Abstract
Due to the recently uncovered health benefits and anti-HIV activities of dicaffeoylquinic acids (diCQAs), understanding their structures and functions is of great interest for drug discovery efforts. DiCQAs are analytically challenging to identify and quantify since they commonly exist as a diverse mixture of positional and geometric (cis/trans) isomers. In this work, we utilized ion mobility spectrometry coupled with mass spectrometry to separate the various isomers before and after UV irradiation. The experimental collision cross sections were then compared with theoretical structures to differentiate and identify the diCQA isomers. Our analyses found that naturally the diCQAs existed predominantly as trans/trans isomers, but after 3 h of UV irradiation, cis/cis, cis/trans, trans/cis, and trans/trans isomers were all present in the mixture. This is the first report of successful differentiation of cis/trans diCQA isomers individually, which shows the great promise of IMS coupled with theoretical calculations for determining the structure and activity relationships of different isomers in drug discovery studies.
Graphical Abstract
Chlorogenic acids (CGAs) are polyphenolic natural products consisting of esters of quinic acid and hydroxycinnamic acids (i.e. caffeic-, ferulic-, sinapic- or coumaric acid). CGAs are found in a wide variety of food and beverage plants and have been linked to reducing obesity and influenza, and preventing cataractogenesis,1–3 therefore making them of great interest for drug discovery efforts. Further, caffeoylquinic acids (CQAs) and dicaffeoylquinic acids (diCQAs), which are two main classes of CGAs, also have reported antioxidant, anti-HIV, antihypertensive, and anti-inflammatory health benefits.4–11 Analyzing CGAs, however, can be very difficult as they normally exist as complex mixtures with various positional and geometric (cis/trans) isomers.12,13 Additionally, exposure to UV light further complicates their analyses as it has been shown to change the trans double bond orientations to cis, and when multiple double bonds exist, cis/cis and combinations of cis/trans isomers are very common.14 Thus, the structure and activity relationship for these cis/trans isomers has not been well understood to date, but the bioactivities of CGAs are known to be affected by the cis/trans isomerism.15,16
As mentioned, diCQAs exhibit anti-HIV-1 activity by binding irreversibly to the HIV-1 DNA integrase (a key enzyme for viral HIV DNA integration) and inhibiting HIV-1 DNA replication.7–9 Related hydroxycinnamic acid derivatives such as L-chicoric acid (dicaffeoyltartaric acid) also possess the same anti-HIV activities, but both the diCQAs and dicaffeoyltartaric acids activities are highly dependent on the structural isomers present.16–18 Structural isomerization analyses for other natural or synthetic drugs have also shown either agonistic or antagonistic effects. For example, an isomeric mixture of conjugated linoleic acids has been shown to possess enhanced biological activities when compared to the pure components,19 indicating biological synergism of the isomers. Also, the unnatural cis isomers of potent proteasome inhibitors have exhibited stronger inhibitory activity than the trans counterparts.20 Thus, understanding the structures and activities of CGAs is essential for their use in drug studies.
The identification and quantification of phytochemicals in plant extracts is challenging due to their structural diversity; therefore, reliable analytical methods capable of differentiating related structures are needed.21 Although great efforts have been put forth, commonly used analytical platforms have shown significant deficiencies with these separations. For instance, LC-MSn approaches developed to discriminate positional isomers of CGAs provide limited differentiation of geometric isomers.22,23 Geometric CGA isomers in previous studies were determined through chromatographic elution orders; however, this can be problematic depending on the chromatographic parameters and their reproducibility.14 Recently it was reported that cis geometrical isomers of diCQAs preferentially bind to alkali metal ions and can be discriminated from the trans isomers using LC-MS.24 However, this method is only capable of discriminating the group of cis isomers from the group of trans isomers, but not identifying individual isomers.
With recent technology advancements, ion mobility spectrometry-mass spectrometry (IMS-MS) has become an appealing tool for the separation and structural characterization of biomolecules.25,26 IMS-MS has also been used in the characterization of positional and geometrical isomers of small molecules27,28 and those due to photoisomerization.29,30 Recently, IMS was applied to separate mono- and diCQAs and differentiate their positional isomers,31,32 but unfortunately the cis/trans isomers were not separated. Here we utilized drift tube ion mobility spectrometry-mass spectrometry (DTIMS-MS) to elucidate the cis/trans isomer structures of diCQAs generated by UV irradiation-induced photoisomerization. Furthermore, a newly developed structures for lossless ion manipulations (SLIM) platform was used for ultrahigh-resolution IMS separations of the cis/trans isomers. SLIM IMS uses traveling waves and a compact serpentine drift path for high ion mobility resolution. A switch can also be utilized in these devices to perform multiple passes in the drift path for even higher IMS resolution.33–38 The work detailed in this manuscript allowed the structure for each cis/trans isomer to be unambiguously identified by comparing the experimental collision cross section (CCS) values with theoretical CCS values obtained from molecular modeling. This comparison enabled the differentiation and identification of isomers from complex natural bioactive phytochemicals, enabling an understanding of their bioactivity.
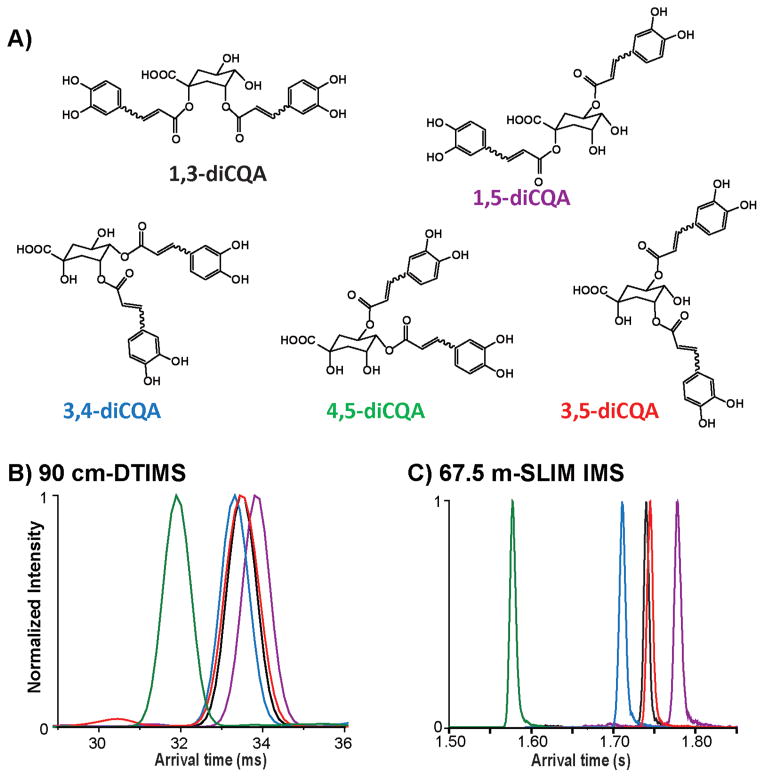
In this study, first we characterized the positional isomers for the five diCQA standards (1,3-diCQA, 1,5-diCQA, 3,4-diCQA, 3,5-diCQA, and 4,5-diCQA; Figure 1A) using a 90 cm DTIMS-MS platform (Agilent 6560 IMS-QTOF MS, Santa Clara, California, USA).39,40 The DTIMS profiles for these diCQA isomers ([M + Na]+, m/z = 539.12) are shown in Figure 1B. Prior to UV irradiation, each diCQA isomer displayed a single IMS peak, consistent with the fact that these compounds naturally exist as trans/trans isomers. The arrival time distributions (ATDs) for these positional isomers were similar; however, 4,5-diCQA eluted first (indicating a smaller structure) and was baseline separated, while 1,5-diCQA had the longest arrival time and the largest structure. The CCS value for each diCQA isomer was measured by DTIMS and is given in Table 1. To better differentiate the positional isomers, the SLIM IMS- MS platform was then used to obtain ultrahigh-resolution IMS separations. As shown in Figure 1C, after five passes of the SLIM IMS-MS platform (67.5 m drift length) 4,5-diCQA, 3,4-diCQA, and 1,5-diCQA were well separated, and 1,3-diCQA and 3,5-diCQA were partially separated. Since the SLIM IMS-MS platform is based upon traveling waves, direct CCS measurement is not possible without calibration, so only the CCS values from DTIMS are noted in Table 1.
Figure 1.
IMS characterization of the dicaffeoylquinic acid positional isomers 1,3-diCQA, 1,5-diCQA, 3,4-diCQA, 3,5-diCQA, and 4,5-diCQA. (A) The chemical structures of each isomer and arrival time distributions (ATDs) using a (B) DTIMS platform and (C) ultrahigh-resolution SLIM IMS.
Table 1.
Experimental Collision Cross Sections Measured by DTIMS in Nitrogen Gas (DTCCSN2, in Å2) for Each diCQA Positional Isomera
| experimental DTCCSN2 (Å2) | experimental CV (%) | |
|---|---|---|
| 1,3-diCQA | 250 | 0.5 |
| 1,5-diCQA | 252 | 0.3 |
| 3,4-diCQA | 249 | 0.1 |
| 3,5-diCQA | 251 | 0.5 |
| 4,5-diCQA | 236 | 0.1 |
The coefficient of variance (CV) determined by triplicate measurements are provided.
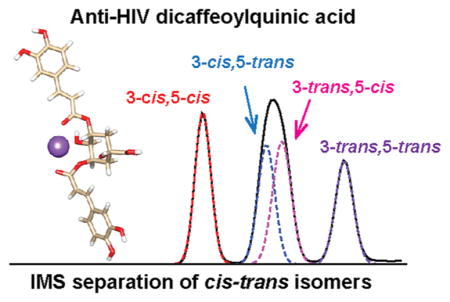
Among these positional isomers, the most abundant form and potent HIV-1 inhibitor 3,5-diCQA was selected to establish an analytical method for cis/trans isomer separation since four possible geometric isomers exist: 3-trans,5-trans-diCQA isomer, 3-cis,5-trans-diCQA, 3-trans,5-cis-diCQA, and 3-cis,5-cis-diCQA. Previous molecular docking studies showed that these isomers have different binding activities with the HIV-1 INT enzyme, a key enzyme for viral HIV DNA integration.41 While these isomers all bind to the catalytic domain of HIV-1 INT enzyme, the cis isomers were found to bind to the metal cofactor of HIV-1 INT, which is related to its antiviral activity. Moreover, 3-trans,5-cis-diCQA interacted with both the LYS156 and LYS159 residues that are significant for viral DNA integration. These docking results also illustrated that different binding activities between the 3,5-diCQA isomers with HIV-1 INT enzyme are synergistic and provide wider inhibition activity than a single isomer. Therefore, understanding the structures of the geometric isomers is important for elucidating their bioactivities.
Upon UV irradiation at 245 nm for 3 h, the photo-isomerization products of 3,5-diCQA were examined by DTIMS-MS (Figure 2). Without UV irradiation, 3,5-diCQA mainly illustrates a single feature which corresponds to the 3-trans,5-trans isomer (Figure 2A). An additional peak at a shorter arrival time however was also observed, indicating a small fraction of 3,5-diCQA exists in other conformations prior to UV irradiation. After 3 h of UV irradiation, several additional features arose with shorter arrival times (Figure 2B). The feature with the shortest arrival time (left) displayed a very narrow distribution similar to the feature with the longest arrival time (right). The middle feature however was twice as broad as the other features and could be fitted with two features, indicating the presence of two conformations with similar abundances.
Figure 2.
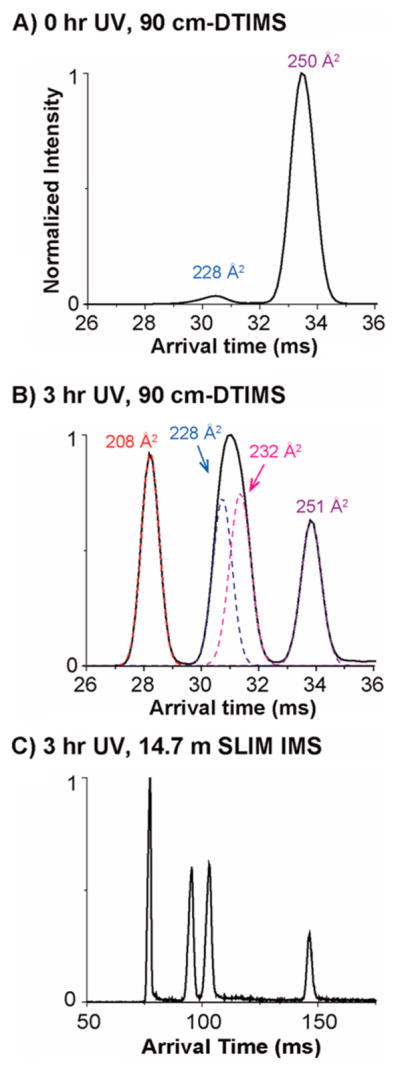
ATDs for the 3,5-diCQAs (A) before UV irradiation and (B) after 3 h UV irradiation using the DTIMS-MS, and (C) the SLIM IMS-MS platforms. The dashed lines in panel B represent the IMS peak shapes expected for single structures.
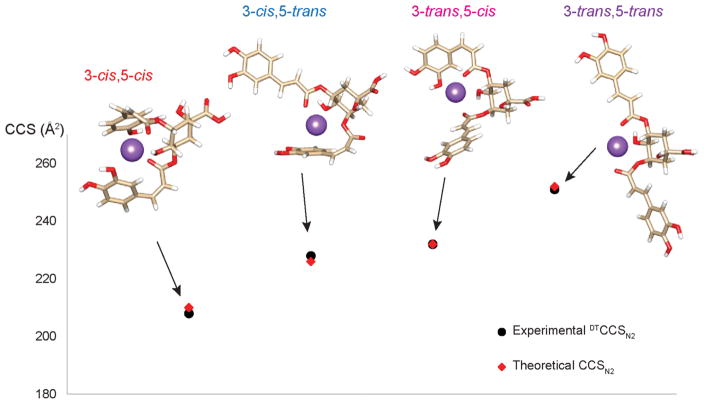
To better differentiate the 3,5-diCQA cis/trans isomers, SLIM IMS-MS measurements were performed for the conformers resulting from UV irradiation. As shown in Figure 2C with only a 14.7 m SLIM IMS separation, the middle feature that was unresolved in Figure 2B is clearly separated into two features, confirming that there are four isomers for 3,5-diCQA after photoisomerization. To assign these cis/trans isomers, theoretical modeling was performed for each isomer allowing the calculation of theoretical CCS values (Figure 3 and Table 2). By comparing the experimental and theoretical CCS values, the four ATD features were assigned (left to right) as 3-cis,5-cis, 3-trans,5-cis, 3-trans,5-cis, and 3-trans,5-trans (Table 2). The theoretical structures also revealed that the 3-cis,5-cis isomer has the most compact structure with the two caffeic acid (CA) groups/moieties collapsed toward each other, while the 3- trans,5-trans isomer was the most extended with the two CA groups widely open and extending in opposite directions. The 3-cis,5-trans and 3-trans,5-cis isomers both adapt a partially open conformation with one CA group collapsing toward the center and the other group extending to the side. The 3-trans,5-cis isomer, however, does have a slightly more open structure than the 3-cis,5-trans isomer resulting in a larger CCS value. Thus, the cis or trans orientation has an important impact on the molecule’s conformation. Moreover, the intensity of the 3-cis,5-trans isomer is comparable to that of the 3-trans,5-cis isomer, indicating the chance of formation for these two isomers appears to be similar. To investigate this occurrence and the conversion pathways between the 3,5-diCQA isomers, we monitored the products formed after 2, 5, 10, 20, and 30 min of UV irradiation. Our analyses found that the cis,cis isomer only forms through the 3-cis,5-trans and 3-trans,5-cis isomers not directly from the trans,trans isomer (see SI Figure S1). To estimate the reaction barriers associated with the transformation pathways from the 3-trans,5-trans isomer to the 3-cis,5-cis isomer, we determined the minimum energy paths using the “string method”42 similar to Yoon et al.43 The reaction energies (ΔE) along the minimum energy paths are depicted in Figure S2. The results illustrate that the conversion from 3-trans,5-trans to 3-trans,5-cis or 3-cis,5-trans is very high, explaining why diCQA exists as trans,trans until it is irradiated. They also suggest that, statistically, the 3-trans,5-cis isomer will form first; however, the energy barriers are similar and the photoisomerization process appears to result in the equivalent formation of 3-trans,5-cis and 3-cis,5-trans. Following the formation of the 3-trans,5-cis and 3-cis,5-trans isomers, the reaction barriers show that the 3-cis,5-cis isomer will preferentially arise from the 3-cis,5-trans isomer. Thus, by coupling the experimental IMS approach and theoretical calculations, we were able to monitor the photoisomerization process and understand the conversions occurring between isomers.
Figure 3.
Theoretical modeling structures for the cis/trans photoisomerization products of 3,5-diCQA and their theoretical CCSN2 values compared to the experimental DTCCSN2 values. The error associated with each measurement is noted in Table 2.
Table 2.
Theoretical and Experimental CCS Values (in Å2) for Each cis/trans 3,5-diCQA Isomer
| 3,5-diCQA | theoretical CCSN2 (Å2) | theoretical CV (%)a | experimental DTCCSN2 (Å2)b | experimental CV (%) |
|---|---|---|---|---|
| 3-cis,5-cis | 210 | 0.2 | 208 | 0.3 |
| 3-cis,5-trans | 226 | 0.2 | 228 | 0.1 |
| 3-trans,5-cis | 232 | 0.1 | 232 | 0.4 |
| 3-trans,5-trans | 252 | 0.2 | 251 | 0.3 |
The theoretical CV is the deviation from the mean resulting from multiple calculations.
DTCCSN2 is the collision cross sections measured by DTIMS in nitrogen buffer gas
This manuscript tackles the difficult problem of identifying and quantifying cis and trans isomers of phytochemicals in natural product and plant extracts, which to date has been extremely challenging due to the vast structural diversity of isomers present in the mixtures. Here IMS-based approaches were successfully utilized to separate complex diCQA isomers and identify the cis/trans isomers with the aid of theoretical modeling. The ultrahigh-resolution SLIM IMS-MS technology enabled baseline separation of the isomers and was a powerful tool for analyzing the complex natural products and elucidating structure-activity relationships. Further, coupling the experiment IMS separations and theoretical calculations provided insight into isomeric conversions, which will be extremely important for drug discovery studies and optimizing specific bioactivities.
METHODS
Materials and Sample Preparations
Authentic standards of trans,trans-dicaffeoylquinic acids (1,3-diCQA, 1,5-diCQA, 3,4-diCQA, 3,5-diCQA, and 4,5-diCQA) were purchased from Phytolab (Vestenbergsgreuth, Germany). Analytical grade methanol was purchased from Romil Ltd. (Cambridge, UK). To obtain the cis isomers of the dicaffeoylquinic acids, a 1 mg/ mL stock solution was prepared in 100% methanol. The samples were then irradiated using a UV lamp (Spectroline, USA) operating at 245 nm with an intensity of 390 μW/cm2. The lamp was not covered with any notch filter. The products of UV irradiation were analyzed by liquid chromatography-mass spectrometry,22 and no other species were observed other than those peaks corresponding to the 3,5-diCQA isomers (LC chromatograms shown in Figure S3). For the time-dependent photoisomerization study, the 3,5-diCQA was UV irradiated for 2, 5, 10, 20, 30, 60, and 180 min and the products were analyzed with IMS-MS.
DTIMS-MS
The chemicals were analyzed using an Agilent 6560 ion mobility-quadrupole time-of-flight mass spectrometry (IM-QTOF MS) platform.39,40 Briefly, for ion mobility measurements, after electrospray ionization, ions were passed through the inlet glass capillary, focused by a high pressure ion funnel, and accumulated in a lower pressure ion funnel trap (IFT). Ions were then pulsed into the 90 cm-long IMS drift tube filled with ~4 Torr of nitrogen gas, where they travel under the influence of a weak electric field (10–20 V/cm). Ions exiting the drift tube were refocused by a rear ion funnel prior to QTOF MS detection, and their arrival time (tA) was recorded. The reduced mobility (the mobility scaled to standard temperature and pressure) can be determined from instrument parameters by plotting tA versus p/V 44
Here L is the drift length, V is the drift voltage, t0 is the time ion spending outside of the drift cell, T is the drift gas temperature, and p is the drift gas pressure. The reduced mobility can be related to the collision cross sections of the analyte using kinetic theory:44
Here, q is the ion charge, N is the buffer gas number density at STP, μ is the reduced mass of the ion-nitrogen collision, and kB is the Boltzmann constant. All the CCS values were measured using seven-stepped field voltages. Each measurement was performed in triplicate, and the coefficient of variance (CV) was below 0.5% in all cases.
SLIM IMS-MS
To achieve ultrahigh-resolution IMS separation of the cis/trans isomers, the UV irradiated products of 3,5-diCQAs were also measured using a structures for lossless ion manipulations IMS-MS platform (SLIM IMS-MS). SLIM IMS uses traveling waves and a compact serpentine ion drift path for efficient ion selection, trapping, and accumulations.33–38 The SLIM IMS-MS platform has a 13 m long serpentine drift path and multipass capability, as recently described.38,45
Theoretical Calculations
Molecular modeling was performed using NWChem (v6.6),46 a high-performance computational chemistry software.47,48 Briefly, 2D structure files (.mol) were analyzed using the Marvin pKa plugin (Marvin 15.9.14, 2015, ChemAxon) for prediction of protonation, deprotonation and adduction sites.49 Initial geometry relaxation was performed using the Merck molecular force field (MMFF94)50 implemented in Avogadro (v1.1.1).51 Density functional theory (DFT) based ab initio molecular dynamics (AIMD),52 as implemented in NWChem, were used to sample 100 conformers for each molecule.52 AIMD calculations were each run for at least 10.2 ps, with conformers sampled every 101.6 fs. The AIMD temperature was maintained using a stochastic velocity rescaling thermostat.53 This was followed by DFT-based frequency calculations for determining the Gibbs free energy of each conformer. The B3LYP exchange-correlation functional was used for all calculations54–57 and Pople basis sets at the 3-21G level (a double-ζ split-valence potential basis set)58,59 and 6-31G* level (a double-ζ valence potential basis set having a single polarization function),60–62 were used for the AIMD and DFT frequency calculations, respectively. All basis sets were obtained from the Environmental Molecular Sciences Laboratory (EMSL) Basis Set Exchange (bse.pnl.gov).63,64 NWChem output files were processed using custom-written Python scripts. Python (v2.7.10), with the NumPy package (v1.9.2),65 was implemented using WinPython (v2.7.10.1, http://winpython.github.io), a free, open-source, and portable full-featured Python-based scientific environment. IPython (v3.2.0),66 an enhanced Python shell, was used within the Scientific Python Development Environment (Spyder v2.3.5.2) for NWChem output data processing. Aided by supercomputers, our molecular modeling approach calculated CCS values with high accuracy, enabling the identification of the geometric isomers. The use of a first-principles theory (DFT) based AIMD approach enables consideration of the electronic structure of each isomer and its role in the conformer geometries. CCS values were calculated for all molecular conformers using the MOBCAL software, modified for the room temperature N2-based trajectory method.67–69 The atom coordinates, radius, and charge distribution of the optimized geometry structures were used as input to the MOBCAL calculations. Final CCS values were obtained by a clustering approach using histogram distributions to determine the most abundant, low-energy, stable conformers. Scatter plots were then generated to show the relative energy of the 100 structures for each molecule (determined by ab initio calculations) versus their corresponding collision cross sections (determined by MOBCAL). An example plot and frequency diagram are shown in Figure S4 for 3-trans,5-cis diCQA. NWChem was also used to calculate the minimum-energy paths using the DFT-based zero temperature “string method”. We employed 40 beads to represent each reaction path, for a total of 80 beads from 3-trans,5-trans diCQA to 3-cis,5-cis diCQA. The B3LYP exchange-correlation functional with the Pople basis set 3-21G was used for the string method with a stepsize of 0.1 and a maximum of 100 iterations.
Supplementary Material
Acknowledgments
Portions of this research were supported by grants from the National Institute of Environmental Health Sciences of the NIH (R01 ES022190), National Institute of General Medical Sciences (P41 GM103493), and the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory for Global Forensic Chemical Exposure Assessment of the Environmental Exposome and Microbes in Transition (MinT) Initiative. This work was also partially funded by the South African National Research Foundation grant (TTK160604167903) awarded to N.E.M. This research utilized capabilities developed by the Panomics program (funded by the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program) and by the National Institute of Allergy and Infectious Diseases under Grant U19AI106772. This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). The NWChem calculations were performed using the Cascade supercomputer at the EMSL. PNNL is operated by Battelle for the DOE under Contract DE-AC05-76RL01830.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.6b03015.
Time-dependent study of the photoisomerization of 3,-5-diCQA by IMS-MS following UV irradiation; the minimum-energy paths from 3-trans,5-trans diCQA through 3-trans,5-cis diCQA and 3-cis,5-trans diCQA to 3-cis,5-cis diCQA using the “string method” as implemented in NWChem; LC chromatograms for 3,5-diCQA before and after UV irradiation; example scatter plot and cluster diagram for 3-trans,5-cis diCQA (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Cho AS, Jeon SM, Kim MJ, Yeo J, Seo KI, Choi MS, Lee MK. Chlorogenic acid exhibits anti-obesity property and improves lipid metabolism in high-fat diet-induced-obese mice. Food Chem Toxicol. 2010;48:937–943. doi: 10.1016/j.fct.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Urushisaki T, Takemura T, Tazawa S, Fukuoka M, Hosokawa-Muto J, Araki Y, Kuwata K. Caffeoylquinic Acids Are Major Constituents with Potent Anti-Influenza Effects in Brazilian Green Propolis Water Extract. Evidence-Based Complementary Altern Med. 2011;2011:1–7. doi: 10.1155/2011/254914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlemi AV, Makri OE, Mermigki PG, Lamari FN, Georgakopoulos CD. Quercetin glycosides and chlorogenic acid in highbush blueberry leaf decoction prevent cataractogenesis in vivo and in vitro: Investigation of the effect on calpains, antioxidant and metal chelating properties. Exp Eye Res. 2016;145:258–268. doi: 10.1016/j.exer.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Clifford MN. Chlorogenic acids and other cinnamates - nature, occurrence, dietary burden, absorption and metabolism. J Sci Food Agric. 2000;80:1033–1043. [Google Scholar]
- 5.Jeng TL, Lai CC, Liao TC, Lin SY, Sung JM. Effects of drying on caffeoylquinic acid derivative content and antioxidant capacity of sweet potato leaves. J Food Drug Anal. 2015;23:701–708. doi: 10.1016/j.jfda.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Könczöl Á, Béni Z, Sipos MM, Rill A, Háda V, Hohmann J, Máthé I, Szántay C, Jr, Keserű GM, Balogh GT. Antioxidant activity-guided phytochemical investigation of Artemisia gmelinii Webb. ex Stechm.: Isolation and spectroscopic challenges of 3, 5-O-dicaffeoyl (epi?) quinic acid and its ethyl ester. J Pharm Biomed Anal. 2012;59:83–89. doi: 10.1016/j.jpba.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Robinson WE, Cordeiro M, Abdel-Malek S, Jia Q, Chow SA, Reinecke MG, Mitchell WM. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: inhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol Pharmacol. 1996;50:846–855. [PubMed] [Google Scholar]
- 8.McDougall B, King PJ, Wu BW, Hostomsky Z, Reinecke MG, Robinson WE. Dicaffeoylquinic and Dicaffeoyltartaric Acids Are Selective Inhibitors of Human Immunodeficiency Virus Type 1 Integrase. Antimicrob Agents Chemother. 1998;42:140–146. doi: 10.1128/aac.42.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu K, Cordeiro ML, Atienza J, Robinson WE, Chow SA. Irreversible Inhibition of Human Immunodeficiency Virus Type 1 Integrase by Dicaffeoylquinic Acids. J Virol. 1999;73:3309–3316. doi: 10.1128/jvi.73.4.3309-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong S, Joo T, Jhoo JW. Antioxidant and anti-inflammatory activities of 3,5-dicaffeoylquinic acid isolated from Ligularia fischeri leaves. Food Sci Biotechnol. 2015;24:257–263. [Google Scholar]
- 11.Abdel Motaal A, Ezzat SM, Tadros MG, El-Askary HI. In vivo anti-inflammatory activity of caffeoylquinic acid derivatives from Solidago virgaurea in rats. Pharm Biol. 2016;54:2864–2870. doi: 10.1080/13880209.2016.1190381. [DOI] [PubMed] [Google Scholar]
- 12.Arantes AA, Fale PL, Costa LCB, Pacheco R, Ascensao L, Serralheiro ML. Inhibition of HMG-CoA reductase activity and cholesterol permeation through Caco-2 cells by caffeoylquinic acids from Vernonia condensata leaves. Rev Bras Farmacogn. 2016;26:738–743. [Google Scholar]
- 13.Souza AHP, Corrêa RCG, Barros L, Calhelha RC, Santos-Buelga C, Peralta RM, Bracht A, Matsushita M, Ferreira ICFR. Phytochemicals and bioactive properties of Ilex paraguariensis: An in-vitro comparative study between the whole plant, leaves and stems. Food Res Int. 2015;78:286–294. doi: 10.1016/j.foodres.2015.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Clifford MN, Kirkpatrick J, Kuhnert N, Roozendaal H, Salgado PR. LC-MSn analysis of the cis isomers of chlorogenic acids. Food Chem. 2008;106:379–385. [Google Scholar]
- 15.Ramabulana T, Mavunda RD, Steenkamp PA, Piater LA, Dubery IA, Madala NE. Secondary metabolite perturbations in Phaseolus vulgaris leaves due to gamma radiation. Plant Physiol Biochem. 2015;97:287–295. doi: 10.1016/j.plaphy.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Healy EF, Sanders J, King PJ, Robinson WE., Jr A docking study of l-chicoric acid with HIV-1 integrase. J Mol Graphics Modell. 2009;27:584–589. doi: 10.1016/j.jmgm.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 17.King PJ, Ma G, Miao W, Jia Q, McDougall BR, Reinecke MG, Cornell C, Kuan J, Kim TR, Robinson WE. Structure-Activity Relationships: Analogues of the Dicaffeoylquinic and Dicaffeoyltartaric Acids as Potent Inhibitors of Human Immunodeficiency Virus Type 1 Integrase and Replication. J Med Chem. 1999;42:497–509. doi: 10.1021/jm9804735. [DOI] [PubMed] [Google Scholar]
- 18.Kyle JE, Zhang X, Weitz KK, Monroe ME, Ibrahim YM, Moore RJ, Cha J, Sun X, Lovelace ES, Wagoner J, et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 2016;141:1649–1659. doi: 10.1039/c5an02062j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavino VC, Gavino G, Leblanc MJ, Tuchweber B. An Isomeric Mixture of Conjugated Linoleic Acids But Not Pure cis-9,trans-11-Octadecadienoic Acid Affects Body Weight Gain and Plasma Lipids in Hamsters. J Nutr. 2000;130:27–29. doi: 10.1093/jn/130.1.27. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura S, Unno Y, List A, Mizuno A, Tanaka M, Sasaki T, Arisawa M, Asai A, Groll M, Shuto S. Potent Proteasome Inhibitors Derived from the Unnatural cis-Cyclopropane Isomer of Belactosin A: Synthesis, Biological Activity, and Mode of Action. J Med Chem. 2013;56:3689–3700. doi: 10.1021/jm4002296. [DOI] [PubMed] [Google Scholar]
- 21.Mncwangi NP, Viljoen AM, Zhao J, Vermaak I, Chen W, Khan I. What the devil is in your phytomedicine? Exploring species substitution in Harpagophytum through chemometric modeling of 1H-NMR and UHPLC-MS datasets. Phytochemistry. 2014;106:104–115. doi: 10.1016/j.phytochem.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Clifford MN, Johnston KL, Knight S, Kuhnert N. Hierarchical Scheme for LC-MSn Identification of Chlorogenic Acids. J Agric Food Chem. 2003;51:2900–2911. doi: 10.1021/jf026187q. [DOI] [PubMed] [Google Scholar]
- 23.Clifford MN, Knight S, Kuhnert N. Discriminating between the Six Isomers of Dicaffeoylquinic Acid by LC-MSn. J Agric Food Chem. 2005;53:3821–3832. doi: 10.1021/jf050046h. [DOI] [PubMed] [Google Scholar]
- 24.Makola MM, Steenkamp PA, Dubery IA, Kabanda MM, Madala NE. Preferential alkali metal adduct formation by cis geometrical isomers of dicaffeoylquinic acids allows for efficient discrimination from their trans isomers during ultra-high-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2016;30:1011–1018. doi: 10.1002/rcm.7526. [DOI] [PubMed] [Google Scholar]
- 25.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Biomolecule Analysis by Ion Mobility Spectrometry. Annu Rev Anal Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 27.Lapthorn C, Pullen F, Chowdhry BZ. Ion mobility spectrometry-mass spectrometry (IMS-MS) of small molecules: Separating and assigning structures to ions. Mass Spectrom Rev. 2013;32:43–71. doi: 10.1002/mas.21349. [DOI] [PubMed] [Google Scholar]
- 28.Baker ES, Hong JW, Gidden J, Bartholomew GP, Bazan GC, Bowers MT. Diastereomer Assignment of an Olefin-Linked Bis-paracyclophane by Ion Mobility Mass Spectrometry. J Am Chem Soc. 2004;126:6255–6257. doi: 10.1021/ja039486k. [DOI] [PubMed] [Google Scholar]
- 29.Adamson BD, Coughlan NJA, Markworth PB, Continetti RE, Bieske EJ. An ion mobility mass spectrometer for investigating photoisomerization and photodissociation of molecular ions. Rev Sci Instrum. 2014;85:123109–112317. doi: 10.1063/1.4903753. [DOI] [PubMed] [Google Scholar]
- 30.Czerwinska I, Kulesza A, Choi C, Chirot F, Simon AL, Far J, Kune C, de Pauw E, Dugourd P. Supramolecular influence on cis-trans isomerization probed by ion mobility spectrometry. Phys Chem Chem Phys. 2016;18:32331–32336. doi: 10.1039/c6cp06168k. [DOI] [PubMed] [Google Scholar]
- 31.Xie C, Yu K, Zhong D, Yuan T, Ye F, Jarrell JA, Millar A, Chen X. Investigation of Isomeric Transformations of Chlorogenic Acid in Buffers and Biological Matrixes by Ultraperformance Liquid Chromatography Coupled with Hybrid Quadrupole/Ion Mobility/ Orthogonal Acceleration Time-of-Flight Mass Spectrometry. J Agric Food Chem. 2011;59:11078–11087. doi: 10.1021/jf203104k. [DOI] [PubMed] [Google Scholar]
- 32.Kuhnert N, Yassin GH, Jaiswal R, Matei MF, Grün CH. Differentiation of prototropic ions in regioisomeric caffeoyl quinic acids by electrospray ion mobility mass spectrometry. Rapid Commun Mass Spectrom. 2015;29:675–680. doi: 10.1002/rcm.7151. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Garimella SVB, Prost SA, Webb IK, Chen TC, Tang K, Tolmachev AV, Norheim RV, Baker ES, Anderson GA, et al. Ion Trapping, Storage, and Ejection in Structures for Lossless Ion Manipulations. Anal Chem. 2015;87:6010–6016. doi: 10.1021/acs.analchem.5b00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen TC, Ibrahim YM, Webb IK, Garimella SVB, Zhang X, Hamid AM, Deng L, Karnesky WE, Prost SA, Sandoval JA, et al. Mobility-Selected Ion Trapping and Enrichment Using Structures for Lossless Ion Manipulations. Anal Chem. 2016;88:1728–1733. doi: 10.1021/acs.analchem.5b03910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamid AM, Garimella SV, Ibrahim YM, Deng L, Zheng X, Webb IK, Anderson GA, Prost SA, Norheim RV, Tolmachev AV, et al. Achieving High Resolution Ion Mobility Separations Using Traveling Waves in Compact Multiturn Structures for Lossless Ion Manipulations. Anal Chem. 2016;88:8949–8956. doi: 10.1021/acs.analchem.6b01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng L, Ibrahim YM, Hamid AM, Garimella SVB, Webb IK, Zheng X, Prost SA, Sandoval JA, Norheim RV, Anderson GA, et al. Ultra-High Resolution Ion Mobility Separations Utilizing Traveling Waves in a 13 m Serpentine Path Length Structures for Lossless Ion Manipulations Module. Anal Chem. 2016;88:8957–8964. doi: 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng L, Ibrahim YM, Garimella SVB, Webb IK, Hamid AM, Norheim RV, Prost SA, Sandoval JA, Baker ES, Smith RD. Greatly Increasing Trapped Ion Populations for Mobility Separations Using Traveling Waves in Structures for Lossless Ion Manipulations. Anal Chem. 2016;88:10143–10150. doi: 10.1021/acs.analchem.6b02678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, Ibrahim YM, Baker ES, Aly NA, Hamid AM, Zhang X, Zheng X, Garimella SVB, Webb IK, Prost SA, et al. Ion Mobility Separations of Isomers based upon Long Path Length Structures for Lossless Ion Manipulations Combined with Mass Spectrometry. ChemistrySelect. 2016;1:2396–2399. doi: 10.1002/slct.201600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, et al. Conformational Ordering of Biomolecules in the Gas Phase: Nitrogen Collision Cross Sections Measured on a Prototype High Resolution Drift Tube Ion Mobility-Mass Spectrometer. Anal Chem. 2014;86:2107–2116. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ibrahim YM, Baker ES, Danielson WF, 3rd, Norheim RV, Prior DC, Anderson GA, Belov ME, Smith RD. Development of a New Ion Mobility (Quadrupole) Time-of-Flight Mass Spectrometer. Int J Mass Spectrom. 2015;377:655–662. doi: 10.1016/j.ijms.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makola MM, Dubery IA, Koorsen G, Steenkamp PA, Kabanda MM, du Preez LL, Madala NE. The Effect of Geometrical Isomerism of 3,5-Dicaffeoylquinic Acid on Its Binding Affinity to HIV-Integrase Enzyme: A Molecular Docking Study. Evidence-Based Complementary Altern Med. 2016;2016:1–9. doi: 10.1155/2016/4138263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.EW, Ren W, Vanden-Eijnden E. String method for the study of rare events. Phys Rev B: Condens Matter Mater Phys. 2002;66:052301–052304. [Google Scholar]
- 43.Yoon M, Han S, Kim G, Lee SB, Berber S, Osawa E, Ihm J, Terrones M, Banhart F, Charlier JC, et al. Zipper Mechanism of Nanotube Fusion: Theory and Experiment. Phys Rev Lett. 2004;92:075504–075507. doi: 10.1103/PhysRevLett.92.075504. [DOI] [PubMed] [Google Scholar]
- 44.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2005. Kinetic Theory of Mobility and Diffusion; pp. 137–193. Sections 5.1–5.2. [Google Scholar]
- 45.Deng L, Ibrahim YM, Hamid AM, Garimella SV, Webb IK, Zheng X, Prost SA, Sandoval JA, Norheim RV, Anderson GA, et al. Ultra-High Resolution Ion Mobility Separations Utilizing Traveling Waves in a 13 m Serpentine Path Length Structures for Lossless Ion Manipulations Module. Anal Chem. 2016;88:8957–8964. doi: 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valiev M, Bylaska EJ, Govind N, Kowalski K, Straatsma TP, Van Dam HJJ, Wang D, Nieplocha J, Apra E, Windus TL, et al. NWChem: A comprehensive and scalable open-source solution for large scale molecular simulations. Comput Phys Commun. 2010;181:1477–1489. [Google Scholar]
- 47.Graham TR, Renslow R, Govind N, Saunders SR. Precursor Ion-Ion Aggregation in the Brust-Schiffrin Synthesis of Alkanethiol Nanoparticles. J Phys Chem C. 2016;120:19837–19847. [Google Scholar]
- 48.Zheng X, Zhang X, Schocker NS, Renslow RS, Orton DJ, Khamsi J, Ashmus RA, Almeida IC, Tang K, Costello CE, et al. Enhancing glycan isomer separations with metal ions and positive and negative polarity ion mobility spectrometry-mass spectrometry analyses. Anal Bioanal Chem. 2017;409:1–10. doi: 10.1007/s00216-016-9866-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Csizmadia JSaF. A method for calculating the pKa values of small and large molecules. American Chemical Society Spring meeting; March 25–29th, 2007. [Google Scholar]
- 50.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 51.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminf. 2012;4:1–17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer SA, Ueltschi TW, El-Khoury PZ, Mifflin AL, Hess WP, Wang HF, Cramer CJ, Govind N. Infrared and Raman Spectroscopy from Ab Initio Molecular Dynamics and Static Normal Mode Analysis: The C-H Region of DMSO as a Case Study. J Phys Chem B. 2016;120:1429–1436. doi: 10.1021/acs.jpcb.5b03323. [DOI] [PubMed] [Google Scholar]
- 53.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101–014107. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 54.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 55.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B: Condens Matter Mater Phys. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 56.Vosko SH, Wilk L, Nusair M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: a critical analysis. Can J Phys. 1980;58:1200–1211. [Google Scholar]
- 57.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J Phys Chem. 1994;98:11623–11627. [Google Scholar]
- 58.Binkley JS, Pople JA, Hehre WJ. Self-Consistent Molecular-Orbital Methods 0.21. Small Split-Valence Basis-Sets for 1st-Row Elements. J Am Chem Soc. 1980;102:939–947. [Google Scholar]
- 59.Gordon MS, Binkley JS, Pople JA, Pietro WJ, Hehre WJ. Self-Consistent Molecular-Orbital Methods 0.22. Small Split-Valence Basis-Sets for 2nd-Row Elements. J Am Chem Soc. 1982;104:2797–2803. [Google Scholar]
- 60.Francl MM, Pietro WJ, Hehre WJ, Binkley JS, Gordon MS, DeFrees DJ, Pople JA. Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for second-row elements. J Chem Phys. 1982;77:3654–3665. [Google Scholar]
- 61.Hariharan PC, Pople JA. The influence of polarization functions on molecular orbital hydrogenation energies. Theor Chim Acta. 1973;28:213–222. [Google Scholar]
- 62.Rassolov VA, Ratner MA, Pople JA, Redfern PC, Curtiss LA. 6-31G* basis set for third-row atoms. J Comput Chem. 2001;22:976–984. [Google Scholar]
- 63.Feller D. The role of databases in support of computational chemistry calculations. J Comput Chem. 1996;17:1571–1586. [Google Scholar]
- 64.Schuchardt KL, Didier BT, Elsethagen T, Sun LS, Gurumoorthi V, Chase J, Li J, Windus TL. Basis set exchange: A community database for computational sciences. J Chem Inf Model. 2007;47:1045–1052. doi: 10.1021/ci600510j. [DOI] [PubMed] [Google Scholar]
- 65.van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput Sci Eng. 2011;13:22–30. [Google Scholar]
- 66.Perez F, Granger BE. IPython: A system for interactive scientific computing. Comput Sci Eng. 2007;9:21–29. [Google Scholar]
- 67.Campuzano I, Bush MF, Robinson CV, Beaumont C, Richardson K, Kim H, Kim HI. Structural Characterization of Drug-like Compounds by Ion Mobility Mass Spectrometry: Comparison of Theoretical and Experimentally Derived Nitrogen Collision Cross Sections. Anal Chem. 2012;84:1026–1033. doi: 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- 68.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural information from ion mobility measurements: Effects of the long-range potential. J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 69.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.