ABSTRACT
Early stage assays that evaluate monoclonal antibody drug-like properties serve as valuable tools for selection of lead candidates. One liability for clinical development, off-target reactivity, is often assessed by binding to a mixture or panel of noncognate proteins. While robust, these mixes are often ill-defined, and can suffer from issues such as lot-to-lot variability. In this study, we discovered in immunoprecipitation experiments that certain chaperones are present in one of these mixtures;we then explored the use of recombinant chaperone proteins as well-characterized agents to predict antibody nonspecificity. Antibody binding to the heat shock proteins HSP70, HSP90, or trigger factor all served as predictors of cross-interaction propensity, with HSP90 providing the greatest ability to predict antibody clearance rates in mouse. Individual chaperone binding correlates surprisingly closely with binding to complex cell extracts, with the exception of a few “false negatives” (assuming a complex cell extract as the “true” value). As defined reagents, these chaperone reagents present advantages for high throughput assays of nonspecificity.
KEYWORDS: cross-interaction, developability, monoclonal antibody, nonspecificity, polyreactivity
The success of therapeutic monoclonal antibody (mAb) development depends both on functional target binding as well as desirable drug-like characteristics. Early stage biophysical assays, including self-association and nonspecificity evaluation, are effective tools that can save development time and costs and ensure only robust molecules are advanced into clinical stages. Early evaluation of nonspecificity, or cross-reactivity, is often assayed by binding of candidate clones to a panel of noncognate antigens,1–3 or a mixture of proteins.4–6 We have previously reported one of these assays, incubation of antibodies with a polyspecificity reagent (PSR) containing a membrane preparation of proteins,5 and have shown its ability to predict systemic clearance rates of such antibodies in mice.6 While this assay is robust, the composition of the reagent is ill-defined, leading to lot-to-lot variability in assay signal magnitude. For this reason, a single protein reagent would be an attractive replacement, in addition to potentially providing insights into the mechanistic sources of nonspecificity. In this report, we explore the use of chaperone proteins as potential single protein replacements for the PSR assay, finding heat shock protein 90 (HSP90) as a particularly good reagent with good correlation to PSR over three antibody data sets.
As a starting point to identify potential single protein candidates, we performed the immunoprecipitation of a soluble cytosolic protein (SCP) preparation from human embryonic kidney (HEK) cells using previously isolated polyreactive single-chain variable fragments (scFv) candidates expressed as scFvs on the surface of yeast. This cytosolic preparation performs nearly identically to the membrane preparation (Fig. S1). Analysis of the resulting pool via mass spectrometry revealed multiple common high abundance proteins, including the chaperone proteins HSP70, HSP 90-β, and the 60 kDa HSP from mitochondria (complete list in Supplemental Data). While other candidate antigens were present, we hypothesized that heat shock proteins might be uniquely appropriate to use as a nonspecificity reagent due to their naturally promiscuous capacity to bind and stabilize folding or misfolded proteins.7–11 To encompass a wide variety of chaperone functions, we selected one chaperone that aids in stabilization of nascent polypeptide chains (Trigger Factor, TF), one non-ribosomal early stage chaperone (HSP70), and one chaperone that aids in later stage folding (HSP90). We selected the human variant in each case except the E. coli protein TF, as the nascent chain is stabilized by a complex of proteins in eukaryotic organisms. We additionally omitted the HSP60 class of chaperones due to their natural formation into large complexes,11 and testing of HSP40 or HSP70/HSP40 complexes revealed promiscuous binding to all antibodies tested (data not shown).
We first assessed binding of each chaperone against a panel of IgGs isolated in a selection campaign against hen egg lysozyme (complete sequences in Supplementary Data). This panel displayed a wide range of scores on the solubilized membrane protein (SMP) assay, which correlated well to the individual chaperone binding assays (Fig. 1). Of the chaperones, HSP90 was most closely correlated (Pearson's r = 0.97), followed by TF (Pearson's r = 0.94) and HSP70 (Pearson's r = 0.92). These high correlations are striking for labeling with three such unrelated chaperones, indicative that some physicochemical property drives the observed binding rather than highly stereospecific complex formation in the classic sense of an antibody/antigen complex.
Figure 1.
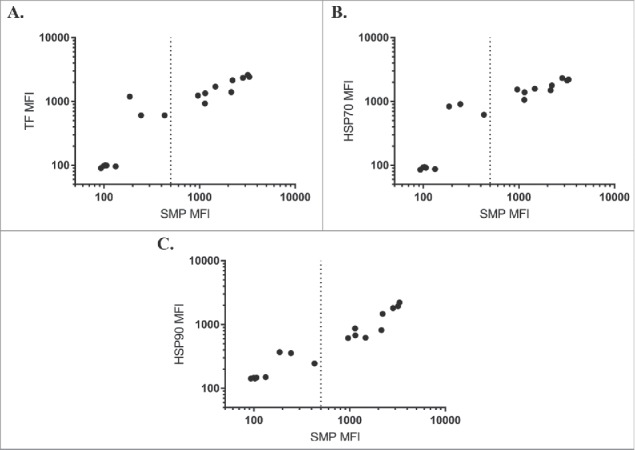
Chaperone binding correlates to SMP binding. SMP binding correlates to binding of TF (A, Pearson's r = 0.94), HSP70 (B, Pearson's r = 0.92), or HSP90 (C, Pearson's r = 0.97) on a panel of antibodies isolated from internal screening campaigns against hen egg lysozyme.
Based on the promising initial results, we next expanded the test to a panel previously used to demonstrate correlation between the PSR assay and clearance rates in mice.6 All three chaperone proteins correlated with mouse clearance rates, albeit slightly less than the multicomponent SMP reagent originally used (Fig. 2). Comparing rank-correlations between clearance and the reagents, HSP90 was most highly correlated (Spearman's ρ = 0.65), followed by HSP70 (Spearman's ρ = 0.60), and finally TF (Spearman's ρ = 0.43). Using a receiver operator curve (ROC) analysis, HSP90 was the most predictive chaperone (ROC Area Under Curve = 0.79), while both HSP70 (ROC AUC = 0.75) and TF (AUC = 0.63) were highly specific but had a higher rate of false negatives and lower overall dynamic range (Fig. 3). Using a median fluorescence intensity (MFI) cutoff of approximately 200, HSP90 (78% specificity, 85% sensitivity) compared well to the SMP assay at a cutoff of 500 (86% specificity, 89% sensitivity), suggesting it could serve as a good single agent replacement. At this cutoff, the sum of the true positive and true negative detection rate was maximized (13 of 16 correctly identified) and the associated maximum likelihood for the odds ratio is 21 [Fisher's Exact Test, 95% confidence interval (1.6 to 266.1)]. The exclusion of 1 in this interval implies statistical significance.
Figure 2.
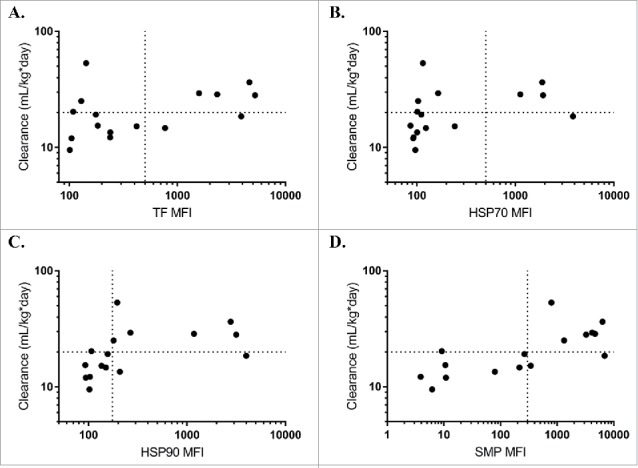
Chaperone proteins predict mouse clearance rates. Median fluorescence intensity (MFI) of trigger factor (A, Spearman's ρ = 0.43), HSP70 (B, Spearman's ρ = 0.60), HSP90 (C, Spearman's ρ = 0.65), or SMP (D, Spearman's ρ = 0.72) binding for each antibody correlates with mouse clearance rates from a previous study comparing SMP binding to mouse pharmacokinetics.6
Figure 3.
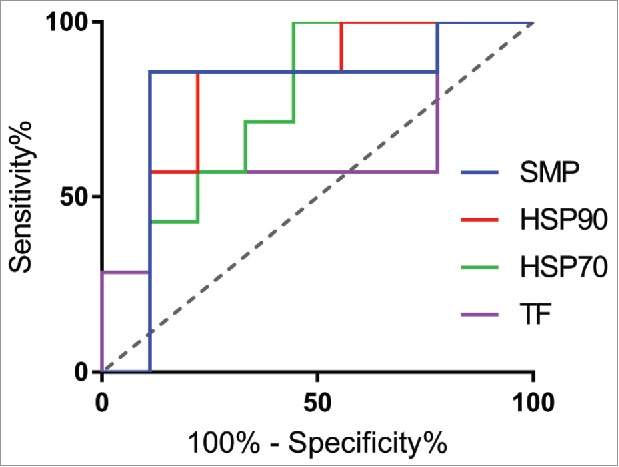
Predictive power of nonspecificity reagents via Receiver Operator Curve analysis. Comparing predictive power of the 3 reagents against SMP using a ROC analysis, HSP90 (red, AUC = 0.79) compared most favorably to SMP (blue, AUC = 0.79), followed by TF (pink, AUC = 0.64) and HSP70 (green, AUC = 0.75).
We next examined a panel of samples corresponding to mAbs in different stages of commercial development, applying the PSR assays to a set of antibodies drawn from a recent survey of biophysical properties of such samples.12 Performance on this larger data set was reflective of the previous panel (Fig. 4), with HSP90 (Pearson's r = 0.88) correlating the most with SMP, followed by TF (Pearson's r = 0.81) and HSP70 (Pearson's r = 0.72). None of the chaperone reagents matched the similarity between the SMP and SCP (Pearson's r = 0.94), with an apparent higher rate of “false negatives” for each of the chaperone reagents. Comparing the reactivity profiles of the chaperones, the majority of candidates display similar binding, including the apparent false negatives (patritumab, etrolizumab). There are a few cases where an antibody preferentially binds to HSP90 (lenzilumab) or TF (brentuximab, olaratumab), suggesting that the binding modalities of the chaperones are not wholly shared.
Figure 4.
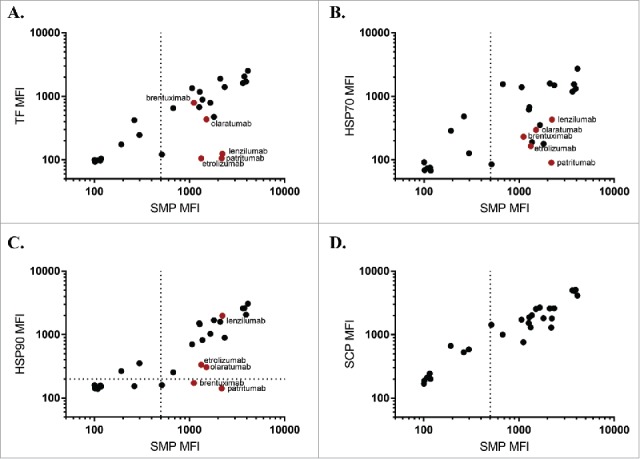
Chaperone binding correlates to SMP binding in approved and preclinical candidates. SMP binding was correlates to binding of TF (A, Pearson's r = 0.81), HSP70 (B, Pearson's r = 0.72), HSP90 (C, Pearson's r = 0.88), or SCP (D, Pearson's r = 0.94) on a subset of a panel of approved and preclinical antibodies. Red points are used to highlight particular clones with interesting binding properties.
To better understand the chaperone-negative clones in this panel, we examined clinical pharmacokinetic data where available. Patritumab, which targets the ErbB family receptor HER3, has a reported terminal half-life of approximately 9 days.13,14 This compares favorably to the SMP negative ErbB family inhibitors cetuximab (7 days15) and panitumumab (9 days16), but poorly to the HER2-targeted trastuzumab (28 days17). In all these cases, target-mediated drug disposition (TMDD) plays a major role in overall clearance and may cause results from different studies and antigens to be incomparable;however, patritumab additionally performs poorly on other nonspecificity metrics including the baculovirus ELISA assay.4,12 Two other chaperone-negative antibodies, etrolizumab (13–15 days18) and olaratumab (11 days19), displayed shorter half-lives than naturally circulating antibodies, but again TMDD likely plays a major role in pharmacokinetic parameters. Considering clinical pharmacokinetics as the reference, then the SMP assay might be construed as a false positive for etrolizumab and patritumab, while each of the three chaperones provides a true negative signal. Such ambiguity motivates application of multiple complementary assays in early-stage developability testing, since no individual metric exhibits perfect predictive capability of systemic clearance rates.
Individual chaperone proteins perform surprisingly well as predictors of nonspecificity, despite being single homogeneous entities. Both HSP90 and TF correlated well to SMP, and HSP70 binding was the weakest predictor. This was consistent across each of the three antibody panels tested in this study. To assess whether SMP or SCP binding could be attributed to the presence of chaperone proteins, we estimated the quantity of HSP90 in the PSR reagents, finding no HSP90 in SMP, and approximately 80 ng per 1μg SCP (Fig. S2A, B). This is a significant portion of the SCP, but cannot explain any observed SMP binding. This suggests that the chaperone-antibody interaction resembles that of the SMP-antibody binding interaction. As the panel of hen egg lysozyme and clinical antibodies (Figs. 1 and 3) all have human IgG1 constant sequence, chaperone binding presumably occurs through sites within the variable domain. We originally hypothesized that chaperones may perform better as a nonspecificity reagent than an average protein due to their ability to bind and stabilize partially misfolded structures and hydrophobic regions, and it appears that this is indeed the case, with a broader binding profile than other individual proteins.1,2 A cocktail of chaperones may perform even better than a single reagent while still allowing for a defined reagent that can be reproducibly manufactured or obtained from commercial sources.
Materials and methods
Yeast display immunoprecipitation and mass spectrometry
All immunoprecipitations were conducted using the yeast display immunoprecipitation technique as described previously.20,21 Briefly, 3 × 109 yeast expressing previously isolated nonspecific scFv clones were prepared by standard yeast display techniques,22,23 and first fixed for one hour using 3% w/v formaldehyde in phosphate-buffered saline (PBS) at room temperature. Next, clones were briefly washed once using PBS, followed by incubation in 1 mg/mL SCP from HEK 293 cells overnight at 4°C. Cells were pelleted, washed, and bound protein was eluted using 50 µL 0.2 glycine-HCl solution (pH 2.0) and precipitated using trichloroacetic acid. Proteins were subsequently resolubilized in 8 M urea, then reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin overnight. Samples were desalted with Millipore C18 Ziptips, then the proteolytic peptides were analyzed via liquid chromatography-mass spectrometry performed on a Thermo LTQ mass spectrometer with an Agilent 1100 series Nanoflow HPLC system. Proteins present in the sample were identified by database search using the Mascot database search software. The data was searched against all human protein sequences present in the Swissprot protein database.
Production of Chaperone proteins
All chaperones were expressed as fusion proteins containing an N-terminal 6x-His tag followed by small ubiquitin-like modifier (SUMO) and the chaperone using the pE-SUMO vector (LifeSensors, Malvern, PA). Proteins were expressed and purified as described previously using TALON metal affinity resin (Clonetech).24 Chaperone sequences were amplified from alternate expression plasmids using primers specific to the N and C-termini with proper 5′ extensions to allow for cloning into the pE-SUMO vector (Fwd: CAGGTCTCAAGGT, Rev: GTTCTAGATTATTA). HSP90 was a gift from William Sessa (Addgene plasmid #22487),25 pcDNA5/FRT/TO HSPA1A was a gift from Harm Kampinga (Addgene plasmid #19456),26 and pNIC28-Bsa4-TF was a gift from Brian Smith (Addgene plasmid #61689).
Antibody reagents
Anti-hen egg lysozyme antibodies were discovered and produced as human IgG1 using the Adimab yeast platform. All clinical stage antibodies except golimumab were expressed as human IgG1 isotype and produced as described previously.12 Golimumab was obtained from myoderm.com.
Chaperone and PSR binding assay
Chaperone and polyspecificity reagent binding were measured as described previously for the PSR assay.5 Briefly, SCP and SMP fractions were prepared from Chinese hamster ovary (CHO) or HEK293 cells. SCP, SMP, and recombinant chaperones were biotinylated with NHS-LC-Biotin reagent (Pierce, ThermoFisher Cat#21336). For binding assays, either 1 μM recombinant chaperone protein or the SCP or SMP poly-specificity reagent was incubated with IgG-presenting yeast. Unbound chaperone or polyspecificity reagent was removed by washing and samples were incubated with a secondary labeling mix (Extravidin-R-PE, goat F(ab')2 anti-human kappa-FITC (Southern Biotech catalog #2062–02), and propidium iodide). Following labeling, samples were analyzed for chaperone or polyspecificity reagent binding using a FACSCanto (BD Biosciences) with HTS sample injector. Binding was determined by analyzing the MFI in the R-PE channel from the flow cytometry data. All measurements were made in triplicate. Complete data, including MFI for all panels against all nonspecificity reagents, can be found in Supplemental data.
Quantitation of HSP90 in PSR reagents
To provide an estimate of the amount of HSP90 in the SCP and SMP reagents, a standard curve of purified HSP90 was prepared, along with dilutions of both SCP and SMP. Known masses of each protein or mixture were loaded onto a Novex NuPAGE 4–12% Bis-Tris protein gel (Life Technologies) for SDS-PAGE analysis. Gels were either developed by silver staining using the Pierce Silver Stain Kit or transferred to a nitrocellulose membrane using the iBlot 2 dry blotting system (Life Technologies) for western blot analysis. For western blots, membranes were first blocked using 5% nonfat milk for two hours, followed by addition of mouse anti-HSP90 (1:100, Novus NB100–1972) overnight. Membranes were washed and subsequently incubated in HRP goat anti-mouse (1:3000, Biolegend 405306) secondary antibody for one hour. Membranes were washed and bands were detected using SuperSignal West Pico PLUS Substrate (Thermo). To estimate quantity of protein, ImageJ was used to quantify intensity of bands and a standard curve was fit in Graphpad Prism. The curve was subsequently used to interpolate quantity of HSP90 in both SMP and SCP reagents.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the biopolymers and proteomics core. This work was supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. R.L.K. was supported by a Siebel scholarship and a graduate fellowship from the National Institute of General Medical Sciences Interdepartmental Biotechnology Training Program at the National Institutes of Health [T32 GM008334–25].
References
- 1.Frese K, Eisenmann M, Ostendorp R. An automated immunoassay for early specificity profiling of antibodies. MAbs. 2013;5(2):279–87. doi: 10.4161/mabs.23539. PMID:23412646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377(5):1518–28. doi: 10.1016/j.jmb.2008.01.093. PMID:18336836 [DOI] [PubMed] [Google Scholar]
- 3.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–7. doi: 10.1126/science.1086907. PMID:12920303 [DOI] [PubMed] [Google Scholar]
- 4.Hotzel I, Theil F–P, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al.. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4(6):753–60. doi: 10.4161/mabs.22189. PMID:23778268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu T–Y, Torrey J, Thomas J, Bobrowicz P, Vásquez M, et al.. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS–based, high–throughput selection and analytical tool. Protein Eng Des Sel. 2013;26(10):663–70. doi: 10.1093/protein/gzt047. PMID:24046438 [DOI] [PubMed] [Google Scholar]
- 6.Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vásquez M, Wittrup KD, et al.. High throughput cross–interaction measures for human IgG1 antibodies correlate with clearance rates in mice. MAbs. 2015;7(4):770–7. doi: 10.1080/19420862.2015.1043503. PMID:26047159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindquist S, Craig EA. The heat–shock proteins. Annu Rev Genet. 1988;22:631–77. doi: 10.1146/annurev.ge.22.120188.003215. PMID:2853609 [DOI] [PubMed] [Google Scholar]
- 8.Lindquist S. The heat–shock response. Annu Rev Biochem. 1986;55:1151–91. doi: 10.1146/annurev.bi.55.070186.005443. PMID:2427013 [DOI] [PubMed] [Google Scholar]
- 9.Mashaghi A, Bezrukavnikov S, Minde DP, Wentink AS, Kityk R, Zachmann–Brand B, Mayer MP, Kramer G, Bukau B, Tans SJ. Alternative modes of client binding enable functional plasticity of Hsp70. Nature. 2016;539(7629):448–451. doi: 10.1038/nature20137. PMID:27783598 [DOI] [PubMed] [Google Scholar]
- 10.Feder ME, Hofmann GE. Heat–shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82. doi: 10.1146/annurev.physiol.61.1.243. PMID:10099689 [DOI] [PubMed] [Google Scholar]
- 11.Kim YE, Hipp MS, Bracher A, Hayer–Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. doi: 10.1146/annurev-biochem-060208-092442. PMID:23746257 [DOI] [PubMed] [Google Scholar]
- 12.Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al.. Biophysical properties of the clinical–stage antibody landscape. Proc Natl Acad Sci. 2017;114(5):944–949. doi: 10.1073/pnas.1616408114. PMID:28096333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu T, Yonesaka K, Hayashi H, Iwasa T, Haratani K, Yamada H, Ohwada S, Kamiyama E, Nakagawa K. Phase 1 study of new formulation of patritumab (U3–1287) Process 2, a fully human anti–HER3 monoclonal antibody in combination with erlotinib in Japanese patients with advanced non–small cell lung cancer. Cancer Chemother Pharmacol. 2017;79(3):489–495. doi: 10.1007/s00280-016-3231-3. PMID:28144730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakui H, Yamamoto N, Nakamichi S, Tamura Y, Nokihara H, Yamada Y, Tamura T. Phase 1 and dose–finding study of patritumab (U3–1287), a human monoclonal antibody targeting HER3, in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73:511–6. doi: 10.1007/s00280-014-2375-2. PMID:24442032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fracasso PM, Burris H, Arquette MA, Govindan R, Gao F, Wright LP, Goodner SA, Greco FA, Jones SF, Willcut N, et al.. A phase 1 escalating single–dose and weekly fixed–dose study of cetuximab: Pharmacokinetic and pharmacodynamic rationale for dosing. Clin Cancer Res. 2007;13:986–93. doi: 10.1158/1078-0432.CCR-06-1542. PMID:17289894 [DOI] [PubMed] [Google Scholar]
- 16.Krens LL, Baas JM, De Jong FA, Guchelaar HJ, Gelderblom H. Pharmacokinetics of panitumumab in a patient with liver dysfunction: A case report. Cancer Chemother Pharmacol. 2014;73:429–33. doi: 10.1007/s00280-013-2353-0. PMID:24258455 [DOI] [PubMed] [Google Scholar]
- 17.Bruno R, Washington CB, Lu JF, Lieberman G, Banken L, Klein P. Population pharmacokinetics of trastuzumab in patients with HER2+ metastatic breast cancer. Cancer Chemother Pharmacol. 2005;56:361–9. doi: 10.1007/s00280-005-1026-z. PMID:15868146 [DOI] [PubMed] [Google Scholar]
- 18.Vermeire S, O'Byrne S, Keir M, Williams M, Lu TT, Mansfield JC, Lamb CA, Feagan BG, Panes J, Salas A, et al.. Etrolizumab as induction therapy for ulcerative colitis: a randomised, controlled, phase 2 trial. Lancet. 2014;384:309–18. PMID:24814090 [DOI] [PubMed] [Google Scholar]
- 19.Lartruvo® [package insert] Eli Lilly and Company, Indianapolis, IN; 2016. https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/761038lbl.pdf
- 20.Wang XX, Cho YK, Shusta E V. Mining a yeast library for brain endothelial cell–binding antibodies. Nat Methods. 2007;4:143–5. doi: 10.1038/nmeth993. PMID:17206151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho Y, Chen I, Wei X, Li L, Shusta E. A yeast display immunoprecipitation method for efficient isolation and characterization of antigens. J Immunol Methods. 2009;341(1–2):117–26. doi: 10.1016/j.jim.2008.11.005. doi: 10.1016/j.jim.2008.11.005. PMID:19041873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006. 1(2):755–68. doi: 10.1038/nprot.2006.94. PMID:17406305 [DOI] [PubMed] [Google Scholar]
- 23.VanDeventer JA, Wittrup KD. Yeast surface display for antibody isolation: Library construction, library screening, and affinity maturation. Methods Mol Biol. 2014;1131:151–81. doi: 10.1007/978-1-62703-992-5_10. PMID:24515465 [DOI] [PubMed] [Google Scholar]
- 24.Traxlmayr MW, Kiefer JD, Srinivas RR, Lobner E, Tisdale AW, Mehta NK, Yang NJ, Tidor B, Wittrup KD. Strong enrichment of aromatic residues in binding sites from a charge–neutralized hyperthermostable Sso7D scaffold library. J Biol Chem. 2016;291:22496–508. doi: 10.1074/jbc.M116.741314. PMID:27582495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García–Cardeña G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392(6678):821–4. doi: 10.1038/33934. PMID:9580552 [DOI] [PubMed] [Google Scholar]
- 26.Hageman J, Kampinga HH. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones. 2009;14:1–21. doi: 10.1007/s12192-008-0060-2. PMID:18686016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


