Abstract
Systemic Lupus Erythematosus (SLE) is a heterogeneous autoimmune disease with heightened disease severity in children. The incomplete understanding of the precise cellular and molecular events that drive disease activity pose a significant hurdle to the development of targeted therapeutic agents. Here, we performed single-cell phenotypic and functional characterization of pediatric SLE patients and healthy controls blood via mass cytometry. We identified a distinct CD14hi monocyte cytokine signature, with increased levels of monocyte chemoattractant protein-1 (MCP1), macrophage inflammatory protein-1β (Mip1β), and interleukin-1 receptor antagonist (IL-1RA). This signature was shared by every clinically heterogeneous patient, and reproduced in healthy donors’ blood upon ex-vivo exposure to plasma from clinically active patients only. This SLE-plasma induced signature was abrogated by JAK1/JAK2 selective inhibition. This study demonstrates the utility of mass cytometry to evaluate immune dysregulation in pediatric autoimmunity, by identification of a multi-parametric immune signature that can be further dissected to delineate the events that drive disease pathogenesis.
Keywords: Pediatric systemic lupus erythematosus, mass cytometry, monocytes, cytokines, MCP1, plasma circulating factors, type I interferons, JAK inhibition, ruxolitinib
INTRODUCTION
SLE is a highly morbid autoimmune disease characterized by heterogeneous clinical presentation and unpredictable disease activity [1]. Up to 20 percent of SLE patients are diagnosed as children younger than 16 years old. These pediatric patients typically have more severe disease than adults [2] underscoring the need to better understand immunopathogenesis in this population.
The immunopathogenesis of SLE mirrors its clinical heterogeneity—variably involving multiple cell types and plasma circulating mediators. Neutrophil death results in extrusion of neutrophil extracellular traps, which represent neoantigens for autoantibody formation [3]. Chromatin-containing immune complexes (ICs), free DNA and RNA, and cellular debris engage Toll-like-receptors (TLRs) 7 and 9 on plasmacytoid dendritic cells (pDCs), resulting in a type I interferon (IFN) signature seen in many pediatric and adult SLE patients [4]. The production of autoantibodies leading to the generation of ICs that co-engage TLRs and the B cell antigen receptor appears to amplify activation of autoreactive B cells [5]. Autoreactive T cells help B cells achieve full activation, differentiation, and isotype switching [6]. Thus, an integrated evaluation of how these apparently dysregulated cellular and molecular processes drive SLE disease activity has the potential to translate into improved therapeutic approaches.
The type I IFN signature has been repeatedly shown to correlate with active SLE [7–10]. This relationship has been suggested primarily by surrogate measures, specifically transcriptomic studies undertaken due to difficulty associated with measuring type I IFN proteins in SLE blood [11]. Findings regarding involvement of other cytokines in SLE have been conflicting. Some studies show that serum TNFα levels are elevated in SLE patients and correlate with disease activity, while others show the opposite, suggesting a protective role for TNFα [12,13]. Similarly, the data on involvement of specific immune cell types in SLE pathogenesis have also been conflicting. For example, in some studies the number of circulating regulatory T cells in SLE patients have been described as decreased, while other studies have shown that the numbers remain the same but the suppressive function is decreased [14,15]. This incomplete picture of SLE pathogenesis may be related to a study design that often focuses on one specific aspect of the immune system (a single cell type or cytokine), or systems-level transcriptomic approaches in the setting of complex cell mixtures. While these studies have been informative, they have provided an assessment of singular cellular and molecular elements, but not within the context of an integrated immune system with single-cell resolution.
To achieve a single-cell systems-level perspective of SLE immunopathogenesis that integrates dysregulated cellular and molecular interactions with clinical outcomes, we leveraged the high-dimensionality of mass cytometry. We simultaneously measured phenotypic and functional (cytokines) perturbations in pediatric SLE whole blood samples to understand how cellular and molecular perturbations may drive SLE disease activity. We applied an unsupervised hierarchical clustering algorithm and regression analysis. The analysis revealed a remarkably common monocyte cytokine signature shared among clinically heterogeneous pediatric SLE patients. To understand the immune mechanisms underlying this signature, we evaluated ex vivo the extent to which this signature was induced by plasma from SLE patients, and abrogated by selective cytokine signaling inhibitors. Plasma from clinically active SLE patients only (and not those in remission) induced the monocyte cytokine signature in healthy donor peripheral blood, to the same extent as seen in those patients’ blood. Selective JAK inhibition abrogated the SLE plasma-induced monocyte cytokine signature, but type I IFN receptor blockade did not. This study represents a proof of principle for the application of mass cytometry and complementary computational tools to understand mechanisms of immune dysregulation in pediatric autoimmune disorders, with potential therapeutic applications.
RESULTS
1. Mass cytometry analysis of newly diagnosed and treatment naive pediatric SLE patients demonstrates a shared distinct monocyte cytokine signature (MCP1/Mip1β/IL1RA)
The underlying immunopathogenesis of SLE involves activation of multiple innate and adaptive cell subsets, and plasma circulating soluble factors such as ICs and pro-inflammatory cytokines, which alter lymphocyte activation and eventually cause organ damage [5,16]. Based on the premise that in SLE immune perturbations involve multiple cell types and cytokines, we used mass cytometry to systematically monitor phenotypic (22 surface markers) and functional (16 cytokines) immune parameters in pediatric SLE patients, with single-cell granularity (Figure 1).
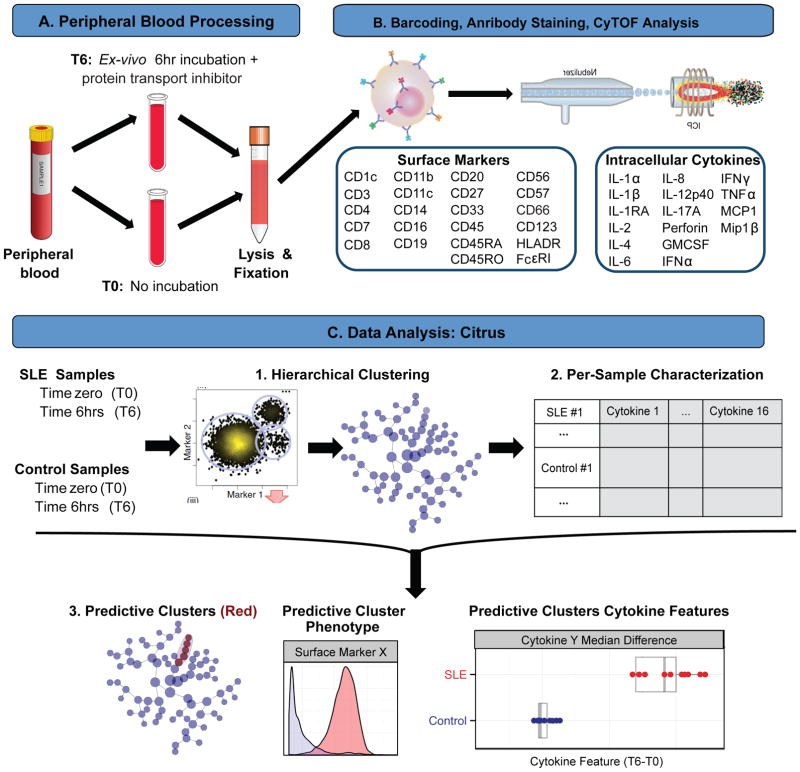
Figure 1. Experimental workflow for mass cytometry analysis of phenotypic and functional immune parameters in pediatric SLE.
A. Peripheral blood samples underwent RBC lysis and fixation immediately following collection (T0), or after incubation at 37°C with a protein transport inhibitor for 6 hours (T6). B. Lysed/fixed samples were barcoded and stained with a panel of antibodies against 22 surface markers and 16 cytokines, and analyzed by CyTOF. C. CyTOF data were analyzed using Citrus (cluster identification, characterization, and regression). CD45+ cells from all samples were combined and clustered by unsupervised hierarchical clustering using 22 cell surface markers (1). Descriptive features (cytokines) of clustered samples are calculated on a per-sample basis, and used in conjunction with additional experimental data (SLE vs. control, T6-T0) to train a regularized regression model predictive of experimental endpoint (SLE vs. control) (2). Predictive clusters are phenotypically characterized based on clustering parameters and plotted as a function of experimental endpoint and descriptive features (3) (adapted from Bruggner et al., 2014).
Peripheral blood samples were collected from 10 newly diagnosed and treatment naïve pediatric SLE patients, and 10 age and gender-matched healthy controls. Every SLE patient met ACR diagnostic criteria [17] (Table 1). Exclusion criteria for SLE patients included prior history of immunosuppression, suspected malignancy or immunodeficiency, and concurrent infection (further details in Methods). Exclusion criteria for healthy controls included chronic medication usage, suspected underlying immunodeficiency, autoimmunity, malignancy, and/or concurrent immunosuppressive therapy (further details in Methods, Table S1).
Table 1.
Summary of laboratory and clinical information for 10 newly diagnosed untreated SLE patients
| SLE Patient |
Gender | Age (Yrs) |
ESR | C3, C4 | ANA | Anti- dsDNA |
Other AutoAbs |
Malar rash |
Photo sens |
Heme | Renal | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #121 | Female | 15 | 58 | Low | >1:1280 | 1:640 | APL, histone, SS-A, SS-B, Smith | Yes | Yes | Platelets, ALC | No | Psychosis |
| #122 | Male | 12 | Normal | Very low | 1:80 | 1:640 | None | No | No | ALC | Class IV | No |
| #123 | Female | 16 | Normal | Normal | 1:80 | Negative | Direct Coombs | No | No | Anemia | No | Cerebritis, Pleuritis, Arthritis |
| #124 | Female | 15 | >140 | Very low | 1:640 | >1:1280 | APL, SS-A, direct Coombs | Yes | No | Platelets, ALC, Anemia | Class IV | Oral ulcers |
| #125 | Female | 12 | 22 | Normal, low | 1:640 | 1:160 | APL | Yes | Yes | No | No | Arthritis |
| #126 | Female | 12 | 40s | Very low | 1:5120 | 1:2560 | SS-A, SS-B, cardiolipin, APL | Yes | Yes | ALC, Anemia | No | Arthritis |
| #127 | Male | 13 | 56 | Very low | 1:320 | >1:1280 | RNP | No | No | No | Class IV | No |
| #128 | Female | 13 | >140 | Very low | 1:320 | 1:80 | SS-A, Smith, RNP, | No | No | ALC, Anemia | Class IV | Pleuritis, colon vasculitis |
| #129 | Male | 9 | >140 | Very low | 1:5120 | 1:160 | Smith | Yes | No | ALC, Anemia | No | Oral ulcers, Arthritis |
| #130 | Female | 12 | 90s | Very low | 1:320 | 1:2560 | Smith | Yes | Yes | No | Class IV | Arthritis |
Abbreviations: ESR, erythrocyte sedimentation rate; C3/C4, complement components 3 & 4; ANA, anti nuclear antibody titer; APL, antiphospholipid antibodies; RNP, ribonucleotide protein; SS-A/Ro, SS-B/La; ALC, absolute lymphocyte count.
Peripheral blood was processed within 30 minutes of collection, to remain as close as possible to in vivo conditions. To internally control for individual variability, for each study participant, peripheral blood underwent red blood cell (RBC) lysis and fixation either immediately following collection (time zero, T0, Figure 1A) or after 6 hours of incubation with a protein transport inhibitor cocktail to prevent cytokine secretion (time 6 hours, T6, Figure 1A). This T6 condition reflects the in vivo “baseline” intrinsic cytokine perturbations in SLE patients’ blood, as no exogenous stimuli were added. Measurements of cell frequency and cytokine production were analyzed relative to the corresponding participant’s time zero measurements (T6-T0), ensuring that each participant served as his/her own control to account for intra- and inter-individual differences. Samples were barcoded using a combination of palladium isotope mass tags to decrease technical variability[18], pooled and stained with antibodies recognizing 22 surface proteins and 16 cytokine proteins, and processed for mass cytometry (Methods, Figure 1B). Our previous studies validated all of the surface marker and cytokine antibodies used in this assay [19].
Surface markers were chosen to delineate lymphoid and myeloid cell subsets previously described in SLE pathology, such as B and T cell subsets [20], and plasmacytoid dendritic cells (pDCs) [21] (Figure S1). Evaluated cytokines included those with a pro-inflammatory role, such as the IL-1 family, IL-6, TNFα [22–24], type I and II IFNs [21,25–27], IFN-regulated chemokines [28,29], and IL-17 [30,31]. Data were analyzed via traditional hand-gated strategies (Figure S1) and with the unsupervised hierarchical clustering algorithm named Citrus (Figure 1C).
Mass cytometry leverages high-dimensional single-cell analysis, which affords the ability to detect phenotypical and functional disease-relevant cells with minimal bias when applying unsupervised computational data analysis tools [32]. To comprehensively explore SLE-induced immune perturbations across multiple immune cell types and cytokines, we analyzed CD45+ cells from every SLE and healthy control blood sample using Citrus [33] (Figure 1C). Citrus distilled multidimensional CyTOF data from every patient and healthy control, from T0 and T6 conditions, to a hierarchy of related clusters based on 22 surface markers. It then split the clustered data into individual sample components and calculated features (arcsinh median differences for cytokines at T6-T0 conditions) that describe each cluster on a per-sample basis. Citrus uses a regularized regression model predictive of the experimental endpoint (PAM, Prediction Analysis of Microarrays) to calculate the minimum number of cluster features that best classify the analyzed samples into the correct category (disease vs. control), based on a false discovery rate (FDR) of <1% (Figure 1C).
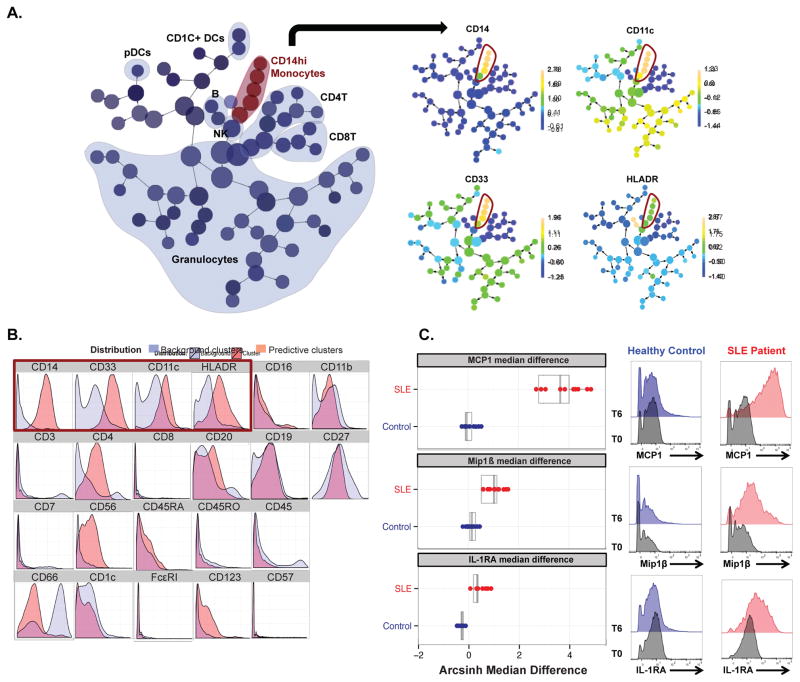
Cluster-specific regularized regression analysis of cytokine changes (T6-T0) identified clusters with phenotypic characteristics of activated CD14hi monocytes (CD66−CD3−CD19−CD7−CD33+CD11c+HLADR+ CD14hiCD16loCD4+) (Figure 2A, 2B) as those demonstrating cytokine features predictive of the SLE disease category. These cytokine features consisted of increased levels of the cytokine signature MCP1/Mip1β/IL-1RA (FDR adjusted q-value <0.01), in SLE patients compared to healthy controls, with median arcsinh T6-T0 differences of 3.851±0.753 (SD) for MCP1, 1.193±0.37 for Mip1β, and 0.32±0.11 for IL-1RA (Figure 2C, left panel). Conversion of arcsinh median differences to absolute fold changes can be found in Table S2 [34]. Cytokine histograms from hand-gated CD14hi monocyte populations (guided by phenotypic characteristics of predictive clusters in Figure 2C) demonstrate the increased production of MCP1, Mip1β, and IL-1RA in one representative SLE patient, but not in the healthy control counterpart (Figure 2C, right panel). Notably, the predictive CD14hi monocyte cluster population frequency did not significantly differ between SLE patients and healthy controls (Figure S2), demonstrating that the increased production of MCP1, Mip1β, and IL-1RA reflect functional differences between SLE and healthy control monocytes.
Figure 2. Clinically heterogeneous pediatric SLE patients share a distinct monocyte cytokine signature (MCP1, Mip1β, and IL-1RA).
A. Visual representation of unsupervised hierarchical clustering, “Citrus tree,” of all CD45+ immune cells from 10 SLE patients and 10 gender-matched control samples at conditions T0 and T6 (n=20 study subjects, 40 samples). (Left) Major immune cell types are contoured based on expression of canonical lineage markers. (Right) CD14hi monocytes phenotypic clusters are delineated. A regularized regression model (pamr) was applied to identify clusters (red contour) with intracellular cytokine features predictive of SLE (FDR adjusted q-value<0.01). B. Phenotypic characteristics of predictive clusters. Histograms for each surface marker show expression within the predictive cluster (red) compared to the background clusters (blue). Histograms enclosed in red box represent lineage markers of CD14hi monocytes. C. (Left) Box plots for MCP1, Mip1β, and IL-1RA median differences (T6-T0) within CD14hi monocytes (clusters in red) are shown as predictive of SLE patient category, when taken together as a signature (FDR adjusted q-value <0.01). Each colored dot represents a single SLE patient (red) or control (blue) and their respective T6-T0 arcsinh median difference for the specified cytokine. (Right) Histograms show expression of MCP1, Mip1β, and IL-1RA at T0 and T6 on manually gated CD14hi monocytes from one representative SLE patient and one gender-matched control.
Citrus analysis of pediatric SLE patient samples produced results consistent with previous literature reports describing monocyte abnormalities in SLE patients [35–37]. These Citrus results expanded our previous work on the study of intracellular cytokine production, which was analyzed via manual gating approach. This cluster-specific regularized regression allowed for the analysis of “multi-cytokine positivity,” as opposed to “single-cytokine” analysis based on the manual gating approach [19]. Our analysis expanded on the identification of monocyte functional alterations, particularly the dysregulated production of MCP1, Mip1β, and IL-1RA at disease “baseline,” in the absence of any exogenous stimuli or immunomodulatory treatment. The MCP1/Mip1β/IL-1RA signature was induced in 10/10 clinically heterogeneous SLE patients studied, but in none of the healthy controls, underscoring the pathogenic role of CD14hi monocytes in dysregulated cytokine networks in SLE patients.
2. Plasma circulating factors from newly diagnosed treatment naive pediatric SLE patients consistently induce the monocyte cytokine signature in healthy donor peripheral blood cells
Peripheral whole blood analysis of pediatric SLE patients via mass cytometry enabled the identification of a CD14hi monocyte cytokine signature in the context of plasma circulating factors. Plasma from SLE patients have been previously shown to have immune activating properties, such as the ability to induce monocyte differentiation into DCs [38], and pDCs production of type I IFNs [39,40]. Hence, we aimed to understand the role of plasma circulating factors from these patients in the induction of the monocyte cytokine signature.
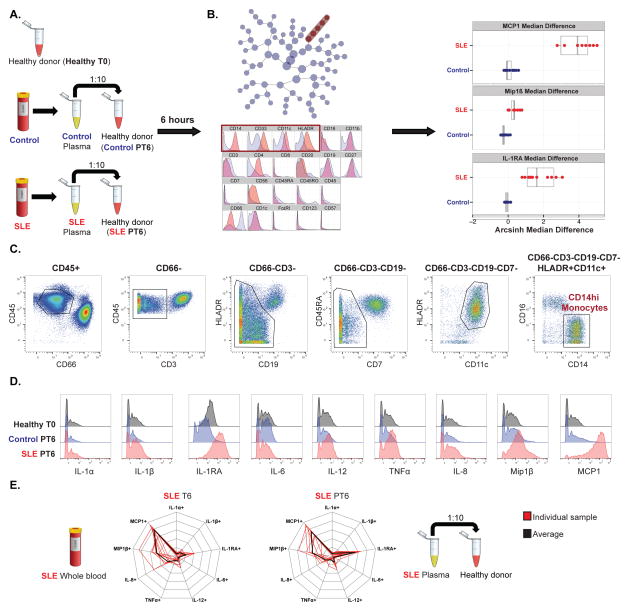
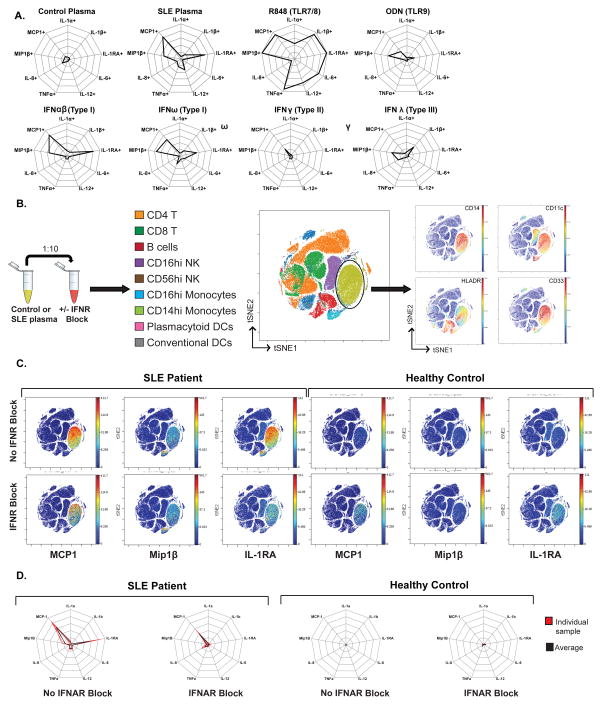
We incubated plasma from each of the 10 pediatric SLE patients and matched controls (separately) with one healthy donor peripheral blood sample (distinct from healthy controls used in Figure 2), and analyzed immune parameters by mass cytometry (Figure 3A). Citrus analysis of intact healthy donor blood (Healthy T0), and healthy donor blood incubated with control or SLE plasma (Control PT6, SLE PT6), resulted in the identification of a set of clusters predictive of SLE disease category, again with phenotypic characteristics of activated CD14hi monocytes with increased production of MCP1 (3.875±1.02), Mip1β (0.375±0.11) and IL-1RA (1.775±1.004) (Figure 3B), as was seen in peripheral blood samples from the same SLE patients (Figure 2B, 2C). Cytokine histograms from the hand-gated CD14hi monocyte population, guided by Citrus predictive clusters phenotypic characteristics (Figure 3C), demonstrated the induction of the MCP1/Mip1β/IL-1RA cytokine signature in healthy donor blood exposed to plasma from the SLE patient only (Figure 3D). The unique monocyte cytokine signature observed in the pediatric SLE patients is consistently reproduced in healthy donor blood exposed to the same patients’ plasma, though IL-1RA was induced to a greater degree by SLE patients’ plasma as compared to the findings in the SLE patients’ peripheral blood (Figure 3E).
Figure 3. Plasma from pediatric SLE patients induces the distinct monocyte cytokine signature (MCP1, Mip1β, and IL-1RA) in healthy donor blood.
A. Healthy donor peripheral blood sample (healthy T0), healthy donor blood sample incubated with plasma from either controls (control PT6) or SLE patients (SLE PT6) for 6 hours (with protein transport inhibitor), were processed and analyzed as described in Figure 1. B. (Left) Unsupervised hierarchical clustering “Citrus tree” based on expression of 22 surface markers constructed from CD45+ cells from healthy donor blood, healthy donor blood incubated separately with plasma from 10 SLE patients and 10 controls (n=20 plasma samples, one negative control, 21 samples). (Right) A regularized regression model (pamr) was applied to identify clusters (red) with intracellular cytokine features predictive of SLE vs. control category. Phenotypic characterization and cytokine features for predictive clusters are shown. C. Gating strategy for identification of predictive clusters of CD14hi monocytes, based on phenotypic features characterized by Citrus in B. D. Histograms showing expression of 9 cytokines by CD14hi monocytes for one healthy donor (healthy T0), and the same healthy donor incubated with plasma from one representative healthy control (Control PT6) and SLE patient (SLE PT6) pair. E. Radar plots showing the unique monocyte cytokine signature. Cytokine signatures are represented as radar plots with 20% radial intervals. Cells demonstrating cytokine production levels higher than the 95th percentile of “baseline” condition (time zero for SLE whole blood, healthy donor blood with no plasma for SLE PT6, respectively) were defined as cytokine positive. (Left) Data derived from SLE patient samples peripheral whole blood. (Right) Data derived from SLE patient plasma samples incubated with healthy donor blood.
Of note, the induction of the monocyte cytokine signature was independent of gender differences between the plasma samples and the healthy donor blood with which they were incubated. Additionally, the signature was reproducible in 4 distinct healthy donors (2 male and 2 female). These healthy donors were different individuals and had a different gender than those from whom plasma samples were acquired, eliminating alloreactivity as an underlying variable in the observed result (Figure S3). Furthermore, the absence (serum) or presence (plasma) of fibrinogen was not a variable factor in the induction of this cytokine signature, as both plasma and serum from the same patients induced the monocyte cytokine signature in healthy donor blood (Figure S4).
While “reverse transcriptomic” approaches measuring global transcriptional responses in healthy donor blood cells exposed to patient plasma in vitro have been done before [10,41], a “reverse single-cell targeted proteomic” approach, evaluating the changes in cytokine protein production across multiple immune cell types, has not. This global single-cell proteomic analytical approach demonstrated a statistically significant and distinct monocyte cytokine signature in pediatric SLE patients’ blood, which was reproduced in healthy donor blood exposed to plasma from these same patients (but not of healthy controls). These data support the previously demonstrated role of CD14hi monocytes in SLE immunopathogenesis [41,42], and also uncover a unique disease-relevant MCP1/Mip1β/IL-1RA cytokine signature induced by plasma circulating factors.
3. The distinct monocyte cytokine signature is induced by plasma from clinically active SLE patients only
To further understand the disease relevance of the monocyte cytokine signature, we evaluated the ability of plasma from patients at different clinical disease states to induce this signature. To this end, we used a retrospectively collected cohort of 18 pediatric SLE patients, at 4 different disease timepoints (n=72). We assessed the capability of these patients’ plasma samples to induce the monocyte cytokine signature in healthy donor blood. The first timepoint was collected at the diagnosis visit prior to any immunosuppressive treatment (new diagnosis or Dx—clinically active state); the succeeding timepoints were collected either at routine follow up visits during clinical inactivity (remission—clinically inactive state) or acute visits for flare episodes (flare—clinically active state). Flare episodes met ACR definition of SLE disease exacerbations [17] (Methods, see Table S3 for clinical details). In the 18-patient cohort, there were 7 patients who had one flare episode. Clinically active disease category included new diagnosis (n=18) and flare (n=7) timepoints (n=25 total). The remainder of the timepoints were collected at clinical remission (n=47). Clinically active and inactive disease status was defined by ACR guidelines (Methods). Gender and age-matched healthy controls (n=10) from one timepoint only were also analyzed (different from initial set of healthy controls in Figures 1–3, see Methods for details on these healthy control plasma).
We incubated the aforementioned plasma samples with healthy donor blood and analyzed them via mass cytometry as in Figure 1 (Figure 4A). We used this approach to assess the presence/absence of the monocyte cytokine signature with active/inactive disease states, given that 1) plasma samples demonstrated different states of disease activity and 2) the reproducibility of the induction of the monocyte cytokine signature by the SLE patients’ plasma (Figure 3). To gain an objective view of the relationship between the monocyte cytokine signature and clinical disease activity of these patients, analysis of the mass cytometry data was initially performed blinded to the patients’ clinical disease status.
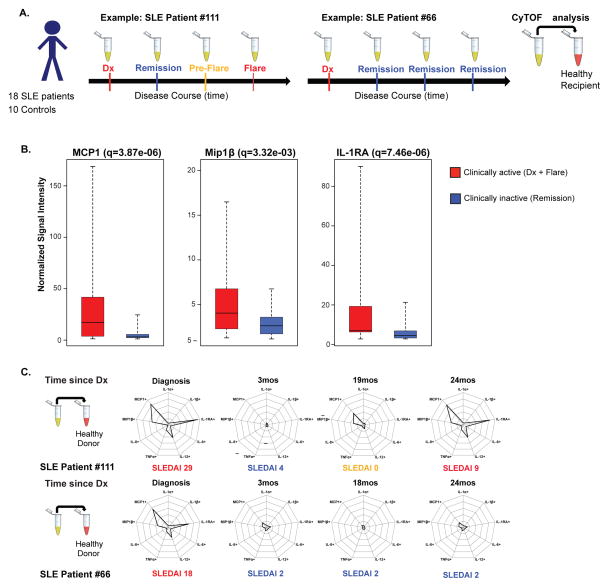
Figure 4. The monocyte cytokine signature (MCP1, Mip1β, and IL-1RA) fluctuates with clinical disease activity.
A. Experimental workflow. SLE patient plasma were collected at diagnosis (no therapy), and every 3–6 months thereafter (on therapy), including flare episodes (patient #111), or only clinical remission timepoints (patient #66). Plasma from 4 timepoints for each SLE patient and 1 timepoint from each healthy age-matched control were incubated separately with healthy donor blood, followed by analysis via CyTOF as in Figure 1. B. For all 18 SLE patients and their timepoints, gated on CD14hi monocytes, box plots represent minimum and maximum (dotted line), median (middle line within box), and first and third quartile signal intensity values (box end values). Values on the Y-axis denote signal intensity normalized against the baseline of healthy donor blood without any plasma incubation (see Table S4 for all values). Clinically active timepoints (new diagnosis and flare) for all 18 patients are grouped (red, 18 new diagnosis and 7 flares, n=25). Clinically inactive timepoints (remission) for all 18 patients are grouped (blue, n=47). Comparison for all 16 cytokines between these two categories was performed using a generalized estimating equation (GEE) approach, and FDR-adjusted q-values were calculated and shown above. C. Cytokine signatures of CD14hi monocytes depicted using radar plots. Each radial axis in the radar plot represents a cytokine, and each radial interval represents 20% positivity for the cytokine, based on 95th percentile threshold at healthy T0. Radar plots depict the monocyte cytokine signature for each timepoint for SLE patients #111 and #66. Associated SLEDAI scores for each timepoint are indicated below each respective radar plot.
The CD14hi monocyte population was hand-gated guided by Citrus predictive clusters phenotypic characteristics (Figure 3C), and all 16 cytokines were evaluated. Comparison of cytokine production between healthy donor blood incubated with control and SLE plasma samples demonstrated differences in MCP1, Mip1β, and IL-1RA production only (see Figure S5 for data on other cytokines). Since Figures 2 and 3 supported statistically significant differences for MCP1, Mip1β, and IL-1RA between newly diagnosed SLE patients and healthy controls, we sought to make an equivalent comparison between the clinically active (red, Dx + Flare) and inactive (blue, Remission) samples from the 18-patient plasma sample cohort. Comparison of the clinically active (n=25) and inactive category (n=47) demonstrated again statistically significant differences for MCP1 (q=3.87e-06), Mip1β (q=3.32e-03), and IL-1RA (q=7.46e-06), corroborating our previous findings (q=FDR-adjusted q-values) (Figure 4B). Medians, first and third quartile, minimum and maximum values for each of the comparisons and cytokines analyzed above can be found in Table S4.
The CD14hi monocyte cytokine signature fluctuates significantly between clinically active and inactive disease status, whether the clinical activity is present at new diagnosis in the absence of any treatment, or during a flare episode while on maintenance treatment. In Figure 4C, analysis of the cytokine signature in two patient samples, one with a flare episode and one without, illustrate the fluctuation of the cytokine signature with clinical disease activity within a single patient. In patient #111, the monocyte cytokine signature has a robust presence at time of diagnosis (treatment naive) and flare (on maintenance treatment), recedes to healthy control levels at time of remission, but re-appears variably at “pre-flare” timepoint independently of SLEDAI score. The emergence (of intermediate values) of this signature during “pre-flare” timepoints was challenging to evaluate given the limited number of flares in this study patient cohort. Future longitudinal studies focusing on patients’ primary peripheral blood cells, with more frequent peripheral blood sampling (every 3 months), and a larger patient cohort are needed to capture increased number of flares.
The data shown in Figure 4 indicate that the magnitude of the monocyte cytokine signature induced in healthy donor blood fluctuated with clinical disease status (active vs. inactive) of the SLE patient from whom the plasma was obtained. This result demonstrates a correlation between the presence of “pathogenic” plasma factors and clinically active disease state, suggesting that these “pathogenic” factors drive disease activity and induce the monocyte cytokine signature. Hence, identification of a strategy for removal or neutralization of the downstream effects (e.g., cytokine induction) of SLE plasma immune activating factors may provide an avenue toward modifying disease activity, as an adjunctive therapy to current immunomodulators. Additionally, analysis of the mechanism by which these “pathogenic” plasma factors induce the monocyte cytokine signature may also provide novel therapeutic insights.
4. Type I IFNs are necessary but not sufficient to induce the monocyte cytokine signature
Plasma from SLE patients contain several circulating factors that have been shown to perpetuate disease pathology; among these are nucleic acids and other cellular debris that trigger pattern recognition receptors such as TLR7 and TLR9 [43–45], pro-inflammatory cytokines such as type I and type II IFNs [21,25–27], and nucleic acid-autoantibody immune complexes [40,46,47]. To evaluate the ability of individual plasma circulating factors to induce the monocyte cytokine signature, we treated healthy donor blood with TLR7/8 agonist (resiquimod, R848) and TLR9 agonist (oligodeoxnucleotide, ODN) to mimic endogenous nucleic acid and cellular debris, and human recombinant type I, II, or III IFNs to mimic pro-inflammatory states. Stimulated blood samples were analyzed via mass cytometry as in Figure 1. TLR7/8 agonist induced multiple pro-inflammatory cytokines including MCP1, Mip1β, and IL-1RA. TLR9 agonist induced MCP1, Mip1β, and IL-1RA to a much lesser degree than SLE plasma. IFNγ induced primarily MCP1, while IFNλ induced Mip1β, IL-1β, and IL-8. Type I IFNs, including IFN α, β, and ω induced a cytokine signature in CD14hi monocytes characteristic of the SLE plasma-induced signature (Figure 5A). This result is consistent with previous reports based on RNA microarray analysis and serum cytokine measurements in SLE patients, which demonstrated that chemokines such as MCP1 and Mip1β are regulated by type I IFN [8,48]. The effect of TLR7/8 agonist R848 in the induction of MCP1, Mip1β, and IL-1RA is likely exerted by direct activation of NF-κB pathway, given that in vitro stimulation of CD14hi isolated monocytes with R848 also induces MCP1 and IL-1RA (Figure S6). While in vitro these observations demonstrate the different (and separate) roles/processes of TLR7/8 agonists and type I IFNs in the activation of CD14hi monocytes and their induction of the monocyte cytokine signature (NF-κB and interferon-stimulated genes, respectively), there is likely a mixed phenomenon in vivo within the patient.
Figure 5. The monocyte cytokine signature is partially abrogated by IFNAR blockade.
A. Plasma from an SLE patient, a matched control, or TLR ligands R848 and ODN2006, recombinant human type I (α,β, ω), type II (γ), and type III (λ) interferons were incubated with healthy donor blood and analyzed via CyTOF as described in Figure 1. Gated on CD14hi monocytes, cytokine signatures are represented as radar plots with 20% radial intervals. Cells demonstrating cytokine production levels higher than the 95th percentile of unstimulated sample (healthy donor blood only) were defined as cytokine positive. All plots obtained from analysis of one healthy donor blood sample. B. Plasma from 10 healthy controls and 10 SLE patients (patient cohort from Figure 2) were separately incubated with healthy donor blood that was either pre-treated or not with IFNAR blocking antibody (20ug/ml). These healthy donor blood samples were analyzed by CyTOF as described in Figure 1. This dataset was analyzed by t-SNE, based on 22 surface markers. Each dot in the visNE map represents an individual cell. visNE maps show distinct clusters representing different cell types, based on signal intensity localization of surface markers defining each cell population. CD14hi monocytes were localized to the area of visNE map (light green) based on CD14, CD11c, HLADR, and CD33 among other markers. In each of these panels, the same visNE map is shown, colored sequentially by the labeling intensity of each of the surface markers indicated on the panel. See Figure S7 for visNE maps for each of the 22 surface markers. One representative donor shown. C. Each visNE map shown demonstrates the signal intensity for each cytokine indicated, with or without IFNAR pre-treatment. The signal intensity for each cytokine indicated is localized to the CD14hi monocyte population, as shown in B. One representative SLE patient and healthy control pair shown. D. Radar plots showing the SLE-plasma induced monocyte cytokine signature and its changes with IFNAR blockade. For individuals (red) and average (black) shown in each plot. Hand-gated on CD14hi monocyte population, cytokine signatures are represented as radar plots with 20% radial intervals. Cells demonstrating cytokine production levels higher than the 95th percentile of “baseline” condition (healthy donor blood with IFNAR) were defined as cytokine positive. Data is derived from SLE patient (Left) or Control (Right) plasma samples incubated with healthy donor blood.
Because type I interferons, particularly IFNα, have been implicated as key pathogenic cytokines in SLE [8,48,49]), several anti-type I IFN therapeutics have been recently developed and are currently being evaluated in clinical trials. The anti-IFNα monoclonal antibodies (mAbs), sifalumab and rontalizumab, have completed phase II clinical trials [50,51]. While the clinical trials revealed that the safety profile was acceptable and that there was successful suppression of the IFN signature, reduction of clinical disease activity was not consistent [52,53]. We evaluated the effect of IFNα inhibition on the monocyte cytokine signature ex vivo, by pre-treatment of plasma from clinically active SLE patients (from the initial newly diagnosed untreated SLE patient cohort) with an anti- IFNα IgA antibody, prior to incubation with healthy donor blood. We found that pre-treatment with anti-IFNα mAb did not consistently abrogate the monocyte cytokine signature (Figure S7). This inconsistent effect is likely related to the mAb’s inability to block all 13 types of IFNα and other type I IFNs such as β, and ω, thus leading to remaining type I IFN activity. This lack of cross reactivity across type I IFNs also likely explains the inconsistent clinical disease activity improvement observed in the clinical trials with anti-IFNα mAbs. To bypass this issue, a mAb against the receptor common to all type I IFNs (IFNAR) is also currently being studied in clinical trials. Anifrolumab has recently entered phase III clinical trials for the treatment of active SLE (clinicaltrials.gov, NCT02446899). We evaluated the effect of IFNAR blockade on the monocyte cytokine signature ex vivo. Healthy donor blood was pre-treated (or not) with an anti-IFNAR mAb prior to incubation with plasma from clinically active SLE patients from the same initial SLE patient cohort. Processed samples were analyzed via mass cytometry as in Figure 1.
We performed analysis with viSNE on cells from healthy donor blood incubated (separately) with SLE plasma, with or without pre-treatment with IFNAR blocking mAb. visNE is a visualization tool based on the t-SNE (t-distributed stochastic neighbor embedding) algorithm that maps multi-parameter relationships of cellular data into two dimensions in an unsupervised manner [54]. Cells were mapped by viSNE using 22 surface markers (Figure 1). As expected, major cell subsets were identified largely based on expression of canonical surface markers (Figure 5B, Figure S8). Based on the clusters assembled by the viSNE graph, we evaluated cytokine production across multiple immune cell subsets. In this manner, we were able to assess the induction of the monocyte cytokine signature by SLE plasma via a single-cell principal component-based analysis algorithm, validating our previous findings via Citrus (Figure 3). Additionally, this analysis also allowed for the evaluation of off-target effects of IFNAR blockade, as it allows exploration of phenotypic and cytokine changes probed by 22 surface markers and 16 cytokines (Figure 1). Indeed, SLE plasma induced the monocyte cytokine signature, with production of MCP1, Mip1β, and IL-1RA by CD14hi monocytes, while the healthy control plasma did not (Figure 5C). The IFN blockade partially abrogated the SLE plasma-induced monocyte cytokine signature with significant decrease in the induction of IL-1RA (65.03%±16.27% with no IFNAR, 5.48%±2.3% with IFNAR) and to a lesser degree MCP1 (73.13%±2.7 with no IFNAR, 40.1%±15.4% with IFNAR) (Figure 5C, 5D). IFNAR blockade did not result in changes to other cytokines induction (Figure S9). Given that several reports have also described the role of type II IFNs in SLE pathogenesis [26,27], we also assessed IFNγ receptor (IFNGR) blockade, which did not abrogate the cytokine signature by itself, but did additively abrogate the monocyte cytokine signature when used in combination with IFNAR (Figure S10). This result indicates that type I IFNs are necessary for the induction of the monocyte cytokine signature, but other plasma circulating factors, such as other cytokines and TLR agonists, also play a role in the induction of this signature.
5. Ruxolitinib abrogates the SLE plasma-induced monocyte cytokine signature
The expression of IFN-induced genes (and hence the production of IFN-inducible chemokines and other proteins) is not only stimulated by Type I IFNs. Engagement of other signaling pathways, including TLR and other patterns recognition receptors such as cytosolic RNA helicases RIG-I, can also induce IFN-dependent genes [55,56]. Additionally, type II IFNs, or IFNγ, have also been implicated in SLE pathogenesis [57,58]. In Figure 5A, while type I IFNs induced the monocyte cytokine signature most closely resembling the signature induced by SLE plasma, TLR 7/8/9 also induced MCP1, Mip1β, and IL-1RA to different extents, and type II and type III IFNs also induced MCP1. This observation suggests that TLR agonists, particularly those that are endoplasmic, and type II and type III IFNs may also play a role in the induction of IFN-stimulated response elements, namely MCP1. Therefore, the blockade of signaling pathways downstream of TLRs and IFNs may provide a more broad abrogation of the SLE plasma-induced monocyte cytokine signature.
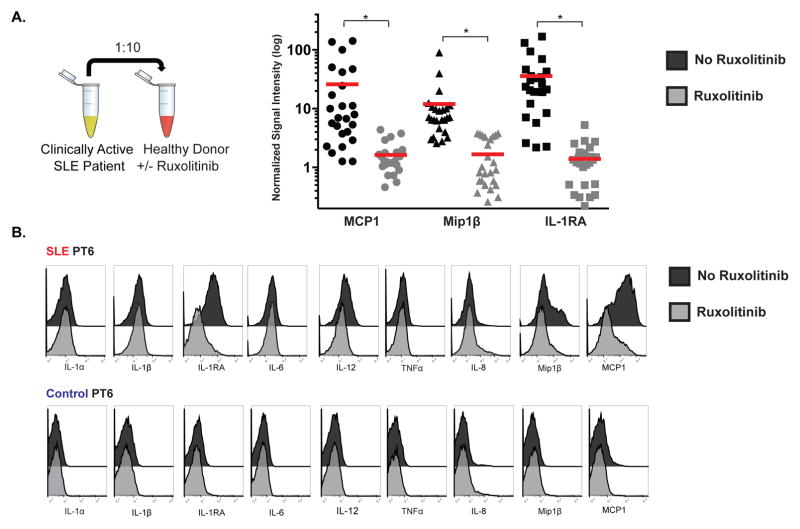
Type I, type II and type III IFNs signal through different receptor complexes, but their downstream signaling pathways overlap. Type I and type III IFNs signal through JAK1/Tyk2, and type II IFNs signal through JAK1/JAK2 [59]. Given that IFNAR blockade only partially abrogated the monocyte cytokine signature, we pursued inhibition of signaling proteins downstream IFNAR, such as JAK1. Additionally, inhibition of this JAK/STAT pathway disrupts signaling of other cytokines involved in SLE pathogenesis. To this end, we evaluated the effect of selective JAK1 and JAK2 inhibition on the monocyte cytokine signature. Healthy donor blood was pre-treated or not with ruxolitinib, an FDA-approved selective JAK1 and JAK2 inhibitor, prior to incubation with plasma from clinically active SLE patients (n=25, from patient cohort in Figure 4). Processed samples were analyzed via mass cytometry as in Figure 1, and CD14hi monocytes were hand-gated as in Figure 3C. SLE plasma samples incubated with healthy donor blood that was pre-treated with ruxolitinib demonstrated significant reductions in the induction of MCP1 (from 47.22±40.02 to 1.48±1.015 with ruxolitinib treatment), Mip1β (from 6.144±4.17 to 2.063±1.147), and IL-1RA (from 19.3±20.74 to 1.161±1.403) (p<0.0001 for each cytokine) (Figure 6A). Additional cytokines measured were not affected (Figure S11). In Figure 6B, a single SLE and healthy control pair is represented, demonstrating that ruxolitinib specifically abrogates the SLE-plasma induced signature in CD14hi monocytes, with no effect on other cytokines analyzed.
Figure 6. Ruxolitinib, a selective JAK1 and JAK2 inhibitor, abrogates the monocyte cytokine signature.
A. Plasma from clinically active timepoints from 18 SLE patients (18 new diagnosis and 7 flares, n=25) were incubated with healthy donor blood that was either pre-treated or not with ruxolitinib (2uM). These healthy donor blood samples were analyzed by CyTOF as described in Figure 1. MCP1, Mip1β, and IL-1RA signal intensity values (normalized against the baseline of healthy donor blood without any plasma incubation) for each sample are plotted in the presence or absence of ruxolitinib treatment (gated on CD14hi monocytes). Scale for signal intensity values is log transformed. p-values were calculated using Mann-Whitney U test for each cytokine category, comparing ruxolitinib untreated (black) vs. treated (grey). p<0.0001 (*) for every comparison. B. Cytokine histogram overlays illustrate the effects of JAK inhibition on MCP1, Mip1β, and IL-1RA expression. Data were from one representative SLE and one matched control. Analyzed cells were gated on CD14hi monocytes.
We used mass cytometry to evaluate IFNAR blockade and JAK inhibitors, emerging therapies for SLE [60–62], using pediatric human primary samples ex-vivo. We provided immunological mechanistic parameters (immunophenotypic and cytokine read outs) to evaluate targeting of IFNAR and JAK/STAT signaling pathways in SLE. In this study, using mass cytometry to evaluate immune dysregulated cytokines in pediatric SLE, we defined a distinct monocyte cytokine signature (MCP1/Mip1β/IL-1RA) that was induced by plasma circulating factors, and abrogated by ruxolitinib, providing a systems immunology platform to study immune dysregulated mechanisms in pediatric SLE pathogenesis.
DISCUSSION
Unsupervised clustering and regression analysis (Citrus) of immune cell frequency distributions and corresponding intracellular cytokine production in pediatric SLE blood samples demonstrated a monocyte cytokine signature characterized by increased production of MCP1, Mip1β, and IL-1RA (Figure 2). Remarkably, despite the variability in organ system involvement, inflammatory marker values, autoantibody profile, and SLEDAI scores in the clinically heterogeneous pediatric SLE patients studied (Table 1), the monocyte cytokine signature (MCP1/Mip1β/IL-1RA) was observed in every patient. Consistent with previous literature, we demonstrated that monocytes from SLE patients and healthy controls did no exhibit phenotypic or population frequency differences [63] (Figure 2, Figure S2).
Evaluation of newly diagnosed, treatment naïve patients allowed us to study immunopathology at the peak of disease, and eliminated confounding effects of therapeutic heterogeneity. While females have a strong predilection for development of SLE compared to males [64], data regarding gender-based differences in disease course, outcomes, and pathogenesis are not as consistent, particularly among pediatric patients [65]. No obvious differences in the monocyte cytokine signature or disease severity were seen in the two males in our study group (data not shown), however with only two male individuals our study was not designed nor powered to detect any differences.
Because cytokine dysregulation is pervasive in SLE pathogenesis, this study and numerous others have focused on cytokine expression. Although the type I IFN signature has been convincingly correlated with active disease [7–10], elevated IFNα can be detected in sera from only about 50% of pediatric SLE patients, while microarray analysis of PBMCs indicated that the majority (>95%) of children with mild to severe disease display a type I IFN signature [8]. This finding may be due to limitations in systemic detection of local production of type I IFNs, quick uptake of these cytokines by effector cells, and limitations in the detection range of ELISA-based assays. Challenges in detection of increased levels of IFNα by intracellular cytokine staining in SLE patients may be due to the low frequency of pDCs in peripheral circulation (0.01%–0.05% of circulating immune cells)[66], and the even lower frequency of pDCs engaged with a pathogenic immune complex via TLR7 and/or TLR9 ligation. Hence, studies focusing on type I IFNs are often conducted via measurement of IFN-induced genes, which also possesses the advantage of evaluating genes induced by all type I IFNs, including different types of IFNα, IFNβ and IFNω. Consistent with previous findings [8], our study did not detect statistically significant elevated levels of IFNα cytokine protein measured intracellularly in pediatric SLE patient blood samples (Figures 2, 3, S5).
While patient PBMCs (peripheral blood mononuclear cells) have been used in many studies to evaluate immune dysregulation in SLE, we chose to use peripheral whole blood samples. Whole blood samples represent a physiologically relevant ‘medium’ to study SLE because whole blood more completely represents the disease-related immune state; whole blood includes non-mononuclear blood cells which are often involved in disease (e.g. neutrophils, platelets), as well as plasma circulating factors (e.g. nucleic acids, ICs, and cytokines) which have immune activating roles in SLE [38–40]. Thus, we examined the immune activating properties of plasma from SLE patients to understand the underlying mechanisms that lead to the MCP1/Mip1β/IL1RA signature. We found that the cytokine signature was induced in CD14hi monocytes from healthy donors after incubation with SLE patient plasma, but not with age and sex-matched control plasma (Figure 3). While stimulation of healthy donor cells with SLE plasma has been previously demonstrated to induce monocyte apoptosis [67], monocyte differentiation into dendritic cells [63], stimulate IL-10 and IL-6 production by PBMCs [22], and induce transcription of IFN responsive genes [21,68], SLE plasma had not been shown to induce MCP1, Mip1β, and IL-1RA production by healthy CD14hi monocytes.
This monocyte cytokine signature induced by plasma from newly diagnosed patients pre-treatment was also induced by plasma from flaring patients on maintenance therapy. Remarkably, we were able to detect the monocyte cytokine signature in both newly diagnosed untreated patients and those who were already on maintenance therapy but were flaring, when we stimulated healthy donor blood with SLE patient plasma (Figure 4B). This signature was extinguished in patients who were clinically inactive. Though we clearly showed that the SLE-plasma induced signature recedes during clinical remission, our study cohort did not contain enough pre-flare and flare episodes to allow for its evaluation as a prognostic indicator. Further longitudinal studies are necessary to rigorously evaluate the flare predictive value of this signature.
To further investigate the mechanism by which SLE plasma induced the monocyte cytokine signature, we evaluated how the immunomodulation of the IFN pathway could abrogate this signature. There are currently several phase II clinical trials investigating IFNα blockade (clinicaltrials.gov, NCT01283139) as well as a phase III trial studying IFNAR blockade (clinicaltrials.gov, NCT02446899) for the treatment of SLE. Examination of the effect of these agents on the monocyte cytokine signature demonstrated that the ex vivo experimental data correlated with the in vivo clinical trials data. We showed that mAbs against IFNα were not consistent in abrogating the signature, while mAb against IFNAR resulted in consistent partial abrogation of the signature (Figure 5). In the clinical trials, anti-IFNα biologics have demonstrated inconsistent improvement in clinical activity, while anti-IFNAR demonstrates a more uniform improvement across patients [52,53].
Given that type I IFNs, TLRs and other pro-inflammatory cytokines involved in SLE pathogenesis signal through the JAK/STAT pathways, we examined the effect of JAK inhibition on the monocyte cytokine signature. JAK inhibitors have recently been incorporated into the therapeutic arsenal employed against autoimmune diseases, particularly rheumatoid arthritis [69]. Tofacitinib, a selective JAK1/JAK3 inhibitor, has been shown to reduce symptoms in active rheumatoid arthritis [70,71]. Pre-treatment of healthy donor blood with ruxolitinib, an FDA-approved selective JAK1/JAK2 inhibitor, before incubation with clinically active SLE plasma, demonstrated complete abrogation of the monocyte cytokine signature (Figure 6). The safety and efficacy of JAK inhibition in SLE are under current investigation (clinicaltrials.gov, NCT02535689, phase Ib); however, our mass cytometry approach provides a physiologically relevant (whole blood) ex vivo analysis platform to test novel therapies such as ruxolitinib, and to evaluate off-target effects, since multiple immune cell types and cytokines are evaluated simultaneously.
In conclusion, the application of mass cytometry to study pediatric SLE immunopathogenesis allowed for an integrated evaluation of cellular and molecular events that drive disease activity. This approach identified a distinct common dysregulated cell type and its “pathogenic” cytokine production, in spite of clinical variability. Further evaluation of the role of this signature in SLE pathogenesis demonstrated that 1) it was induced by plasma circulating factors of clinically active patients only, and 2) it was abrogated by selective JAK inhibition, providing a mechanistic insight with potential therapeutic implications.
MATERIALS AND METHODS
1. Study participants
SLE patients with presentation prior to age 18 years old who met the American College of Rheumatology revised criteria for classification of SLE[72] were recruited at the Pediatric Rheumatology Clinic at Stanford Children’s Health and Lucile Packard Children’s Hospital Stanford. Age appropriate consent and assent were obtained. Gender-matched controls were recruited from the Pediatric Rheumatology, and Allergy and Immunology clinics at Stanford Children’s Health. Patients who were clinically ruled out for autoimmune, immunodeficient, or allergic disorders; and who were not treated with anti-histamines or other immunomodulatory medications at the time of initial clinical evaluation, were considered for study enrollment as controls. Inclusion and exclusion criteria for gender-matched controls can be found in Table S1. All human donors were enrolled under a study protocol approved by the Institutional Review Board of the Research Compliance Office at Stanford University.
Data from Figures 2–3 were generated from prospective enrollment of 10 pediatric SLE patients and 10 age and gender-matched healthy controls. Peripheral blood (for CyTOF analysis and plasma isolation) was collected at initial diagnosis prior to the initiation of any immunosuppressive treatment. Patient demographics and characteristics at time of collection are available in Table 1. Patient exclusion and inclusion criteria are detailed in Table S1.
Data from Figures 4–6 was generated from plasma samples retrospectively collected from 18 pediatric SLE patients (IRB protocol number 13952), and purchased age- and gender- matched healthy controls plasma samples from Biodesign International Inc. (Saco, ME, USA) (n = 10). Peripheral blood samples were collected for plasma isolation only at multiple timepoints starting from diagnosis and every 3–6 months thereafter during routine follow-up clinical visits and/or acute sick visits. Patient demographics and clinical characteristics, including complement levels, anti-double-stranded DNA titers, nephritis status, and modified SELENA (Safety of Estrogen in Lupus Erythematosus National Assessment) SLEDAI scores were collected (Table S3). SELENA SLEDAI scores were calculated including serology and complement data as per Petri et al., 1999 [17]. Flare episodes were defined as a change in SLEDAI ≥3 points, new or worsening clinical manifestations, increased prednisone dose or additional therapy, physician’s global assessment score ≥1.0 or hospitalization for SLE, based on the SELENA trial definition of flare [17].
2. Whole blood sample processing and stimulations
SLE patient or gender-matched control peripheral whole blood was collected into heparinized vacutainers (BD). Blood volume was divided for CyTOF analysis and plasma isolation. For CyTOF analysis, blood samples were fixed with Phosflow lyse/fix buffer (BD 558049) either immediately after collection (T0); or after incubation at 37C, mixed 1:1 with RPMI 1640 (Gibco 21870076) plus protein transport inhibitor cocktail (eBioscience 00-4980-93), for 6 hours (T6). Lysed/fixed cells were stored at −80C, and were thawed on the day of barcoding and staining. To decrease technical variability, palladium isotopes were used in different combinations for mass tag barcoding of separate samples, pooled in sets of 20, surface stained in a single tube with a metal-labeled antibody panel, then permeabilized with Perm/Wash buffer I (BD 558050) to facilitate intracellular staining. Barcoding methodology was adapted from Zunder and Finck et al, 2015 [18]. Protocols for intracellular cytokine staining (ICS) assays were adapted from previous studies [73,74].
For experiments involving incubation of plasma with healthy donor experiments (Control PT6, SLE PT6), four different healthy donor blood samples from Stanford Blood Center were used, demonstrating comparable results (Figure S3). Plasma isolated from SLE patients and healthy controls were separately incubated with healthy donor blood (mixed 1:1 with RPMI) at a 1:10 (plasma to blood+RPMI) volume ratio for 6 hours at 37C with a protein transport inhibitor cocktail. Blood samples were lysed/fixed, barcoded, and stained as indicated above. For IFNα blockade experiments, plasma from clinically active SLE patients was either untreated or pre-treated with an IgA neutralizing monoclonal antibody against human IFNα at several concentrations of 100ng/ml, 1ug/ml and 10ug/ml (Invivogen maba-hifna-3) for 60 minutes at 37C, prior to plasma transfer to healthy donor blood protocol as described above. For the IFNAR1 blockade experiments, the plasma transfer protocol described above was performed with healthy donor blood that was either untreated, or pre-treated with a blocking monoclonal antibody against human interferon alpha/beta receptor 1 (20 μg/mL; PBL 21385-1) for 90 minutes at 37C prior to plasma transfer. Blocking anti-IFNAR1 antibody titration was performed at 10 μg/mL and 20 μg/mL concentrations (data not shown). For the IFNGR blockade experiments, the plasma transfer protocol described above was performed with healthy donor blood that was either untreated, or pre-treated with a blocking monoclonal antibody against human interferon gamma receptor 1 (2 μg/ml, R&D Systems # MAB6732-100) for 90 minutes at 37C prior to plasma transfer. Blocking anti-IFNGR antibody titration was performed at 0.5 μg/ml, 1 μg/ml, and 2 μg/ml concentrations (data not shown). For ruxolitinib treated experiments, the plasma transfer protocol described above was performed with healthy donor blood that was either untreated, or pre-treated with 2μM ruxolitinib for 30 minutes at 37C prior to plasma transfer. Ruxolitinib dosage titration was performed at 0.5, 2, 5, and 10 μM concentrations (Figure S12).
For IFN and TLR ligand stimulation experiments, healthy donor blood samples from Stanford Blood Center were treated separately with each of the following: recombinant human IFNα (25,000 units/ml; PBL 11100–1), IFNβ (25,000 units/ml; PBL 11410–2), IFNω (100 ng/ml, 1 μg/ml, and 10 μg/ml; Sigma SRP3061-100 UG), IFNγ (1 μg/ml), and IFNλ (50 ng/ml, 500 ng/ml, 5 μg/ml; Sigma SRP3059-20UG), R848 (1 μg/ml; Invivogen tlrl-r848), and ODN2006 (50 μM; Invivogen tlrl-2006) by incubation for 6 hours at 37C in a mixed ratio of 1:1 with RPMI plus protein transport inhibitor cocktail. Blood samples were lysed/fixed, and processed as indicated above.
3. Mass cytometry analysis
Clone, vendor, and conjugation information for all mAbs used in these studies are shown in Table S5. This antibody panel was previously validated in O’Gorman and Hsieh et al., 2015 [19]. Barcoding reagents were prepared according to the procedures described in Bodenmiller et al., 2012 [75]. Timepoints from the same patient were barcoded and processed simultaneously for antibody staining. To protect against potential batch effects, all findings were quantified as relative changes between time points (all reported values were normalized to a signal from its same barcode plate). All decisions regarding which patients were barcoded together, and all staining, mass cytometry analysis, and quantification of changes were performed blinded to the patients’ clinical disease activity. In addition, immune features used for regression analysis predictive of experimental endpoint (SLE vs. control) were derived blinded to clinical disease activity.
Stained cells were analyzed on a mass cytometer (CyTOF, Fluidigm) at an event rate of 400 to 500 cells per second. To make all samples maximally comparable, data were acquired using internal metal isotope bead standards and normalized as previously described [76]. Files were debarcoded using the Matlab Debarcoder Tool. Gating was performed using Flowjo (Treestar) and Cytobank. An inverse hyperbolic sine transformation (arcsinh) was applied to analyze intracellular cytokine differences between T6 and T0, and PT6 and T0, as previously described [34].
4. Citrus analysis
For Figures 2 and 3, regularized regression analyses of molecular features derived from mass cytometry data and clinical disease activity were performed using Citrus, a method for unsupervised identification of cellular responses associated with a clinical outcome [33].
4.1 Clustering
Hierarchical clustering using Ward’s linkage and Euclidean distance was performed as described by Bruggner et al. [33], 2014, on CD45+ cells using R. Cells were clustered on the basis of the expression of CD1c, CD3, CD4, CD7, CD8, CD11b, CD11c, CD14, CD16, CD19, CD20, CD27, CD33, CD45RA, CD45RO, CD56, CD57, CD66, CD123, HLADR, and FcERI. Clusters were based on 10,000 events from each patient sample. Clusters containing at least 2% of all clustered cells are graphically displayed. Cluster plots (“Citrus tree”) depict the clustering hierarchy; nodes are scaled on the basis of frequency of cells in that cluster. For Figure 2, data from SLE patients and matched controls, at T0 and T6, were included in the same PAM analysis, and were clustered together to enable comparison of clusters between conditions. For Figure 3, data from SLE patients and matched controls plasma transfers with healthy donor, at healthy T0 (no plasma incubation), SLE PT6, and Control PT6, were included in the same PAM analysis, and were clustered together to enable comparison of clusters between conditions. Repeated runs of the analysis with identical parameters confirmed that results were reproducible.
4.2
Regularized and shrinkage method analysis of molecular parameters and class prediction for SLE disease vs. control significant changes in cell frequency and cytokine production were inferred with PAM[77], using the “pamr” package in R. PAM is a statistical technique for class prediction using nearest shrunken centroids. The method of nearest shrunken centroids identifies subsets of features that best characterize each class, based on an estimate of FDR derived from permuting the observed data. This test was selected because it considers multiple hypotheses testing, makes few assumptions about the distribution of the underlying data, and has been validated for use on high-dimensional biological data. Significance was inferred for an FDR <1% (q < 0.01).
5. visNE analysis
viSNE analyses were performed using the Cytobank implement (https://www.cytobank.org/). Each file was pre-gated on CD45+CD66− cells. Subsampling of CD45+CD66− events was performed per visNE algorithm, analyzing 25,000 events per sample. CD1c, CD3, CD4, CD7, CD8, CD11b, CD11c, CD14, CD16, CD19, CD20, CD27, CD33, CD45RA, CD45RO, CD56, CD57, CD66, CD123, HLADR, and FcERI were used to create the t-sNE axes of the visNE map. Expression levels of each surface marker protein were normalized by the maximum value of the channel within each donor (Figure S8). CD14himonocytes were gated as CD66−CD3−CD19−CD7−CD33+CD11bhiCD11c+HLADR+ CD14hiCD16lo (Figure 5B, S8).
6. Statistical analyses of monocyte cytokine signature
For Figures 4, 5A, 5D and 6, we used data from the hand-gated CD14hi monocyte population only (Figure 3C). In Figure 4B, for the CD14hi monocyte population, for each of the 16 cytokines, we tested whether there is any difference in the signal intensity between the clinically active (new diagnosis and flare) and clinically inactive (remission) patients. A generalized estimating equation (GEE) approach was used for this purpose. The GEE approach was chosen because it can reflect the longitudinal nature of the study without very stringent parametric assumptions [78,79]. A gamma family was chosen due to the non-negative signal intensity values and positive skewing in the data. The robust variance estimation was used. We controlled for multiple hypothesis testing using the control of false discovery rate (FDR) at 5%. In Figures 5D and 6A, significant changes in cytokine production were calculated using two-tailed Mann-Whitney U test on hand-gated CD14hi monocyte data (Figure 3C). Mann-Whitney U test was selected because it tests the null hypothesis that two samples come from the same population against an alternative hypothesis; however, unlike the t-test, the Mann-Whitney U test does not require a normal distribution (non-parametric).
Supplementary Material
HIGHLIGHTS.
Mass cytometry analysis of immune dysregulation in pediatric SLE identifies a distinct commonly shared monocyte cytokine signature, despite clinical heterogeneity
The monocyte cytokine signature is induced by plasma circulating factors from clinical active SLE patients only
The monocyte cytokine signature is partially abrogated by type I interferon receptor blockade and completely abrogated by selective JAK1/JAK2 inhibition
Acknowledgments
We would like to thank David B. Lewis, Joseph D. Hernandez, and Aimee Pugh-Bernard for their intellectual input and helpful comments.
1. Funding
E.W.Y.H. was a fellow of the Pediatric Scientist Development Program while she was at Stanford University. She was supported by award number K12-HD000850 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the Lucile Packard Foundation for Children’s Health, Stanford CTSA UL1 TR001085, the ITI Young Investigator award, and Child Health Research Institute of Stanford University. She is currently supported by the Boettcher Foundation Webb-Waring Early Investigator Award. I.M.B. was supported by the NIH grant K08 AI080945, the Stanford Child Health Research Institute, and the Arthritis Foundation Postdoctoral Fellowship. This work is supported by funds from NIH grants U19AI057229, U19AI090019, U54CA149145, T32AI007290, N01HV00242, 1U19AI100627, 5R01AI07372405, R01CA184968, 1R33CA183654, R33CA183692; NIH-Baylor Research Institute 41000411217; NIH-Northrop Grumman Corp 7500108142; California Institute for Regenerative Medicine DR101477; Food and Drug Administration HHSF223201210194C; Bill and Melinda Gates Foundation OPP 1017093; the European Commission HEALTH.2010.1.2-1; Alliance for Lupus Research; Entertainment and Industry Foundation NWCRA grant; the U.S. Department of Defense OC110674, 11491122; and Howard Hughes Medical Institute.
2. Competing interests
Funding sources had no involvement in study design, collection, analysis or interpretation of data; in writing, or in the decision to submit the article for publication.
ABBREVIATIONS
- TLR
Toll like receptor
- IC
Immune complexes
- JAK/STAT
Janus kinase/Signal transducer and activator of transcription
- ODN
oligodeoxynucleotide
- R848
Resiquimod
- MCP1
Monocyte chemotactic protein 1
- Mip1β
Macrophage inflammatory protein 1β
- IL-1RA
IL-1 receptor antagonist
- TNFα
Tumor necrosis factor α
- IFNα
Interferon α
- mAb
Monoclonal antibody
Footnotes
AUTHOR CONTRIBUTIONS
W.E.O and E.W.Y.H. conceived the study, developed the reagents, performed experiments, analyzed data, and wrote the manuscript. D.S.K. performed experiments and assisted in data analysis. E.W.Y.H. and I.M.B. recruited SLE patients and healthy controls. I.M.B. also provided intellectual guidance and expertise in immunobiology and autoimmunity. C.R. B., P.R., and D.G. assisted with statistical analysis. M.M.D. and G.P.N. provided intellectual guidance on this project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Haddon DJ, Diep VK, Price JV, Limb C, Utz PJ, Balboni I. Autoantigen microarrays reveal autoantibodies associated with proliferative nephritis and active disease in pediatric systemic lupus erythematosus. Arthritis Res Ther. 2015;17:162. doi: 10.1186/s13075-015-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Science Translational Medicine. 2011;3:73ra19–73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obermoser G, Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19:1012–1019. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings DJ, Schwartz MA, Jackson SW, Meyer-Bahlburg A. Integration of B cell responses through Toll-like receptors and antigen receptors. Nat Rev Immunol. 2012;12:282–294. doi: 10.1038/nri3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busser BW, Adair BS, Erikson J, Laufer TM. Activation of diverse repertoires of autoreactive T cells enhances the loss of anti-dsDNA B cell tolerance. J Clin Invest. 2003;112:1361–1371. doi: 10.1172/JCI18310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USa. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and Granulopoiesis Signatures in Systemic Lupus Erythematosus Blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A Modular Analysis Framework for Blood Genomics Studies: Application to Systemic Lupus Erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiche L, Jourde-Chiche N, Pascual V, Chaussabel D. Disease Mechanisms in Rheumatology-Tools and Pathways: Current Perspectives on Systems Immunology Approaches to Rheumatic Diseases. Arthritis & Rheumatism. 2013;65:1407–1417. doi: 10.1002/art.37909. [DOI] [PubMed] [Google Scholar]
- 11.Morimoto AM, Flesher DT, Yang J, Wolslegel K, Wang X, Brady A, et al. Association of endogenous anti-interferon-α autoantibodies with decreased interferon-pathway and disease activity in patients with systemic lupus erythematosus. Arthritis & Rheumatism. 2011;63:2407–2415. doi: 10.1002/art.30399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez D, Correa PA, Gómez LM, Cadena J, Molina JF, Anaya JM. Th1/Th2 cytokines in patients with systemic lupus erythematosus: is tumor necrosis factor alpha protective? Semin Arthritis Rheum. 2004;33:404–413. doi: 10.1016/j.semarthrit.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Lit LCW. Raised plasma concentration and ex vivo production of inflammatory chemokines in patients with systemic lupus erythematosus. Annals of the Rheumatic Diseases. 2006;65:209–215. doi: 10.1136/ard.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venigalla RKC, Tretter T, Krienke S, Max R, Eckstein V, Blank N, et al. Reduced CD4+,CD25− T cell sensitivity to the suppressive function of CD4+,CD25high,CD127 −/low regulatory T cells in patients with active systemic lupus erythematosus. Arthritis & Rheumatism. 2008;58:2120–2130. doi: 10.1002/art.23556. [DOI] [PubMed] [Google Scholar]
- 15.Chavele KM, Ehrenstein MR. Regulatory T-cells in systemic lupus erythematosus and rheumatoid arthritis. FEBS Lett. 2011;585:3603–3610. doi: 10.1016/j.febslet.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 16.Li TT, Zhang T, Chen GM, Zhu QQ, Tao JH, Pan HF, et al. Low level of serum interleukin 27 in patients with systemic lupus erythematosus. J Investig Med. 2010;58:737–739. doi: 10.231/JIM.0b013e3181d88f7b. [DOI] [PubMed] [Google Scholar]
- 17.Petri M, Buyon J, Kim M. Classification and definition of major flares in SLE clinical trials. Lupus. 1999;8:685–691. doi: 10.1191/096120399680411281. [DOI] [PubMed] [Google Scholar]
- 18.Zunder ER, Finck R, Behbehani GK, Amir EAD, Krishnaswamy S, Gonzalez VD, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nature Protocols. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Gorman WE, Hsieh EWY, Savig ES, Gherardini PF, Hernandez JD, Hansmann L, et al. Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol. 2015;136:1326–1336. doi: 10.1016/j.jaci.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Good-Jacobson KL, Song E, Anderson S, Sharpe AH, Shlomchik MJ. CD80 expression on B cells regulates murine T follicular helper development, germinal center B cell survival, and plasma cell generation. The Journal of Immunology. 2012;188:4217–4225. doi: 10.4049/jimmunol.1102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.BLANCO P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 22.Rönnelid J, Tejde A, Mathsson L, Nilsson-Ekdahl K, Nilsson B. Immune complexes from SLE sera induce IL10 production from normal peripheral blood mononuclear cells by an FcgammaRII dependent mechanism: implications for a possible vicious cycle maintaining B cell hyperactivity in SLE. Annals of the Rheumatic Diseases. 2003;62:37–42. doi: 10.1136/ard.62.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripley BJM, Rahman MA, Isenberg DA, Latchman DS. Elevated expression of the Brn-3a and Brn-3b transcription factors in systemic lupus erythematosus correlates with antibodies to Brn-3 and overexpression of Hsp90. Arthritis & Rheumatism. 2005;52:1171–1179. doi: 10.1002/art.21000. [DOI] [PubMed] [Google Scholar]
- 24.Mageed RA, Isenberg DA. Tumour necrosis factor alpha in systemic lupus erythematosus and anti-DNA autoantibody production. Lupus. 2002;11:850–855. doi: 10.1191/0961203302lu306rr. [DOI] [PubMed] [Google Scholar]
- 25.Rönnblom LE, Alm GV, Oberg KE. Autoimmunity after alpha-interferon therapy for malignant carcinoid tumors. Ann Intern Med. 1991;115:178–183. doi: 10.7326/0003-4819-115-3-178. [DOI] [PubMed] [Google Scholar]
- 26.Harigai M, Kawamoto M, Hara M, Kubota T, Kamatani N, Miyasaka N. Excessive production of IFN-gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B. The Journal of Immunology. 2008;181:2211–2219. doi: 10.4049/jimmunol.181.3.2211. [DOI] [PubMed] [Google Scholar]
- 27.Karonitsch T, Feierl E, Steiner CW, Dalwigk K, Korb A, Binder N, et al. Activation of the interferon-gamma signaling pathway in systemic lupus erythematosus peripheral blood mononuclear cells. Arthritis & Rheumatism. 2009;60:1463–1471. doi: 10.1002/art.24449. [DOI] [PubMed] [Google Scholar]
- 28.Bauer JW, Petri M, Batliwalla FM, Koeuth T, Wilson J, Slattery C, et al. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: A validation study. Arthritis & Rheumatism. 2009;60:3098–3107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Q, Chen X, Cui H, Guo Y, Chen J, Shen N, et al. Association of elevated transcript levels of interferon-inducible chemokines with disease activity and organ damage in systemic lupus erythematosus patients. Arthritis Res Ther. 2008;10:R112. doi: 10.1186/ar2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crispín JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. The Journal of Immunology. 2008;181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doreau A, Belot A, Bastid J, Riche B, Trescol-Biemont MC, Ranchin B, et al. Interleukin 17 acts in synergy with B cell-activating factor to influence B cell biology and the pathophysiology of systemic lupus erythematosus. Nature Immunology. 2009;10:778–785. doi: 10.1038/ni.1741. [DOI] [PubMed] [Google Scholar]
- 32.Aghaeepour N, Nikolic R, Hoos HH, Brinkman RR. Rapid cell population identification in flow cytometry data. Cytometry A. 2011;79:6–13. doi: 10.1002/cyto.a.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci USa. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendall SC, Simonds EF, Qiu P, Amir EAD, Krutzik PO, Finck R, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu MF, Li JS, Weng TH, Lei HY. Differential expression and modulation of costimulatory molecules CD80 and CD86 on monocytes from patients with systemic lupus erythematosus. Scand J Immunol. 1999;49:82–87. doi: 10.1046/j.1365-3083.1999.00452.x. [DOI] [PubMed] [Google Scholar]
- 36.Katsiari CG, Liossis SNC, Sfikakis PP. The pathophysiologic role of monocytes and macrophages in systemic lupus erythematosus: a reappraisal. Semin Arthritis Rheum. 2010;39:491–503. doi: 10.1016/j.semarthrit.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Alzawawy A, Zohary M, Ablordiny M, Eldalie M. Estimation of monocyte-chemoattractantprotein-1 (Mcp-1) level in patients with lupus nephritis. Int J Rheum Dis. 2009;12:311–318. doi: 10.1111/j.1756-185X.2009.01429.x. [DOI] [PubMed] [Google Scholar]
- 38.BLANCO P, PALUCKA A, Pascual V, Banchereau J. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine & Growth Factor Reviews. 2008;19:41–52. doi: 10.1016/j.cytogfr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lövgren T, Eloranta ML, Båve U, Alm GV, Rönnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis & Rheumatism. 2004;50:1861–1872. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 40.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, et al. Toll-like receptor 9–dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nature Immunology. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 41.Joo H, Coquery C, Xue Y, Gayet I, Dillon SR, Punaro M, et al. Serum from patients with SLE instructs monocytes to promote IgG and IgA plasmablast differentiation. Journal of Experimental Medicine. 2012;209:1335–1348. doi: 10.1084/jem.20111644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone RC, Feng D, Deng J, Singh S, Yang L, Fitzgerald-Bocarsly P, et al. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis & Rheumatism. 2012;64:788–798. doi: 10.1002/art.33395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nature Immunology. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 45.Pawar RD, Patole PS, Ellwart A, Lech M, Segerer S, Schlöndorff D, et al. Ligands to nucleic acid-specific toll-like receptors and the onset of lupus nephritis. Journal of the American Society of Nephrology. 2006;17:3365–3373. doi: 10.1681/ASN.2006030263. [DOI] [PubMed] [Google Scholar]
- 46.Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. Journal of Experimental Medicine. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody–DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauer JW, Baechler EC, Petri M, Batliwalla FM, Crawford D, Ortmann WA, et al. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med. 2006;3:e491. doi: 10.1371/journal.pmed.0030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dall’era MC, Cardarelli PM, Preston BT, Witte A, Davis JC. Type I interferon correlates with serological and clinical manifestations of SLE. Annals of the Rheumatic Diseases. 2005;64:1692–1697. doi: 10.1136/ard.2004.033753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khamashta M, Merrill JT, Werth VP, Furie R, Kalunian K, Illei GG, et al. Sifalimumab, an anti-interferon-α monoclonal antibody, in moderate to severe systemic lupus erythematosus: a randomised, double-blind, placebo-controlled study. Annals of the Rheumatic Diseases. 2016;75:1909–1916. doi: 10.1136/annrheumdis-2015-208562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalunian KC, Merrill JT, Maciuca R, McBride JM, Townsend MJ, Wei X, et al. A Phase II study of the efficacy and safety of rontalizumab (rhuMAb interferon-α) in patients with systemic lupus erythematosus (ROSE) Annals of the Rheumatic Diseases. 2016;75:196–202. doi: 10.1136/annrheumdis-2014-206090. [DOI] [PubMed] [Google Scholar]
- 52.Merrill JT, Wallace DJ, Petri M, Kirou KA, Yao Y, White WI, et al. Safety profile and clinical activity of sifalimumab, a fully human anti-interferon α monoclonal antibody, in systemic lupus erythematosus: a phase I, multicentre, double-blind randomised study. Annals of the Rheumatic Diseases. 2011;70:1905–1913. doi: 10.1136/ard.2010.144485. [DOI] [PubMed] [Google Scholar]
- 53.McBride JM, Jiang J, Abbas AR, Morimoto A, Li J, Maciuca R, et al. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis & Rheumatism. 2012;64:3666–3676. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- 54.Amir EAD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature Biotechnology. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Y, King NJC, Kesson AM. Major histocompatibility complex class I (MHC-I) induction by West Nile virus: involvement of 2 signaling pathways in MHC-I up-regulation. J Infect Dis. 2004;189:658–668. doi: 10.1086/381501. [DOI] [PubMed] [Google Scholar]
- 56.Jia T, Leiner I, Dorothee G, Brandl K, Pamer EG. MyD88 and Type I interferon receptor-mediated chemokine induction and monocyte recruitment during Listeria monocytogenes infection. The Journal of Immunology. 2009;183:1271–1278. doi: 10.4049/jimmunol.0900460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welcher AA, Boedigheimer M, Kivitz AJ, Amoura Z, Buyon J, Rudinskaya A, et al. Blockade of interferon-γ normalizes interferon-regulated gene expression and serum CXCL10 levels in patients with systemic lupus erythematosus. Arthritis Rheumatol. 2015;67:2713–2722. doi: 10.1002/art.39248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gottschalk TA, Tsantikos E, Hibbs ML. Pathogenic Inflammation and Its Therapeutic Targeting in Systemic Lupus Erythematosus. Front Immunol. 2015;6:550. doi: 10.3389/fimmu.2015.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amezcua-Guerra LM, Ferrusquía-Toriz D, Castillo-Martínez D, Márquez-Velasco R, Chávez-Rueda AK, Bojalil R. Limited effectiveness for the therapeutic blockade of interferon α in systemic lupus erythematosus: a possible role for type III interferons. Rheumatology. 2015;54:203–205. doi: 10.1093/rheumatology/keu020. [DOI] [PubMed] [Google Scholar]
- 60.Markopoulou A, Kyttaris VC. Small molecules in the treatment of systemic lupus erythematosus. Clin Immunol. 2013;148:359–368. doi: 10.1016/j.clim.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan CH, Fang C, Qiao Y, Yarilina A, Prinjha RK, Ivashkiv LB. BET bromodomain inhibition suppresses transcriptional responses to cytokine-Jak-STAT signaling in a gene-specific manner in human monocytes. Eur J Immunol. 2015;45:287–297. doi: 10.1002/eji.201444862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson SW, Jacobs HM, Arkatkar T, Dam EM, Scharping NE, Kolhatkar NS, et al. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. Journal of Experimental Medicine. 2016;213:733–750. doi: 10.1084/jem.20151724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Pla A, Patel P, Maecker HT, Rossello-Urgell J, Baldwin N, Bennett L, et al. IFN priming is necessary but not sufficient to turn on a migratory dendritic cell program in lupus monocytes. The Journal of Immunology. 2014;192:5586–5598. doi: 10.4049/jimmunol.1301319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu LJ, Wallace DJ, Ishimori ML, Scofield RH, Weisman MH. Review: Male systemic lupus erythematosus: a review of sex disparities in this disease. Lupus. 2010;19:119–129. doi: 10.1177/0961203309350755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartzman-Morris J, Putterman C. Gender differences in the pathogenesis and outcome of lupus and of lupus nephritis. Clin Dev Immunol. 2012;2012:604892–9. doi: 10.1155/2012/604892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 67.Klint C, Truedsson L, Andreasson A, Johansson I, Sturfelt G. Toxic effects of SLE serum on normal monocytes in vitro: cell death induced by apoptosis related to complement dysfunction. Lupus. 2000;9:278–287. doi: 10.1191/096120300680198999. [DOI] [PubMed] [Google Scholar]
- 68.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis & Rheumatism. 2006;54:1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 69.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fleischmann R, Cutolo M, Genovese MC, Lee EB, Kanik KS, Sadis S, et al. Phase IIb dose-ranging study of the oral JAK inhibitor tofacitinib (CP-690,550) or adalimumab monotherapy versus placebo in patients with active rheumatoid arthritis with an inadequate response to disease-modifying antirheumatic drugs. Arthritis & Rheumatism. 2012;64:617–629. doi: 10.1002/art.33383. [DOI] [PubMed] [Google Scholar]
- 71.Lee EB, Fleischmann R, Hall S, Wilkinson B, Bradley JD, Gruben D, et al. Tofacitinib versus methotrexate in rheumatoid arthritis. N Engl J Med. 2014;370:2377–2386. doi: 10.1056/NEJMoa1310476. [DOI] [PubMed] [Google Scholar]
- 72.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis & Rheumatism. 1997;40:1725. doi: 10.1002/1529-013119970940:9<1725::AID-ART29>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 73.Jansen K, Blimkie D, Furlong J, Hajjar A, Rein-Weston A, Crabtree J, et al. Polychromatic flow cytometric high-throughput assay to analyze the innate immune response to Toll-like receptor stimulation. Journal of Immunological Methods. 2008;336:183–192. doi: 10.1016/j.jim.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Corbett NP, Blimkie D, Ho KC, Cai B, Sutherland DP, Kallos A, et al. Ontogeny of Toll-Like Receptor Mediated Cytokine Responses of Human Blood Mononuclear Cells. PLoS ONE. 2010;5:e15041. doi: 10.1371/journal.pone.0015041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nature Biotechnology. 2012;30:857–866. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. Normalization of mass cytometry data with bead standards. Cytometry. 2013;83A:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, et al. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Series B Stat Methodol. 2012;74:245–266. doi: 10.1111/j.1467-9868.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 79.Guo X, Pan W, Connett JE, Hannan PJ, French SA. Small-sample performance of the robust score test and its modifications in generalized estimating equations. Stat Med. 2005;24:3479–3495. doi: 10.1002/sim.2161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.