Abstract
The Health Insurance Portability and Accountability Act (HIPAA) Safe Harbor method requires removal of 18 types of protected health information (PHI) from clinical documents to be considered “de-identified” prior to use for research purposes. Human review of PHI elements from a large corpus of clinical documents can be tedious and error-prone. Indeed, multiple annotators may be required to consistently redact information that represents each PHI class. Automated de-identification has the potential to improve annotation quality and reduce annotation time. For instance, using machine-assisted annotation by combining de-identification system outputs used as pre-annotations and an interactive annotation interface to provide annotators with PHI annotations for “curation” rather than manual annotation from “scratch” on raw clinical documents. In order to assess whether machine-assisted annotation improves the reliability and accuracy of the reference standard quality and reduces annotation effort, we conducted an annotation experiment. In this annotation study, we assessed the generalizability of the VA Consortium for Healthcare Informatics Research (CHIR) annotation schema and guidelines applied to a corpus of publicly available clinical documents called MTSamples. Specifically, our goals were to (1) characterize a heterogeneous corpus of clinical documents manually annotated for risk-ranked PHI and other annotation types (clinical eponyms and person relations), (2) evaluate how well annotators apply the CHIR schema to the heterogeneous corpus, (3) compare whether machine-assisted annotation (experiment) improves annotation quality and reduces annotation time compared to manual annotation (control), and (4) assess the change in quality of reference standard coverage with each added annotator’s annotations.
Keywords: Natural language processing, Medical informatics, Confidentiality, Patient data privacy, De-identification, Anonymization, Electronic health records, Clinical corpora
1. Introduction
In most electronic medical record (EMR) systems, a great deal of relevant clinical information is stored in clinical documents. Clinical documents and other medical records data are rich in protected health information (PHI). Preserving a patient’s privacy and confidentiality of PHI is fundamental to the physician-patient relationship. In order to use patient medical records for purposes other than providing health care (e.g. clinical research), informed consent from the patient is required. Indeed, use of patient medical record data is subject to the ethical and legal considerations defined by the Health Insurance Portability and Accountability Act (HIPAA) codified as 45 CFR §160 and 164 and the Common Rule [1]. However, obtaining the informed consent of a large population of patients, especially for retrospective research is difficult, time-consuming, and costly. This requirement can be waived if clinical documents are de-identified (i.e., all information identifying the patient has been redacted). Although de-identification of clinical documents remains a significant challenge, fulfilling these ethical and legal requirements is often a necessary step prior to using them for clinical research. However, manually de-identifying clinical documents represents a considerable expense in terms of time and human workload.
Automated methods that apply natural language processing (NLP) techniques may reduce the time and effort required to manually de-identify clinical documents, especially for large-scale projects applied to tens of thousands of patient records in which manual redaction of PHI is impractical. An NLP de-identification system must accurately remove the 18 types of PHI identifiers specified under the HIPAA Safe Harbor method for clinical documents to be considered “de-identified”. NLP systems that de-identify clinical documents are readily available [2–17], but are often developed and evaluated using specific document types. The approaches used by these systems may not be generalizable to all document types due to document specific formatting, clinical sublanguages, and prevalence of PHI [2]. Indeed, there is always the possibility that even with “de-identified” documents a PHI identifier may slip by and not be removed by all review methods [18]. A combined approach may reduce the likelihood of missing PHI identifiers and achieve acceptable coverage for certain PHI types by combining the efforts of many human reviewers with the outputs of an NLP system used as pre-annotations [19–21]. By leveraging NLP system outputs, this approach could offer a lower cost solution by pre-annotating potential PHI identifiers that human annotators review i.e., modifying existing, adding missing, or deleting spurious machine annotations. However, with any human review task relying on understanding of guidelines and tools, the cost of manual effort is high and may produce marginal returns of improved coverage as additional reviewers are added. The number of judges required to achieve acceptable coverage may also correlate with the risk of re-identification for different PHI types.
In this study, we evaluate the effects of a combined machine pre-annotation plus interactive annotation interface used to de-identify clinical documents from a publicly accessible document corpus called MTSamples. This heterogeneous clinical document corpus was selected for this study because it is a publicly available data source that could be easily obtained without a rigorous institutional data release process and contains replaced PHI mentions in context (“Dr. Sample Doctor…”) that are useful for de-identification research. We first describe the MTSamples corpus. We then describe an annotation experiment including the annotation scheme used and training process. Finally, we further detail our annotation training, experiment, and evaluation approaches and assess the effects of combining machine pre-annotation plus an interactive annotation interface used to de-identify clinical documents.
2. Background
Creating a reference standard that adequately identifies all HIPAA PHI identifier types and provides accurate training and evaluation data is imperative for developing rule-based or machine-learning-based de-identification systems. A few NLP researchers have championed efforts to facilitate the creation of state-of-the-art de-identifications for clinical documents and evaluate such systems against a standard corpus [16]. In 2006, NLP researchers from the University of Albany and MIT CSAIL sponsored the 2006 i2b2 Challenge task for automatic de-identification of clinical documents. A corpus of 889 discharge summaries from Partners Healthcare was annotated in two phases. In phase 1, PHI of eight types – patient names, doctor names, hospital names, IDs, dates, locations, phone numbers, and ages – were pre-annotated using an automated de-identification system that applied machine learning approaches [17]. In phase 2, three annotators sequentially annotated each report using a serial review method and achieved consensus after each review round. The inter-annotator agreement (IAA) between annotators and the performance of the NLP de-identification system was not reported as part of the 2006 i2b2 Challenge [16].
In contrast to the 2006 i2b2 Challenge, the goal for our manual de-identification task was to estimate the effects of machine pre-annotations and an interactive annotation interface on human annotator performance and quality of generated data for a heterogeneous clinical document corpus. We compare and contrast between annotators and the generated reference standard using IAA and standard performance metrics (i.e. recall, precision, and F1-measure) to assess annotator task consistency and accuracy. The effects of pre-annotation on the quality of annotated data has been investigated in many studies that include annotation of medical literature [20], POS tagging [19], named entity recognition (NER) [22] and clinical named entities [23,24], as well as common PHI types [25]. Other studies have employed semi-automated annotation methods that produce machine-generated candidate spans presented in such a way that the human reviewer must either modify incorrect annotations, delete spurious annotations, or add missed annotations [26–28]. It was our goal to produce a corpus of clinical documents annotated for PHI that maximized annotation quality while minimizing annotation effort.
3. Methods
We begin by describing the annotated MTSamples corpus. Next, we describe an annotation experiment including the annotation schema and training process. We further detail our annotation training, experiment, and evaluation approaches.
3.1. MTSamples corpus
A document sample consisting of 2,330 unique clinical documents was obtained from a publicly available resource of clinical documents called MTSamples (Medical Transcription Samples at www.mtsamples.com). These clinical documents were originally created to train medical coders and transcriptionists. The sample corpus contains document samples from 40 different medical specialties – consults, discharge summaries, and specialized medical services – including some uncommon formats. Although the MTSamples corpus does include data representing most of the 18 types of PHI identifiers specified under the HIPAA regulation, names and dates that remain have been changed (or removed) to preserve confidentiality of the users providing the data.
3.2. Annotation schema
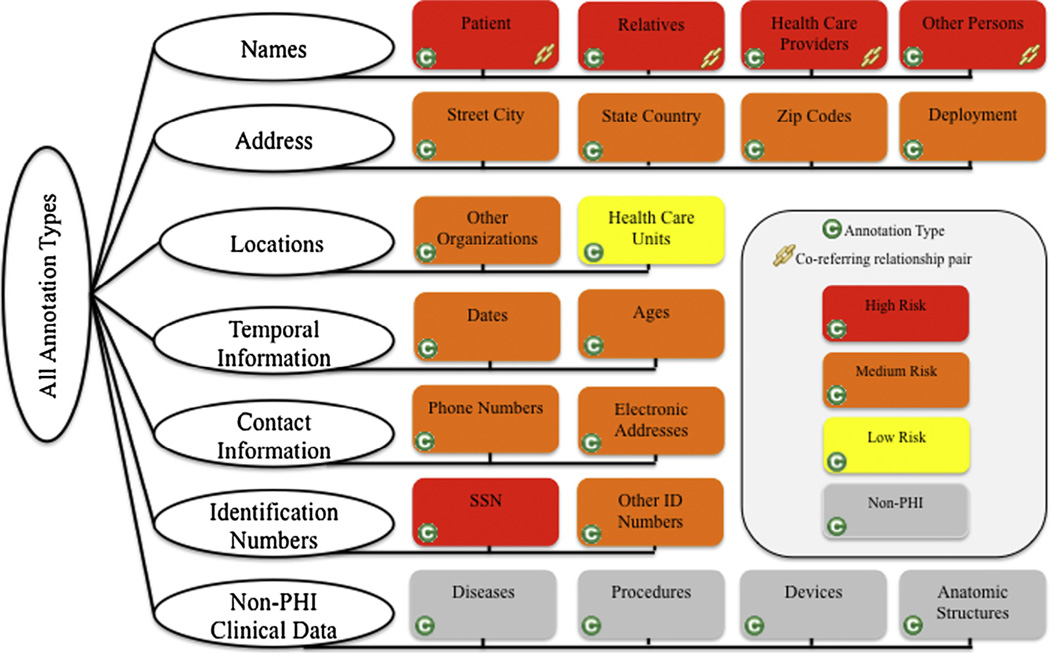
We build upon previous efforts [29] by expanding PHI types defined as part of the 2006 i2b2 challenge [16] and definitions for the Veteran Affair’s (VA) setting using an annotation schema and guidelines originally developed as part of the VA Consortium for Healthcare Informatics Research (CHIR) De-identification project [8,11]. These annotation guidelines go beyond the PHI types annotated from the 2006 i2b2 Challenge. We include annotation types representing clinical eponyms, organization names, military deployments, health care units, and co-referring-paired relationships between annotations for person names (Table 1). For example, “Patient Joe Smith… and Mr. Smith…”, “Joe Smith” and “Smith” might refer to the same person, in which case they would be linked in a paired relationship.
Table 1.
Annotation type definitions between i2b2 and extended CHIR Schema. Annotation types having co-referring relationships.
| i2b2 PHI types [16] |
Definitions | Extended CHIR Annotation Types [8,11] |
Definitions |
|---|---|---|---|
| Dates | All elements of a date except for the year | Dates | Date, including year and/or time, and specific time of day. Ex. “clinic on July 4, 2001@01:00”. This does not include mentions of durations. Ex. “2 h”, “5 days”, “day 1”, “x2”. |
| Patients | First and last names of patients, their health proxies, and family members | Patient Names | Patient’s first name, last name, middle name, and initials excluding salutations. Ex. “Mr. Smith complains of cough” |
| Relative Names | Proper name of relatives. Ex. “patient’s daughter Jennifer” | ||
| Other Person Names | Other persons mentioned or patient proxy. Ex. “lived in his friend Mike’s place” | ||
| Doctors | Medical doctors and other practitioners as well as transcriber’s name and initials | Health Care Provider Names | Health care worker’s first name, last name, middle name, and initials excluding salutations Ex. “JONES, JANE MD” |
| Ages | Ages above 89 | All mentions of age | Expanded to include all mentions of age. Ex. “52-year-old man” |
| IDs | Any combination of numbers, letters, and special characters identifying medical records, patients, doctors, or hospitals | Other ID Numbers | All combinations of numbers and letters that could represent a medical record number, lab test number, or other patient or provider identifier such as driver’s license number. Ex. “Driver’s license: S-012-34567” |
| Electronic Addresses | Electronic mail addresses and references to personal Websites, Facebook pages, Twitter. Ex. “CC: smarty@yahoo.com” | ||
| Social Security Numbers | Numbers and/or characters, that could represent a social security reference. Ex. “SSN is 000-00-0000” | ||
| Locations | Geographic locations such as cities, states, street names, zip codes, building names, and numbers | Street City | Street or city names excluding name as part of organization name. Ex. “lived on 5 Main Street” |
| State Country | State or country. Ex. “lived in Alaska” | ||
| Zip codes | All digits acting as a zipcode. Ex. “works in 08536 area” | ||
| Deployments | A specific geographic location, or mention of unit, battalion, regiment, brigade etc. Ex. “deployed with the 81st infantry unit” | ||
| Hospitals | Names of medical organizations and of nursing homes where patients are treated and may also reside including room numbers of patients, and buildings and floors related to doctors’ affiliations | Other Organization Names | Affiliation with companies such as employment that are not related to health care. Ex. “employed by WalMart” |
| Health Care Unit Names | Sub-specialty clinics, consults or referral to services, or recommendations from services where health care was or will be provided to a patient. Ex “Care provided at VA SALT LAKE CITY HCS” | ||
| Phone Numbers | Telephone, pager, and fax numbers | Phone Numbers | Phone/fax/pager numbers including phone number extensions. Ex. “Fax No: 381-7777” |
| Non-PHI | Not annotated as part of i2b2 | Clinical eponyms as part of medical procedure names | Medical procedures that contain proper names of persons, places, or locations. Ex. “DeLuca criteria was used” |
| Non-PHI | Not annotated as part of i2b2 | Clinical eponyms as part of medical device names | Medical devices that contain proper names of persons, places, or locations excluding brand names. Ex. “Foley catheter” |
| Non-PHI | Not annotated as part of i2b2 | Clinical eponyms as part of disease names | Diseases that contain proper names of persons, places, or locations. Ex. “history of Crohn’s disease” |
| Non-PHI | Not annotated as part of i2b2 | Clinical eponyms as part of anatomic structures | Anatomic structures contain proper names of persons, places, or locations. Ex. “Achilles tendon” |
Our motivation to include annotation of clinical eponyms was twofold. First, we wished to measure human performance identifying clinical information that machine systems may misclassify as PHI. Second, we wished to enrich available data sources for training classifiers and methods to identify these information types. Human reviewers more easily identify this type of information than machines because the reviewer can take into account contextual cues that may not be integrated with machine learned systems. We show a logical representation of these annotation types in Fig. 1.
Fig. 1.
Logical representation of the annotation schema. Annotation types color-coded by PHI privacy risk ranking: red (high risk), orange (medium risk), yellow (low risk), and gray (non-PHI). Co-referring paired relationships were created between annotations for person names. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For each annotation type, we developed detailed guidelines specifying inclusion and exclusion criteria regarding what to mark and not mark, which tokens to include, and what type of information should be marked. Annotations were defined using a contiguous span, beginning at the start of a phrase and ending at the completion of the phrase to capture instances rather than individual word tokens.
3.3. Experimental design
Manual annotation can be a slow, laborious process. We performed an experiment to determine the effects of combining machine pre-annotations with an interactive annotation interface. It was our goal to maximize annotation quality while minimizing manual annotation effort. We also wished to limit confusion or uncertainty related to annotator training on the guidelines, schema, and tool while maximizing the number of documents annotated from the original 2,330 MTSamples document corpus. This was achieved by separating annotation of the MTSamples corpus into annotator training and experiment. A stratified random sample was obtained for both training and experiment based on document type and the number of lines, words and tokens found in each clinical document. During the annotator training, seven reviewers annotated a random sample of 350 documents divided into 15 batches of 20–25 documents. Annotator training continued until a reviewer either exhausted their supply of training documents/ batches or achieved a pre-defined IAA performance threshold of 75% or greater when compared to other annotators on the training corpus. During the annotator experiment, the same seven reviewers annotated another random sample of 1,535 documents divided into 15 batches of 35 documents. Each annotator reviewed a total of 525 documents with 1,229 (80%) of these annotated by two or more reviewers. For both annotator training and experiment, annotators applied the same guidelines and annotation schema using an annotation tool called the extensible Human Oracle Suite of Tools (eHOST) [27]. Following the annotator training and experiment, a final reference standard was created after adjudicating discrepancies and consensus review of the resulting annotations from all reviewers.
During the annotator experiment, we employed two types of machine-assisted annotation: (1) machine pre-annotations (pre-annotations generated using an out-of-the-box de-identification system) and (2) interactive annotation (interactive annotation interface integrated with the annotation tool). We hypothesized that a combined approach using machine pre-annotations and an interactive annotation interface would reduce the time required for manual annotation of annotation types defined by our schema and found in clinical texts and would not reduce the quality of the data annotated. We used an “out-of-the-box” version of a de-identification system called BoB to generate pre-annotations, and a function integrated with the eHOST tool called “the Oracle” to provide the interactive annotation interface.
3.3.1. BoB: machine-generated pre-annotations from a de-identification system
One automated de-identification system designed for clinical documents is the Veterans Affairs “Best-of-Breed” (BoB) de-identification system [8]. BoB is a hybrid system that integrates known high-performing approaches specific to each particular PHI type from existing rule-based and machine learning systems. BoB is developed on a UIMA framework and processes documents using two main components: a high-sensitivity extractor and a false positive filterer. The high-sensitivity extractor applies a conditional random field classifier and rules to identify all potential PHI annotations maximizing recall. The false positive filterer leverages a support vector machine classifier to reclassify incorrectly tagged PHI annotations maximizing precision. For instance, the filterer may reclassify clinical eponyms such as those mentioned within Anatomical Structures e.g., “Circle of Willis” as non-PHI. We processed the MTSamples corpus using an out-of-the-box version of BoB originally trained on VA document types to generate pre-annotations provided to annotators during the experiment. Under these conditions, the annotation task is modified slightly and the human annotator accepts correct pre-annotations, modifies incorrect spans and deletes incorrect pre-annotations. We evaluate how helpful the system outputs could be without additional training on the MTSamples document corpus, a reality most researchers face when obtaining any open-source, de-identification software. We report recall, precision and F1-measure and provide baseline “out-of-the-box” performance of BoB without domain adaptation on the MTSamples corpus.
3.3.2. The eHOST Oracle: machine-assisted interactive annotation interface
One function integrated with the eHOST tool is a module called “the Oracle”. When enabled, the Oracle provides new annotation suggestions to the annotator based on an exact string match of the last human reviewer-produced annotation corresponding with that annotation type. For instance, if an annotator spans “Jane” as a Patient Name, the Oracle can search either the current document or across an entire batch of documents for other spans of “Jane” and then present these as potential candidate spans representing Patient Name. The annotator can choose to accept or reject these candidate PHI annotations. Annotators completed the annotator training using the eHOST Oracle module and were accustomed to its functionality before starting the annotator experiment discussed later. We report annotator utilization of the eHOST Oracle module in comparison to the total number of annotations generated during the experiment.
3.4. Annotation prevalence
We characterized the final reference standards generated from the annotator training and experiment. We report prevalence and performance metrics for PHI types specified by HIPAA and all annotation types defined in our annotation scheme. We further stratify these analyses according to the following re-identification risk ranking in the case where PHI is potentially missed.
High Risk: Social Security Numbers, Patient Names, Relative Names, Other Person Names, and Health Care Provider Names.
Medium Risk: Dates, Street City, State Country, Zip codes, Deployments, Other Organization Names, Other ID Numbers, Phone Numbers, Electronic Addresses and Ages.
Low Risk: Health Care Unit Names.
Non-PHI: Clinical eponyms (Anatomic Structures, Devices, Diseases, Procedures) and Person Relations.
3.5. Annotator performance metrics
We evaluated inter-annotator agreement (IAA) using F1-measure as a surrogate for Kappa since the number of document strings not annotated as a PHI annotation or true negatives (TN) are unknown [30]. We applied three types of annotator comparisons using standard performance metrics:
BoB-Reference Standard: compare BoB-generated pre-annotations and the reference standard generated during the annotator training using average exact performance metrics (recall, precision, F1-measure).
Annotator–Annotator: compare average paired exact and partial IAA between annotators.
Annotator-Reference Standard: compare average exact and partial performance metrics (i.e. recall, precision, F1-measure) between annotators and the annotator experiment reference standard.
F1-measure was calculated using the harmonic mean of recall (TP/(TP + FN)) and precision (TP/(TP + FP)), defined as 2 ((recall * − precision)/(recall + precision)). For example, during Annotator-Reference Standard comparisons we defined:
True Positive (TP): an annotation that exactly (exact) or partially (partial) overlapped a reference standard annotation for the same annotation type.
False Positive (FP): an annotator’s annotation that did not occur as a reference standard annotation.
False Negative (FN): a reference standard annotation that did not occur in the annotator’s annotations.
3.6. Annotation experiment
For the experiment, we assessed whether human annotators provided with machine pre-annotations and an interactive interface could generate similar quality data for span and classification of annotation type than without machine pre-annotations plus an interactive interface. We created two versions of the corpus – one with and one without BoB pre-annotated machine annotations and two versions of the eHOST tool – one with and one without the eHOST Oracle module. Annotators were randomly assigned 7 batches with pre-annotations plus the interactive annotation interface and 8 batches without (Fig. 2). For each annotation type and PHI risk of re-identification ranking, we evaluated whether human annotators receiving pre-annotations plus the interactive interface (experiment = BoB + eHOST Oracle) were able to generate data of similar quality with human annotators that did not receive the experiment (control = raw annotation). For this evaluation, we used the Wilcoxon Rank Sum test (Mann–Whitney U) [31]. The Wilcoxon Rank Sum test is a non-parametric test equivalent to a parametric 2-sample t-test for determining whether median F1-measures for the experiment are different than medians of the control (calculated for each annotation type and stratified by PHI risk ranking). For significance testing independent t-tests were used to determine if there were differences in averaged F1-measure between control and experiment for each annotator on each clinical document. For all statistical analyses, we used a null hypothesis stating there was no difference between the control and experiment using a significance level of 0.05.We calculated statistics (mean, median, and quartiles) and significance tests using SAS version 9.3.
Fig. 2.
Annotation experimental conditions.
3.7. Time comparison
Next, we hypothesized that human annotators receiving the experimental condition (BoB + eHOST Oracle) could produce annotations in less time (seconds) than human annotators under the control condition (annotation on raw clinical documents). We compared the average time per annotation for the experiment and control conditions. These calculations were made using the mean time between annotation spans using the timestamp for each annotation type classification within each document.
3.8. Coverage differences with added annotators
Finally, since our goal was to maximize annotation quality while minimizing annotation effort, we wanted to estimate how adding additional reviewers would affect recall, precision, and F1-measure. During the annotator training, we assessed the effects of annotation coverage as a function of adding additional reviewers. All seven reviewers annotated the same 350 documents. Discrepant annotations were adjudicated and a final consensus review was conducted to create a reference standard after the completion of both annotator training and experiment.
4. Results
We characterized prevalence of each PHI risk ranking and annotation type by training and experiment. For the annotator experiment, we report performance metrics for BoB compared with the reference standard generated for annotator training. We also report averaged IAA for annotators during the annotator experiment, and performance metrics for annotators compared with the reference standard generated at the completion of the annotator experiment. We compared distributions of the final annotations produced by experimental and control conditions and report the effects of the experiment applying the Wilcoxon Rank Sum test. Time-savings introduced by the experiment were also calculated. We also determined the coverage differences for each added annotator based on the annotation training reference standard. Finally, we report the distribution of each annotation type generated during the training and experiment using the complete annotated corpus.
4.1. Annotation prevalence
The majority of documents were annotated during the experiment. Discordant annotations generated from the training and experiment were adjudicated and subjected to a final consensus review. We characterized the prevalence of annotations by PHI risk category and annotation type found in the final reference standard in Table 2. PHI categorized as medium risk had the highest prevalence for both annotator training and experiment; PHI categorized as high risk had the lowest prevalence for training and experiment. Counts are expanded by annotation type for each collapsed risk ranking. For each PHI risk ranking, the most common PHI types represented Health Care Provider Names for high risk, Dates for medium risk, and Healthcare Unit Names for low risk. The most prevalent clinical eponyms were medical Devices. It is important to note that paired relations between person relations were quite common (5% within the entire annotated corpus); the most prevalent were Health Care Provider Names and Patient Names. Some PHI types, Social Security Numbers, Zip codes, and Electronic Addresses, did not occur in the MTSamples data.
Table 2.
Prevalence of annotation types and PHI risk category by annotator training and experiment for the final reference standard.
| Annotation prevalence training and experiment | ||||
|---|---|---|---|---|
| Annotator training | Annotator experiment | |||
| N | % | N | % | |
| Documents Reviewed | 350 | 18.6 | 1,535 | 81.43 |
| Annotation Type | ||||
| High Risk | 311 | 12.8 | 1,135 | 11.2 |
| Social Security Numbers | – | – | – | – |
| Patient Names | 86 | 3.5 | 248 | 2.5 |
| Health Care Provider Names | 204 | 8.4 | 860 | 8.5 |
| Relative Names | 17 | <1.0 | 12 | <1.0 |
| Other Person Names | 4 | <1.0 | 15 | <1.0 |
| Medium Risk | 1,220 | 50.2 | 4,357 | 43.2 |
| Dates | 630 | 26.0 | 2,305 | 22.8 |
| Street City | 24 | 1.0 | 119 | 1.2 |
| State Country | 33 | 1.4 | 95 | 1.0 |
| Zip codes | – | – | – | – |
| Phone Number | 2 | <1.0 | 6 | <1.0 |
| Deployments | 2 | <1.0 | 1 | <1.0 |
| Other Organization Names | 49 | 2.0 | 109 | 1.1 |
| Electronic Addresses | – | – | – | – |
| Other ID Numbers | 4 | <1.0 | 178 | 1.8 |
| Ages | 476 | 19.6 | 1,544 | 15.3 |
| Low Risk | 110 | 9.0 | 469 | 4.6 |
| Health Care Unit Names | 110 | 9.0 | 469 | 4.6 |
| Non-PHI | 661 | 27.1 | 2762 | 27.4 |
| Clinical Eponyms | 661 | 27.1 | 2762 | 27.4 |
| Anatomic Structures | 44 | 1.8 | 164 | 1.6 |
| Devices | 412 | 16.9 | 1,622 | 16.1 |
| Diseases | 48 | 2.0 | 263 | 2.6 |
| Procedures | 157 | 6.5 | 713 | 7.1 |
| Person Relations | 129 | 5.3 | 456 | 4.5 |
| Health Care Provider Names relations | 66 | 2.7 | 287 | 2.8 |
| Patient Names relations | 61 | 2.5 | 167 | 1.66 |
| Relative Names relations | 2 | <1.0 | 2 | <1.0 |
| Total Annotations | 2,431 | 19.4 | 10,091 | 80.6 |
| Overall | 12,522 | |||
Bold is provided for super categories of annotation classes only and overall numbers within these tables.
4.2. BoB-reference standard performance metrics
Baseline performance for out-of-the-box BoB pre-annotations on the MTSamples experiment corpus using standard performance metrics (recall, precision and F1-measure) was low when micro-averaged across all annotation types (0.20, 0.42, 0.27), moderate on medium risk (0.44 0.48, 0.46), but very low for high risk (0.17, 0.04, 0.07) and low risk (0.10, 0.76, 0.18) PHI types. This is in contrast to the published overall micro-averaged performance of BoB trained on VA clinical documents averaged across all PHI types (0.92, 0.86, 0.86) [8]. Highest performance on BoB pre-annotations on the MTSamples corpus was for Dates (0.78, 0.80, 0.79), followed by Other ID Numbers (0.34, 0.25, 0.29) and State Country (0.85, 0.18, 0.29). BoB’s lowest performance was on Other Person Names (1.0, 0.04, 0.09). There were a total of 8,181 BoB pre-annotations provided to annotators across the experiment document corpus and over half of these were false positive annotations 67% with only 16% (2,899) of these left unmodified prior to final adjudication and consensus review. Indeed, human annotators were more likely to delete BoB pre-annotations than modify or accept them. The majority of false positive annotations introduced by BoB pre-annotations were clinical eponyms that were incorrectly classified as Health Care Unit Names 21% (1,740) and Other Person Names 27% (2,237). The majority of false negative annotations corresponded with Ages 10% (850) and Dates 3.5% (285).
Annotators used the eHOST Oracle for only 640 (3.6%) annotations out of the total 17,643 annotations generated by all 7 annotators in the experiment. Out of these annotations the eHOST Oracle was used to mark 243 clinical eponyms, 145 Ages, 120 proper names of persons, and 104 Dates, which is not surprising since these types of annotations can easily be found using exact string matching and some are highly prevalent (Clinical Eponyms, Ages, Dates) in the MTSamples corpus. The eHOST Oracle produced only 16 false positive annotations (<1%), on those annotations where it was used.
4.3. Annotator–annotator agreement
For all annotation types (Table 3), agreement was moderate for exact IAA (control 0.75; experiment 0.66) and slightly higher for partial IAA (control 0.79; experiment 0.69). For each PHI risk ranking, both exact and partial IAA was higher for annotation on raw documents, ranging from moderate IAA for low risk PHI to high IAA for medium and high risk PHI. For Person Relations, the experiment condition produced higher IAA than the control. Inter-annotator agreement on raw document annotation ranged from low (Other ID Numbers, Deployments, and Other Person Names) to moderate (Phone Numbers, Other Organization Names, Health Care Unit Names, and all clinical eponyms) to high (all other types). Agreement on experiment documents ranged from low (Relative Names, Phone Numbers, Other Organization Names, and Other Person Names) to moderate (Street City, State Country, Other ID Numbers, Health Care Unit Names, and most clinical eponyms) to high (all other types). It is worth noting that both exact (control 0.60; experiment 0.91) and partial (control 0.62; experiment 0.95) IAA was higher for person relations generated under the experimental condition.
Table 3.
Inter-annotator agreement for the experiment.
| Inter-annotator agreement (IAA) (experiment) | ||||
|---|---|---|---|---|
| Exact (IAA) | Partial (IAA) | |||
| Control: raw annotation | Experiment: BoB + eHOST Oracle | Control: raw annotation | Experiment: BoB + eHOST Oracle | |
| Annotation Type | 0.75 | 0.66 | 0.79 | 0.69 |
| High Risk | 0.90 | 0.73 | 0.95 | 0.75 |
| Social Security Numbers | – | – | – | – |
| Patient Names | 0.87 | 0.40 | 0.91 | 0.80 |
| Health Care Provider Names | 0.90 | 0.91 | 0.95 | 0.92 |
| Relative Names | 0.8 | 0 | 0.8 | 0 |
| Other Person Names | 0.33 | 0.10 | 0.33 | 0.11 |
| Medium Risk | 0.81 | 0.76 | 0.85 | 0.60 |
| Dates | 0.84 | 0.75 | 0.86 | 0.76 |
| Street City | 0.82 | 0.44 | 0.84 | 0.44 |
| State Country | 0.78 | 0.35 | 0.79 | 0.46 |
| Zip codes | – | – | – | – |
| Phone Numbers | 0.50 | 0 | 0.50 | 0 |
| Deployments | 0.33 | – | 0.33 | – |
| Other Organization Names | 0.61 | 0.30 | 0.64 | 0.39 |
| Electronic Addresses | – | – | – | – |
| Other ID Numbers | 0.07 | 0.60 | 0.15 | 0.60 |
| Ages | 0.84 | 0.83 | 0.92 | 0.89 |
| Low Risk | 0.50 | 0.50 | 0.54 | 0.55 |
| Health Care Unit Names | 0.50 | 0.50 | 0.54 | 0.55 |
| Non-PHI | 0.64 | 0.63 | 0.67 | 0.65 |
| Clinical Eponyms | 0.64 | 0.63 | 0.67 | 0.65 |
| Anatomic Structures | 0.67 | 0.55 | 0.68 | 0.59 |
| Devices | 0.68 | 0.76 | 0.72 | 0.77 |
| Diseases | 0.62 | 0.67 | 0.65 | 0.67 |
| Procedures | 0.55 | 0.40 | 0.56 | 0.45 |
| Person Relations | 0.60 | 0.91 | 0.62 | 0.95 |
Bold is provided for super categories of annotation classes only and overall numbers within these tables.
4.4. Annotator-reference standard performance metrics
We report performance metrics (recall, precision, and F1-measure) using the reference standard generated during the annotation experiment (Table 4). We observed high exact recall (control 0.82, experiment 0.80), precision (control 0.91, experiment 0.81), and F1-measure (control 0.86, experiment 0.81) between annotators, with improved partial recall (control 0.84, experiment 0.84), precision (control 0.94, experiment 0.85), and F1-measure (control 0.89, experiment 0.84). For each PHI risk category, similar to Annotator–Annotator performance, both exact and partial metrics were higher when annotating on raw clinical documents. These differences were statistically significant for all annotation types when comparing averaged exact F1-measure for each annotator and annotated clinical document between control (0.84, ±0.211) and experiment (0.81, ±0.255), t(3.13) = 1363.5, p = 0.0018.
Table 4.
Performance metrics for control (raw annotation) and experimental (BoB + eHOST Oracle) conditions.
| Performance metrics annotator (experiment) | |||||
|---|---|---|---|---|---|
| Exact (recall, precision, F1-measure) | Partial (recall, precision, F1-measure) | ||||
| Control: raw annotation | Experiment: BoB + eHOST Oracle | Control: raw annotation | Experiment: BoB + eHOST Oracle | ||
| Annotation Type | 0.82, 0.91, 0.86 | 0.80, 0.81, 0.81 | 0.84, 0.94, 0.89 | 0.84, 0.85, 0.84 | |
| High Risk | 0.94, 0.96, 0.95 | 0.87, 0.74, 0.80 | 0.96, 0.98, 0.97 | 0.93, 0.78, 0.85 | |
| Social Security Numbers | – | – | – | – | |
| Patient Names | 0.95, 0.98, 0.96 | 0.78, 0.85, 0.81 | 0.96, 0.99, 0.98 | 0.91, 0.99, 0.95 | |
| Health Care Provider Names | 0.94, 0.96, 0.95 | 0.90, 0.96, 0.93 | 0.97, 0.98, 0.97 | 0.93, 0.99, 0.96 | |
| Relative Names | 0.82, 0.93, 0.88 | 0.50, 0.50, 0.50 | 0.88, 1.0, 0.94 | 1, 1, 1 | |
| Other Person Names | 0.50, 0.80, 0.62 | 0.69, 0.06, 0.11 | 0.50, 0.80, 0.62 | 0.81, 0.07, 0.13 | |
| Medium Risk | 0.85, 0.92, 0.88 | 0.82, 0.86, 0.84 | 0.88, 0.96, 0.92 | 0.86, 0.91, 0.88 | |
| Dates | 0.86, 0.95, 0.90 | 0.84, 0.93, 0.88 | 0.88, 0.97, 0.92 | 0.86, 0.94, 0.90 | |
| Street City | 0.88, 0.92, 0.90 | 0.92, 0.50, 0.65 | 0.89, 0.93, 0.91 | 0.93, 0.51, 0.66 | |
| State Country | 0.80, 0.94, 0.86 | 0.83, 0.50, 0.62 | 0.80, 0.95, 0.87 | 0.96, 0.57, 0.72 | |
| Zip codes | – | – | – | – | |
| Phone Numbers | 0.50, 0.71, 0.59 | 1, 1, 1 | 0.70, 1.0, 0.82 | 1, 1, 1 | |
| Deployments | 0.67, 0.67, 0.67 | – | 0.67, 0.67, 0.67 | – | |
| Other Organization Names | 0.69, 0.81, 0.74 | 0.61, 0.53, 0.57 | 0.72, 0.84, 0.77 | 0.67, 0.58, 0.62 | |
| Electronic Addresses | – | – | – | – | |
| Other ID Numbers | 0.37, 0.46, 0.41 | 0.36, 0.54, 0.44 | 0.54, 0.69, 0.61 | 0.53, 0.80, 0.64 | |
| Ages | 0.90,0.93,0.91 | 0.89, 0.93, 0.91 | 0.94, 0.98, 0.96 | 0.93, 0.98, 0.95 | |
| Low Risk | 0.69, 0.75, 0.72 | 0.76, 0.54, 0.63 | 0.73, 0.80, 0.76 | 0.83, 0.59, 0.69 | |
| Health Care Unit Names | 0.69, 0.75, 0.72 | 0.76, 0.54, 0.63 | 0.73, 0.80, 0.76 | 0.83, 0.59, 0.69 | |
| Non-PHI | 0.75, 0.89, 0.81 | 0.74, 0.84, 0.96 | 0.76, 0.91, 0.83 | 0.75, 0.86, 0.96 | |
| Clinical Eponyms | 0.75, 0.89, 0.81 | 0.74, 0.84, 0.96 | 0.76, 0.91, 0.83 | 0.75, 0.86, 0.96 | |
| Anatomic Structures | 0.77, 0.83, 0.80 | 0.64, 0.82, 0.72 | 0.78, 0.84, 0.81 | 0.65, 0.83, 0.73 | |
| Devices | 0.77, 0.91, 0.83 | 0.79, 0.88, 0.83 | 0.79, 0.94, 0.86 | 0.81, 0.91, 0.85 | |
| Diseases | 0.76, 0.87, 0.81 | 0.81, 0.79, 0.80 | 0.79, 0.91, 0.84 | 0.83, 0.81, 0.82 | |
| Procedures | 0.69, 0.85, 0.76 | 0.62, 0.73, 0.67 | 0.69, 0.85, 0.76 | 0.63, 0.75, 0.68 | |
| Person Relations | 0.75, 0.93, 0.83 | 0.74, 0.89, 0.81 | 0.76, 0.94, 0.84 | 0.74, 0.90, 0.81 | |
Bold is provided for super categories of annotation classes only and overall numbers within these tables.
4.5. Annotation experiment
We evaluated whether annotators provided with machine pre-annotations plus an interactive interface (experiment) produced annotations and annotation type classification of similar quality as compared to annotators reviewing raw clinical texts (control). In Table 5, we show summary statistics for the control and experimental conditions by annotation type stratified by PHI risk category computed from the Wilcoxon Rank Sum test. Significant differences were observed when comparing raw annotation (control) and annotation using BoB + eHOST Oracle (experiment). Annotation on raw clinical documents provided higher quality data for Patient Names, Other Person Names, Relative Names, Street City, State Country, Other Organization Names, and Health Care Unit Names.
Table 5.
Experimental effects estimated using the wilcoxon rank sum test.
| Wilcoxon rank sum test | |||||
|---|---|---|---|---|---|
| Control: raw annotation | Experiment: BoB + eHOST Oracle | Significance | |||
| Median F1-measure | N | Median F1-Measure | N | Pr>|Z| | |
| All Annotation Types | 0.91 | 1156 | 0.91 | 741 | 0.296 |
| High Risk | 1 | 365 | 1 | 274 | <0.0001a |
| Patient Names | 1 | 78 | 1 | 32 | 0.0389a |
| Health Care Provider Names | 1 | 338 | 1 | 201 | 0.278 |
| Relative Names | 1 | 8 | 0.5 | 2 | 0.553 |
| Other Person Names | 0.5 | 11 | 0 | 106 | <0.0001a |
| Medium Risk | 1 | 879 | 1 | 579 | 0.0748 |
| Street City | 1 | 68 | 0 | 72 | <0.0001a |
| State Country | 0.96 | 48 | 0 | 64 | 0.0009a |
| Zip codes | – | – | – | – | – |
| Deployments | 0.33 | 2 | – | – | – |
| Other Organization Names | 0.5 | 72 | 0 | 65 | 0.0319a |
| Dates | 1 | 533 | 1 | 342 | 0.195 |
| Ages | 1 | 764 | 1 | 493 | 0.992 |
| Phone Numbers | 0.58 | 4 | 1 | 2 | 0.140 |
| Electronic Addresses | – | – | – | – | – |
| Other ID Numbers | 0.20 | 47 | 0 | 37 | 0.553 |
| Low Risk | 0.667 | 277 | 0 | 221 | 0.0002a |
| Health Care Unit Names | 0.667 | 277 | 0 | 221 | 0.0002a |
| Non-PHI | 0.857 | 729 | 0.995 | 459 | 0.7103 |
| Clinical Eponyms | |||||
| Anatomic structures | 0.933 | 101 | 0.8 | 61 | 0.600 |
| Devices | 0.872 | 485 | 1 | 303 | 0.103 |
| Diseases | 0.919 | 116 | 1 | 66 | 0.784 |
| Procedures | 0.667 | 347 | 0.667 | 211 | 0.929 |
| Person Relations | 1 | 141 | 1 | 72 | 0.458 |
Bold is provided for super categories of annotation classes only and overall numbers within these tables.
Control condition generated significantly higher quality data than the experimental condition.
4.6. Time comparison
There was no statistically significant difference when comparing times for annotation of raw clinical documents compared with annotation of documents annotated under the experimental conditions across all annotation types. Observed mean time in seconds per annotation was 13.7 s for annotation on raw clinical documents and 13.6 s for documents annotated using BoB + eHOST Oracle. Although these differences were not significant across all annotation types, the mean time between annotations generated using only the eHOST Oracle was 5.24 s.
4.7. Coverage differences with added annotators
In Fig. 3, we show the change in performance metrics as logical combinations of reviewers are compared. Recall ranged from 0.66 (1 reviewer) plateauing at a high of 0.92 (7 reviewers). Alternatively, precision decreased from 0.82 (1 reviewer), to a low of 0.61 for the union of all seven judges. F1-measures ranged from 0.73 (1 reviewer), 0.79 (2), 0.78 (3), 0.77 (4), 0.75 (5), 0.74 (6), and 0.73 (7 reviewers). Document level F1-measure (not shown) by PHI risk ranking ranged from 0.20 to 1.00 (mean = 0.96, std = 0.12) for high risk, 0.11–1.00 (mean = 0.89, std = 0.17) for medium risk, and 0.07–1.0 (mean = 0.81, std = 0.22) for low risk.
Fig. 3.
PHI coverage differences as a function of annotator number.
5. Discussion
5.1. Annotation prevalence
For the annotator training and experiment the most prevalent PHI category included those PHI types categorized as medium risk and the least prevalent PHI types were those in the high risk. At the corpus level, these prevalence estimates are difficult to compare with other published studies [8,14,16,20,27] due to the large variety of report types used in our study and the differences in annotation schema between studies. Our average prevalence of 4.0 PHI annotated per document for the MTSamples corpus is lower than those reported using other clinical corpora such as 26 per document from the VA [8], 22 from the 2006 i2b2 De-identification challenge [16], 8.79 per document from the 2012 Deleger et al. study [14], 49 from the 2013 Hanauer et al. study [20], and 7.9 per document from a 2006 study by Dorr et al. [32]. This difference could be due to the varied PHI types and prevalence of document types annotated in these other studies. For instance, a general clinical document containing instructions for how a patient should continue to treat “Athlete’s foot” would not contain PHI.
We should note that our corpus does contain clinical information that can be mistaken for PHI. Clinical eponyms are one such example accounting for 27.3% of the total corpus, which is significantly higher than the 3.5% previously observed using the same annotation schema on VA clinical documents [33].
5.2. BoB-reference standard performance metrics
We observed low to moderate F1-measure for predicting low to high risk PHI mentions. This is not surprising since this performance is based on pre-annotations produced by BoB previously trained using VA clinical documents and not on MTSamples documents. Even though the performance of the baseline pre-annotation system was poor we would expect (particularly for medium and low risk PHI types), that a combined approach using machine-generated pre-annotations plus the interactive annotation interface would result in improvements in the quality of annotated data and result in gains in annotation efficiency. This expectation was not borne out for the majority of annotation types in our study. Furthermore, annotators used the eHOST Oracle for only a small proportion of their annotations because they found it easier to annotate on raw clinical texts without the interactive machine suggestions. It is not clear whether this preference was due to the high number of false positives introduced by BoB or related to usability issues with the eHOST Oracle. However, annotator usability ratings of the eHOST Oracle based on the system usability scale (SUS) [34] were slightly above average. Despite this preference the number of false positives introduced using the eHOST Oracle was very small compared with the number of false positives introduced by BoB. Although the Oracle was not specifically designed for relations it was used to annotate one or both entities in co-referring relation pairs for annotation types representing proper names of persons. This is interesting since identifying a co-referring pair first involves identifying the entities that should be linked.
5.3. Annotator–annotator agreement
When viewed in aggregate for each PHI risk category, raw annotation on clinical texts produced the highest inter-annotator agreement. The combination of BoB + eHOST Oracle introduced false positives producing less reliable annotation between annotators. However, these false positives were introduced in the majority of cases due to the low baseline performance of the BoB outputs used as pre-annotations and not via annotator interaction with the eHOST Oracle. Moreover, even though not statistically significant, use of the eHOST Oracle produced higher quality data when building relation pairs between person names. Person relations IAA was higher where BoB + eHOST was used due to the high IAA for Health Care Provider Names and Patient Names (particularly for partial IAA) and their high prevalence in the corpus. We are not surprised to observe less prevalent PHI types like Relative Names and Deployments had the lowest IAA. Introducing more training instances could boost IAA performance for these types.
5.4. Annotator performance: annotator-reference standard performance metrics
Standard performance metrics demonstrated similar results with the control condition producing higher quality data among all PHI risk categories as demonstrated in Table 4 and the Wilcoxon Rank Sum Test in Table 5.
5.5. Annotation experiment
There are several lessons we learned from integrating a combined approach using outputs from an untrained de-identification system along with an interactive interface. First, the experimental condition did not introduce significant gains in recall, precision, and F1-measure. This is surprising since particular annotation types including clinical non-PHI can easily and consistently be found using the eHOST Oracle since they follow standard naming conventions and were often flagged as false positive BoB pre-annotations (i.e. clinical eponyms and Other Organization Names). Annotation on raw clinical texts produced higher quality data across all annotation types when compared with the experiment. For some annotation types (i.e. Other Person Names, Health Care Unit Names), annotator agreement remained lower than expected throughout the experiment and never plateaued. In the best of all possible experiments annotators would train until their agreement meets or exceeded some pre-defined threshold. This brings us to several remaining questions reserved for future experimentation. First, we did not explore how applying a “tag a little, learn a little” approach could be implemented in a practical way [20]. Second, we did not explore “how high” system performance should be to optimize annotator performance e.g., would higher performing pre-annotation with precision and/or recall greater than 50% produce better results instead of the out-of-the-box application of BoB?
The methods used for this annotation task could be modified to fit annotation of other types of information commonly found in clinical texts including clinical entities. However, caution should be used when pre-annotation or machine-assisted methods are employed as a means to improve the quality of generated data or reduce the time required to generate annotated data. This is particularly true when an untrained system is used out-of-the-box to produce pre-annotations with no domain adaptation. On the one hand, providing pre-annotated information derived from system outputs may result in human annotators either trusting the pre-annotations too much in the case where system outputs are highly precise or missing incorrect annotations when system outputs produce results of high recall. This is a limitation in the way BoB outputs were used as pre-annotations in our study since they are derived using both rules and machine learning approaches. High performing machine learning based systems usually require training on similar documents to those being de-identified [20].
5.6. Time comparison
Across all annotation types, we observed no statistical differences for annotation times between the experiment and control conditions. Lack of time difference may be due to time added for deleting false positives that could equate to the same amount of time required to identify a PHI span in the same document that is not reviewed using BoB + eHOST. This result is contrary to a study by Fort and Saggot [19] that used machine pre-annotations for POS tagging in which significant reduction in time was observed for the experiment, as well as a more recent study by Lingren et al. [24] in which machine pre-annotation was employed to annotate clinical named entities resulting in significant reduction in annotation time and no effect on IAA or standard performance metrics. However, our experimental results are congruent with findings by Ogren and colleagues [23] that outputs generated from a third-party system used as pre-annotations decreased efficiency and produced little gain in data quality.
Although annotations using only the eHOST Oracle were generated faster than the control condition alone, the lack of time difference between experimental and control conditions may be a consequence of combining pre-annotations with the interactive annotation interface. Higher quality pre-annotations may introduce efficiencies compared with annotation on raw clinical texts. On the other hand lower quality pre-annotations certainly do not offer a net gain in efficiency or annotator performance due to the added task of modifying existing, adding missed, or deleting spurious annotations. It is likely that the ratio of correct to incorrect pre-annotations must be small in order for there to be any efficiency gains offered by the machine-assisted approach [35].
5.7. Coverage differences with added annotators
The number of annotators needed to achieve adequate recall and precision may be dependent on various factors that should be explored in future annotation studies. First, different clinical documents may require more reviewers as compared with fewer. Second, a privacy risk ranking of PHI types should be one consideration for these tasks. Third, there are policy implications for the redaction of PHI from clinical texts that extend beyond simply removing personally identifiable information. A reference standard generated by human reviewers is never perfect and the ability of humans to reliably annotate for PHI and generate an accurate reference standard is a difficult goal to achieve. Even though annotators trained on the de-identification task and tools until they achieved a pre-defined performance threshold in the training, IAA never plateaued across either annotator training or experiment for both control and experimental conditions for some annotation types in our study. This indicates human annotators were still “learning” to correctly identify and classify some annotation types through both the training and experiment. There are two tasks that must go on simultaneously in the reviewers mind, first the reviewer must read the clinical text and second, the reviewer must apply the guidelines and annotation schema. This observation speaks to the complexity of a manual de-identification task, the difficulty of providing enough examples of each annotation type, and the ability of human annotators to consistently apply an annotation schema written in the spirit of the HIPAA Safe Harbor method.
6. Conclusions
We have demonstrated the generalizability of a manual de-identification task on a publicly available, heterogeneous corpus of clinical documents, MTSamples, using an annotation schema and guidelines originally developed for a similar annotation effort on VHA clinical documents. Based on this schema and the resulting annotations, we determined most PHI annotations represent expressions of medium risk of re-identification Overall, we observed that PHI classes can be annotated with high average inter-annotator agreement. In this experiment, machine-assisted annotation did not improve annotation quality for most PHI classes and did not provide statistically significant time-savings compared to manual annotation of raw documents. However, we determined that two annotators perform PHI annotation with highest F1-measure and observed diminishing PHI coverage with each added annotator. This could be an important finding for institutions creating a de-identification service where humans would be hired to manually redact PHI from clinical texts. Finally, we have produced a de-identified clinical document corpus and a reference standard that can be used for future experimentation on NLP de-identification methods.
In the case of building a reference standard that will be used to train automated systems for de-identification, it is better to err on the side of high recall considering the implications and negative impacts of HIPAA violations on the institution providing the data. These issues should be considered in the context of patient privacy, potential information loss, and the workload associated with manual de-identification of clinical texts. Balancing the expectations of existing ethical and legal responsibility with practicality and the burdens of human review is paramount for any sound implementation of automated machine methods used for clinical text de-identification. This study contributes to the ongoing analysis of human review methods used for de-identification of clinical texts.
Acknowledgments
Development of the eHOST annotation tool was supported by the VA Consortium for Healthcare Informatics Research (CHIR), VA HSR HIR 08-374. Annotation of the MTSamples corpus was supported by NIH Grant U54 HL 108460 for integrating Data for Analysis, Anonymization and Sharing (iDASH), NIGMS 7R01GM090187. We also wish to acknowledge the efforts of all annotators involved in pilot testing annotation schema and annotating this document corpus. The fully de-identified MTSamples document corpus is available from iDASH via a data use agreement.
References
- 1.GPO USA. GPO; 2008. Oct 1, CFR Title 45 Subtitle A Part 46: Protection of Human Subjects [Internet] < http://www.access.gpo.gov/nara/cfr/waisidx_08/45cfr46_08.html>. [Google Scholar]
- 2.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aberdeen J, Bayer S, Yeniterzi R, Wellner B, Clark C, Hanauer D, et al. The MITRE identification scrubber toolkit: design, training, and assessment. Int J Med Inform. 2010;79(12):849–859. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc. 2008;15(5):601–610. doi: 10.1197/jamia.M2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckwith BA, Mahaadevan R, Balis UJ, Kuo F. Development and evaluation of an open source software tool for de-identification of pathology reports. BMC Med Inform Decis Mak. 2006;6:12. doi: 10.1186/1472-6947-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner J, Xiong L. HIDE: an integrated system for health information DE-identification. IEEE Symp Comput Med Syst Proc. 2008:254–259. [Google Scholar]
- 7.Neamatullah I, Douglass MM, Lehman LW, Reisner A, Villarroel M, Long WJ, et al. Automated de-identification of free-text medical records. BMC Med Inform Decision Making. 2008;8(1):32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrández O, South BR, Shen S, Friedlin FJ, Samore MH, Meystre SM. BoB, a best-of breed automated text de-identification system for VHA clinical documents. J Am Med Inform Assoc. 2013;20(1):77–83. doi: 10.1136/amiajnl-2012-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. Proc AMIA Annu Fall Symp. 1996:333–337. [PMC free article] [PubMed] [Google Scholar]
- 10.Taira RK, Bui AA, Kangarloo H. Identification of patient name references within medical documents using semantic selectional restrictions. AMIA Annu Symp Proc. 2002:757–761. [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrández O, South BR, Shen S, Friedlin FJ, Samore MH, Meystre SM. Evaluating current automatic de-identification methods with Veteran’s health administration clinical documents. BMC Med Res Methodol. 2012;12(1):109. doi: 10.1186/1471-2288-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman JJ. Concept-match medical data scrubbing. How pathology text can be used in research. Arch Pathol Lab Med. 2003;127(6):680–586. doi: 10.5858/2003-127-680-CMDS. [DOI] [PubMed] [Google Scholar]
- 13.Ruch P, Baud RH, Rassinoux AM, Bouillon P, Robert G. Medical document anonymization with a semantic lexicon. AMIA Ann Symp. 2000:729–733. [PMC free article] [PubMed] [Google Scholar]
- 14.Deleger L, Molnar K, Savova G, Xia F, Lingren T, Li Q, et al. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform Assoc. 2012 doi: 10.1136/amiajnl-2012-001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GPO USA. GPO; 2008. Oct 1, CFR 45 Subtitle A Part 164: Security and Privacy [Internet] < http://www.access.gpo.gov/nara/cfr/waisidx_08/45cfr164_08.html>. [Google Scholar]
- 16.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sibanda T, Uzuner Ö. Role of local context in de-identification of ungrammatical. Fragmented Text NAACL-HLT. 2006:65–73. [Google Scholar]
- 18.Carrel D, Malin B, Aberdeen J, Bayer S, Clark C, Wellner B, et al. Hiding in plain sight: use of realistic surrogates to reduce exposure of protected health information in clinical text. J Am Med Inform Assoc. 2013;20(2):342–348. doi: 10.1136/amiajnl-2012-001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fort K, Sagot B. Influence of pre-annotation on pos-tagged corpus development. Proceedings of the fourth linguistic annotation workshop, LAW IV. 2010:56–63. [Google Scholar]
- 20.Hanauer D, Aberdeen J, Bayer S, Wellner B, Clark C, Zheng K, et al. Bootstrapping a de-identification system for narrative patient records: cost-performance tradeoffs. Int J Med Inform. 2013;82:821–831. doi: 10.1016/j.ijmedinf.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proceedings of the AMIA symposium: American medical informatics association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 22.Ganchev K, Pereira F, Mandel M, Carrol S, White P. Proceedings of the linguistic annotation workshop. Prague, Czech Republic: Association for, Computational Linguistics; 2007. Semi-automated named entity annotation; pp. 53–56. [Google Scholar]
- 23.Ogren PV, Savova GK, Chute CG. Constructing evaluation corpora for automated clinical named entity recognition. Proceedings of the sixth international conference on language resources and evaluation LREC. 2008:3143–3150. [Google Scholar]
- 24.Lingren T, Deleger L, Molnar K, Zhai H, Meinzen-Derr J, Kaiser M, Stoutenborough L, Li Q, Solte I. Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements. J Am Med Inform Assoc. doi: 10.1136/amiajnl-2013-001837. http://dx.doi.org/10.1136/amiajnl-2013-001837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.South BR, Shen S, Friedlin FJ, Samore M, Meystre SM. Enhancing annotation of clinical text using pre-annotation of common PHI; Proceedings of the AMIA annual symposium; 2010. p. 1267. [Google Scholar]
- 26.Stenetorp P, Pyysalo S, Topic G, Ananiadou S, Tsujii J. BRAT: a web-based tool for NLP-assisted text annotation. EACL. 2012;2012:102. [Google Scholar]
- 27.South B, Shen S, Leng J, Forbush T, DuVall S, Chapman WW. Proceedings of the 2012 workshop on biomedical natural language processing. BioNLP ‘12. Stroudsburg, PA, USA: Association for Computational Linguistics; 2012. A prototype tool set to support machine-assisted annotation; pp. 130–139. [Google Scholar]
- 28.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics. 2005;21:3191–3192. doi: 10.1093/bioinformatics/bti475. [DOI] [PubMed] [Google Scholar]
- 29.Mayer JM, Shen S, South BR, Meystre S, Friedlin FJ, Ray WR, et al. Inductive creation of an annotation schema and a reference standard for de-identification of VA electronic clinical notes. AMIA Annu Symp Proc. 2009;14(2009):416–420. [PMC free article] [PubMed] [Google Scholar]
- 30.Hripcsak G, Rothschild A. Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc. 2005 May-Jun;12(3):296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilcoxon Rank Sum Test. [Accessed 21.08.13]; < http://analytics.ncsu.edu/sesug/2004/TU04-Pappas.pdf>. [Google Scholar]
- 32.Dorr DA, Phillips WF, Phansalkar S, Sims SA, Hurdle JF. Assessing the difficulty and time costs of de-identification in clinical narratives. Method Inform Med. 2006;45(3):246–252. [PubMed] [Google Scholar]
- 33.South B, Shen S, Maw M, Ferrández O, Friedlin FJ, Meystre S. Prevalence estimates of clinical eponyms in de-identified clinical documents. AMIA Summits Transl Sci Proc CRI. 2012:136. [Google Scholar]
- 34.Brooke J. SUS: A “quick and dirty” usability scale. In: Jordan PW, Thomas B, Weerdmeester BA, McClelland, editors. Usability evaluation in industry. London UK: Taylor & Francis; 1996. pp. 189–194. [Google Scholar]
- 35.Felt P, Ringger E, Seppi K, Heal K, Haertel R, Londsdale D. Proceedings of the tenth international conference on language resources and evaluation. LREC; 2012. First results in a study evaluating pre-annotation and correction propagation for machine-assisted syriac morphological analysis; pp. 878–885. [Google Scholar]