Abstract
Macroalgae are a diverse group of organisms. Marine macroalgae, in particular, have numerous medicinal and industrial applications. Molecular studies of macroalgae require suitable concentrations of DNA free of contaminants. At present, numerous protocols exist for DNA extraction from macroalgae. However, they are either time consuming, expensive or work only with few species. The method described in this study is rapid and efficient and applicable to different types of marine macroalgae. This method yields an average of 3.85 µg of DNA per 50 mg of algal tissue, with an average purity of 1.88. The isolated DNA was suitable for PCR amplification of universal plastid region of macroalgae.
Keywords: Chlorophyta, DNA extraction, Genomic DNA, Macroalgae, Phaeophyceae, Rhodophyta
Introduction
Macroalgae are a vital part of the marine ecosystem. They contribute to the food web and provide a habitat for various marine organisms (Klinger 2015). Macroalgae or seaweed is also important from an industrial and medical perspective. They have been used for biorefinery and as feedstock for biofuel production (Bruhn et al. 2011; Baghel et al. 2014). Marine macroalgae contain sulphated polysaccharides such as fucans, carrageenans and ulvans, which have been reported to have antioxidant, antitumour, immunostimulatory, anticoagulant, and antimicrobial properties (Patel 2012). Studies have shown that macroalgal species like Laminaria japonica contain essential oils, which have antioxidant and antibacterial properties (Patra et al. 2015). Hydromethanolic extracts of the brown seaweed Padina tetrastromatica have been proven to have antihyperglycemic and antihyperlipidemic effects on rat models on a high-calorie diet (Mohan et al. 2014).
Various molecular studies have been reported for macroalgae including DNA barcoding (Kazi et al. 2013; Zhao et al. 2013), microsatellite library construction (Varela-Álvarez et al. 2006), phytoplankton community composition (Wallace and Gobler 2015) and whole genome sequencing (Cock et al. 2010). Good quality DNA is a prerequisite for most molecular studies involving macroalgae. The extraction of genomic DNA from marine algae is a challenging task owing to the various contaminants that are co-extracted with the DNA such as polysaccharides and polyphenols (Hoarau et al. 2007). These contaminants can inhibit the action of enzymes such as Taq polymerase, rendering the DNA useless for downstream applications (Jin et al. 1997). Macroalgae have a complicated cellular structure composed of various polysaccharides such as cellulose, sulphated fucans, laminarins and alginates (Mabeau et al. 1990; Michel et al. 2010), which hinder the DNA extraction process. Their morphology is also an important factor to consider during DNA extraction because homogenization of algal tissues is a tough task. Hence, an efficient homogenization method applicable for morphologically different algae is essential for efficient DNA extraction. A number of homogenization methods have been reported to disrupt the complicated algal cell wall structure including treatment with an enzyme cocktail consisting of β-glucuronidase, β-glucanase, xylanase, cellulase and agarase (Joubert and Fleurence 2005), grinding with liquid nitrogen (Shivji et al. 1992), and bead beating (Greco et al. 2014). However, a few of these methods require a large amount of biomass, and some methods are time-consuming and expensive, and most of the methods have been standardized for a single organism or only for a few species.
In our recent study on metagenomic DNA extraction, cell lysis was performed using sterile glass powder (Devi et al. 2015). The method was found to be rapid and efficient. In the current study, we have used glass powder for the homogenization of representative species of Rhodophyta, Phaeophyceae and Chlorophyta along with other components for DNA extraction. This is the first report on using glass powder as a homogenization agent for marine macroalgae, and the developed method was found to be efficient, rapid and suitable for other downstream applications.
Materials and methods
Sample collection and morphological identification
Fresh samples of representative species of Rhodophyta, Chlorophyta and Phaeophyceae were collected from Nochiurani (N09°16′16.0″; E79°01′02.0″) and Rameswaram (N09°09′09.6″; E78°39′39.5″) along the Gulf of Mannar region of Tamil Nadu, India. The collected samples were morphologically identified at Central Salt and Marine Research Institute (CSMCRI), Mandapam, Tamil Nadu, India (Fig. 1). The samples were stored at − 80 °C for further studies.
Fig. 1.
Representative species of macroalgae used in this study. a Gracilaria corticata, b Acanthophora spicifera, c Champia parvula, d Sargassum tenerrimum, e Padina tetrastromatica, f Sargassum wightii, g Sargassum polycystum, h Caulerpa scalpelliformis, i Caulerpa racemosa, j Chaetomorpha aerea
Genomic DNA extraction
Sterile glass powder was prepared by crushing broken borosilicate glass to a fine powder using a mortar and pestle. Extraction buffer was prepared having the following composition 100 mM Tris–HCl, 20 mM EDTA, 1.5 M NaCl, 1% sarkosyl, 2% PVP and 0.2% β mercaptoethanol (added fresh just before extraction). Algal tissue (50 mg) was homogenized in a mortar and pestle with extraction buffer and sterile glass powder (50 mg). The ground mixture was centrifuged to remove cell debris at 14,200×g, 4 °C, for 10 min. Absolute ethanol (1/9 volume) and 3 M potassium acetate (pH 4.8) (1/4 volume) were added to the supernatant to remove polysaccharide contamination. One volume of chloroform: isoamyl alcohol (24:1) was added to the solution. The tube was vortexed vigorously for a few seconds and incubated at − 20 °C for 20 min with constant mixing. The tube was then centrifuged at 14,200×g, 4 °C for 20 min. RNase A (50 µg) was added to the aqueous phase, and the tube was gently mixed by inversion. The tube was incubated at 37 °C for 30 min. An equal volume of chloroform: isoamyl alcohol was added; the tube was vortexed vigorously for few seconds and incubated at − 20 °C for 20 min with constant mixing. The tube was centrifuged at 14,200×g, 4 °C for 20 min. Isopropanol (0.8 volume), 3 M sodium acetate (pH 5.2) (0.1 volume), and β mercaptoethanol (0.2%) were added to the aqueous phase. The tube was kept at − 80 °C for 1 h and centrifuged at 14,200×g, 4 °C for 20 min. The pellet was washed with 1 ml of 70% ethanol and centrifuged at 14,200×g, 4 °C for 10 min. The pellet was air-dried and dissolved in 50 µl of sterile distilled water.
Agarose gel electrophoresis of DNA
The extracted DNA was loaded on an agarose gel (1% w/v), stained with ethidium bromide (10 µg/mL) and subjected to electrophoresis at 100 V. After electrophoresis, the gel was visualized using gel documentation system (Gelstan 1012, Mediccare, India) to check the quality and integrity of the DNA.
Assessment of yield and purity of the extracted DNA
Concentration of the extracted DNA was determined using Biophotometer (Eppendorf, NY). The total yield of DNA was calculated using the following equation:
The purity of DNA was determined by measuring the absorbance ratio at A 260/A 280.
PCR amplification of universal plastid amplicon region
The extracted DNA was used as template for PCR amplification to assess its suitability for downstream applications. Universal Plastid Amplicon [UPA] (400 bp) region was selected as a reference gene for PCR amplification. To amplify the UPA sequences, the primers p23S1 (forward, 5′-GGACAGAAAGACCCTATGAA-3′) and p23S2 (reverse, 5′-TCAGCCTGTTATCCCTAGAG-3′) were used (Zhao et al. 2013). The PCR reaction was carried out using the following reaction conditions: initial denaturation of 94 °C for 2 min, followed by 35 cycles of 94 °C for 20 s, 55 °C for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 10 min. PCR products were run on an agarose gel (1% w/v) and visualized using gel documentation system (Gelstan 1012, Mediccare, India) after staining with ethidium bromide (10 µg/mL). The reaction without the template served as a non-template control (NTC).
Results
Genomic DNA extraction
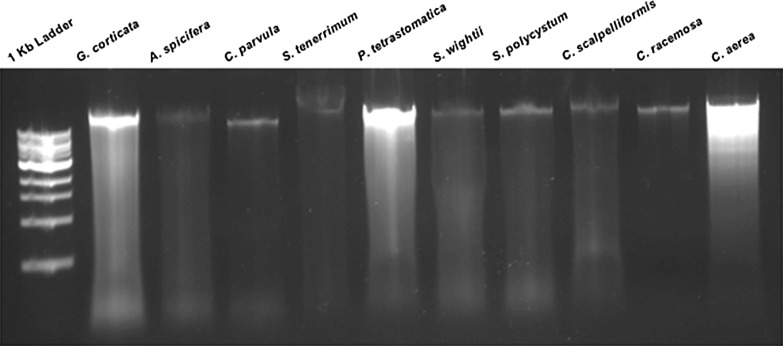
Genomic DNA was extracted using the developed method from representative samples of red, brown and green macroalgae. A distinct, intense band was seen on the agarose gel for all the species tested in this study (Fig. 2).
Fig. 2.
Genomic DNA isolated using the developed method from various macroalgal samples run on a 1% agarose gel
Yield and purity of genomic DNA
The concentration and purity of the extracted DNA was determined by spectrophotometric method. The total yield of DNA was calculated (Table 1). DNA yield varied between different genera of macroalgae. The DNA extracted from the red algae ranged from 1.73 to 6.5 µg/50 mg of biomass. Brown algae yielded DNA ranging from 2.2 to 5.4 µg/50 mg of biomass. DNA extracted from green algae ranged between 2.9 and 6.0 µg/50 mg of biomass. The purity of the extracted DNA ranged from 1.78 to 2.05 for all the samples.
Table 1.
Yield and purity of DNA isolated using the developed method from various macroalgal samples
| S. no. | Macroalgae | Classification | Concentration of DNA (ng/µL) | Purity A 260/A 280 | Yield (μg/50 mg of sample) |
|---|---|---|---|---|---|
| 1 | Gracilaria corticata | Rhodophyta | 130.6 ± 9.29 | 1.86 ± 0.01 | 6.5 ± 0.4 |
| 2 | Acanthophora spicifera | Rhodophyta | 34.6 ± 4.16 | 1.81 ± 0.03 | 1.73 ± 0.2 |
| 3 | Champia parvula | Rhodophyta | 37.33 ± 4.5 | 1.83 ± 0.10 | 1.86 ± 0.2 |
| 4 | Sargassum tenerrimum | Phaeophyceae | 45.33 ± 3.21 | 1.9 ± 0.05 | 2.2 ± 0.1 |
| 5 | Padina tetrastromatica | Phaeophyceae | 109.6 ± 6.5 | 1.80 ± 0.06 | 5.4 ± 0.32 |
| 6 | Sargassum wightii | Phaeophyceae | 87.66 ± 3.05 | 1.78 ± 0.01 | 4.38 ± 0.15 |
| 7 | Sargassum polycystum | Phaeophyceae | 81.6 ± 3.51 | 1.86 ± 0.09 | 4.0 ± 0.17 |
| 8 | Caulerpa scalpelliformis | Chlorophyta | 72 ± 3.6 | 1.95 ± 0.02 | 3.6 ± 0.18 |
| 9 | Caulerpa racemosa | Chlorophyta | 59.6 ± 2.88 | 2.05 ± 0.04 | 2.9 ± 0.14 |
| 10 | Chaetomorpha aerea | Chlorophyta | 120.5 ± 22.5 | 1.98 ± 0.05 | 6.0 ± 0.1 |
PCR amplification
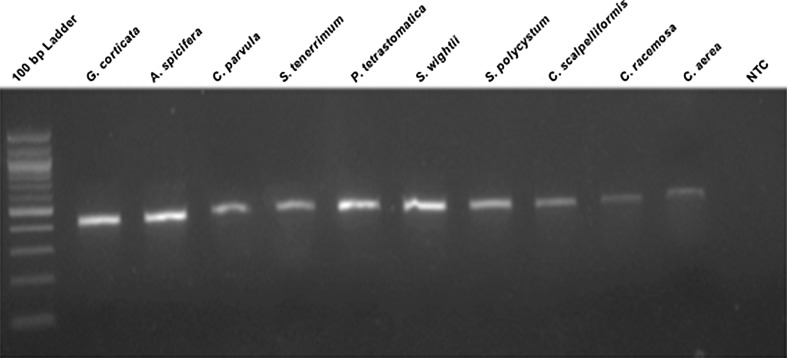
The extracted DNA was amplified using specific primers designed for the UPA region. The amplified products were separated and visualized on an agarose gel (1% w/v). The amplicons were seen as a distinct band at 400 bp from all the species of macroalgae. The non-template control showed no band (Fig. 3).
Fig. 3.
PCR amplification of UPA region of macroalgae samples run on a 1% agarose gel. NTC—non-template control
Discussion
Methods such as bead beating, detergent-based cell lysis, treatment with enzymes and grinding with liquid nitrogen have been employed for homogenization of macroalgae for DNA extraction and have proven to be effective (Joubert and Fleurence 2005; Wang et al. 2005; Hoarau et al. 2007; Greco et al. 2014). However, most enzymes are expensive and hence cannot be used for a large number of samples. Bead beating requires a thermomixer/bead beater and is a time-consuming process. Glass powder can be made from waste glassware in any laboratory and is economical when compared to the use of enzymes. Additionally, glass grinding is rapid and greatly shortens the time required for homogenization. Glass powder acts as an abrasive agent, resulting in complete maceration of algal tissue. In this study, glass powder together with extraction buffer was used during the homogenization step to enhance cell lysis, leading to the release of DNA into solution.
The method described in this study is rapid and comparable to other fast methods while yielding higher concentrations of DNA (Varela-Álvarez et al. 2006; Hoarau et al. 2007). It also requires just 50 mg of fresh biomass while other protocols require substantial quantities of biomass (Joubert and Fleurence 2005; Varela-Álvarez et al. 2006). Furthermore, our method has been standardized to work for different types of macroalgae, with representatives from Rhodophyta, Chlorophyta and Phaeophyceae.
The DNA extracted using this method yields an average of 3.36 µg of DNA for red algae, 3.9 µg for brown algae and 4.1 µg for green algae per 50 mg of biomass. The yield of DNA obtained was higher compared to several methods proposed previously (Wang et al. 2005; Varela-Álvarez et al. 2006). The variation in the yield of DNA could be attributed to the variation in morphological structure and chemical composition of the algae. The species which possess a soft, fleshy/leafy thallus are easily homogenized and hence released more DNA when ground with glass powder. The average purity of the DNA extracted is 1.88. These values suggest that the DNA is of good quality with low levels of contamination.
Although there was a distinct band of DNA observed on the gel image, there was also some amount of smearing which could be a result of shearing of the DNA. This could be attributed to the harshness of the method. However, the extracted DNA was still applicable for downstream applications. The UPA region of the DNA extracted from all the species was successfully amplified. These results suggest that the isolated DNA is of good quality and can be used for routine molecular biology experiments.
Acknowledgements
We would like to acknowledge the facility given by SRM University, Tamil Nadu, India, to carry out this project. The author Gautham Subramaniam Ramakrishnan acknowledges the GATE fellowship from SRM University for financial support.
Author contributions
Conceived and designed the experiments: MR. Performed the experiments: GSR and AAF. Analyzed the data: GSR, MR. Wrote the paper: GSR, MR.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Baghel RS, Reddy CRK, Jha B. Characterization of agarophytic seaweeds from the biorefinery context. Bioresour Technol. 2014;159:280–285. doi: 10.1016/j.biortech.2014.02.083. [DOI] [PubMed] [Google Scholar]
- Bruhn A, Dahl J, Bangsø H, Nikolaisen L, Rasmussen BM, Markager S, Olesen B, Arias C, Jensen PD. Bioenergy potential of Ulva lactuca: biomass yield, methane production and combustion. Bioresour Technol. 2011;102:2595–2604. doi: 10.1016/j.biortech.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GY, Coelho SM, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper FC, Lang D, Le Bail A, Leblanc C, Lerouge P, Lohr M, Lopez PJ, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli CA, Nelson DR, Nyvall-Collén P, Peters AF, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing SA, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder DC, Ségurens B, Strittmatter M, Tonon T, Tregear JW, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y, Wincker P. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Devi SG, Fathima AA, Radha S, Arunraj R, Curtis WR, Ramya M. A rapid and economical method for efficient DNA extraction from diverse soils suitable for metagenomic applications. PLoS One. 2015;10(7):1–16. doi: 10.1371/journal.pone.0132441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco M, Sáez C, Brown MT, Bitonti MB. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model brown alga. PLoS One. 2014;9(5):e96470. doi: 10.1371/journal.pone.0096470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G, Coyer J, Stam WT, Olsen JL. A fast and inexpensive DNA extraction/purification protocol for brown macroalgae: technical article. Mol Ecol Notes. 2007;7:191–193. doi: 10.1111/j.1471-8286.2006.01587.x. [DOI] [Google Scholar]
- Jin HJ, Kim JH, Sohn CH, DeWreede RE, Choi TJ, Towers GHN, Hudson JB, Hong Y. Inhibition of Taq DNA polymerase by seaweed extracts from British Columbia, Canada and Korea. J Appl Phycol. 1997;9:383–388. doi: 10.1023/A:1007925202219. [DOI] [Google Scholar]
- Joubert Y, Fleurence J. DNA isolation protocol for seaweeds. Plant Mol Biol Rep. 2005;23:197. doi: 10.1007/BF02772712. [DOI] [Google Scholar]
- Kazi MA, Reddy CRK, Jha B. Molecular phylogeny and barcoding of caulerpa (bryopsidales) based on the tufA, rbcL, 18S rDNA and ITS rDNA genes. PLoS One. 2013;8(12):e82438. doi: 10.1371/journal.pone.0082438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger T. The role of seaweeds in the modern ocean. Perspect Phycol. 2015;2:31–39. doi: 10.1127/pip/2015/0024. [DOI] [Google Scholar]
- Mabeau S, Kloareg B, Joseleau JP. Fractionation and analysis of fucans from brown algae. Phytochemistry. 1990;29:2441–2445. doi: 10.1016/0031-9422(90)85163-A. [DOI] [Google Scholar]
- Michel G, Tonon T, Scornet D, Cock JN, Kloareg B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 2010;188:82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- Mohan DS, Saraswathy M, Kurup M, Kurup G. Attenuation of hyperglycemia and hyperlipidemia in high calorie fed/streptozotocin—treated rats by hydromethanolic extract of Padina tetrastromatica. Bangladesh J Pharmacol. 2014;9(1):37–42. doi: 10.3329/bjp.v9i1.17153. [DOI] [Google Scholar]
- Patel S. Therapeutic importance of sulfated polysaccharides from seaweeds: updating the recent findings. 3. Biotech. 2012;2(3):171–185. [Google Scholar]
- Patra J, Das G, Baek KH. Chemical composition and antioxidant and antibacterial activities of an essential oil extracted from an edible seaweed, Laminaria japonica L. Molecules. 2015;20:12093–12113. doi: 10.3390/molecules200712093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji M, Rogers S, Stanhope M. Rapid isolation of high molecular weight DNA from marine macroalgae. Mar Ecol Prog Ser. 1992;84:197–203. doi: 10.3354/meps084197. [DOI] [Google Scholar]
- Varela-Álvarez E, Andreakis N, Lago-Lestón A, Pearson AG, Serrao EA, Procaccini G, Duarte CM, Marba N. Genomic DNA isolation from green and brown algae (Caulerpales and Fucales) for microsatellite library construction. J Phycol. 2006;42:741–745. doi: 10.1111/j.1529-8817.2006.00218.x. [DOI] [Google Scholar]
- Wallace RB, Gobler CJ. Factors controlling blooms of microalgae and macroalgae (Ulva rigida) in a eutrophic, urban estuary: Jamaica Bay, NY, USA. Estuar Coasts. 2015;38:519–533. doi: 10.1007/s12237-014-9818-1. [DOI] [Google Scholar]
- Wang G, Li Y, Xia P, Duan D. A simple method for DNA extraction from sporophyte in the brown alga Laminaria japonica. J Appl Phycol. 2005;17(1):75–79. doi: 10.1007/s10811-005-5557-9. [DOI] [Google Scholar]
- Zhao X, Pang S, Shan T, Liu F. Applications of three DNA barcodes in assorting intertidal red macroalgal flora in Qingdao, China. J Ocean U China. 2013;12:139–145. doi: 10.1007/s11802-013-2052-9. [DOI] [Google Scholar]