Transient junction anisotropies orient annular cell polarization in the Drosophila airway tubes. Nat. Cell Biol. 17, 1569-1576. 10.1038/ncb3267 [DOI] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N. and Mohr S. E. (2011). An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. D., Dubnicoff T., Liaw G. J., Bai Y., Valentine S. A., Shirokawa J. M., Lengyel J. A. and Courey A. J. (1995). Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev. 9, 3177-3189. 10.1101/gad.9.24.3177 [DOI] [PubMed] [Google Scholar]
- Johnson W. A., McCormick C. A., Bray S. J. and Hirsh J. (1989). A neuron-specific enhancer of the Drosophila dopa decarboxylase gene. Genes Dev. 3, 676-686. 10.1101/gad.3.5.676 [DOI] [PubMed] [Google Scholar]
- Junell A., Uvell H., Davis M. M., Edlundh-Rose E., Antonsson A., Pick L. and Engstrom Y. (2010). The POU transcription factor Drifter/Ventral veinless regulates expression of Drosophila immune defense genes. Mol. Cell. Biol. 30, 3672-3684. 10.1128/MCB.00223-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. and McGinnis W. (2011). Phosphorylation of Grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc. Natl. Acad. Sci. USA 108, 650-655. 10.1073/pnas.1016386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. and Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Y. and Adler P. N. (2004). The grainy head transcription factor is essential for the function of the frizzled pathway in the Drosphila wing. Mech. Dev. 121, 37-49. 10.1016/j.mod.2003.11.002 [DOI] [PubMed] [Google Scholar]
- Lehmann R. and Tautz D. (1994). In situ hybridization to RNA. Methods Cell Biol. 44, 575-598. 10.1016/S0091-679X(08)60933-4 [DOI] [PubMed] [Google Scholar]
- Li X., Erclik T., Bertet C., Chen Z. Q., Voutev R., Venkatesh S., Morante J., Celik A. and Desplan C. (2013). Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456-462. 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw G. J., Rudolph K. M., Huang J. D., Dubnicoff T., Courey A. J. and Lengyel J. A. (1995). The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 9, 3163-3176. 10.1101/gad.9.24.3163 [DOI] [PubMed] [Google Scholar]
- Llimargas M. and Casanova J. (1997). ventral veinless, a POU domain transcription factor, regulates different transduction pathways required for tracheal branching in Drosophila. Development 124, 3273-3281. [DOI] [PubMed] [Google Scholar]
- Luschnig S., Bätz T., Armbruster K. and Krasnow M. A. (2006). serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 16, 186-194. 10.1016/j.cub.2005.11.072 [DOI] [PubMed] [Google Scholar]
- Mace K. A., Pearson J. C. and McGinnis W. (2005). An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308, 381-385. 10.1126/science.1107573 [DOI] [PubMed] [Google Scholar]
- Matsuda R., Hosono C., Saigo K. and Samakovlis C. (2015a). The intersection of the extrinsic hedgehog and WNT/wingless signals with the intrinsic Hox code underpins branching pattern and tube shape diversity in the drosophila airways. PLoS Genet. 11, e1004929 10.1371/journal.pgen.1004929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R., Hosono C., Samakovlis C. and Saigo K. (2015b). Multipotent versus differentiated cell fate selection in the developing Drosophila airways. Elife 4, e09646 10.7554/eLife.09646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C., Cheng L. and Gould A. P. (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891-902. 10.1016/j.cell.2008.03.034 [DOI] [PubMed] [Google Scholar]
- Page-McCaw A., Serano J., Santé J. M. and Rubin G. M. (2003). Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev. Cell 4, 95-106. 10.1016/S1534-5807(02)00400-8 [DOI] [PubMed] [Google Scholar]
- Paré A., Kim M., Juarez M. T., Brody S. and McGinnis W. (2012). The functions of grainy head-like proteins in animals and fungi and the evolution of apical extracellular barriers. PLoS ONE 7, e36254 10.1371/journal.pone.0036254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier D., Davie K., Hulselmans G., Naval Sanchez M., Haagen L., Huynh-Thu V. A., Koldere D., Celik A., Geurts P., Christiaens V. and Aerts S. (2014). Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell Rep. 9, 2290-2303. 10.1016/j.celrep.2014.11.038 [DOI] [PubMed] [Google Scholar]
- Pyrgaki C., Liu A. M. and Niswander L. (2011). Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev. Biol. 353, 38-49. 10.1016/j.ydbio.2011.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y., Parekh V., Wilanowski T., Hislop N. R., Auden A., Ting S. B., Cunningham J. M. and Jane S. M. (2010). Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 345, 237-245. 10.1016/j.ydbio.2010.07.017 [DOI] [PubMed] [Google Scholar]
- Saramäki A., Diermeier S., Kellner R., Laitinen H., Vaisanen S. and Carlberg C. (2009). Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3. J. Biol. Chem. 284, 8073-8082. 10.1074/jbc.M808090200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillos S., Espinosa-Vázquez J. M., Foglia F., Hu N. and Hombría J. C.-G. (2010). An efficient approach to isolate STAT regulated enhancers uncovers STAT92E fundamental role in Drosophila tracheal development. Dev. Biol. 340, 571-582. 10.1016/j.ydbio.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubbe G., Popp C., Schmidt A., Pauli A., Ringrose L., Beisel C. and Paro R. (2011). Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. USA 108, 5572-5577. 10.1073/pnas.1007916108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson L. E. and Beitel G. J. (2006). Tubulogenesis: an inside job. Curr. Biol. 16, R51-R53. 10.1016/j.cub.2006.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiklová K., Senti K. A., Wang S., Graslund A. and Samakovlis C. (2010). Epithelial septate junction assembly relies on melanotransferrin iron binding and endocytosis in Drosophila. Nat. Cell Biol. 12, 1071-1077. 10.1038/ncb2111 [DOI] [PubMed] [Google Scholar]
- Tiklová K, Tsarouhas V. and Samakovlis C. (2031). Control of airway tube diameter and integrity by secreted chitin-binding proteins in Drosophila. PLoS ONE 8, e67415 10.1371/journal.pone.0067415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S. B., Wilanowski T., Auden A., Hall M., Voss A. K., Thomas T., Parekh V., Cunningham J. M. and Jane S. M. (2003a). Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat. Med. 9, 1513-1519. 10.1038/nm961 [DOI] [PubMed] [Google Scholar]
- Ting S. B., Wilanowski T., Cerruti L., Zhao L.-L., Cunningham J. M. and Jane S. M. (2003b). The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem. J. 370, 953-962. 10.1042/bj20021476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S. B., Caddy J., Hislop N., Wilanowski T., Auden A., Zhao L. L., Ellis S., Kaur P., Uchida Y., Holleran W. M. et al. (2005). A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308, 411-413. 10.1126/science.1107511 [DOI] [PubMed] [Google Scholar]
- Tonning A., Hemphala J., Tang E., Nannmark U., Samakovlis C. and Uv A. (2005). A transient luminal chitinous matrix is required to model epithelial tube diameter in the Drosophila trachea. Dev. Cell 9, 423-430. 10.1016/j.devcel.2005.07.012 [DOI] [PubMed] [Google Scholar]
- Tonning A., Helms S., Schwarz H., Uv A. E. and Moussian B. (2006). Hormonal regulation of mummy is needed for apical extracellular matrix formation and epithelial morphogenesis in Drosophila. Development 133, 331-341. 10.1242/dev.02206 [DOI] [PubMed] [Google Scholar]
- Tsarouhas V., Senti K. A., Jayaram S. A., Tiklová K., Hemphälä J., Adler J. and Samakovlis C. (2007). Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev. Cell 13, 214-225. 10.1016/j.devcel.2007.06.008 [DOI] [PubMed] [Google Scholar]
- Tsarouhas V., Yao L. and Samakovlis C. (2014). Src-kinases and ERK activate distinct responses to Stitcher receptor tyrosine kinase signaling during wound healing in Drosophila. J. Cell Sci. 127, 1829-1839. 10.1242/jcs.143016 [DOI] [PubMed] [Google Scholar]
- Tuckfield A., Clouston D. R., Wilanowski T. M., Zhao L.-L., Cunningham J. M. and Jane S. M. (2002). Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol. Cell. Biol. 22, 1936-1946. 10.1128/MCB.22.6.1936-1946.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uv A. E., Harrison E. J. and Bray S. J. (1997). Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol. Cell. Biol. 17, 6727-6735. 10.1128/MCB.17.11.6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A., Johnson D. S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R. M. and Sidow A. (2008). Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods 5, 829-834. 10.1038/nmeth.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Mahavadi P., Sasikumar S., Cushing L., Hyland T., Rosser A. E., Riccardi D., Lu J., Kalin T. V., Kalinichenko V. V. et al. (2014). Grainyhead-like 2 (GRHL2) distribution reveals novel pathophysiological differences between human idiopathic pulmonary fibrosis and mouse models of pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 306, L405-L419. 10.1152/ajplung.00143.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K., McManus H. R., Mello C. C., Smith T. F. and Hansen U. (2003). Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 31, 4304-4316. 10.1093/nar/gkg644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Q., Tsarouhas V., Xylourgidis N., Sabri N., Tiklová K., Nautiyal N., Gallio M. and Samakovlis C. (2009). The tyrosine kinase Stitcher activates Grainy head and epidermal wound healing in Drosophila. Nat. Cell Biol. 11, 890-895. 10.1038/ncb1898 [DOI] [PubMed] [Google Scholar]
- Wilanowski T., Tuckfield A., Cerruti L., O'Connell S., Saint R., Parekh V., Tao J., Cunningham J. M. and Jane S. M. (2002). A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech. Dev. 114, 37-50. 10.1016/S0925-4773(02)00046-1 [DOI] [PubMed] [Google Scholar]
- Wilanowski T., Caddy J., Ting S. B., Hislop N. R., Cerruti L., Auden A., Zhao L.-L., Asquith S., Ellis S., Sinclair R. et al. (2008). Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 27, 886-897. 10.1038/emboj.2008.24 [DOI] [PMC free article] [PubMed] [Google Scholar]






