Abstract
Filamentous fungi have been of great interest because of their excellent ability as cell factories to manufacture useful products for human beings. The development of genetic transformation techniques is a precondition that enables scientists to target and modify genes efficiently and may reveal the function of target genes. The method to deliver foreign nucleic acid into cells is the sticking point for fungal genome modification. Up to date, there are some general methods of genetic transformation for fungi, including protoplast-mediated transformation, Agrobacterium-mediated transformation, electroporation, biolistic method and shock-wave-mediated transformation. This article reviews basic protocols and principles of these transformation methods, as well as their advantages and disadvantages.
Keywords: Filamentous fungi, Protoplast-mediated transformation, Agrobacterium-mediated transformation, Electroporation, Biolistic method, Shock-wave-mediated transformation
Background
Fungi exist widely in different environments, such as soil, biological wastes and plants. Some of them have been utilized by humans for over 1000 years. In nature, fungi play a vital role in numerous degradation processes. In agriculture, many species of fungi are used for control of plant pests and diseases [1, 2]. In medicine, fungi are utilized to produce antibiotics for the treatment of diseases. For example, penicillin is a product of Penicillium chrysogenum and cephalosporin of Cephalosporium acremonium. Due to their good capacity in manufacturing valuable proteins and secondary metabolites, fungi are important economic contributors. With the explosion and exploration of fungal genomic sequence information, mycology is coming into a new era of functional studies [3].
Filamentous fungi, molds, grow well and rapidly on simple and inexpensive media and thus are preferred cell factories due to their outstanding capacity in expression and secretion of heterologous proteins with post-translational processing. They have also been used in the production of a wide variety of products, such as citric acid, kojic acid and other organic acids, secondary metabolites like penicillin, cephalosporin, as well as cellulase, amylase, glucanase, rennet, lipase, laccase and unsaturated fatty acids, soy sauce, fermented soya beans.
The development of genetic transformation techniques is a breakthrough in our attempt to genetically modify fungal strains. This technique enables scientists to target and modify genes efficiently to reveal functions of targeted genes, or to insert new genetic elements into the genome of the strain such as promoters to modify the expression of endogenous genes [4].
Conceptually, fungal biotechnology is expected to be transformed by the application of genetic transformation technologies. In practice, genetic transformation of fungi meets with many difficulties. First, although many articles of fungal transformation have been published each team established their own protocols without sufficient details. Second, because of the huge number of fungal species and their complex cell wall structures, different transformation methods are required for different fungal species. Thus, species specific transformation protocols must be optimized for each strain. Even completely new methods must be established for efficient transformation of some strains [5].
In this review, we summarize all major methods of genetic transformation for fungi, including the protoplast-mediated transformation method and the Agrobacterium-mediated transformation method, which are common methods used for many years, the electroporation method and the biolistic method, which are not currently popular but are particularly suitable in some specific circumstances, and the emerging shock-wave-mediated transformation technology.
Common filamentous fungi and their applications
Aspergillus niger is a typical species used for producing glucoamylase and citric acid. In recent years, using fungi, such as A. niger, to produce cellulase to degrade inexpensive cellulosic materials into glucose with high efficiency has become one of research hotspots. Aspergillus oryzae, with a safe application history of over 1000 years, can be applied in producing protease, amylase, glucoamylase, cellulose, and phytase. As pointed out by Kubodera [6], A. oryzae, which has been one of the most important workhorses in Japanese fermentation industry, was used for the production of tempeh, one of the oldest fermented soy products in China [7]. A. oryzae can secrete a variety of enzymes, including amylase, protease and esterase, etc. Protein in beans can be hydrolyzed into soluble nitrogenous compounds by protease hydrolysis in the fermentation process. In addition, Kojic acid, which has an antibacterial effect on aerobic microbes, is an organic acid produced by A. oryzae. Because of its nontoxic property, Kojic acid has important applications such as being used as food additives, or in cosmetics, pharmaceuticals, etc. [8]. Aspergillus nidulans has been one of the most widely studied species in terms of genetics and biochemistry [9, 10]. It is often used as model organism in the identification of gene function and protein interaction studies [11, 12]. Penicillium is one of the most widely distributed fungi in nature. In medicine, penicillin also is the earliest clinical application of antibiotics. Additionally, Penicillium species have the capability of degrading lignocellulose. Monascus purpureus Went is mainly used traditionally for making wine, vinegar, food coloring, and meat preservation. In 1979, Brooklyn K (Monacolin K), an agent with activity to lower cholesterol levels [13], was isolated from Monascus.
Transformation methods
The establishment of genetic transformation systems has enabled scientists to transform foreign DNA into filamentous fungi and thus obtained the desired strains for industrial purposes. We now can take full advantage of the superior secretory power of fungi and their excellent efficiency in manufacturing valuable metabolites.
Protoplast-mediated transformation (PMT)
PMT is the most commonly used fungal transformation method, which relies on a large number of competent fungal protoplasts. The principle is to use some commercially available enzymes to remove fungal complex cell wall components for generating protoplasts. Subsequently, some chemical reagents (such as PEG) are used to promote the fusion of exogenous nucleic acids and protoplasts, as described in more detail below. The components of the fungal cell wall are highly variable among different strains. Even components of the spore coat are significantly different from that of hyphae from the same strain [14]. Thus, there is no universal transformation method that can be applied to different fungal strains. Preparation of protoplast can hardly be standardized. Part of the difficulties comes from our limited knowledge of cell wall hydrolases. Development of an optimized PMT method for fungi still requires significant effort.
PMT is a routinely used transformation method. The method has been under constant improvement to achieve higher efficiency for genetic transformation and for targeting suitable gene loci through gene editing. Preparation of protoplasts requires removing the cell wall, which is mainly achieved through enzyme treatment. Non-enzyme methods to prepare protoplasts have also been reported, such as physical methods including grinding and supersonic wave shock [15]. However, they are not widely used because of practical inconvenience and the low yield of protoplasts. A summary of protoplast-mediated transformation protocols for different fungal species are provided in Table 1.
Table 1.
Summary of protoplast-mediated transformation protocols for different fungal species
| Strain | Starting material | Number of protoplasts | Name of vectors | The concentration of DNA | Selective markers | Transformation efficiency (transformants/μg DNA) | References |
|---|---|---|---|---|---|---|---|
| Trichoderma reesei QM 9414 and VTT-D-79125 | Mycelium | 5 × 107 to 5 × 108 | pSa143 containing the A. nidulans argB gene | 2–5 μg of transforming DNA | argB | 600 transformants | [16] |
| T. reesei QM 9414 | Mycelium | 5 × 107–108 | pRLMEX30 (5.5 kb) | 1–6 μg of transforming DNA | hygB | 1800–2500 transfomants | [17, 18] |
| N. crassa 74A | Mycelium | 1 × 107 | pVK88 | 60 μg/mL | qa-2 + | 5-30 transformants | [19] |
| A. oryzae NRRL 492 | Mycelium | 2 × 106 | pILJ16 | 10 μg | argB | 5–10 transformants | [20, 21] |
| M. purpureus NRRL 1596, M. ruber NRRL 1597 and M. purpureus IBCC1 | Mycelium | 1.8 × 108 | pULJL43 (4.6 kb) | 15 μg | Sh ble | M. purpureus NRRL 1596 (870 transformants), M. ruber NRRL1597 (137 transformants) and M. purpureus IBCC1 (43 transformants) | [22] |
| Aspergillus giganteus IfGB15/0903 | Germinated conidia | 1 × 107 to 2.5 × 108 | pAN7-1 | 10 μg | hygB | 55 transformants per 1 × 107 protoplasts | [23] |
Basic steps of the PMT method
PMT was first applied to Saccharomyces cerevisiae. Researchers [24] prepared protoplasts with the commercial snailase, and used sorbitol to preserve protoplasts. Later, such method was applied to filamentous fungi, such as Neurospora crassa [19], and A. nidulans [25]. Although the transformation methods have been improved, the basic steps remain essentially the same. Basic steps of the PMT method are provided in Fig. 1.
Fig. 1.
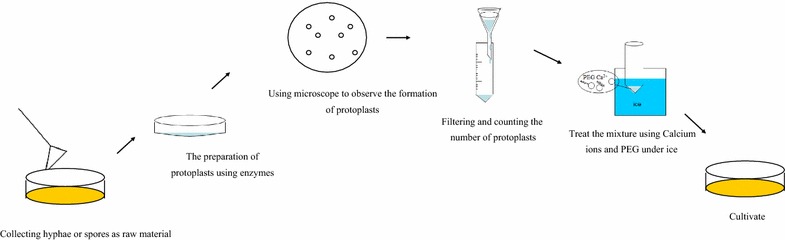
Basic steps of the protoplast-mediated transformation
Preparation of the protoplasts
The first step in protoplast preparation is the removal of cell wall through enzymatic digestion. The fungal cell wall is comprised of glucan, mannan and chitin. The structure of the fungal cell wall is highly dynamic, and the cell wall varies during the cell division and growth of fungi, as well as in spore germination, hyphal branching and formation of the diaphragm. The cell wall components are also different in different fungal species, therefore, various enzymes should be used in combination. It has been reported that the selection of an appropriate enzyme mix is a key factor in protoplast preparation [26].
In general, the hyphae are sensitive to a suitable enzyme which hydrolyzes its cell wall during the logarithmic phase. In the PMT procedure of Neurospora, the protoplasts are prepared by hydrolyzing the newly born hyphae (culture for 4–6 h under 25–30 °C) [27, 28]. Similarly, protoplasts can also be prepared with conidiospores. For example, for Aspergillus and Penicillium, one can choose germinal spores or thalli [29].
Protoplasts are sensitive to osmotic pressure, care should be taken to maintain a stable osmotic pressure to keep the protoplasts intact during the enzymolysis of cell walls. Thus, osmotic stabilizers (such as sorbitol, sodium chloride, and potassium chloride) should be included in all of the buffers for protoplast preparation to avoid rupture of cells. For instance, sorbitol solution with a concentration of 0.8–1.2 M is used in the protoplast preparation of N. crassa [19], Aspergillus sp. [30] and Trichoderma sp. [17] to maintain the osmotic stability of protoplasts. A summary of protoplast preparation parameters for some common fungal species are provided in Table 2.
Table 2.
Summary of protoplast preparation parameters for some common fungal species
| Strain | Starting material | Enzyme | Enzymolysis conditions | Osmotic stabilizer | Protoplast yield (protoplasts/mL) | References |
|---|---|---|---|---|---|---|
| A. niger K10 | NA | Lyophilized snail | Incubate at 31 °C for 3 h on a laboratory reciprocal shaker (78 strokes/min, amplitude 3 cm) | 0.7 M NaCl | 8.9 × 106 | [31] |
| A. niger N583 | Mycelium | Lysing enzymes, chitinase griseus and β-glucuronidase | Incubate in a 100 mL glass bottle for 2 h at 37 °C and 130 rpm | 0.8 M sorbitol | NA | [32] |
| Aspergillus nidulans A89 | Mycelium | Yatalase, Kitalase | Incubate 6 h at 30 °C with constant shaking (60–80 rpm) | 1 M sorbitol | NA | [33] |
| Aspergillus nidulans G191 | Mycelium | Novozym 234 | Incubate 1.5 h at 30 °C | 0.6 M KCl | NA | [34] |
| Trichoderma atroviride T11 | Mycelium | Lysing enzymes from Trichoderma. harzianum | Incubate at 30 °C for 2 h with shaking at 80 rpm | 1 M sorbitol | 1–5 × 106 | [26] |
| Penicillium simplicissimum | Mycelium | Driselase, caylase, cellulase, enzyme cocktail II from Merck company | Incubate at 30 °C for 20 or 180 min | 1.2 M sorbitol | 2 × 109 | [26] |
| Trichoderma reesei QM 9414 | Mycelium | Lysing enzymes from T. harzianum | Incubate at 28–30 °C for 2 h | 1 M sorbitol | NA | [17] |
| Trichoderma reesei QM 9414 and VTT-D-79125 | Mycelium | Novozym 234 | Incubate at 28 °C for approx. 1.5 h | 1 M and 1.2 M sorbitol | 5 × 107 to 5 × 108 | [16] |
| N. crassa M246 (qa-2 mutation) and M6-11 (arom-9 mutation) | Mycelium | Proteinase K | Incubate at 1 h at room temperature | 1 M sorbitol | 1 × 107 | [19] |
| Aspergillus fumigatus 2085 | Mycelium | β-glucuronidase | Incubate at 37 °C for 1 h with occasional shaking | 0.7 M KCl | 1 × 106 | [35] |
| A. oryzae NRRL 492 | Mycelium | Novozyme 234 and β-glucuronidase | Incubate at 30 °C for 1.5 h with shaking at 80 rpm | 0.6 M sorbitol and 1 M sucrose | Each 400 mL culture yielded 5–10 × 107 protoplasts | [20, 21] |
| Rhizopus oryzae AS 3.819 | Mycelium | Helicase, cellulase, lyticase | Incubate at 35 °C for 140 min with gentle shaking | 0.6 M sorbitol | 1 × 107 | [36] |
| Monascus purpureus | Mycelium | Glucanex, lysing enzyme from T. harzianum β-glucuronidase and Caylase | Incubate at 28 °C for 4 h with orbital shaking (100 rpm) | 0.5 M KCl and 0.1 M MgSO4 | 1 × 109 | [22] |
| Penicillium expansum | Mycelium | 0.6% cellulase and 0.6% snailase | Incubate at 25–35 °C for 3–3.5 h | 0.6 M NaCl | 2.36 × 108 | [37] |
Uptake of exogenous DNA
The solution used to suspend protoplasts usually contains calcium ions and osmotic stabilizers. Calcium is thought to open channels in the cytomembrane, which facilitates entry of exogenous DNA into the cell, while osmotic stabilizer are necessary for maintaining the morphology of the protoplasts. Usually, certain amount of polyethylene glycol (PEG) is added together with purified DNA (which can either be the circular double-stranded DNA or the linearized DNA). PEG is a commonly used cell fusion promoter [38]. It can form the molecular bridge between cells or between cytomembrane and DNA, and thus promotes adhesion. In addition, it can also induce disordered charges on the cytomembrane surface, alter the membrane permeability, and facilitate entry of exogenous nucleic acids into cells [39].
PEG is a crucial agent enhancing transformation efficiency. Low transformation efficiency in most cases can be improved by adding more PEG. Under normal conditions, the performance of low-molecular-weight PEG (like PEG3000) is superior to that of high-molecular-weight PEG (like PEG8000). However, this needs to be optimized for various species [40].
Transformation efficiency is also influenced by temperature. Generally, the DNA and protoplast mixture should be placed on ice for 15–30 min, so that the DNA can adhere to the surface of protoplasts [41].
Regeneration of protoplasts
In order to guarantee good recovery of viable protoplasts, protoplasts are allowed to recuperate on the plate with no selection pressure for a certain amount before they are transferred to a selective plate. An osmotic stabilizer should be included in the regeneration culture. Stable osmotic pressure is the key factor for protoplast to regenerate cell wall. Only the protoplasts that carry exogenous nucleic acids can grow on the selective medium.
Comments on the PMT method
Protoplast transformation method is simple and effective with no need for expensive equipment. But the protocol involves many steps and critical reagents. Each step needs to be optimized and the quality of the reagents needs to be critically tested. The growth status of fungi being transformed needs to be carefully monitored. Experience is critical for the successful implementation of this method.
Agrobacterium -mediated transformation (AMT)
Agrobacterium is a Gram-negative bacterium commonly found in soil. Agrobacterium tumefaciens can infect injured plants. The tumor-inducing plasmid of > 200 kb, which is also referred to as the Ti plasmid, could be isolated in the early stage of infection. When A. tumefaciens infects a plant, it enters the plant through the wound, and integrates part of the Ti plasmid into the genome of the infected plant cells. The integrated DNA fragment from the Ti plasmid is commonly referred to as transfer DNA or T-DNA. The T-DNA inserts into the plant genome randomly as monoclone. The T-DNA is flanked by two directional imperfect repeats (called left and right border) and contains genes that encode enzymes responsible for the formation of plant hormones, which cause tumor growth [42]. A binary vector was designed to have the target gene inserted in between the left and right T-DNA borders, and the recombinant plasmid was transformed into the Agrobacterium tumefaciens. The positive Agrobacterium clone was used as a vehicle to integrate the target gene into the fungal genome. The specific steps will be discussed in detail below.
AMT method has been shown to be more stable and efficient than conventional transformation methods since the first paper [43] reported that this method could be applied to fungal transformation. The AMT method was first applied to transform S. cerevisiae [28]. A plasmid carrying a hygromycin-resistance gene is commonly used [43] to transform Aspergillus awamori [44]. The AMT method has been applied to many Ascomycetes, including the Aspergillus [43], and Monascus purpureus [22]. The basic steps of AMT method are provided in Fig. 2. A summary of Agrobacterium-mediated transformation protocol for different fungal species is provided in Table 3.
Fig. 2.
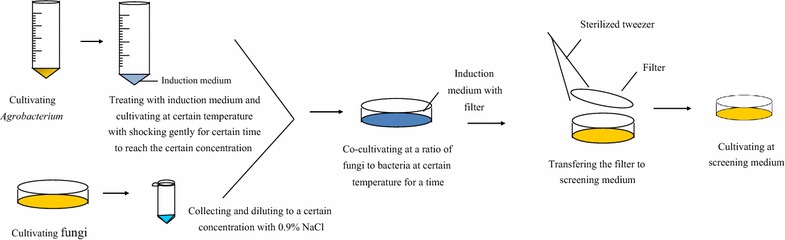
The basic steps of the Agrobacterium–mediated transformation
Table 3.
Summary of Agrobacterium-mediated transformation protocols for different fungal species
| Fungus | Agrobacterium | Starting material | The number of fungal cells | Pre-culturing of Agrobacterium | The conditions of co-culturing | Name of vector (size) | Selective marker | Transformation efficiency | References |
|---|---|---|---|---|---|---|---|---|---|
| Aspergillus awamori | A. tumefaciens LBA1100 | Conidiospores | 1 × 106 | Incubate at 28 °C 4–5 h with 0.2 M AS and 100 rpm to OD600 nm of 0.8 | Incubate the plates for 3 days at 22.5 °C | NA | hygB | 200–250 transformants per 106 conidiospores | [45] |
| Aspergillus fumigatus B-5233 | A. tumefaciens EHA105 | Conidia | 1 × 107 | Further incubation at 28 °C for 24 h to a ratio of 1:10 (conidia to bacteria) | Incubated at 37 °C for 3 days | pDHt/hph (NA) | hygB | 100 transformants per 107 conidia | [46] |
| Trichoderma reesei QM 9414 | A. tumefaciens AGL-1 | Protoplasts and conidia | 1 × 107 | Further incubation for 6 h at 28 °C to OD600 nm of 0.15 | Incubation at 25 °C for 48 h | pBI-hph (15846 bp) | hygB | Start from protoplasts: 2000 to 9000 transformants per 107 protoplasts. Start from conidia: 200–500 transformants per 107 condia | [47] |
| Mucor circinelloides ATCC1216b | A. tumefaciens AGL-1 | Spores | 1 × 105 spores per mL and | Incubate for 4 h at 20 °C to OD600 nm of 0.6 | Incubation at 15 °C for 6 days | pTiBo542 (NA) | hygB | 2000–9000 transformants per 107 sporangiospores | [48] |
| Aspergillus giganteus IfGB15/0903 | A. tumefaciens LBA1100 | Germinated conidia | 5 × 107 | Incubate for 6 h at 30 °C with 200 μM AS to OD 600 nm of 2.5 | Incubation at 24 °C for 3 days | pUR5750 (NA) | hygB | 500 to 7900 transformants per 108 conidia | [23] |
Factors that influence the AMT efficiency
Many factors affect the AMT efficiency, including the type of starting fungal material (protoplast, spore, hypha, and fruit body tissue), concentration of the acetosyringone, ratio of fungus to Agrobacterium, and the condition for co-culturing.
The type of starting fungal material The AMT method can use the protoplasts, spores, hyphae, and fruit body tissue of fungi as the recipient. Appropriate starting materials should be selected for different strains. For instance, the AMT method only works for the protoplasts of Rhizopus. oryzae and Mucor circinelloides, while spores or germinal spores would not produce transformants [49].
The concentration of acetosyringone (AS) AS acts on two stages during the AMT process. One is the induction process, and the other is the transformation process. AS is generally used to induce the expression of the Vir domain of T-DNA, and the gene in the Vir domain activate the transfer of T-DNA. Numerous studies have demonstrated that an appropriate amount of AS was necessary during the transformation process. However, the addition of AS is not absolutely necessary during the pre-culturing stage of the Agrobacterium, which could reduce transformation efficiency for some strains. The concentration of AS is an important factor affecting the transformation efficiency during the fungus-Agrobacterium co-culturing process in the AMT of Aspergillus awamori [45].
The ratio of fungi to Agrobacterium Within certain limits, the transformation efficiency will reach the maximum level with the increase of amount of fungus or Agrobacterium. An optimal ratio for the AMT for different fungi must be empirically determined. Ratio of fungal to that of bacterial cells should be optimized for different fungus-Agrobacterium transformation systems.
The condition for co-culturing The conditions for co-culturing are an important factor in the AMT method. This includes culture time, temperature, pH, and the selection of filter. The temperature and time for co-culturing are the key factors among the AMT steps. In the fungus-Agrobacterium transformation, an appropriate condition to start is a temperature of 20–28 °C and a co-culturing time of 16–96 h. A lower temperature (20–25 °C) is usually beneficial for the AMT method. The filter, which is hydrophilic and serves as support for fungus-Agrobacterium co-culturing, facilitates the transfer of single colonies to the screening plate. A nitrocellulose membrane, nylon membrane, filter paper, cellophane and polyvinylidene fluoride (PVDF) membrane can be used as the filter [3].
Comments on the Agrobacterium-mediated transformation
The AMT method opens up a new avenue for those fungi recalcitrant to transformation by conventional methods. The AMT method is especially suitable for generating knock-in mutations in fungi because T-DNA randomly inserts into the genome as a single copy. In addition, AMT can achieve high homologous recombination efficiency in various gene targeting experiments [3].
Major advantages of the AMT method include: firstly, diversified transformation recipients, including protoplasts, hyphae, and spores; secondly, the ability to integrate exogenous genes into the genome to form stable transformants; and thirdly, high transformation efficiency resulting in a large number of transformants [3].
The AMT method requires binary vectors, which are tedious to prepare. Multiple factors need to be taken into consideration in the optimization of the transformation process. This is a major limitation of the AMT method [50, 51].
Electroporation transformation
Electroporation is a simple, rapid and efficient transformation method for filamentous fungi. In electroporation, electric charges are stored in a capacitor to build a high voltage, the sample is struck by the impulse voltage, and the exogenous nucleic acid can be transferred instantly into cells. Usually, square waves or exponential decay waves are used in the transformation of fungi [52, 53]. Exponential-decay pulses are generated simply by charging and discharging a capacitor. The electric field declines exponentially from the peak value. A square wave is a non-sinusoidal periodic waveform (which can be represented as an infinite summation of sinusoidal waves), in which the amplitude alternates at a steady frequency between fixed minimum and maximum values. Different waveforms of electroporation are utilized for different species. A summary of waveforms used in electroporation for different species is provided in Table 4.
Table 4.
Summary of waveforms used in electroporation of different species
| Strain | Waveform | References |
|---|---|---|
| Saccharomyces cerevisiae | Exponential decay waveform | [54] |
| Pichia pastoris | Exponential decay waveform | [55] |
| A. niger ATCC 20739 | Exponential decay waveform | [52] |
| Rhizopus oryzae | Square wave | [53] |
| Aspergillus nidulans | Exponential decay waveform | [56] |
| Trichoderma harzianum | Exponential decay waveform | [57] |
When a cell is exposed to the electric field, the structure of the cytomembrane will be changed with a voltage induced between the cytomembrane. Micropores can be formed in the cytomembrane after electric shock. The induced cell wall permeability is reversible within the thresholds of the voltage and the duration, otherwise, it will cause irreversible injury to the cells. Therefore, the micropores in the cytomembrane appear to have two patterns after electric shock, the reversible and the irreversible pattern. The lipid and protein molecules in the cytomembrane can restore the original structure when an appropriate field intensity is applied, while the irreversible electric shock will give rise to irreparability or extremely slow recovery, which eventually leads to cell death [58]. Exogenous DNA can be transferred into the bacterium [59], plant protoplast [60], animal cell [61] and filamentous fungi [62] through electroporation. This method has been successfully applied to multiple fungi. Ozeki et al. discovered that germinal spores are more amenable to transformation by electroporation [52]. In recent years, electroporation has become a reliable method for gene transformation of some common strains [63]. A summary of electroporation-mediated transformation protocols for different fungal species is provided in Table 5.
Table 5.
Summary of electroporation-mediated transformation protocols for different fungal species
| Strain | Waveform | Instrument | Electroporation parameters | Raw material | Number of cell | Name and size of vectors | Amount of DNA | Selective markers | Transformation efficiency (per μg DNA) | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Rhizopus oryzae AS 3.819 | NA | Multiporator (Eppendorf) | 12 kV/cm | Protoplasts | 1 × 107 | pBS-hygro-ldhA (8533 bp) | 5 μg | hygB | 10.2 transformants | [36] |
| R. oryzae AS 3.819 | NA | Multiporator (Eppendorf) | 15 kV/cm | Germinated spores | 1 × 107 | pBS-hygro-ldhA (8533 bp) | 5 μg | hygB | 8.8 transformants | [36] |
| A. niger ATCC 20739 | Exponential decay waveform | Electro Gene Transfer Unit | 6 kV/cm and 3 ms time constant | Germinated spores | 1 × 107/0.4 mL | pXbal92 (5.7 kb) and pBXba2 (6.0 kb) | 1–10 μg | argB − | 1.2 transformants for integrative vector and 100 colonies for plasmid DNA. | [52] |
| Penicillium urticae NRRL 2159A | Exponential decay waveform | Gene Pulser (Bio-Rad) Multiporatr (Eppendorf) | 12.5 kV/cm, 8.6 ms time constant and 25 μF | Germinated conidia | 8 × 106 | pCSN44(NA) | 1–5 μg | hygB | 1.8 × 103 transformants | [64] |
| A. oryzae ATCC 14895 | Exponential decay waveform | Gene Pulser (Bio-Rad) Multiporatr (Eppendorf) | 11–12.5 kV/cm, 4.6–4.8 ms time constant and 25 μF | Germinated conidia | 2.5 × 106 | pBEN(NA) | 1–5 μg | BenomylR | 2.6 × 103 transformants | [64] |
| Leptosphaeria maculans “Virulent” | Exponential decay waveform | Gene Pulser (Bio-Rad) Multiporatr (Eppendorf) | 12.5 kV/cm, 4.8 ms time constant and 25 μF | Germinated conidia | 1.2 × 106 | pCSN44(NA) | 1-5 μg | hygB | 1–6 × 102 transformants | [64] |
| N. Crassa R-206A | Exponential decay waveform | Gene Pulser (Bio-Rad) Multiporatr (Eppendorf) | 12.5 kV/cm, 5 ms time constant and 25 μF | Germinated conidia | 3–6 × 106 | Bsqa(NA) | 1–5 μg | hygB | 5.7 × 103 transformants | [64] |
Factors that influence electroporation transformation
Electroporation parameters
Electric field intensity Electric field intensity is the most important factor that influences the electroporation efficiency. When the applied electric field intensity reaches the magnitude of kV/cm and the pulse width of μs–ms scale, the cytomembrane will be changed and many micropores will be generated on the cell walls [65]. High electric field intensity is associated with high uptake rate of exogenous nucleic acids and lower cell survival rate. However, various types of cells require different electric field intensities due to differences in cytomembrane components [66]. Few micropores are formed when the electric field intensity does not exceed the required threshold. In the contrary, excessive electric field intensity will result in irreversible damage to the cytomembrane, leading to cell death [58].
Capacitance During the electroporation process, variation in electric charges and the electric field intensity applied to the cell suspension depend on the capacitance and the pulse duration. The intensity and duration of the pulse are also influenced by the capacitance, therefore, greater capacitance has better transformation effects [67].
Pulse duration and frequency The duration of perforation on the cytomembrane, which directly related to electroporation transformation efficiency, is influenced by pulse duration and frequency [68].
Electroporation environment and external factors
Buffer solution The buffer solution provides an important environment for electroporation of cells, and the pH value of the electric shock buffer solution is of great importance. Normally, a buffer of pH 7.0 is used. Cells are easily punctured and killed at pH higher than 7.0 [52].
Temperature A large amount of heat is produced during the electroporation process, which will be released into the buffer solution. Therefore, a reduced temperature (0–4 °C) is recommended for better effect [66]. Furthermore, ice-bathing the pre-electric-shock mixture can also improve the electric shock efficiency.
Concentration of exogenous nucleic acid Overall, the electroporation efficiency increases with the concentration of exogenous nucleic acid. Compact superhelix DNA more easily enters the cells through the cytomembrane. In 1995, one study reported that every 1 μg of plasmid DNA could generate 100 transformants for A. niger [52].
Comments on the electroporation method
The electroporation method has been extensively applied to numerous types of cells, including prokaryotes and eukaryotes. This technology has the potential to be the method of choice for the transformation of unexplored fungal species. Compared with the PMT method, where complicated steps are involved, electroporation is simple and more convenient. However, the mechanism of electroporation still remains unclear. The perforation rate of the cytomembrane is dependent on many parameters of the electric field. And also, it requires suitable buffer conditions to be optimally effective.
Biolistic transformation
Biolistic transformation is also known as particle bombardment. Its principle is that foreign DNA is adsorbed on the surface of tungsten or gold particles. Under the push of high pressure, the particles are injected into host cells. Particle bombardment can realize both stable and transient transformation.
Various factors affect the efficiency of bombardment in patterns of complex interactions [69]. Biological parameters (cell type, growth condition, and cell density) and instrumental settings (particle type and size, vacuum and pressure level, target distance) are important variables [70].
Particle bombardment is the most powerful among all the genetic transformation methods. It is not subject to the limitations of cell types of host or species. For fungi, particle bombardment is sufficiently efficient for those organisms that are difficult to culture or from which protoplasts are hard to prepare. Particle bombardment is easy and convenient to operate. However, instruments and consumables for particle bombardment are expensive. It will only be considered in the case where other methods fail to work. At present, particle bombardment has been utilized to successfully transform A. nidulans and T. reesei [71, 72], etc.
Shock-wave-mediated transformation (SWMT)
SWMT utilizes the principle of energy transformation and transmission to generate transient pressure disturbance and twisting force across cells to form transient cavitation effect [73]. This method has been applied in medical treatment such as orthopedics and crushing kidney stones [74, 75]. SWMT changes the permeability of cell membranes through acoustic cavitation resulting in the uptake of exogenous nucleic acid into cells. The method has been successfully used in introducing exogenous nucleic acid into Escherichia coli, Pseudomonas aeruginosa and Salmonella typhimurium [76–78]. In 2013, Denis Magaña-Ortíz et al. first reported the application of SWMT for fungi, including A. niger, Fusarium oxysporum, and Phanerochaete chrysosporium [79]. In this article, three advantages of the SWMT method were noted. Firstly, compared with conventional transformation methods, this method is able to directly act on spores but not protoplasts. Secondly, the physical parameters were easily controllable, only the number of spores, energy and speed of the shockwave needed to be precisely controlled. Thirdly, the transformation efficiency was excellent. Results of Denis Magaña-Ortíz et al. indicated that, compared with the Agrobacterium transformation method, SWMT method could enhance the transformation efficiency by 5400 fold for A. niger [79].
But some limitations in this transformation method were also notable. Since a large proportion of DNA is damaged in the shock wave treatment, the transformation efficiency as determined by the ratio of DNA to cells was quite low [80]. However, for the number of cells involved, the transformation efficiency was significantly higher [52, 81]. In evaluating the efficiency, one needs to consider two aspects: the amount of DNA and the number of cells. For example, in the experiment performed by Magana-Ortiz et al. [79], in general, plasmid DNA used in protoplast transformation and electroporation is about 1–10 μg [16–19, 23, 36, 52, 64]. It is expensive and inconvenient to produce such huge amount of plasmid in the laboratory for the SWMT method. Furthermore, shockwave sources and instruments are expensive because they are primarily designed for medical purpose. This turns out to be a major obstacle to adopting this method in a microbiology laboratory with limited resources.
Prospects
Filamentous fungi are a category of organisms widely existing in nature. Their secondary metabolite products are used in all aspects of our daily life. Their important economic value is self-evident. However, although filamentous fungi can produce beneficial secondary metabolites, some filamentous fungi also produce secondary metabolites that are toxic to humans and animals, such as mycotoxins. Mycotoxins seriously threaten the health and safety of livestock, poultry, and even rodents and humans. Most mycotoxins have strong acute toxicity. Some types of mycotoxins are strong carcinogens and mutagens and can damage liver, kidney, and other organs [82]. With the development of the genetic transformation method for fungi, these toxic genes could be removed and fungi can be made much safer or even harmless to humans and animals.
Transformation technology is a premise for modifying the genome of filamentous fungi. At present, the PMT method, electroporation transformation method and AMT method are mainly utilized in the study of fungal transformation. Compared with electroporation transformation, the protocols of PMT or AMT have been well developed and routinely used. At present, no universal fungal transformation methods works for every fungal species and one must find a specific protocol for the species of interest. In mammalian cell research, some fast, effective and simple transformation methods such as viral vectors-mediated transformation, nanomaterial-mediated transformation or liposome-mediated transformation are well established. Unfortunately, these methods are not suitable for fungi. There has been no significant breakthrough in fungal transformation for almost over 20 years.
In recent years, genome editing technology has been rapidly developed. The existing technologies mainly include Zinc-finger nucleases (ZFNs) [83, 84] technology, transcription activator-like effector nucleases (TALENs) [85] and clustered regularly interspersed short palindromic repeats (CRISPR) technology [86]. ZFNs have been widely used, but may results in a high off-targeting rate and high level of cytotoxicity. TALENs is similar but superior to ZFNs, which is much more cumbersome and expensive [87]. The combination of fungal transformation technology and effective genome editing technology are expected to realize the modification of fungal genomes to improve the protein production capacity of industrial fungi, to reduce the level of mycotoxins of certain strains, and to develop new strains for protein expression. However, application of genome editing technology to filamentous fungi is currently at an early stage. Numerous issues await exploration.
Authors’ contributions
DL, YT, JL and WC wrote this paper. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This work was financially supported by National Natural Science Foundation of China (31301537, 31371287), The Critical Patented Project of The Science & Technology Bureau of Fujian Province, China (2013YZ0002-1, 2013YZ0002-2) and Science Development Foundation of Fuzhou University (2013-XY-17).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dandan Li, Email: 254304727@qq.com.
Yu Tang, Email: 449177238@qq.com.
Jun Lin, Phone: +008659122863805, Email: jun@fzu.edu.cn.
Weiwen Cai, Phone: +008659122863805, Email: caiww@fzu.edu.cn.
References
- 1.Huang Y. Transformation in filamentous fungi mediated by Agrobacterium tumefaciens. Biotechnol Bull. 2007;17:111–114. [Google Scholar]
- 2.Fernándezbodega MA, Mauriz E, Gómez A, Martín JF. Proteolytic activity, mycotoxins and andrastin A in Penicillium roqueforti strains isolated from Cabrales, Valdeón and Bejes-Tresviso local varieties of blue-veined cheeses. Int J Food Microbiol. 2009;136:18–25. doi: 10.1016/j.ijfoodmicro.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Michielse CB, Hooykaas PJJ, van den Hondel CA, Ram AFJ. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- 4.Martín JF. Fungal transformation: from protoplasts to targeted recombination systems. Berlin: Springer International Publishing; 2015. [Google Scholar]
- 5.van den Berg MA, Maruthachalam K. Genetic transformation systems in fungi. Switzerland: Springer; 2015. pp. 3–4. [Google Scholar]
- 6.Kubodera T, Yamashita N, Nishimura A. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci Biotechnol Biochem. 2000;64:1416–1421. doi: 10.1271/bbb.64.1416. [DOI] [PubMed] [Google Scholar]
- 7.Zhao D. Evolution and development of soy sauce production technology in China. Chin Brew. 2009;9:15–17. [Google Scholar]
- 8.Zhao LF, Ya-Jun XU. Research progress on the application of Aspergillus oryzae. Chin Brew. 2006;3:8–10. [Google Scholar]
- 9.Malavazi I, Lima JF, de Castro PA, Savoldi M, de Souza Goldman MH, Goldman GH. Genetic interactions of the Aspergillus nidulans atmAATM homolog with different components of the DNA damage response pathway. Genetics. 2008;178:675–691. doi: 10.1534/genetics.107.080879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamazaki H, Tanaka A, Kaneko J, Ohta A, Horiuchi H. Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet Biol. 2008;45:963–972. doi: 10.1016/j.fgb.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 11.David H, Ozcelik IS, Hofmann G, Nielsen J. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genom. 2008;9:163. doi: 10.1186/1471-2164-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes MJ, Murray SL, Khew GS, Davis MA. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics. 2008;178:1355–1369. doi: 10.1534/genetics.107.085795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo A. Monacolin K, a new hypocholesterolemic agent produced by a Monascus species. J Antibiot (Tokyo). 1979;32:852–854. doi: 10.7164/antibiotics.32.852. [DOI] [PubMed] [Google Scholar]
- 14.Martin JF, Nicolas G, Villanueva JR. Chemical changes in the cell walls of conidia of Penicillium notatum during germination. Can J Microbiol. 1973;19:789–796. doi: 10.1139/m73-128. [DOI] [PubMed] [Google Scholar]
- 15.Tan WH, Li YP, Xu Y. Factors affect the formation and regeneration of protoplasts of microorganism. Mod Food Sci Technol. 2006;22:263–265. [Google Scholar]
- 16.Penttila M, Nevalainen H, Ratto M, Salminen E, Knowles J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987;61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- 17.Gruber F, Visser J, Kubicek CP, de Graaff LH. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr Genet. 1990;18:71–76. doi: 10.1007/BF00321118. [DOI] [PubMed] [Google Scholar]
- 18.Mach RL, Schindler M, Kubicek CP. Transformation of Trichoderma-Reesei based on hygromycin-B resistance using homologous expression signals. Curr Genet. 1994;25:567–570. doi: 10.1007/BF00351679. [DOI] [PubMed] [Google Scholar]
- 19.Case ME, Schweizer M, Kushner SR, Giles NH. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc Natl Acad Sci USA. 1979;76:5259–5263. doi: 10.1073/pnas.76.10.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahm YT, Batt CA. Genetic Transformation of an argB Mutant of Aspergillus oryzae. Appl Environ Microbiol. 1988;54:1610–1611. doi: 10.1128/aem.54.6.1610-1611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yelton MM, Hamer JE, Timberlake WE. Transformation of Aspergillus nidulans by using a Trpc plasmid. Proc Natl Acad Sci USA. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campoy S, Perez F, Martin JF, Gutierrez S, Liras P. Stable transformants of the azaphilone pigment-producing Monascus purpureus obtained by protoplast transformation and Agrobacterium-mediated DNA transfer. Curr Genet. 2003;43:447–452. doi: 10.1007/s00294-003-0417-0. [DOI] [PubMed] [Google Scholar]
- 23.Meyer V, Mueller D, Strowig T, Stahl U. Comparison of different transformation methods for Aspergillus giganteus. Curr Genet. 2003;43:371–377. doi: 10.1007/s00294-003-0406-3. [DOI] [PubMed] [Google Scholar]
- 24.Hutchison HT, Hartwell LH. Macromolecule synthesis in yeast spheroplasts. J Bacteriol. 1967;94:1697–1705. doi: 10.1128/jb.94.5.1697-1705.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilburn J, Scazzocchio C, Taylor GG, Zabicky-Zissman JH, Lockington RA, Davies RW. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- 26.Gallmetzer M, Burgstaller W, Schinner F. An optimized method for the isolation of protoplasts from Penicillium simplicissimum to produce sealed plasma membrane vesicles. Mycologia. 1999;91:206–212. doi: 10.2307/3761209. [DOI] [Google Scholar]
- 27.Vollmer SJ, Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci USA. 1986;83:4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fincham JR. Transformation in fungi. Microbiol Rev. 1989;53:148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azizi M, Yakhchali B, Ghamarian A, Enayati S, Khodabandeh M, Khalaj V. Cloning and Expression of Gumboro VP2 Antigen in Aspergillus niger. Avicenna J Med Biotechnol. 2013;5:35. [PMC free article] [PubMed] [Google Scholar]
- 31.Musílková M, Fencl Z. Some factors affecting the formation of protoplasts in Aspergillus niger. Folia Microbiol. 1968;13:235–239. doi: 10.1007/BF02871040. [DOI] [PubMed] [Google Scholar]
- 32.De BC, Wiebenga A, Aguilar G, Wösten HA. An enzyme cocktail for efficient protoplast formation in Aspergillus niger. J Microbiol Method. 2008;76:305–306. doi: 10.1016/j.mimet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Kuwano T, Shirataki C, Itoh Y. Comparison between polyethylene glycol- and polyethylenimine-mediated transformation of Aspergillus nidulans. Curr Genet. 2008;54:95–103. doi: 10.1007/s00294-008-0204-z. [DOI] [PubMed] [Google Scholar]
- 34.Ballance DJ, Buxton FP, Turner G. Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene of Neurospora crassa. Biochem Biophys Res Commun. 1983;112:284–289. doi: 10.1016/0006-291X(83)91828-4. [DOI] [PubMed] [Google Scholar]
- 35.Archer DB. Chitin biosynthesis in protoplasts and subcellular fractions of Aspergillus fumigatus. Biochem J. 1977;164:653–658. doi: 10.1042/bj1640653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Sun Q, Li H. Advances in genetic modification technologies. Chin J Biotechnol. 2015;31:1162. [PubMed] [Google Scholar]
- 37.Lin J, Shi B, Shi Q, Yun-Xia H, Wang MZ. Rapid improvement in lipase production of Penicillium expansum by Genome shuffling. Chin J Biotechnol. 2007;23:672–676. doi: 10.1016/S1872-2075(07)60044-2. [DOI] [PubMed] [Google Scholar]
- 38.Kao KN, Michayluk MR. A method for high-frequency intergeneric fusion of plant protoplasts. Planta. 1974;115:355–367. doi: 10.1007/BF00388618. [DOI] [PubMed] [Google Scholar]
- 39.Fincham JR. Transformation in fungi. Microbiol Rev. 1991;55:148. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker DM, Lundblad V. Introduction of DNA into yeast cells. Curr Protoc Mol Biol. 2001;13(7):1. doi: 10.1002/0471142727.mb1307s27. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Díez B. Strategies for the transformation of filamentous fungi. J App Microbiol. 2002;92:189–195. doi: 10.1046/j.1365-2672.2002.01516.x. [DOI] [PubMed] [Google Scholar]
- 42.Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, et al. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 2007;9:9–20. doi: 10.1111/j.1462-5822.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- 43.de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 44.Gouka RJ, Gerk C, Hooykaas PJ, Bundock P, Musters W, Verrips CT, et al. Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nat Biotechnol. 1999;17:598–601. doi: 10.1038/9915. [DOI] [PubMed] [Google Scholar]
- 45.Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat Protoc. 2008;3:1671–1678. doi: 10.1038/nprot.2008.154. [DOI] [PubMed] [Google Scholar]
- 46.Sugui JA, Chang YC, Kwon-Chung KJ. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl Environ Microbiol. 2005;71:1798–1802. doi: 10.1128/AEM.71.4.1798-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong YH, Wang XL, Wang TH, Jiang Q. Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Fungal Genet Biol. 2007;73:1348–1354. doi: 10.1007/s00253-006-0603-3. [DOI] [PubMed] [Google Scholar]
- 48.Nyilasi I, Acs K, Papp T, Nagy E, Vágvölgyi C. Agrobacterium tumefaciens-mediated transformation of Mucor circinelloides. Folia Microbiol (Praha). 2005;50:415–420. doi: 10.1007/BF02931423. [DOI] [PubMed] [Google Scholar]
- 49.Michielse CB, Salim K, Ragas P, Ram AF, Kudla B, Jarry B, et al. Development of a system for integrative and stable transformation of the zygomycete Rhizopus oryzae by Agrobacterium-mediated DNA transfer. Mol Genet Genom. 2004;271:499–510. doi: 10.1007/s00438-004-1003-y. [DOI] [PubMed] [Google Scholar]
- 50.Hazell BW, Te’o VS, Bradner JR, Bergquist PL, Nevalainen KM. Rapid transformation of high cellulase-producing mutant strains of Trichoderma reesei by microprojectile bombardment. Lett Appl Microbiol. 2000;30:282–286. doi: 10.1046/j.1472-765x.2000.00715.x. [DOI] [PubMed] [Google Scholar]
- 51.Klein RM, Wolf ED, Wu R, Sanford JC. High-velocity microprojectiles for delivering nucleic acids into living cells. 1987. Biotechnology. 1987;24:384–386. [PubMed] [Google Scholar]
- 52.Ozeki K, Kyoya F, Hizume K, Kanda A, Hamachi M, Nunokawa Y. Transformation of intact Aspergillus niger by electroporation. Biosci Biotechnol Biochem. 1994;58:2224–2227. doi: 10.1271/bbb.58.2224. [DOI] [PubMed] [Google Scholar]
- 53.Rai GA. Transformation of enzyme-treated Rhizopus Oryzae germlings by square-wave electroporation. In: Fungal Genetics Conference Proceedings; 2003.
- 54.Neumann E. Guide to electroporation and electrofusion. San Diego: Academic Press; 1992. [Google Scholar]
- 55.Wu S, Letchworth GJ. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques. 2004;36:152–154. doi: 10.2144/04361DD02. [DOI] [PubMed] [Google Scholar]
- 56.Richey MG, Marek ET, Schardl CL, Smith DA. Transformation of filamentous fungi with plasmid DNA by electroporation. Phytopathology. 1989;79:844–847. doi: 10.1094/Phyto-79-844. [DOI] [Google Scholar]
- 57.Goldman GH, Geremia R, Montagu MV, Herrera-Estrella A. Molecular genetics of the biocontrol agents Trichoderma spp. developments in agricultural & managed forest ecology. Amsterdam: Elsevier; 1991. pp. 175–180. [Google Scholar]
- 58.Weaver JC. Electroporation of biological membranes from multicellular to nano scales. IEEE Trans Dielectr Electr Insul. 2003;10:754–768. doi: 10.1109/TDEI.2003.1237325. [DOI] [Google Scholar]
- 59.Calvin NM, Hanawalt PC. High-efficiency transformation of bacterial cells by electroporation. J Bacteriol. 1988;170:2796–2801. doi: 10.1128/jb.170.6.2796-2801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohyama K, Gamborg OL, Miller RA. Uptake of exogenous DNA by plant protoplasts. Can J Bot. 2011;50:2077–2080. doi: 10.1139/b72-267. [DOI] [Google Scholar]
- 61.Zerbib D, Amalric F, Teissie J. Electric field mediated transformation: isolation and characterization of a TK+ subclone. Biochem Biophys Res Commun. 1985;129:611–618. doi: 10.1016/0006-291X(85)91935-7. [DOI] [PubMed] [Google Scholar]
- 62.Chakraborty BN, Kapoor M. Transformation of filamentous fungi by electroporation. Nucleic Acids Res. 1990;18:6737. doi: 10.1093/nar/18.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lakrod K, Chaisrisook C, Skinner DZ. Expression of pigmentation genes following electroporation of albino Monascus purpureus. J Ind Microbiol Biotechnol. 2003;30:369–374. doi: 10.1007/s10295-003-0058-9. [DOI] [PubMed] [Google Scholar]
- 64.Chakraborty BN, Patterson NA, Kapoor M. An electroporation-based system for high-efficiency transformation of germinated conidia of filamentous fungi. Can J Microbiol. 1991;37:858–863. doi: 10.1139/m91-147. [DOI] [PubMed] [Google Scholar]
- 65.Weaver JC. Electroporation of cells and tissues. IEEE Trans Plasma Sci. 2000;28:24–33. doi: 10.1109/27.842820. [DOI] [Google Scholar]
- 66.Chu G, Hayakawa H, Berg P. Electroportion for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987;15:1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potter H. Electroporation in biology: methods, applications, and instrumentation. Anal Biochem. 1988;174:361–373. doi: 10.1016/0003-2697(88)90035-8. [DOI] [PubMed] [Google Scholar]
- 68.Liang H, Purucker WJ, Stenger DA, Kubiniec RT, Hui SW. Uptake of fluorescence-labeled dextrans by 10T 1/2 fibroblasts following permeation by rectangular and exponential-decay electric field pulses. Biotechniques. 1988;6:550–552. [PubMed] [Google Scholar]
- 69.Sanford JC, Smith FD, Russell JA. Optimizing the biolistic process for different biological applications. Methods Enzymol. 1993;217:483–509. doi: 10.1016/0076-6879(93)17086-K. [DOI] [PubMed] [Google Scholar]
- 70.Gouka RJ, Punt PJ, vanden Hondel CA. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl Microbiol Biotechnol. 1997;47:1–11. doi: 10.1007/s002530050880. [DOI] [PubMed] [Google Scholar]
- 71.Barcellos FG, Fungaro MH, Furlaneto MC, Lejeune B, Pizziranikleiner AA, de Azevedo JL. Genetic analysis of Aspergillus nidulans unstable transformants obtained by the biolistic process. Can J Microbiol. 1998;44:1137–1141. doi: 10.1139/w98-213. [DOI] [PubMed] [Google Scholar]
- 72.Herzog RW, Daniell H, Singh NK, Lemke PA. A comparative study on the transformation of Aspergillus nidulans by microprojectile bombardment of conidia and a more conventional procedure using protoplasts treated with polyethyleneglycol. Appl Microbiol Biotechnol. 1996;45:333–337. doi: 10.1007/s002530050692. [DOI] [Google Scholar]
- 73.Ogden JA, Toth-Kischkat A, Schultheiss R. Principles of shock wave therapy. Clin Orthop Relat Res. 2001;387:8–17. doi: 10.1097/00003086-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 74.Haupt G. Use of extracorporeal shock waves in the treatment of pseudarthrosis, tendinopathy and other orthopedic diseases. J Urol. 1997;158:4–11. doi: 10.1097/00005392-199707000-00003. [DOI] [PubMed] [Google Scholar]
- 75.Loske AM. Shock wave physics for urologists. México: Universidad Nacional Autónoma de México; 2007. [Google Scholar]
- 76.Jagadeesh G, Nataraja KN, Udayakumar M. Shock waves can enhance bacterial transformation with plasmid DNA. Curr Sci India. 2004;87:734–735. [Google Scholar]
- 77.Loske AM, Campos-Guillen J, Fernandez F, Castano-Tostado E. Enhanced shock wave-assisted transformation of Escherichia coli. Ultrasound Med Biol. 2011;37:502–510. doi: 10.1016/j.ultrasmedbio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 78.Divya PG, Anish RV, Jagadeesh G, Chakravortty D. Bacterial transformation using micro-shock waves. Anal Biochem. 2011;419:292–301. doi: 10.1016/j.ab.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 79.Magana-Ortiz D, Coconi-Linares N, Ortiz-Vazquez E, Fernandez F, Loske AM, Gomez-Lim MA. A novel and highly efficient method for genetic transformation of fungi employing shock waves. Fungal Genet Biol. 2013;56:9–16. doi: 10.1016/j.fgb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 80.Campos-Guillén J, Fernández F, Pastrana X, Loske AM. Relationship between plasmid size and shock wave-mediated bacterial transformation. Ultrasound Med Biol. 2012;38:1078–1084. doi: 10.1016/j.ultrasmedbio.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 81.Lorito M, Hayes CK, Di Pietro A, Harman GE. Biolistic transformation of Trichoderma harzianum and Gliocladium virens using plasmid and genomic DNA. Curr Genet. 1993;24:349–356. doi: 10.1007/BF00336788. [DOI] [PubMed] [Google Scholar]
- 82.Zhang ZP, Nan GU, Zhao GH. Research progress on biological detoxification of mycotoxins. Cereals Oils. 2012;1:43–45. [Google Scholar]
- 83.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 85.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 86.Zhang M, Jiang S, Zheng J, Zheng Z, Li X, Pan L, et al. Construction of an integration vector carrying hygromycin B resistance gene and its genetic transformation in Rhizopus oryzae. Chin J Biotechnol. 2015;31:1203. [PubMed] [Google Scholar]
- 87.Yu T, Li L, Lin J. Advances in transcription activator-like effectors–a review. Chin J Biotechnol. 2015;31:1024. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


