Abstract
Structure-switching sensors utilize recognition elements that undergo a conformation change upon target binding that is converted into a quantitative signal. Electrochemical, aptamer-based sensors achieve detection of analytes through a conformation change in an electrode-bound, oligonucleotide aptamer by measuring changes in electron transfer efficiencies. The analytical performance of these sensors is related to the magnitude of the conformation change of the aptamer. The goal of the present work is to develop a general method to predictably tune the analytical performance (sensitivity and linear range) of electrochemical, aptamer-based sensors by utilizing a mixture of rationally-designed aptamer sequences that are specific for the same target but with different affinities on the same electrode surface. To demonstrate control over sensor performance, we developed heterogeneous sensors for two representative small molecule targets (adenosine triphosphate; tobramycin). We demonstrate that mixtures of modified sequences can be used to tune the affinity, dynamic range, and sensitivity of the resulting sensors predicted by a bi-Langmuir-type isotherm.
Graphical abstract
Introduction
The use of nucleic acid aptamers in the development of chemical and biochemical sensors is a rapidly expanding field. While the list of sensing methods coupled with aptamers is long, electrochemical 1–12 and optical methods13–15 dominate the literature. The specificity of aptamer-based sensors is afforded by the recognition abilities of the nucleic acid aptamer sequence to its binding target, which can range from ions, proteins, cells, and small molecules.3,4,6,8,16,17,18 The aptamer-target interaction typically involves a single binding site with a combination of shape complementarity, hydrogen bonding, hydrophobic-hydrophobic, stacking, and electrostatic interactions.19 The analytical figures of merit of the resulting sensors are a function of the signal transduction methodology, as well as the intrinsic binding abilities, or affinity, of the aptamer-target pair.8,20,21 When these aptamers are immobilized on a sensor surface (e.g., a sensing electrode), the resulting sensors exhibit single-site binding isotherms, similar to the Langmuir binding isotherm.5,11,22 Fitting data to this binding isotherm enables quantitative characterization and benchmarking of the analytical figures of merit, including the maximum signal, sensitivity, dynamic range, and observed binding affinity.
Electrochemical, aptamer-based (E-AB) sensors utilize surface immobilized aptamers to achieve sensitive, specific, and reusable analyte detection.1,3,4,8,10,17,23 The signaling abilities of an E-AB sensor is based on the target-induced conformation change of the electrode-bound aptamer.3,6,8,22,24,25 Target binding changes the conformation of the aptamer and alters the electron transfer efficiency between a covalently-altered redox reporter and the electrode surface.21 As mentioned above, the sensor response typically follows a Langmuir-like binding isotherm. Using this sensing method, E-AB sensors have been reported to achieve dynamic ranges encompassing one to three orders of magnitude.10,21,26
Various techniques exist to optimize the response and analytical figures of merit of E-AB sensors. Specifically, modifying the packing density of DNA or RNA on an electrode surface by changing the concentration of the nucleic acid probe used to fabricate sensors can affect the observed binding affinities, maximum signal change, and sensitivity.22,27 These parameters, however, have only modest effects. For example, White et al. utilized the cocaine aptamer at various packing densities and obtained binding affinities ranging from 327 μM to 101 μM.22 Other methods have been utilized to tune the sensing abilities of E-AB sensors. For example, optimizing the potential waveforms used to voltammetrically interrogate the sensor surface can control the magnitude and polarity of the signal change upon target addition, as well as the observed binding affinity.20,21,27,28 A more radical technique to improve sensors responses is to modify aptamer sequences to undergo larger conformation changes upon target binding, resulting in increased signaling with consequential changes in the observed affinity.9,21 In all of these examples, the sensing attributes are still limited by the employed aptamer sequence and its binding interactions. For the detection of complementary DNA with structure-switching sensors, Kang et al. introduced the concept of heterogeneous sensor surfaces with multiple DNA probes for the same complementary DNA strand, but with different binding affinities.24 They accomplished this by designing multiple stem-loop probes for the same complementary target with stems of various stabilities. They then combined these on the electrode surface at various ratios to control the dynamic range of the resulting sensors.24 This strategy of altering stem-loop DNA probes for tuned binding affinities is relatively straightforward and has been reported for optical sensors as well.29
Here, we present for the first time, heterogeneous surfaces with rationally designed aptamer sequences to control the dynamic range and sensitivity of resulting EAB sensors. We design two different representative E-AB sensors employing DNA-based ATP aptamers20 and RNA-based aminoglycoside aptamers7,21,30 to demonstrate that the technique is general. Using a combination of in-house designed mutant aptamer sequences with different binding abilities to the same targets we developed mixed ratio sensor surfaces that exhibit predicted analytical responses. Specifically, we demonstrate control over the dynamic range and sensitivity of the resulting sensors, as well as provide quantitative predictions of this performance. Finally, while we demonstrate this control using electrochemical-based detection, the aptamer design guidelines we present should be applicable to any type of structure-switching aptamer-based sensing strategy.
Materials and Methods
Materials
Sodium chloride, Trizma® base (2-amino-2-(hydroxymethyl)-1,3-propanediol – here called Tris), magnesium chloride, tris-2-carboxyethyl-phosphine (TCEP), 6-mercapto-1-hexanol (99%) (Sigma Aldrich) were used as received. Buffer solutions were prepared using ultrapure water (Mili-Q Ultrapure Water Purification, Milipore, Billerica, MA). Buffer used for RNA-based sensor fabrication was autoclaved prior to use. RNA and DNA sequences (Tables 1 and 2) were synthesized and purified using dual-HPLC (Biosearch Technologies, Inc. Novato, CA). All the probes were aliquoted at 0.2 mM in 0.01 M EDTA aqueous solution (autoclaved for the RNA sequences) at pH 8.0 (Sigma Aldrich) and stored at −20°C until use.
Table 1.
ATP Aptamer Sequences
| Sequence Name | Sequence |
|---|---|
| Full Length | 5′-HSC6H12-ACCTGGGGGAGTATTGCGGAGGAAGGTT-MB-3′ |
| Mut | 5′-HSC6H12-CTGGGGGAGTATTGCGGAGGAAA-MB-3′ |
| Parent | 5′-HSC6H12-ACCTGGGGGAGTATTGCGGAGGAAGGTTTTTTTCTTC-MB-3′ |
Underlined sequence was conserved in the mutant
Table 2.
Aminoglycoside Aptamer Sequences
| Sequence Name | Sequence |
|---|---|
| Parent | 5′-HSC6H12-GGGACUUGGUUUAGGUAAUGAGUCCC-MB-3′ |
| Mut | 5′-HSC6H12-CUUGGUUUAGGUAAUGAG-MB-3′ |
Fabrication of Electrochemical Aptamer-Based (E-AB) Sensors
All sensors were fabricated on 2 mm diameter polycrystalline gold electrodes (CH Instruments, Austin, TX). Electrode modification was performed as previously described.21 Briefly, the electrodes were hand polished in a circular fashion on microcloth (Buehler) in a 1 μm diamond suspension and then they were polished in an alumina oxide slurry (Buehler). The electrodes were then rinsed and sonicated for 5 min. Sonication was followed by electrochemical cleaning of the electrodes via a series of voltammeric scans in sodium hydroxide and sulfuric acid solutions as previously described.31 After cleaning the electrode surfaces, each electrode was incubated in a 200 nM probe solution for 1h diluted in 20 mM Tris buffer with 100 mM sodium chloride and 5 mM magnesium chloride at pH 7.4, which was autoclaved for the RNA probe solutions. For the mixed monolayer sensors the total concentration of aptamer was kept constant at a total oligonucleotide concentration of 200 nM to allow for constant aptamer packing density. Prior to probe modification, the aptamer was reacted with 4 μL of 10 mM TCEP for 1h to reduce the disulfide bond at the 5′ end of the aptamer sequence, resulting from probe synthesis. The sensors were then incubated in 3 mM 6-mercapto-1-hexanol solution diluted with Tris buffer for 1h.
Electrochemical Measurements
Electrochemical measurements were completed utilizing a CH Instruments 660D Electrochemical Workstation (CH Instruments, Austin, TX). The measurements were performed in a three-electrode cell with a Ag/AgCl (3 M NaCl) reference electrode and a platinum auxiliary electrode. The square wave voltammetry were as follows: pulse amplitude of 25 mV, frequency of 900 Hz for the RNA-based aminoglycoside sensors and 200 Hz for the DNA ATP-based sensors, and a step width of 1 mV.
Results and Discussion
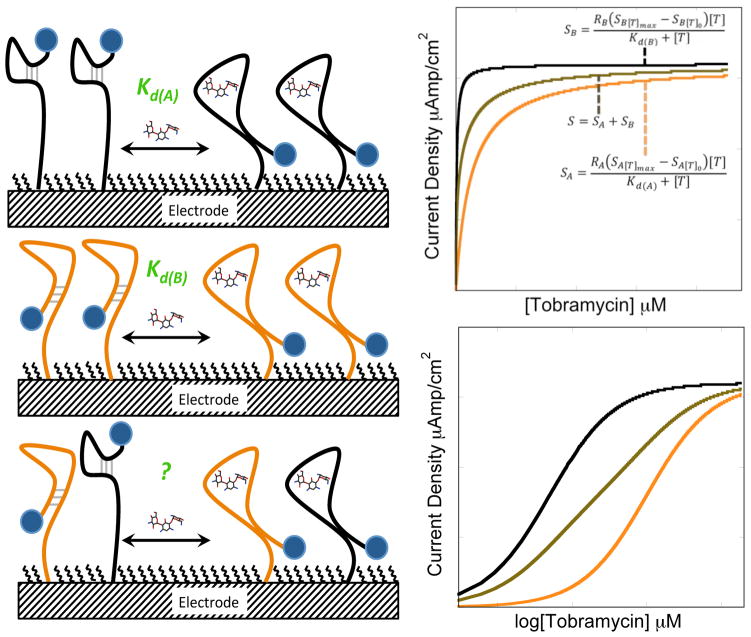
In this manuscript we demonstrate that heterogeneous sensor surfaces employing several aptamer mutants, designed to bind the same target with different affinities, can be used to tune the analytical response characteristics of the resulting sensor (Fig. 1). We have previously reported that rational modifications to an existing aptamer sequence to create larger target-induced conformation changes significantly enhances the observed binding affinity and sensitivity of the resulting sensor.21 In this report, we add a new layer of control by utilizing multiple aptamers with different affinities for the same target molecule on a sensor surface for better control over the analytical figures of merit. We achieve this control by varying the ratio of the different aptamers used during sensing monolayer formation. To demonstrate the generality of our approach we successfully develop tunable sensors for two representative targets employing aptamer sequences for tobramycin and adenosine triphosphate. In addition, we present a general quantitative expression to describe the analytical performance of the resulting aptamer-based sensors that takes into account the variation in current densities of both the target-bound and unbound state of each sensor, employing various aptamer architectures.
Figure 1.
Aptamers engineered to undergo different conformation changes (top left and middle) in the presence of the same target results in different analytical figures of merit for the resulting sensor, including sensitivity and dissociation constants (Kd(A) and Kd(B)). Sensors fabricated with mixtures of aptamers with different affinities for the same target (bottom left) produce sensors with tunable analytical performance based on the ratio of the two aptamers. For example, the predicted sensor response of a sensor employing a 50:50 mixture of high- and low-affinity aptamers falls right in between the responses of sensors fabricated with 100% high-affinity or low affinity aptamers.
Quantitative Binding Isotherms for Heterogeneous Sensor Surfaces
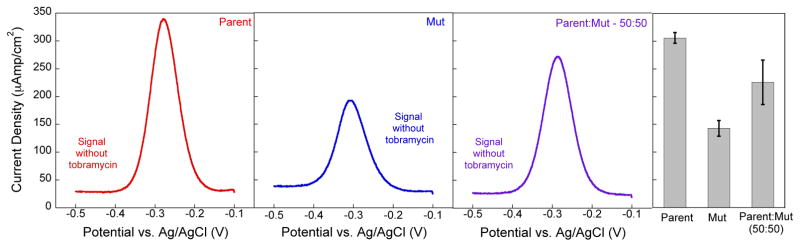
A surface with two distinct non-cooperative binding sites for the same target should exhibit a response that follows a combined binding isotherm or bi-Langmuir isotherm (Fig. 1).24,32 To quantitatively predict heterogeneous E-AB sensor response, we derived a bi-Langmuir isotherm written in terms of signal (S – absolute current density in units of A/cm2) to provide a general expression for sensor response (Eqs. 1–3). To properly describe the heterogeneous sensors, we take into account that the individual aptamer architectures will exhibit different minimum current densities (Smin) when no target is present and different maximum current densities (Smax) at saturating target concentrations. These values are a result of the apparent electron transfer rates given the different aptamer architectures. When mixed on the surface we observe a weighted algebraic average of the signals. Consistent with this, we find sensors employing either the Parent or Mut tobramycin aptamers exhibit Smin values for each architecture are 306 ± 10 μA/cm2 and 143 ± 14 μA/cm2 respectively. When mixed at different ratios (Ri – where R is the fraction of aptamer i) the current density represents the weighted sum of the contributions of both aptamers. For example, a 50:50 mixture of Parent and Mut give a Smin of 226 ± 40 μA/cm2 (see Fig. 2 – of note, no appreciable faradaic signal is observed with the aptamer is not present – Fig. S1). The quantitative binding isotherm for the mixed monolayer can thus be described by the following:
| (1) |
| (2) |
| (3) |
Figure 2.
The absolute signal (current density) measured at a heterogeneous surface in the absence of target molecule represents the weighted algebraic sum of the contributions of each aptamer. At constant surface coverage this results in minimum current densities (signal without target) of 306 ± 10 μA/cm2 and 143 ± 14 μA/cm2 for the homogenous Parent and Mut sensors respectively. When mixed at a 50:50 ratio, the current density with no target present is 226 ± 40 μA/cm2. The voltammograms show representative data and the bar graph and error bars represent the average of at least three independently fabricated sensors.
In eq. 1 the heterogeneous sensor signal (S) is the sum of the contributions of the signal from aptamers A and B employed on the sensor surface. By definition, RA + RB = 1. Eq. 2 and 3 represent the individual binding isotherms for sensors fabricated with aptamers A and B where [T] represents the concentration of free target in solution (M) and Kd is the dissociation constant of the sensor using the respective aptamer (Fig. 1). We proceeded to use the equations above for constructing calibration curves for mixed aptamer coated sensor surfaces. Of note, when plotted as a calibration curve, all data is normalized to the initial current density (signal without target present). This is done to better visualize the change in current density as a function of target concentration for the various mixtures as this change is what is important in quantifying target.
Heterogeneous Sensor Surfaces Allow Tunable Sensor Response
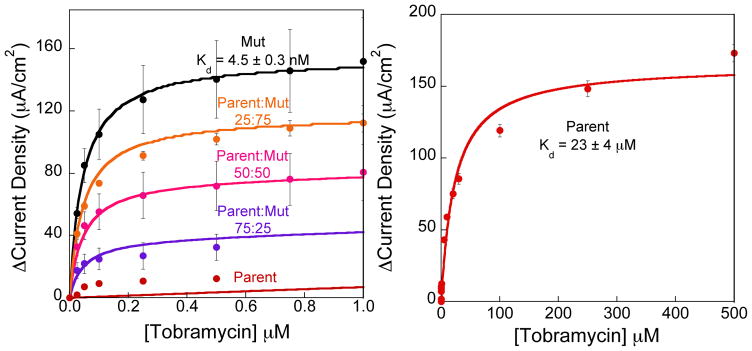
Heterogeneous, electrochemical, aptamer-based sensors with varying ratios of two different aptamers with different affinities for the same target quantitatively respond to target as predicted by our bi-Langmuir isotherm model. Specifically, we investigated sensors for the detection of two representative small molecule targets tobramycin and ATP. To demonstrate rational control over the signaling properties of the heterogeneous sensors, we tested multiple Parent:Mut ratios, including 75:25, 50:50, and 25:75. As a baseline measurement, sensors fabricated with a homogenous surface comprising only the Parent aminoglycoside aptamer exhibit a high dissociation constant (Kd = 23 ± 4 μM). Conversely, sensors comprising only the Mut aptamer exhibit a low dissociation constant (Kd = 0.045 ± 0.003 μM) (Fig. 3). All data points and error bars represent the mean and standard deviation of at least 3 independently fabricated sensors. As expected, skewing the ratio in favor of the Parent aptamer on the sensor surface results in a signal that more closely resembles that of sensors employing the Parent aptamer. For example, sensors fabricated with the Parent aptamer display a 12 ± 1 μA/cm2 signal change at 0.5 μM tobramycin, while, if 75% of the surface is modified with the Parent aptamer, we observe 33 ± 8 μA/cm2 current density at the same concentration. Thus, the heterogeneous sensor responses followed the predicted bi-Langmuir isotherm described above. All data is plotted as a change in current density.
Figure 3.
Heterogeneous sensor surfaces exhibit predictable bi-Langmuir binding properties. (Left) Specifically when mixed in varying ratios on the electrode surface, the sensors perform as a weighted sum of the performance of the individual aptamer architectures. The lines are calculated with our derived combined binding isotherm using the dissociation constant values (Kd) and current densities at saturating concentrations. (Right) Because sensors built with the Parent aptamer show minimal change at low concentrations, a titration curve using higher concentrations is used to calculate the dissociation constant. All data points and error bars represent the mean and standard deviation of at least 3 independently fabricated sensors.
We find that heterogeneous sensor responses fabricated with the Parent and Mut ATP aptamer-based sensors follow the same trend as the aminoglycoside sensors – and the bi-Langmuir isotherm (Fig. 4). For example, the Mut ATP aptamer sensors display a current density of 27 ± 2 μA/cm2 at 2.5 mM ATP, where the sensors fabricated with 75% Mut aptamer exhibit 21 ± 4 μA/cm2 (Fig. 4). Also, the sensors employing Parent aptamers shows a current density of 7.1 ± 0.3 μA/cm2 at 2.5 mM and when the sensors are modified with 75% Parent the signal is 12 ± 1 μA/cm2. When the sensors are employing 50% of each aptamer the sensor response is 17 ± 3 μA/cm2 at 2.5 mM ATP, which is in the middle of the sensor responses of Mut and Parent. The strategy of heterogeneous sensor surfaces is generally applicable to DNA and RNA aptamer-based sensing elements.
Figure 4.
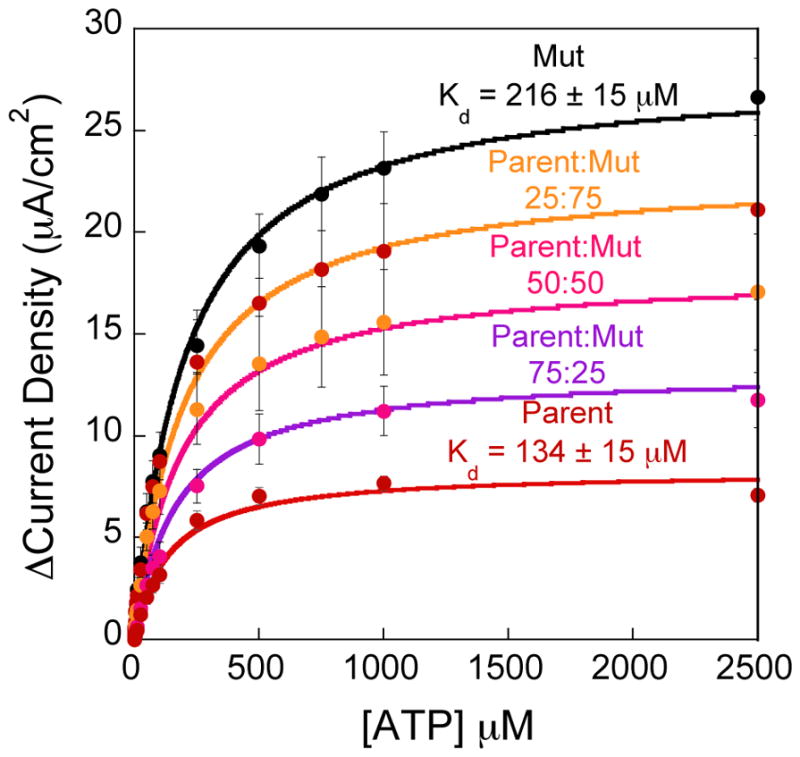
Heterogeneous sensors fabricated with two variations on the ATP aptamer exhibit predictable sensor performance based on the ratio of the two aptamers on the sensor surface. Much like the tobramycin sensors, the ATP sensors are fabricated with a high gain (Mut) and low gain (Parent) sequence. The sensor performance follows the bi-Langmuir isotherm described here as indicated by the solid lines. All data points and error bars represent the mean and standard deviation of at least 3 independently fabricated sensors.
Tuning Affinity, Sensitivity, and Dynamic Range of Electrochemical Sensors by Using Mixed Ratio of Aptamers
Sensors fabricated with mixed ATP DNA and aminoglycoside RNA aptamers have tunable affinities determined by the ratio of the parent and mutated aptamers employed on the sensor surface. To quantify heterogeneous sensor performance we use the concentration at which the signal is half of the maximum signal (Km - Table 3). As boundary values, the dissociation constants of the Parent and Mut ATP DNA-based sensors are 134 and 206 μM, respectively. Because these dissociations are close to each other, the values of Km for sensors comprising mixtures of 75:25, 50:50, and 25:75 (Parent:Mut) fall in between these boundaries and are 148, 162, and 171 μM respectively (Table 3). Conversely, the Km values for the heterogeneous aminoglycoside sensors are very similar to the dissociation constant of the sensors fabricated with 100% Mut aptamer (~0.045 μM). This is a result of the mismatch in the magnitude of signal change at low tobramycin concentrations between the Mut and Parent sequences.21 As such, the presence of Mut aptamer presumably dominates the signaling at low concentrations <1 μM. This observation indicates a quantitative limit to the tunability of sensor performance based on mutant sequences.
Table 3.
Km Values for Heterogeneous Sensor Surfaces
| DNA (Parent:Mut) | μM* | RNA (Parent:Mut) | μM† |
|---|---|---|---|
| 100:0 | 134 | 100:0 | 23 |
| 75:25 | 148 | 75:25 | 0.052 |
| 50:50 | 162 | 50:50 | 0.045 |
| 25:75 | 171 | 25:75 | 0.042 |
| 0:100 | 206 | 0:100 | 0.045 |
The estimated concentration of the signal at half maximum for sensor responses at assuming the maximum response occurs at 2500 μM ATP.
The estimated concentration of the signal at half maximum for sensor responses at assuming the maximum response occurs at 1 μM tobramycin, with the exception of 100% Parent aptamer (assumed maximum at 500 μM tobramycin).
In addition to tunable affinities, the heterogeneous aptamer-based sensors have tunable dynamic ranges and sensitivities are again dictated by the ratio of the two aptamers employed on the sensor surface. The dynamic range for each sensor is calculated using the limit of detection and the limit of linearity. While the limits of detection calculated for the various ATP sensors were all ~2 μM (Table 4), the limit of linearities followed the expected trend based on the ratio of the aptamers used. The same is true for the sensitivities – defined as the slope of the linear portion of the calibration curve. Specifically, sensors fabricated with the parent aptamer have the narrowest dynamic range, hitting an upper limit of 82 μM with a sensitivity of 0.0427 μAμM/cm2. Sensors fabricated with the Mut ATP aptamer display a limit of linearity of 123 μM and the best sensitivity of 0.0948 μAμM/cm2. Heterogeneous sensors fall in between these limits of linearity and sensitivities according to the ration of Parent:Mut (Table 4). Conversely, the RNA aminoglycoside Parent and Mut sensors have large disparity between their signaling, where their limits of detection varied by a factor of ~650 (260 nM and 0.395 nM respectively).20 The heterogeneously coated RNA sensors have limits of detection (LOD) that trended similarly with mixes of the two aptamers, but weighted towards the mutant sensor properties, as did the limits of linearity. The sensitivities also exhibited a similar trend with the mutant being the most sensitive (2381 μAμM/cm2) and the parent being the least sensitive (5 μAμM/cm2) and the mixes falling in between, which were weighted by the ratio of the aptamer sequences employed (Table 4).
Table 4.
The Dynamic Ranges and Sensitivities of the Heterogeneous Sensors
| DNA (Parent:Mut) | Dynamic Range* (μM) | Sensitivity† (μAμM/cm2) | RNA (Parent:Mut) | Dynamic Range* (μM) | Sensitivity† (μAμM/cm2) |
|---|---|---|---|---|---|
| 100:0 | 3.1–82 | 0.0427 | 100:0 | 0.26–13 | 5 |
| 75:25 | 2.1–98 | 0.0555 | 75:25 | 0.0040–0.0276 | 579 |
| 50:50 | 1.0–111 | 0.0681 | 50:50 | 0.0014–0.0272 | 1192 |
| 25:75 | 2.4–118 | 0.0814 | 25:75 | 0.0010–0.0271 | 1788 |
| 0:100 | 2.1–123 | 0.0948 | 0:100 | 0.000395–0.0271 | 2381 |
The lower limits here represent limit of detection calculations and the upper limits are within 10% deviation of the linear fit.
The sensitivities are the slopes of the linear fits of the dynamic ranges.
The ability to quantitatively tune the analytical figures of merit based on mixtures of aptamers with different affinities to the same target is achieved via quantitative control over the ratio of the aptamers attached to the electrode surface. The magnitude of tunability of each figure of merit depends on the signaling of the individual aptamer components. Specifically, the difference between the magnitude of the signal change observed (Smax-Smin) with each aptamer and the dissociation constants (Kd) can bias, or weight, the performance favoring one aptamer over the other as is seen in the RNA-based aminoglycoside sensors. Nonetheless, the ability to quantitatively control sensor performance is general.
Conclusion
In this manuscript we demonstrate, for the first time, that the use of a family of rationally designed aptamer sequences with different affinities for the same target can be combined on a single sensor surface in order to quantitatively and predictably control sensor performance. Specifically, we demonstrate the ability to control the sensitivity, dynamic range, and dissociation constant of the resulting sensors by varying the ratio of a high-affinity and low-affinity aptamer for the same target on a single sensor surface. To demonstrate the generality of our approach we successfully developed tunable sensors using several DNA aptamers for ATP detection and RNA aptamers for tobramycin detection. In addition we present a quantitative binding isotherm that takes into account the absolute signaling of each aptamer sequence in both the target-free and target-bound state. We find that this model accurately predicts the experimental observations of heterogeneously coated sensors. A caveat to this method is the algebraic weighting of sensor signaling in favor of one mutant over the other. The magnitude of signal change (Smax-Smin) and the difference in the dissociation constants limit the tunability of the sensor analytical figures of merit. Nonetheless, this variability is quantitatively predicted using the bi-Langmuir isotherm presented above.
The parameters we outline here to tune the sensitivity, dynamic range, and dissociation constants for electrochemical aptamer-based sensors should be applicable to optimize the performance of any E-AB sensor using relatively simple sequence modifications. Furthermore, while we demonstrate the feasibility of this approach towards the development of electrochemical-based sensors, the approach is broadly applicable to structure-switching, aptamer-based sensors, regardless of the signal transduction mechanism.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by UMBC Startup and the National Institute of Mental Health of the National Institutes of Health under award number R21MH101692. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Footnotes
Supporting Information Available
Background voltammograms using sensors fabricated without aptamers to show the electrochemical response in the absence and presence of tobramycin can be found in Supporting Information. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. J Am Chem Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 2.Cheng AKH, Sen D, Yu HZ. Design and Testing of Aptamer-Based Electrochemical Biosensors for Proteins and Small Molecules. Bioelectrochemistry. 2009;77:1–12. doi: 10.1016/j.bioelechem.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Ferapontova EE, Olsen EM, Gothelf KV. An RNA Aptamer-Based Electrochemical Biosensor for Detection of Theophylline in Serum. J Am Chem Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 4.Lai RY, Plaxco KW, Heeger AJ. Aptamer-Based Electrochemical Detection of Picomolar Platelet-Derived Growth Factor Directly in Blood Serum. Anal Chem. 2007;79:229–233. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Tuleouva N, Ramanculov E, Revzin A. Aptamer-Based Electrochemical Biosensor for Interferon Gamma Detection. Anal Chem. 2010;82:8131–8136. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radi AE, O’Sullivan CK. Aptamer Conformational Switch as Sensitive Electrochemical Biosensor for Potassium Ion Recognition. Chem Commun. 2006;32:3432–3434. doi: 10.1039/b606804a. [DOI] [PubMed] [Google Scholar]
- 7.Rowe AA, Miller Ea, Plaxco KW. Reagentless Measurement of Aminoglycoside Antibiotics in Blood Serum via an Electrochemical, Ribonucleic Acid Aptamer-Based Biosensor. Anal Chem. 2010;82:7090–7095. doi: 10.1021/ac101491d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swensen JS, Xiao Y, Ferguson BS, Lubin Aa, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Continuous, Real-Time Monitoring of Cocaine in Undiluted Blood Serum via a Microfluidic, Electrochemical Aptamer-Based Sensor. J Am Chem Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White RJ, Plaxco KW. Engineering New Aptamer Geometries for Electrochemical Aptamer-Based Sensors. Proc Soc Photo Opt Instum Eng. 2009;7321:732105–732116. doi: 10.1117/12.820419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, Lubin Aa, Heeger AJ, Plaxco KW. Label-Free Electronic Detection of Thrombin in Blood Serum by Using an Aptamer-Based Sensor. Angew Chem Int Ed Engl. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Uzawa T, White RJ, DeMartini D, Plaxco KW. On the Signaling of Electrochemical Aptamer-Based Sensors: Collision- and Folding-Based Mechanisms. Electroanalysis. 2009;21:1267–1271. doi: 10.1002/elan.200804564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Morris MD, Macazo FC, Schoukroun-Barnes LR, White RJ. The Current and Future Role of Aptamers in Electroanalysis. J Electrochem Soc. 2014;161:H301–H313. [Google Scholar]
- 13.Stojanovic MN, de Prada P, Landry DW, De Prada P, Landry DW. Aptamer-Based Folding Fluorescent Sensor for Cocaine. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 14.Zhou C, Jiang Y, Hou S, Ma B, Fang X, Li M. Detection of Oncoprotein Platelet-Derived Growth Factor Using a Fluorescent Signaling Complex of an Aptamer and TOTO. Anal Bioanal Chem. 2006;384:1175–1180. doi: 10.1007/s00216-005-0276-2. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Bao L, Liu Z, Pang DW. Aptamer Biosensor Based on Fluorescence Resonance Energy Transfer from Upconverting Phosphors to Carbon Nanoparticles for Thrombin Detection in Human Plasma. Anal Chem. 2011;83:8130–8137. doi: 10.1021/ac201631b. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Kwa T, Revzin A. Simultaneous Detection of Cell-Secreted TNF-α and IFN-γ Using Micropatterned Aptamer-Modified Electrodes. Biomaterials. 2012;33:7347–7355. doi: 10.1016/j.biomaterials.2012.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferapontova EE, Gothelf KV. Effect of Serum on an RNA Aptamer-Based Electrochemical Sensor for Theophylline. Langmuir. 2009;25:4279–4283. doi: 10.1021/la804309j. [DOI] [PubMed] [Google Scholar]
- 18.Huizenga DE, Szostak JW. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 19.Hermann T, Patel DJ. Adaptive Recognition by Nucleic Acid Aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 20.White RJ, Rowe AA, Plaxco KW. Re-Engineering Aptamers to Support Reagentless, Self-Reporting Electrochemical Sensors. Analyst. 2010;135:589–594. doi: 10.1039/b921253a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoukroun-Barnes LR, Wagan S, White RJ. Enhancing the Analytical Performance of Electrochemical RNA Aptamer-Based Sensors for Sensitive Detection of Aminoglycoside Antibiotics. Anal Chem. 2014;86:1131–1137. doi: 10.1021/ac4029054. [DOI] [PubMed] [Google Scholar]
- 22.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Optimization of Electrochemical Aptamer-Based Sensors via Optimization of Probe Packing Density and Surface Chemistry. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferapontova EE, Gothelf KV. Optimization of the Electrochemical RNA-Aptamer Based Biosensor for Theophylline by Using a Methylene Blue Redox Label. Electroanalysis. 2009;21:1261–1266. [Google Scholar]
- 24.Kang D, Vallée-Bélisle A, Porchetta A, Plaxco KW, Ricci F. Re-Engineering Electrochemical Biosensors to Narrow or Extend Their Useful Dynamic Range. Angew Chem Int Ed Engl. 2012;51:6717–6721. doi: 10.1002/anie.201202204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang D, Zuo X, Yang R, Xia F, Plaxco KW, White RJ. Comparing the Properties of Electrochemical-Based DNA Sensors Employing Different Redox Tags. Anal Chem. 2009;81:9109–9113. doi: 10.1021/ac901811n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshizumi J, Kumamoto S, Nakamura M, Yamana K. Target-Induced Strand Release (TISR) from Aptamer-DNA Duplex: A General Strategy for Electronic Detection of Biomolecules Ranging from a Small Molecule to a Large Protein. Analyst. 2008;133:323–325. doi: 10.1039/b719089c. [DOI] [PubMed] [Google Scholar]
- 27.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Effect of Molecular Crowding on the Response of an Electrochemical DNA Sensor. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Z, Lai RY. Effect of Signaling Probe Conformation on Sensor Performance of a Displacement-Based Electrochemical DNA Sensor. Anal Chem. 2013;85:3340–3346. doi: 10.1021/ac3037987. [DOI] [PubMed] [Google Scholar]
- 29.Vallée-bélisle A, Ricci F, Plaxco KW. Engineering Biosensors with Extended, Narrowed, or Arbitrarily Edited Dynamic Range. J Am Chem Soc. 2012;20:2876–2879. doi: 10.1021/ja209850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Rando RR. Specific Binding of Aminoglycoside Antibiotics to RNA. Chem Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Y, Lai RY, Plaxco KW. Preparation of Electrode-Immobilized, Redox-Modified Oligonucleotides for Electrochemical DNA and Aptamer-Based Sensing. Nat Protoc. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 32.Umpleby RJ, Baxter SC, Rampey AM, Rushton GT, Chen Y, Shimizu KD. Characterization of the Heterogeneous Binding Site Affinity Distributions in Molecularly Imprinted Polymers. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;804:141–149. doi: 10.1016/j.jchromb.2004.01.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.