Abstract
To investigate immune escape during breast tumor progression, we analyzed the composition of leukocytes in normal breast tissues, ductal carcinoma in situ (DCIS), and invasive ductal carcinomas (IDC). We found significant tissue and tumor subtype-specific differences in multiple cell types including T cells and neutrophils. Gene expression profiling of CD45+CD3+ T cells demonstrated a decrease in CD8+ signatures in IDCs. Immunofluorescence analysis showed fewer activated GZMB+CD8+ T cells in IDC than in DCIS, including in matched DCIS recurrent IDC. TCR clonotype diversity was significantly higher in DCIS than in IDCs. Immune checkpoint protein TIGIT expressing T cells were more frequent in DCIS whereas high PD-L1 expression and amplification of CD274 (encoding PD-L1) was only detected in triple negative IDCs. Co-amplification of 17q12 chemokine cluster with ERBB2 subdivided HER2+ breast tumors into immunologically and clinically distinct subtypes. Our results show co-evolution of cancer cells and the immune microenvironment during tumor progression.
INTRODUCTION
The importance of tissue infiltrating leukocytes in breast tumor development and therapeutic responses is widely accepted, but the mechanisms underlying their effects and alterations of leukocyte composition during tumor progression are still poorly understood (1). Leukocytes are one of the most dynamic cell populations present within tumors and they also play a role in normal breast tissue remodeling during pregnancy and involution (2,3). Tumor-associated macrophages (TAMs) are known to facilitate angiogenesis, extracellular matrix (ECM) degradation, and tumor invasion, and high frequency of TAMs is associated with poor clinical outcome (3,4). In contrast, higher frequency of tumor-infiltrating lymphocytes (TILs) and especially more CD8+ and fewer FOXP3+ regulatory T cells within tumors is associated with better outcome (1). The numbers and composition of TILs within tumors seem to be especially relevant in HER2+ and triple negative breast cancer (TNBC) where tumors with higher TIL fractions have better response to HER2-targeted therapies and chemotherapy, respectively (1).
In DCIS high leukocyte density has been observed in a subset of tumors with enrichment of leukocytes at sites of focal myoepithelial cell layer disruptions (5), suggesting that they might play a role in invasive progression. In DCIS, cancer cells are still physically separated from the stroma and direct leukocyte-cancer cell contact is rarely detected. With invasive progression cancer cells and leukocytes are intermingled, and only cancer cells that can survive and proliferate in this environment will contribute to disease progression. Thus, the transition from in situ to invasive carcinoma might be a critical tumor progression step for immune escape in breast cancer and deciphering its mechanism would aid the design of immunotherapies for both advanced and early stage disease. Most prior analyses of leukocytes in breast tumors, especially in DCIS, have been limited to inferring leukocyte composition from gene expression profiles of bulk tumors (6–10) and to the testing of a handful of markers in archived tissue samples (11–16). Thus, our understanding of immune-related changes in early stages of breast tumorigenesis are still rather limited.
Here we used a combination of global profiling and single cell methods for the cellular and molecular characterization of tissue-infiltrating leukocytes, with particular emphasis on T cells, in normal breast tissues, pure DCIS (no histologic evidence of invasion), and in HER2+ and triple negative (TN) invasive ductal breast carcinomas. We also characterized genetic alterations in cancer cells that might impact the tumor immune microenvironment and disease progression. Our goal was to gain insights into the co-evolution of tumor and immune cell compartments during the in situ to invasive carcinoma transition. We focused on HER2+ and triple negative tumor subtypes as these DCIS have a higher risk of invasive recurrence and the resulting invasive tumors are also more likely to progress to metastatic disease.
RESULTS
Leukocyte composition of human breast tissues
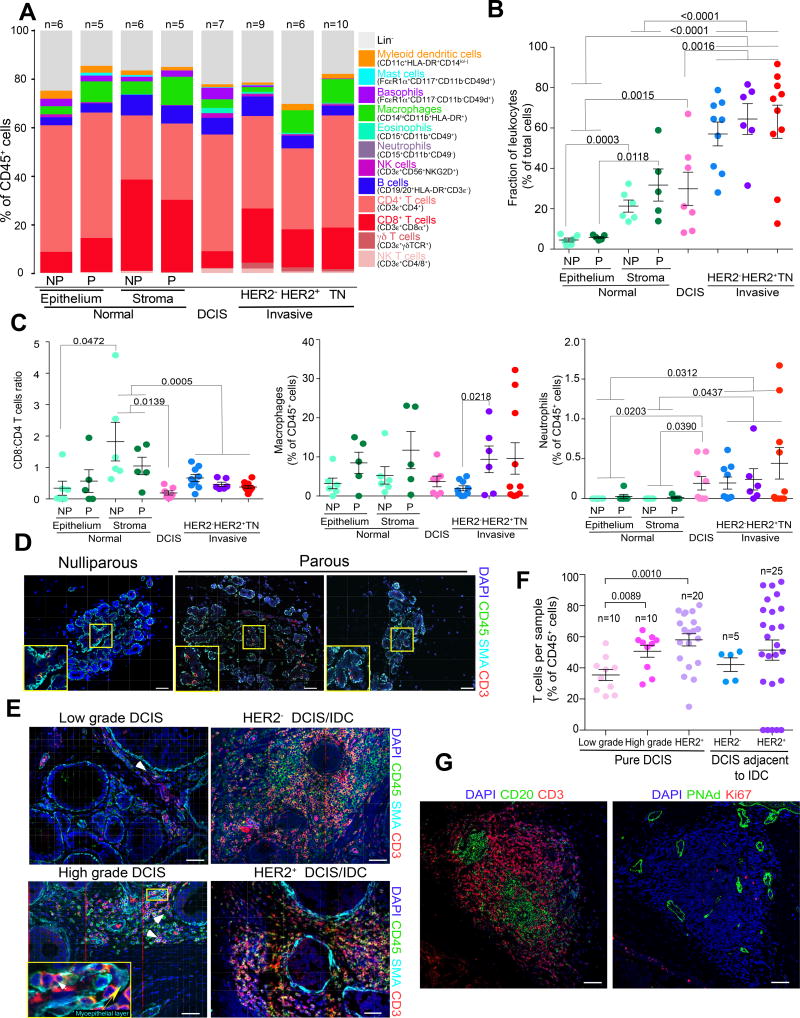
We first characterized the composition of tissue-infiltrating leukocytes in normal and neoplastic breast tissues using a polychromatic FACS, that allows for the quantitative assessment of all major leukocyte cell populations (14) (Fig.1A and Supplementary Fig. S1A). Quantification based on FACS was reproducible and accurate as confirmed by the analysis of the same tumor stained and profiled separately and by comparing it to histologic examination of tissue slides (Supplementary Fig. S1B,C). We analyzed normal breast tissues from nulliparous and parous women, including BRCA1 and BRCA2 mutation carriers, as well as DCIS and IDCs of different subtypes (Supplementary Table S1). In normal breast tissues, we analyzed epithelial and stromal fractions separately to detect potential differences between intra-epithelial and stromal leukocytes. We found that DCIS and IDC contained significantly (p=0.0015 and p<0.0001, respectively) higher numbers of leukocytes, compared to normal breast, whereas in normal tissues more leukocytes were in the stromal than in the epithelial fraction (Fig.1B). We also observed significant differences in the relative frequencies of several CD45+ cell types including increased neutrophils and decreased CD8+/CD4+ T cell ratios in tumors compared to normal stroma (Fig.1A,C and Supplementary Table S2). The relative fraction of CD8+ and CD4+ T cells showed significant inverse correlation in normal breast tissues and DCIS, and in DCIS higher fractions of CD8+ T cells were associated with significantly higher frequency of macrophages (Supplementary Fig. S1D). The frequencies of dendritic cells and T cells showed significant inverse correlation in TN IDCs (Supplementary Fig. S1D), while the relative proportion of γδT cells was significantly higher in IDCs compared to normal breast tissues (p=0.008) and DCIS (p=0.0476) (Supplementary Fig. S1E). This latter result is potentially interesting in light of recent findings in animal models showing that γδT cells promote breast cancer metastasis via their recruitment of neutrophils (17). Corroborating this, we detected higher fraction of neutrophils in IDCs compared to normal breast tissues (Fig.1C). However, the clinical relevance of γδT cells in human breast tumors is inconclusive; some studies suggest that their frequency is associated with good prognosis (18) whereas others show that their higher frequency indicates poor outcome (19). Overall, these changes in leukocyte composition imply a switch to a more immunosuppressive and metastasis-promoting environment during breast tumor progression. We focused our subsequent analyses on T cells due their central role in immune responses and being the most dominant CD45+ cell population in both normal and cancerous breast tissues.
Figure 1.
Leukocyte populations in normal and neoplastic breast tissues. In all panels: NP and P indicates normal breast of nulliparous and parous women, respectively, TN – triple negative, data are represented as mean ± S.E.M., and p-values are calculated using paired two-tailed T test. A, Summary of polychromatic FACS analyses of leukocyte populations in normal breast tissues and breast tumors. Results are shown as a percent of total CD45+ cells. B, Percentage of leukocytes in normal breast tissues and breast tumors relative to all cells. C, Ratio of CD8+/CD4+ T cells, and relative fraction of macrophages and neutrophils shown as percentage of total CD45+ cells. D–E, Immunofluorescence staining for CD45 pan-leukocyte, CD3 T cell, and smooth muscle actin (SMA) myoepithelial cell markers, and DAPI to denote nuclei of normal breast tissues (D) and DCIS and IDC (E). Images are a montage of nine fields captured from one area of the tissue. Yellow boxes on montage image magnify regions where leukocytes are located in the myoepithelium. White and yellow arrows indicate CD3+ cells and myoepithelial cell layer, respectively. Scale bar, 50 µm. F, Frequencies of T cell populations in pure DCIS and in DCIS regions of invasive cancers quantified based on immunofluorescence images. G, Examples of potential tertiary lymphoid structures in DCIS. Scale bar, 150 µm.
Next, we performed multicolor immunofluorescence for CD45 pan-leukocyte and CD3 T cell markers to assess the spatial distribution of leukocytes within normal and neoplastic breast tissues. We also stained for smooth muscle actin (SMA) to mark myoepithelial cells surrounding the ducts; intact myoepithelial cell layer and basement membrane are detected in normal breast and in DCIS but lost in invasive breast carcinomas. In normal breast tissues there were relatively few T cells, with the exception of mastitis or other benign inflammation, and minor differences were observed between nulliparous and parous women (Fig.1D and Supplementary Fig. S1F,G). In DCIS, we found significantly higher fraction of T cells in high-grade compared to low-grade DCIS (p<0.0001), and HER2+ compared to HER2− DCIS (p<0.0001), and DCIS adjacent to IDC also contained significantly (p=0.0002) higher frequency of T cells than pure DCIS (Fig.1E,F). However, the spatial distribution of T cells was highly variable in DCIS; some areas of the tumor had very high leukocyte and T cell density whereas others contained almost none (Fig. 1E). In several DCIS we even detected probable tertiary lymphoid structures (TLS) (20) characterized by tight clusters of B cells surrounded by T cells and presence of peripheral-node addressin-positive (PNAd+) endothelial venules (Fig.1G and Supplementary Fig. S1H). In normal breast tissues T cells surrounded the ducts and intra-epithelial T cells were frequently detected despite their numbers being relatively low (Fig. 1D), in contrast to DCIS where T cells were rarely detected within the ducts but rather localized in the stroma and at sites of focal myoepithelial cell layer disruption (Fig. 1E). These findings suggest that in DCIS there is limited interaction between T cells and cancer cells, thus, immune escape is most likely to occur during in situ to invasive carcinoma transition.
Gene expression profiles of CD3+ T cells
To begin dissecting the properties of T cells infiltrating normal and neoplastic breast tissues, we performed RNA-seq on purified CD45+CD3+ T cells from normal breast tissues (n=12), DCIS (n=11), and IDC (n=12), focusing on HER2+ (n=5) and TN (n=6) IDC cases (Supplementary Table S1). In normal breast tissues, parity and BRCA status did not appear to influence T cell gene expression profiles thus, we considered all normal samples as one group. Principal Component Analysis (PCA) of T cell gene expression profiles demonstrated separation of normal samples from DCIS. Whilst the HER2+ IDC were more similar to normal cases, TN IDCs clustered with DCIS (Fig. 2A,B). Unsupervised hierarchical clustering of the samples using differentially expressed immune-related genes showed a clear separation of T cells from normal breast, DCIS, and HER2+ and TN IDC, indicating that these groups have largely distinct T cell expression profiles (Fig. 2B and Supplementary Table S3). The most abundantly expressed genes were related to T cell receptor (TCR) signaling (e.g., NFATC1, NFKB1) and antigen presentation (e.g., HLA-A, HLA-C), which appeared to be more highly expressed in normal samples. Transcriptional regulators of T cells (e.g., TBX21, GATA3) were somewhat consistently expressed among samples with the exception of EOMES (eomesodermin), which had low expression in T cells from TN IDC, and FOXP3, which was more abundant in T cells from IDCs (both HER2+ and TN cases). IL18R1, CXCL13, CXCL1, and CTLA-4 were also more abundant in T cells from IDCs than in DCIS, whereas the expression of ILF3 and NFKBIA, several interleukin receptors (e.g., IL2RA, IL10RA), was higher in T cells in DCIS and normal breast than in IDCs. TNFRSF25 and other TNF receptors were more highly expressed in DCIS-associated T cells compared to normal and IDCs, whereas LCK kinase, which is involved in T cell development and TCR signaling, showed the opposite pattern; these results were also confirmed by immunofluorescence analysis (Supplementary Fig. S2A).
Figure 2.
Gene expression profiles of T cells. In all panels: TN-triple negative, IDC – invasive ductal carcinoma, NP and P indicates normal breast of nulliparous and parous women, respectively. A, 3-D Principal Component Analysis plots of RNA-seq data. B, Heatmap depicting clustering of samples based on the expression of top differentially expressed immune-related genes with highest variance defined by both edgeR and DESeq2. C, Frequency of enrichment of a particular cell type following GSEA using the Immune c7 compendium. Significance was determined using proportionality test with multiple testing correction. D, Network of enriched immune c7 cell-type specific gene sets. Node size is reflective of number of times a T cell type appears as significantly enriched and arrow thickness is reflective of number of significant gene sets involved in a particular comparison. E, Frequency of T cell populations calculated using CIBERSORT based on RNA-seq expression data from bulk T cell samples, with difference in populations computed using ANOVA.
Gene set enrichment analysis (GSEA) using the canonical gene set compendium (21) showed that, as expected, immune-related gene sets were enriched in our list of significantly differentially expressed genes (Supplementary Fig. S2B and Supplementary Table S4). This included a downregulation of genes in the IL4 signaling pathway in IDC and normal samples compared to DCIS. Furthermore, the Th1Th2 and CTLA-4 signaling pathways were upregulated in IDC compared to DCIS and the complement pathway was higher in normal compared to DCIS samples. Differences between HER2+ and TN IDC included the upregulation of pathways signaling for IL6, IL7, CTLA-4, and antigen-presentation by MHC I in TN cases (Supplementary Table S4). Also enriched were gene sets corresponding to cell signaling pathways, transcription and translation, and cell cycle in the HER2+/TN IDC comparison, suggesting potential T cell expansion. Overall, these results suggest a more inflammatory and more immunosuppressive environment in IDCs compared to DCIS, especially in TN tumors.
To obtain a more detailed view of the specific activities of T cells, GSEA was conducted using ImmuneSigDB (21), a compendium of gene signatures summarizing differences between two given groups of immune cells. First, we summarized enriched gene sets according to the frequency of a particular immune lineage and compared to the overall frequency of that given cell type appearing in the compendium (Fig. 2C). Gene sets relating to thymocytes were not as common as expected when comparing DCIS to IDC and HER2+ to TN IDC samples. Additionally, gene sets relating to CD8+ T cells were enriched when comparing HER2+ to TN IDCs, and NKT cells when comparing both DCIS and IDC to normal cases (Proportionality test, BH-adjusted p<0.05) suggesting that differences between these groups are driven by their activation state rather than lineage (Fig. 2C). Enriched gene sets from ImmuneSigDB were collapsed into network diagrams to highlight the directionality within each comparison (Fig. 2D). When comparing T cells from DCIS to normal cases, multiple enriched gene sets suggest that the expression profiles of T cells from DCIS samples are more similar to CD4+ regulatory T cells (Tregs) or an undifferentiated CD4+ T cell state. In contrast, the expression profile of T cells from normal samples was more varied. Comparison to IDC samples showed that DCIS T cells had a CD8+ or CD4+Th17 phenotype, whereas IDC samples were more similar to Tregs, suggesting even stronger immunosuppression in IDC compared to DCIS. Comparison between HER2+ and TN IDCs suggested activation of T helper cells, including Th17 and Tregs in TN IDCs, whereas T cells from HER2+ IDCs were enriched for Th1 and Th2 cells (Fig. 2D and Supplementary Fig. S2C). Since T cells from HER2+ and TN IDC showed very distinct profiles (Fig. 2A), we compared T cells from DCIS to those of HER2+ IDC alone (Fig. 2D). T cells from HER2+ IDC showed a higher enrichment of Th1, Th2, and Tregs compared to undifferentiated CD4+ T cell gene sets. Conversely, DCIS T cells more closely resembled CD8+ T cells when compared to Treg or CD4+ T cells. Deconvolution of the composition T cell populations within each sample using CIBERSORT (22) supported this trend, with samples having the greatest CD8 or CD4 naïve content being in normal or DCIS. Differences in γδT cells, Tregs and CD4+ memory activated T cells was noted between the different groups, with higher proportions of activated CD4+ memory cells and γδ T cells in HER2+ IDC and lower proportions of Tregs in normal samples (Fig. 2E). Overall, these data suggest that compared to normal tissues both DCIS and IDC have more Tregs, T cells in DCIS are enriched for cytotoxic CD8+ and undifferentiated/naive CD4+ T cell signatures, whereas IDCs have lower proportion of CD8+ T cells but more activated CD4+ T helper cells and Tregs. Although inference of cellular composition based on deconvolution of RNA-seq data has limitations and ideally best would be to characterize each T cell subpopulation by single cell RNA-seq, it provides estimates that agree well with FACS profiles for more frequent cell types (Supplementary Fig. S2D). Nevertheless, further experimental validation of these predictions is needed.
Activation of CD8+ T cells in DCIS
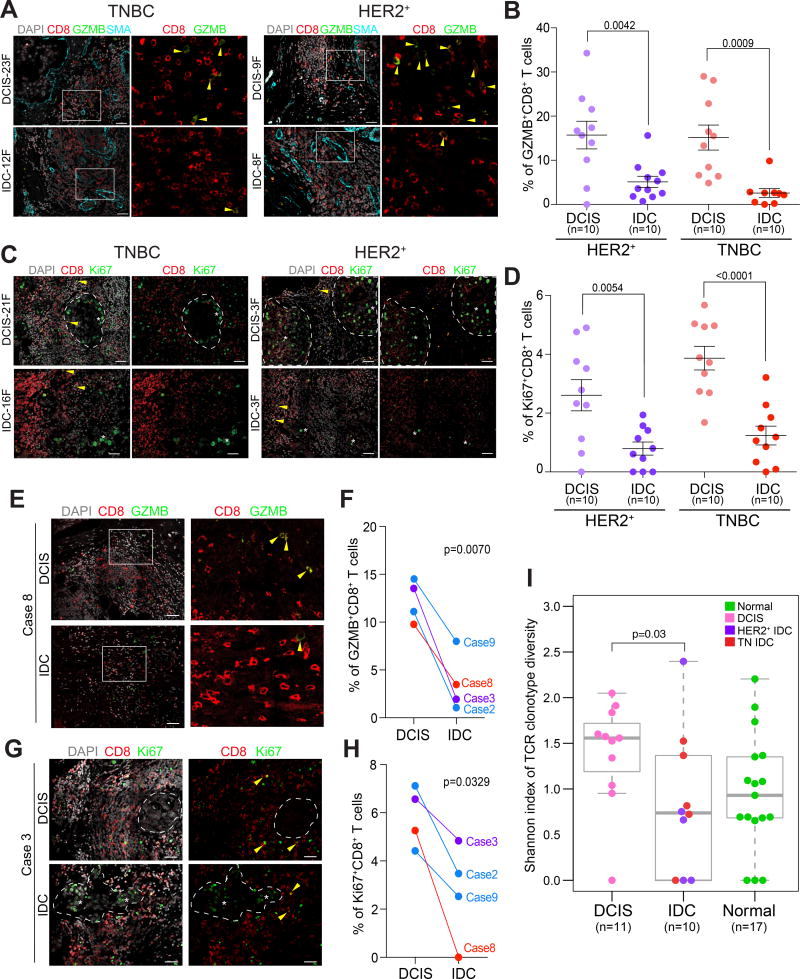
The enrichment of DCIS T cell expression profiles for CD8+ T cell signatures and the predicted higher frequency of CD4+ memory activated T cells in HER2+ IDC suggest potential activation and clonal expansion of T cells in DCIS and HER2+ IDCs. In line with this observation, the expression of MKI67, a proliferation marker, in our RNA-seq data was highly variable across samples. All HER2+ IDCs and a subset of DCIS displayed high MKI67 levels suggesting T cell expansion in these cases (Supplementary Fig. S3A,B). The expression of granzyme B (GZMB), a marker of activated CD8+ T cells, was also high in MKI67high T cells in DCIS (Supplementary Fig. S3B). Moreover, gene signatures specific for naïve and cytotoxic T cells showed a positive correlation in MKI67low T cells but not in MKI67high T cells (Supplementary Fig. S3C and Supplementary Table S5), suggesting that MKI67high T cells reflect expansion of activated CD8+ T cells. Immunofluorescence analysis of markers of activated and effector T cells, including CD8, GZMB, Ki67, and IFNγ, in an independent cohort (Supplementary Table S1) confirmed the predictions of the RNA-seq data as we detected significantly higher fractions of GZMB+, Ki67+, and IFNγ+ CD8+ T cells in DCIS than in IDC in both HER2+ and TN subtypes (Fig. 3A–D and Supplementary Fig. S3D). Furthermore, the relative fractions of Ki67+CD8+ and GZMB+CD8+ T cells showed significant positive correlation in both DCIS and IDCs (Supplementary Fig. S3E). Importantly, analysis of matched DCIS and IDC samples in a cohort of patients who were diagnosed with pure DCIS and underwent lumpectomy, but years later recurred locally with IDC demonstrated the same results (Fig. 3E–H), confirming that a decrease in activated CD8+ T cells is a feature of in situ to invasive breast carcinoma progression.
Figure 3.
Activation status of CD8+ T cells. Dotted lines indicate clusters of tumor epithelial cells, yellow arrows mark double positive T cells, whereas white stars mark epithelial cells positive for the marker. A–D Immunofluorescence analysis of granzymeB (A) and Ki67 (C) expression in CD8+ T cells. SMA staining was used to mark the myoepithelial cell layer in DCIS. Images are a montage of nine fields captured from one area of the tissue. Graphs depict the frequencies of granzymeB+ (B) and Ki67+ (D) CD8+ T cells in multiple regions of ten samples per group. Error bars, S.E.M. Scale bar 50 µm. P-values are calculated using two-tailed T test. E–H, Immunofluorescence analysis of granzymeB (E) and Ki67 (G) expression in CD8+ T cells in matched DCIS and locally recurrent IDC samples. Graphs depict the frequencies of granzymeB+ (F) and Ki67+ (H) CD8+ T cells in multiple regions. Error bars, S.E.M. Scale bar 50 µm. P-values are calculated using paired two-tailed T test. I, Boxplot depicting Shannon index of TCR clonotype diversity in normal breast tissues, DCIS, and IDC. Significance of the difference between tissue types was calculated using Wilcoxon rank sum test.
To further explore the potential clonal expansion of T cells in DCIS, we aligned our RNA-seq data to a library of known variable human complementarity-determining regions (CDR) to determine the frequency of unique T cell receptor (TCR) segments in each sample (23). Samples were normalized based on the relative fraction of T cells. The TCR is rearranged during T cell maturation and a population of T cells that have the same unique TCR sequence is defined as a TCR clonotype. TCR clonotype repertoire can be quantitatively assessed using the Shannon index of diversity (24). DCIS cases overall appeared to have more clonally expanded T cells than IDCs (Supplementary Fig. S4A), and TCR clonotype diversity measured by the Shannon index was significantly (p=0.03) higher in DCIS compared to IDCs (Fig. 3I). The correlation between MKI67 read counts and Shannon index of TCR clonotype diversity was the highest in TN IDCs implying the expansion of multiple T cell clones in these tumors (Supplementary Fig. S4B). Similar positive correlation was observed in TN IDCs between the expression of T cell activation markers (e.g., GZMB, PRF1, and CCL4) and TCR clonotype diversity (Supplementary Fig. S4C) further implying the presence of polyclonal activated T cells. In contrast, the generally lower TCR clonotype diversity in HER2+ cases might reflect expansion of a limited set of T cell clones. As an alternative method to assess the TCR repertoire in DCIS, we performed immunoSEQ that defines TCR clonotype diversity based on DNA sequence (25). ImmunoSEQ also showed polyclonality in all DCIS, although a few dominant clones were also detected, and the top 10 most abundant clones in each case revealed differences among samples (Supplementary Fig. S4D,E). Similarly, clustering of samples based on normalized frequencies of TCR v and j genes highlighted several similarities and differences among samples (Supplementary Fig. S4F,G). The two samples for which we had both RNA-seq and immunoSEQ data (DCIS15 and DCIS17) appeared to have the highest fraction of expanded clones based on TCR and were also highly MKI67+ and GZMB+. Interestingly, we found a few TCR clones that were shared (at the amino acid level) among different tumors and represented relatively more abundant clones (Supplementary Fig. S4H). For one of these TCRs we were able to predict the putative antigen as EBNA-3, an Epstein-Barr virus protein based on prior TCR repertoire analyses (26). The presence of shared, relatively prominent clones, in combination with our RNA-seq data, support the possibility of an adaptive immune response in a subset of DCIS and a potential decline of this after progression to IDC.
Immune checkpoint proteins
Our observation that IDCs, particularly TN IDCs, contained relatively more TILs but fewer in the activated state than DCIS implied potential exhaustion and immunosuppression in IDCs. To investigate this hypothesis, we analyzed our RNA-seq data for genes characteristic of exhausted state including immune checkpoint proteins (e.g., CTLA-4, PD-1, TIGIT, Supplementary Table S5). We found a positive correlation between exhausted and cytotoxic gene signatures in both MIKI67high and MKI67low T cells, which was expected as these markers are commonly co-expressed in the same T cell population and exhaustion is usually due to chronic activation. On the other hand, the expression of inhibitory and activating immune checkpoint proteins showed positive correlation only in MKI67high T cells (Supplementary Fig. S5A,B). Similarly, the expression of T cell activation and dysfunction related genes (27) was significantly different in tumor-associated MKI67high and MKI67low T cells (Supplementary Fig. S5C).
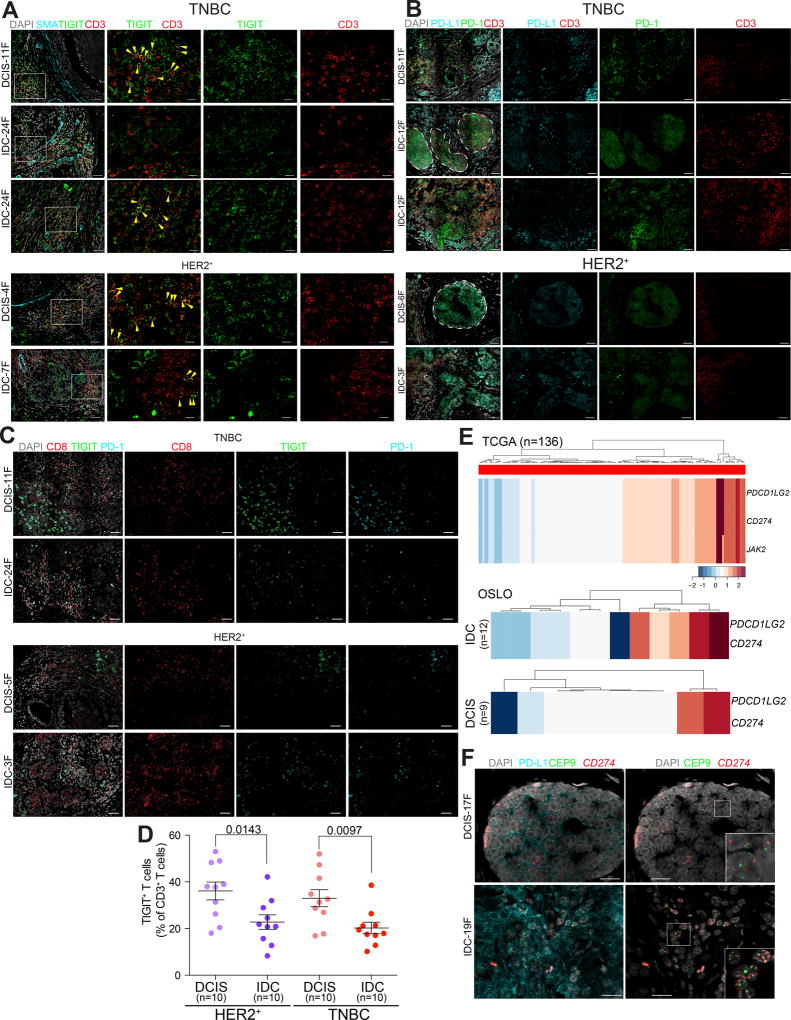
To explore the expression of immune checkpoint proteins in further detail, we performed immunofluorescence analysis for TIGIT, PD-1, and PD-L1 in DCIS and IDCs, since these proteins have been previously implicated in breast cancer with clinical trials targeting them currently ongoing (Fig. 4A–C). TIGIT was expressed mostly in CD3+ T cells but also in other leukocyte populations (Fig. 4A, and Supplementary Fig. S5D). In contrast, PD-L1 expression was more wide-spread with high expression in various immune and in a subset of tumor epithelial cells (Fig. 4B and Supplementary Fig. S5E). In a luminal A IDC with extensive in situ region, PD-L1 was highly expressed in DCIS-associated myoepithelial cells but not in tumor cells, whereas in the invasive areas, the tumor epithelial cells were highly PD-L1-positive (Supplementary Fig. S5F). This finding implies that in DCIS myoepithelial cells might play a role in immune suppression and tumor epithelial cells that gain this phenotype have selective advantage during invasive progression. TIGIT and PD-1 were also frequently co-expressed in CD8+ T cells in both DCIS and IDCs (Fig. 4C), similar to what was described in melanoma (28). Quantification of the signal revealed slightly but significantly more TIGIT+CD3+ T cells in DCIS compared to IDC in both HER2+ and TN cases (Fig. 4D), whereas the relative fraction of PD-1+CD3+ T cells was not significantly different among tumors (Supplementary Fig. S5G). In a few cases we analyzed the expression of TIGIT by both FACS and IF to confirm our results, and we detected similar TIGIT levels by both techniques (Supplementary Fig. S5H). We also analyzed the spatial distribution of TIGIT+ or PD-1+ CD3+ T cells to see potential differences associated with histology or tumor subtype. For this, we digitized the immunofluorescence images by recording spatial coordinates and marking tumor cells and T cells positive or negative for TIGIT or PD-1 and assessed the relative distribution of the various cell types (Supplementary Fig. S6A). We found that the fraction of T cells invading the tumor cells was significantly higher in IDC compared to DCIS only in TN cases (p=0.19 in HER2+ and 0.006 in TN based on t-test), implying that there is more intermixing between T cells and cancer cells in the TN but not in the HER2+ cases (Supplementary Fig. S6B). Interestingly, we also found that in TN DCIS adjacent to IDC, PD-1+CD3+ T cells can commonly be observed within the ducts in the epithelial compartment, whereas intraepithelial T cells were rarely present in HER2+ DCIS adjacent to IDC (Fig. 4B). These results imply potential differences in the microenvironment such as the extracellular matrix (ECM) or differences in local myeloid complexity of HER2+ and TNBCs that could influence the ability of T cells to invade into tumors and enter the ducts. Correlating with this finding, collagen trichrome staining showed some differences between HER2+ and TNBCs, especially in DCIS, with HER2+ DCIS displaying a denser fibrous ECM potentially forming a physical barrier (Supplementary Fig. S6C).
Figure 4.
Expression patterns of immune checkpoint proteins in breast tumors. A–C, Immunofluorescence analysis of the expression of TIGIT, CD3, and SMA (A), PD-L1, CD3, and PD-1 (B), and TIGIT, PD-1, and CD8 combined (C). White rectangles indicate selected areas enlarged in the adjacent panels. Yellow arrows mark T cells positive for both TIGIT and CD3. Dotted lines demarcate clusters of tumor epithelial cells. Scale bar, 50 µm. D, Quantification of TIGIT+CD3+ T cells. Multiple regions of ten samples per group were quantified. Error bars, S.E.M. P-values are calculated using two-tailed T test. E, Copy number gain for 9p24 amplicon genes in basal-like breast tumors in the TCGA and Oslo cohorts. F, Immuno-FISH analysis of PD-L1 protein levels and CD274 (encoding PD-L1) copy number in triple negative DCIS and IDC. CEP9 probe was used as control and nuclei were stained with DAPI. Scale bar 20 µm. Insets are approximately 20×20 µm. Throughout, images are a montage of nine fields captured from one area of the tissue.
The high expression of PD-L1 in tumor epithelial cells in TN IDCs (Fig. 4B) raised the possibility of specific upregulation of PD-L1 in these tumors due to copy number gain, as we and others previously described that a 9p24 amplicon including CD274 (encoding for PD-L1), PDCD1LG2 (encoding PD-L2), and JAK2 only occurs in basal-like breast tumors where ∼30% of cases have a high copy number gain of this region (29,30). To investigate this hypothesis in more detail, we examined the frequency of gains and losses of the CD274 locus in the TCGA cohort composed of invasive breast cancers, and found an enrichment of gains in the basal subtype of ∼40% (P<0.001 chi-square test, Fig. 4E and Supplementary Fig. S7A–C). 9p24 copy number gain was not associated with any differences and cytotoxic T cell gene signatures, probably due to the expression of PD-L1 by many stromal cell types besides tumor epithelial cells (Supplementary Fig. S7D). 9p24 gain was also independent of neoantigen load and estimated immune content, but was associated with overall copy number aberration (CNA) status only in TN cases (Supplementary Table S6). Similar comparisons in the Oslo cohort (9) that included both DCIS and IDCs reported a higher frequency of CD274 and PDCD1LG2 gain in IDC samples (6 of 12 patients) compared to DCIS (2 of 9 patients), which had basal-like expression profiles (Fig. 4E). To confirm the preferential gain of CD274 and overexpression of PD-L1 in TN IDCs compared to DCIS, we developed an immuno-FISH protocol that allows for the combined analysis of PD-L1 protein levels and CD274 copy number status at the single cell level in archived samples. Testing of ten TN IDC revealed CD274 copy number gain and PD-L1 overexpression in 3/10 cases, whereas none of the ten TN DCIS showed gain of this locus and had relatively low expression of PD-L1 (Fig. 4F). These results suggest that a possible mechanism for the in situ to invasive breast carcinoma transition in TN through the selection for tumor cells with higher expression of PD-L1 due to increased copy number for CD274. The clinical relevance of these findings as they relate to the likelihood of response to PD-L1 blockade remains to be determined.
17q12 chemokine amplicon
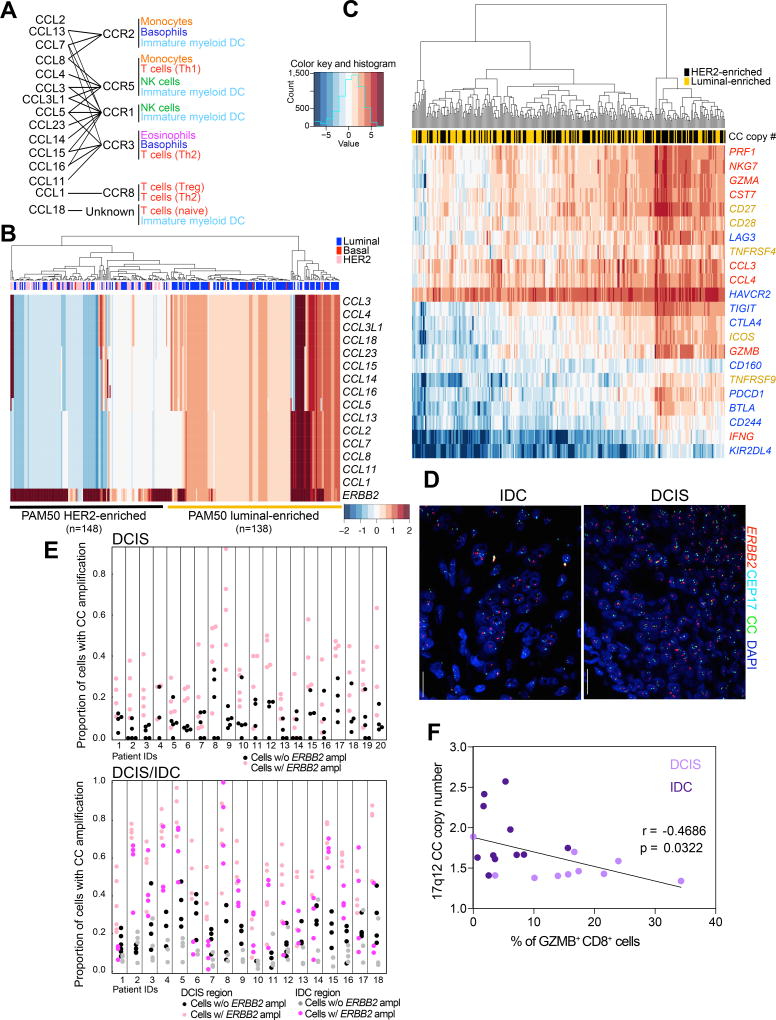
To explore whether other genomic regions encoding for genes involved in immune regulation may also display differential copy number gain between DCIS and IDCs, we analyzed known breast cancer amplicons for the presence of such genes. We found that the 17q12 chromosomal region that contains a cluster of genes encoding chemokines is located proximal to the ERBB2 amplicon, making it prone to possible co-amplification (Fig. 5A). Interestingly, in the TCGA cohort chemokine cluster (CC) gain was associated with ERBB2 amplicon status independent of immune content and overall CNA but negatively associated with neoantigen load, suggesting co-amplification as an alternate mechanism for immune activation (Supplementary Table S6). An enrichment of CC gains was observed in HER2+ tumors defined either by PAM50 classification or ERBB2 amplification (50% compared to 2%, P<0.001 chi-square test, Supplementary Fig. S7E). Interestingly, these HER2+ tumors can roughly be divided into two groups: those which are HER2+ER+ luminal-like and display evidence of co-amplification, and those which are HER2+ER− as defined by PAM50 which do not exhibit co-amplification and in fact suggest loss of this locus (Fig. 5B). To assess whether the copy number gain of this locus is associated with changes in leukocyte composition, we analyzed the expression of immune activation and exhaustion-related genes in these samples and found higher expression of both activation and inhibition-related genes in tumors that lack CC gain (Fig. 5C). Cytotoxic gene scores were associated with CC gain particularly in HER2+ER− tumors (Supplementary Table S6). These results suggest that HER2+ER− tumors with loss of the CC region are more likely to have T cell infiltrates than HER2+ER+ tumors with gain of the CC region.
Figure 5.
Leukocytes and tumor cell heterogeneity. A, Chemokine cluster (CC) on 17q12. Genes within amplicon encoding chemokines and their receptors and responsive cell populations. B, Copy number gain for 17q12 genes and ERBB2 in HER2+ breast tumors. C, Expression of immune activation (yellow), cytotoxic (red), inhibitory and exhaustion (blue) related genes in HER2+ breast tumors with CC copy number gain (yellow) or loss (black). D, Multicolor FISH for ERBB2, Cep17, and 17q12 CC in DCIS and IDC. E, Proportion of cells with chemokine cluster amplification stratified by HER2 status (+/−) in each region of each patient from DCIS (upper panel) and DCIS/IDC (lower panel) cohorts. Cells with HER2 amplification have overall significantly higher proportion of chemokine cluster amplification (Supplementary Table S7). The magnitudes of elevation of probability of chemokine cluster amplification in HER2+ cells vary from patient to patient. F, Plot depicting correlation between 17q12 CC copy number gain and frequency of GZMB+CD8+ T cells.
Next, we developed a multicolor FISH assay to evaluate both the 17q12 CC and ERBB2 copy number gain in HER2+ IDCs and DCIS at the single cell level (Fig. 5D and Supplementary Table S7). Adjacent slides were used for immunofluorescence to assess T cell frequency and activation. We found that cancer cells with ERBB2 gain had significantly higher chance of gaining the CC locus, especially in pure DCIS cases (Fig. 5E and Supplementary Table S7). Analysis of the intratumor spatial distribution of cells with or without CC gain did not reveal any specific pattern in DCIS nor in IDC (Supplementary Fig. S7F,G). The Shannon index of diversity for CC and ERBB2 copy numbers did not display significant correlation with frequencies of CD3+ T cells and CD45+ leukocytes neither in pure DCIS nor in DCIS/IDC cases (Supplementary Fig. 7H). However, CC amplification showed significant inverse correlation with the frequency of GZMB+CD8+ T cells within tumors (Fig. 5F). Moreover, we found a relatively strong positive correlation between the Shannon index and patient age at diagnosis in HER2+ IDC cases but not in pure DCIS (Supplementary Fig. S7I). To investigate whether the decreased T cell activation in tumors with CC gain might be due to higher frequencies of myeloid-derived suppressor cells (MDSC), we stained adjacent slides for CD68, CD33, and HLA-DRB1 that allow for the detection of macrophages (CD68+HLA-DR+CD33+), and monocytic (CD68+HLA-DR+CD33+) and granulocytic (CD68−HLA-DR+CD33+) MDSCs (Supplementary Fig. S7J). However, the numbers of macrophages and MDSCs did not correlate with CC copy number or fraction of GZMB+CD8+ T cells (Supplementary Fig. S7K,L) suggesting other mechanisms underlying the “immune cold” status of these tumors. Lastly, a generalized linear model predicting CC co-amplification (Supplementary Table S7) suggested an association between ERBB2 and ER, consistent with our findings from the TCGA cohort (Fig. 5B). These results imply potential selection for cancer cells that have lost the 17q12 CC amplicon during DCIS to IDC progression only in HER2+ER− tumors but retention of co-amplification in HER2+ER+ IDCs, and some of this selection might influence the immune microenvironment of the tumors.
DISCUSSION
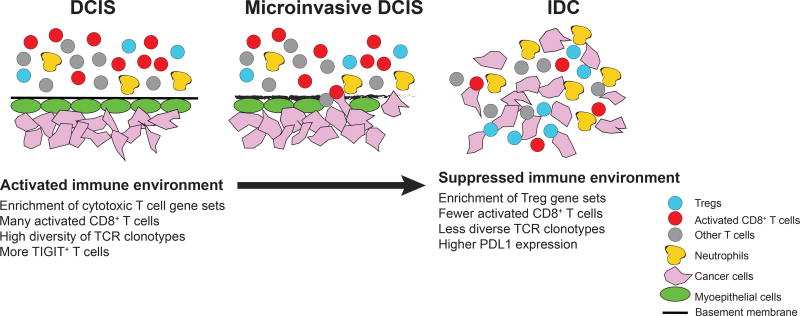
Evasion of immune surveillance is a necessary step in tumor evolution. Despite its importance, our understanding of mechanisms of immune escape in human tumors is rather limited. In DCIS the tumor cells are relatively protected from the immune system due to intact myoepithelial cell layer and basement membrane, and intraductal T cells are rarely detected (Fig. 6). In contrast, in invasive disease, cancer and immune cells are intermingled. Thus, the in situ to invasive carcinoma transition might be a particularly important bottleneck for immune escape and tumor evolution and the analysis of this could identify novel targets for immunotherapies, which has had limited success so far in breast cancer patients (1,31). One reason for this limited success might be that different tumors evade the immune system via different mechanisms, and thus, the identification and characterization of immune escape mechanisms is critical for the design of novel and more effective immunotherapies.
Figure 6.
Schematic model. Major changes in cell types and their activity during DCIS to IDC progression.
Here, we investigated potential mechanisms of immune escape in breast cancer by analyzing the composition and molecular profiles of leukocytes, with special emphasis on T cells, in normal breast tissues, DCIS, and IDC. Using clinical samples from normal and neoplastic breast tissues, we performed FACS analyses and RNA-seq on sorted T cells. Since tissue dissociation can influence the number of viable cells recovered and may also impact the detection of some antigens (the relatively high fraction of Lin− cells in our FACS may be due to this), we also tested selected genes by immunofluorescence on intact tissue sections. Using these approaches we determined that T cells in DCIS are enriched in activated effector CD8+ T cells characterized by the expression of GZMB and MKI67, but the frequencies of these cells decreases in invasive disease. This was particularly evident in DCIS cases that recurred locally as IDC, implying that decreased immune activity may be necessary for invasive progression. Currently there is no clinically useful biomarkers that would predict the risk of invasive progression of DCIS and considering that ∼40,000 women are diagnosed annually with DCIS in the USA alone, this is an important clinical problem. Our data suggest that the frequency of activated CD8+ T cells may predict which DCIS is likely to progress to invasive disease. Analysis of the TCR repertoire also displayed significantly higher clonotype diversity in DCIS compared to IDCs, and we identified several more prominent shared clones among cases. Interestingly one of these TCRs was predicted to react to an epitope in EBNA3, an Epstein-Barr virus protein. This finding does not necessarily imply a casual role for EBV in breast cancer, but it is consistent with a hypothesis that some T cells reactive to commensal microflora and common infectious agents may cross-react with some tumor neoantigens (32). However, further studies are required to test this hypothesis.
Several immune checkpoint proteins displayed significant differences between DCIS and IDC, particularly in TN cases. TIGIT+ T cells were more common in DCIS than in IDC, especially in TN subtype, whereas the expression of PD-L1 was almost undetectable in DCIS tumor epithelial cells but increased to higher levels in IDC with the amplification of the CD274 locus encoding PD-L1 in a subset (∼30%) of the cases. In Hodgkin’s lymphoma the presence of this same 9p24.1 amplicon is highly predictive of response to PD-1 blockade (33) raising the possibility that this might also be the case in TNBCs, but this remains to be explored. The expression of CTLA-4 was also higher in T cells from IDCs compared to DCIS further supporting the development of a suppressive immune microenvironment during invasive progression. The changes in the expression of these immune checkpoint proteins during DCIS to IDC transition suggest that the application of immunotherapies can be and may actually be more effective at earlier stages compared to metastatic disease. This hypothesis is supported by findings in animal models showing that the most efficient anti-tumor immune response was achieved when primary tumor and draining lymph nodes were still present (34,35). Thus, it would be useful to test whether immunotherapy could be more effective in breast cancer when applied in the neoadjuvant phase in combination with chemotherapy.
Most prior studies analyzing leukocytes and the prognostic value of TILs in breast cancer have focused on invasive tumors whereas DCIS and premalignant lesions have been relatively neglected. This is in part due to difficulties with obtaining fresh tissue samples from these small pre-invasive lesions that is required for comprehensive profiling studies and also due to the focus on advanced stage disease that is subject to systemic therapies including immunotherapy. Previous genomic characterization of DCIS cases have identified gene expression signatures implying the presence of activated T cells in a subset of tumors (36), but this was not confirmed by any other method. Prior studies have also shown that the frequencies of Tregs increased during tumor progression suggesting that this could be used to predict risk of invasive progression (13). However, in our experience the frequency of FOXP3+ T cells is very low in DCIS (<10% of T cells) and their topologic distribution is highly variable within tumors making their accurate assessment inaccurate in thin tissue sections (see Supplementary Methods). In contrast, activated GZMB+CD8+ and Ki67+CD8+ T cells were very common within DCIS implying that they could potentially be better biomarkers for predicting risk of invasive progression. Our observation that the frequencies of these cells is decreased in local invasive recurrence of DCIS supports this hypothesis, but further testing in large, uniformly-treated cohorts with long-term follow up would be required to determine this. It would also be useful to analyze each T cell subset in detail in both DCIS and IDC, possibly by single cell RNA-seq, as inferring the composition of T cells based on expression profiles has limitations.
We have also found evidence for co-evolution of cancer cells and leukocytes as exemplified by the negative association between the co-amplification of 17q12 chemokine cluster with ERBB2 in HER2+ breast tumors and presence of activated T cells within tumors. This co-amplification was more common in PAM50 luminal HER2+ IDCs compared to PAM50 HER2-enriched subset that may reflect different evolutionary paths for luminal, commonly ER+, and non-luminal ER− HER2+ tumors. Interestingly, the copy number gain was relatively low in a subset of luminal HER2+ tumors for both ERBB2 and the CC locus, which could imply higher intratumor heterogeneity in these tumors that can contribute to their lower response rate to HER2-targeted therapies. The lower frequency of TILs and particularly fewer activated GZMB+CD8+ T cells in these luminal HER2+ IDCs could further increase the probability of treatment resistance as the presence of these cells is associated with better response to both chemo and HER2-targeted therapies. Due to the high number of chemokines in this 17q12 cluster and their diverse function, dissecting the mechanism by which co-amplification of this locus impacts T cell activity would require future more detailed functional studies.
Immunoediting is a key step in tumor evolution that can be divided into elimination, equilibrium, and escape phases (37). Our data suggest that in DCIS, due to mostly intact basement membrane and myoepithelial cell layer relatively few cancer cells are exposed to immune cells and these can be mostly eliminated keeping the tumor in the intraductal “equilibrium” stage. Due to subsequent selection for cancer cells that have lost (or never had) neoepitopes and/or upregulated immunosuppressive mechanisms, tumor cells escape from immune surveillance and invasive and subsequent metastatic progression occurs. Therefore, the in situ to invasive breast carcinoma transition appears to be the most critical tumor progression step both from clinical and immunology standpoints. A better understanding of cellular and molecular changes that occur during DCIS to IDC transition could improve the design of immunotherapies for the treatment of both early and advanced stage disease. At the same time, our results also suggest that assessing the frequencies of activated CD8+ T cells in DCIS may identify patients with high risk of invasive progression.
METHODS
Human tissues and FACS
Fresh and archival formalin fixed paraffin embedded tissue samples were obtained from Brigham and Women’s Hospital/DF/HCC, Sutter Health, Washington University (St. Louis, MO), Yonsei University Medical College Gangnam Severance Hospital (Seoul, Korea), and Seoul National University Bundang Hospital (Seoul, Korea). Samples were de-identified prior to transfer to the lab. All Institutional Review Boards approved the protocol and waived the informed consent requirement. Fresh human normal and neoplastic breast tissues were dissociated as described (38). Polychromatic FACS was performed essentially as described (14). Briefly, single cell suspensions were blocked in PBS with 0.5%BSA, and 2 mM EDTA and stained at 4°C for 30 minutes with antibodies listed in Supplementary Table S8. Live-Dead Aqua (Invitrogen) was used to eliminate dead cells from the analysis and isotype control antibodies and unstained cells were used as negative controls. Single antibody staining was used for gating controls. Cells were analyzed and sorted using BD LSR Fortessa cell analyzer and BD FACSAria II SORP UV (Becton Dickinson), respectively. For RNA-seq, EpCAM+ cells were gated out to avoid epithelial contamination and CD45+CD3+ double positive cells were sorted into PBS or lysis buffer (1% 2-mercaptoethanol in RLT plus buffer, Qiagen). Associations between different cell populations was computed using Spearman’s correlation.
Histology and immunofluorescence analyses
Histology and multicolor immunofluorescence analyses were performed using 5µ sections of formalin-fixed paraffin-embedded (FFPE) tissues essentially as described (39). Briefly, slides were deparaffinized in xylene and hydrated in a series of descending ethanol. After heat-induced antigen retrieval in either citrate (pH=6) or TRIS-EDTA (pH=9) buffer, the samples were permeabilized with 0.5% TritonX-100, blocked with 5% goat serum-phosphate-buffered saline (PBS) and sequentially co-stained with antibodies as indicated in Supplementary Table S8. For TIGIT and CD33 staining, TSA indirect kit was used following manufacturer’s instructions (PerkinElmer). Image analysis was performed on 3×3 montage images acquired by Nikon Ti microscope attached to a Yokogawa spinning-disk confocal unit, 40× Plan Apo objective, and OrcaER camera controlled by the Andor iQ software. Masson’s trichrome staining was performed as indicated by the manufacturer (American MasterTech). Myeloid suppressor cells classification and staining was performed as recommended by Bronte and colleagues (40).
Multicolor FISH and immuno-FISH
FISH to analyze ERBB2 and 17q12 chemokine cluster was performed using whole sections of FFPE breast tumor samples as described (39). Briefly, 5µM FFPE tissue sections on silylated glass slides were baked overnight at 70°C, dewaxed in xylene (Leica Microsystems), washed in 100%, 70%, and 50% ethanol (Leica Microsystems), rinsed with H2O and air dried for 1h. Tissue digestion was performed by treating the samples with 0.08% Pepsin in PBS (DAKO) at 37°C for 15–22 minutes. Afterwards, the slides were washed in H2O and dehydrated in increasing concentrations of ethanol (50%, 70%, 85%, 100%), and air-dried. A FISH probe mix, containing ERBB2 BAC probe (RP11-94L15, provided by Drs. Hege Russness and Inga Rye, Oslo, Norway; labeled with SpectrumOrange dUTP by Nick Translation Kit; Abbott Molecular, according to the manufacturer’s recommendations), CEP17Aqua probe (Abbott Molecular) and 17q12 chemokine cluster BAC probes (equal concentration of RP11-791G14 and RP11-278F22 cDNA; labeled with SpectrumGreen by Nick Translation Kit) was then applied to the slides, covered with coverslip, and sealed with rubber cement. Hybridization was performed for 7 minutes at 75°C followed by overnight incubation at 37°C in a humid chamber. Next, the slides were washed in 0.4× SSC with 0.3% NP-40 for 2 minutes at room temperature, in 0.4× SSC with 0.3% NP-40 for 2 minutes at 74°C, then in 2× SSC with 0.1% NP-40, in 2× SSC, and in PBS. Nuclear counterstaining was performed by 10 min. incubation at room temperature with 1 µM To-Pro-3 in PBS (Molecular Probes, Life Technologies). After two additional washes in PBS and one in H2O, the slides were dried and mounted with Vectashield Mounting Medium (Vector Laboratories), covered with coverslips and stored overnight to 3 days at −20°C. Different immunofluorescence images from multiple areas of each sample were acquired with a Nikon Ti microscope attached to a Yokogawa spinning-disk confocal unit, 40× plan apo objective, and OrcaER camera controlled by Andor iQ software.
For the detection of CD274 (encoding PD-L1) gene and PD-L1 protein, slides were deparaffinized and digested with Proteinase K for 20min at 37°C before staining with PD-L1 antibody as described before. Co-ordinates of imaged areas were recorded. After imaging, the same procedure as used for multicolor FISH was followed using CEP9 probe (Abbott Molecular) and BAC probe 599H20 (Life Technologies) labeled with SpectrumOrange dUTP by Nick Translation Kit (Abbott Molecular). After hybridization and washes, slides were mounted using Vectashield with DAPI (Vector labs) and imaged using the set of co-ordinates from the first round of imaging. Images were then overlaid using R to compute overlaps and crop images.
RNA-seq and data analysis
RNA was isolated from purified CD45+CD3+ cells by cell sorting using antibodies described above. After approximately two thousand cells were sorted into 96-well plates with 2.5 µl PBS (containing 0.5 µl RNaseOut (Life Technologies) and 0.5 µl dithiothreitol (DTT, Life Technologies), cytoplasmic RNA was isolated as described previously (41). Briefly, 2.5 µl of 2× selected cytoplasm lysis buffer (SCLB) was added and the cells were lysed by pipetting. Lysates were centrifuged at 8,000 rpm for 5 min. at 4°C. Supernatants (∼5 µl) containing the total cytoplasmic RNA were transferred to PCR tubes. 5′-phosphorylated oligo-GdT24 (pGdT24) primer was used for reverse transcription by Superscript Reverse Transcriptase III (Life Technologies) followed by second strand synthesis. cDNAs were purified with Genomic DNA Clean & Concentrator kit (Zymo Research) followed by DNA blunt ending, 5′-end phosphorylation and ligation with End-It DNA End-Repair Kit (Epicentre) and T4 DNA ligase (Epicentre). Products were directly amplified using REPLI-g UltraFast Mini Kit (QIAGEN) and purified using Genomic DNA Clean & Concentrator kit (Zymo Research Corp, Irvine, CA). Products were fragmented to 200–500 bp by a Bioruptor Sonicator (Diagenode). Fragment sizes were validated using High Sensitivity DNA kit (Agilent Technologies) and Agilent 2100 Bioanalyzer. Functional concentrations were determined by qPCR with standards for Illumina sequencing libraries.
RNA-Seq libraries were (single-end) sequenced using the Illumina NextSeq 500 Next Gen Sequencer. Datasets were aligned to the human reference GRCh37/hg19 genome using the STAR RNA-Seq aligner (version STAR_2.5.1b) (42). Two-pass mapping was performed using the following modified parameters: --outSAMstrandField intronMotif, --outFilterMultimapNmax 20, --alignSJoverhangMin 8, --alignSJDBoverhangMin 1, --outFilterMismatchNmax 999, --outFilterMismatchNoverLmax 0.1, --alignIntronMin 20, --alignIntronMax 1000000, --alignMatesGapMax 1000000, --outFilterType BySJout, --outFilterScoreMinOverLread 0.33, --outFilterMatchNminOverLread 0.33, --limitSjdbInsertNsj 1200000, --chimSegmentMin 15, --chimJunctionOverhangMin 15, --twopassMode Basic. The read counts for individual genes were generated using the htseq-count script of the HTSeq framework (version 0.6.1p1) (43) using modified parameters (--stranded no) and the hg19 refGene annotation file available at the UCSC Genome Browser. Genes were filtered to retain only those with at least 45 counts across all samples. Differential gene expression analysis was performed with both edgeR (44) and DESeq2 (45) using design models which take into account batch effects. For PCA and heatmap visualizations, counts were converted to log2(counts per million) and batch adjusted using limma (46). Heatmap visualization was performed using the intersect of genes defined as differentially expressed using both edgeR and DESeq2 which also appear in the ImmPort database of immune related genes (47). Estimation of the relative proportions of different T cells was performed using CIBERSORT (22), using the author’s gene signatures for members of the T cell lineage and 100 permutations. Testing for significant differences among groups was performed using ANOVA.
Gene Set Enrichment Analysis
Gene set enrichment analysis (48) was performed using the HTSAnalyzeR package (49) using a Benjamini & Hochberg corrected p-value cut off of 0.05 to define significant gene sets. Two gene set collections from MSigDB were assessed: (i) curated canonical pathways (c2) to determine whether immune-related gene sets were differentially expressed and (ii) immunologic signatures (c7) reflecting genetic perturbations or differences in cell states (21). The c7 compendium contains 389 curated studies comparing differences in gene expression between cells of different lineages, or cells before and after treatment with specific chemokines. Thus, the multiple appearance of a gene set (e.g., Treg vs. CD4+ T cell) in independent studies would strongly support a difference in phenotype. Enriched gene sets were summarized by: (i) the frequency of specific immune cell types (e.g., CD4+, CD8+, thymocytes) appearing as differentially expressed, with significance calculated using proportionality testing with correction for multiple hypothesis testing and (ii) the corresponding directionality of these enriched gene sets in network diagrams. Node sizes in network diagrams were determined by the frequency of a given cell lineage appearing in the list of enriched gene sets. Edge weights were computed by the net number of gene sets supporting a link between cell types. For each network link, arrows are colored to highlight which group the test sample is more similar to (e.g., pink arrow in direction of CD4 T cell suggests DCIS T cells are more similar to this phenotype)
Immune signatures
A list of genes characteristic of T-cell activation, checkpoint inhibition, exhaustion, naïve T Cells, activation-specific and dysfunction-specific were manually curated from the literature (23,27,50) (Supplementary Table 4) A score for cytotoxicity, exhaustion, naïve, checkpoint activation and checkpoint inhibition was obtained for each sample by computing the mean logCPM value. To take into account differences in cell cycle, patients were grouped into ‘high’ and ‘low’ categories using a logCPM Ki67 threshold of −0.5.
TCR alignment and identification of clonotypic T cell repertoires
The clonotypic repertoire of T cells have been identified by aligning the RNA-seq reads to the human CDR regions using MiXCR (default parameters), a tool specific for immunoglobulin (IG) and T-cell receptor profiling (51). Only clonotypes with at least 2 reads were retained for further analyses. The clonotypes were then normalized using the number of sorted CD45+CD3+ T cells used for the RNA-Seq. The diversity of individual sample based on the number and abundance of different clonotypes is represented using Shannon index (24). The significance of the difference between DCIS, IDC, and normal samples based on clonotype diversity was calculated using Wilcoxon rank sum test. For a comparison of the Shannon index of individual samples and also their subtypes to the expression of selected cytotoxic genes, the raw read counts of the genes have been normalized based on library size using DESeq2 (45).
Copy number analyses
Copy number log2 ratios were obtained from publicly available data from TCGA (52) and from the Oslo cohort (9) using Affymetrix SNP6 arrays with logR ratios adjusted for ploidy and tumor percentage using the allele specific copy number analysis for tumors (ASCAT) algorithm (53). Copy number data was truncated to lie within a [-2, 2] range. To allow for intratumor heterogeneity, a relaxed log2 ratio of ±0.3 was used to define gains and losses. In the TCGA cohort, HER2+ patients were defined as either positive by PAM50 classification or by ERBB2 log2 Ratio of at least 0.3. In both cohorts, triple negative patients were defined using PAM50 Basal classification. Significance was determined using chi squared test.
Generalized linear models
Associations copy number gain were estimated using a linear model taking into account parameters including PAM50 subtype, neoantigen load (54), overall copy number aberration, immune content (55) and patient age.
Spatial, diversity, and statistical analyses
Association between DCIS/IDC cell type, ERBB2 and chemokine cluster amplification
The association between the probability of a cell carrying the 17q12 chemokine cluster (CC) amplification and ERBB2 amplification (together with the cell type (DCIS or IDC cells) in the DCIS/IDC cohorts) was estimated using logistic hierarchical model (or logistic mixed effects regression) (56) to account for the correlation of cells within each sample and of samples within each patient. In addition, we adjusted for important clinical covariates collected in this study including patients’ ages, ER status, and others. The selection of covariates from those collected was justified using the Akaike Information Criterion (AIC) (57). The models with the lowest AIC in both cohorts (AIC = 4412.2 in pure DCIS and 10909.7 in DCIS/IDC) were selected as the final model to interpret (Supplementary Table S5).
Diversity estimation from single cell FISH data
To compute the diversity from the multicolor FISH data, we first categorized each cell into the following four types based on copy number for ERBB2 and the chemokine cluster (CC): ERBB2+CC+, ERBB2+CC−, ERBB2−CC+, and ERBB2−CC−. Next, the number of cells belonging to each category was determined and the Shannon’s entropy (24) was thereby calculated for each sample of each patient. The association between the diversity and patients’ age was evaluated for both the pure DCIS and DCIS-IDC cohorts (Supplementary Fig. S7I). Interestingly, in the DCIS-IDC cohort, diversity was significantly associated with patients’ age (p-value = 0.0183 for DCIS cell type and p-value = 0.0146 for IDC cell type), whereas there was no such association supported by statistical significance in pure DCIS cohort. Furthermore, we also studied the association between diversity and CD3 and CD45 counts, and found no significant correlation between these two factors in both cohorts (Supplementary Fig. S7H).
Spatial analyses
We then considered another layer of information contained in the immuno-FISH images describing the spatial distribution of single cells. We sought to investigate the observed patterns relative to the null hypothesis – complete spatial randomness. Such analysis can help inform future evolutionary modeling of tumor development and intratumor heterogeneity. Here our definition of complete spatial randomness closely follows (58) (1) regardless of the type of cells, the cells are randomly localized on the 2D slice of a tumor by a spatial uniform Poisson process with an intensity parameter estimated from all cells on the image; (2) for each single cell, we randomly assign one of the four mutation types defined in the previous section based on the relative frequency of each mutation type again estimated from the image. This approach is also called marked spatial Poisson process (59). To test such complete spatial randomness, we first used Monte Carlo simulation to generate a number of simulated samples under the null hypothesis, and calculated the so-called “cross” K function (a function of the radius of the neighborhood of each cell) (58) using both the simulated samples and the patient sample. With a collection of simulated samples, we then constructed a 95% confidence band of the K function and determined if the K function computed from the patient data lies within the 95% confidence band under complete spatial randomness, to investigate whether the spatial distribution of different cell types in 2D deviates from the null hypothesis. Supplementary Fig. S7F shows that most of the patient samples do not significantly deviate from the complete spatial randomness, demonstrated by the K function of the data (black curve) lying within the 95% confidence band of the K function of the simulated datasets (grey area).
Analysis of invasiveness of tumor cells and T cells
Due to the relatively small number of cells subdivided by the TIGIT or PD-1 expression levels, we only considered the comparison between tumor cell populations and T cell populations in the image. In Supplementary Fig. S6A, we used different colors to distinguish T cells (black for TIGIT+ or PD-1+, red for TIGIT− or PD-1−) and tumor cells (green for TIGIT+ or PD-1+, gold for TIGIT− or PD-1−). We use k-means clustering (k = 2) to classify the cells on the image into two clusters based only on their coordinate information (or spatial distribution on the image) with an unsupervised approach. We then we computed the percentages of T cells (or equivalently the tumor cells) classified into the two clusters and chose the lower value as the “invasive percentage”, because intuitively, the more invasive the T cells are into the tumor cells, the percentage of T cells belonging to the two clusters should be closer to 50%, and hence there should be a higher “invasive percentage”. Consistent with our expectation (Supplementary Fig. S6B), in the “TIGIT” images (left panel), we observed a higher invasive percentage in “HER2+ IDC” than in “HER2+ DCIS” (p-value = 0.047) and also a higher invasive percentage in “TN IDC” than in “TN DCIS” (p-value = 0.003). However, the differences in “HER2+ IDC” and “HER2+ DCIS” are relatively weak. Similarly, in the “PD-1” images (right panel), we observed a higher invasive percentage in “TN IDC” than in “TN DCIS” (p-value = 0.039), but no significant difference between “HER2+ IDC” and “HER2+ DCIS” (p-value = 0.52).
All code for transcriptomic, spatial and statistical analysis can be found at github.com/polyak-lab/LeukocyteDCISIDC
Supplementary Material
SIGNIFICANCE.
The design of effective cancer immunotherapies requires the understanding of mechanisms underlying immune escape during tumor progression. Here we demonstrate a switch to a less active tumor immune environment during the in situ to invasive breast carcinoma transition, and identify immune regulators and genomic alterations that shape tumor evolution.
Acknowledgments
We thank John F. Daley in the Dana-Farber Cancer Institute Flow Cytometry and Lisa Cameron in Confocal and Light Microscopy Core facilities for their outstanding services and technical support. We thank Drs. Jane Grogan and Patrick Caplazi (Genentech) for their advice on TIGIT immunohistochemistry and Dr. Drew Pardoll (Johns Hopkins University) for his critical reading of our manuscript. We thank Dr. Maria Grazia Daidone, Fondazione IRCCS Instituto Nazionale dei Tumori, Milan, Italy, for a subset of the data included in the Oslo cohort.
Grant Support: This work was supported by the National Cancer Institute F32CA156991 (S.J.H.), R35CA197623 (K.P.) and U54CA193461 (K.P., F.M., R.F.), Susan G. Komen Foundation (Y.S.), and the Breast Cancer Research Foundation (K.P.).
Footnotes
ACCESSION CODES
Gene Expression Omnibus: RNA-Seq datasets have been deposited to GEO with accession number GSE87517. Reviewers’ link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87517
Disclosure of Potential Conflicts of Interest: The authors declare no competing financial interests.
References
- 1.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–41. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 2.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 3.Schedin P, O'Brien J, Rudolph M, Stein T, Borges V. Microenvironment of the involuting mammary gland mediates mammary cancer progression. J Mammary Gland Biol Neoplasia. 2007;12:71–82. doi: 10.1007/s10911-007-9039-3. [DOI] [PubMed] [Google Scholar]
- 4.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Man YG, Sang QX. The significance of focal myoepithelial cell layer disruptions in human breast tumor invasion: a paradigm shift from the “protease-centered” hypothesis. Exp Cell Res. 2004;301:103–18. doi: 10.1016/j.yexcr.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 6.Abba MC, Gong T, Lu Y, Lee J, Zhong Y, Lacunza E, et al. A Molecular Portrait of High-Grade Ductal Carcinoma In Situ. Cancer Res. 2015;75:3980–90. doi: 10.1158/0008-5472.CAN-15-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent-Salomon A, Lucchesi C, Gruel N, Raynal V, Pierron G, Goudefroye R, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956–65. doi: 10.1158/1078-0432.CCR-07-1465. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen VN, Vaske CJ, Ursini-Siegel J, Van Loo P, Nordgard SH, Sachidanandam R, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1108781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesurf R, Aure MR, Mork HH, Vitelli V, Oslo Breast Cancer Research C. Lundgren S, et al. Molecular Features of Subtype-Specific Progression from Ductal Carcinoma In Situ to Invasive Breast Cancer. Cell Rep. 2016;16:1166–79. doi: 10.1016/j.celrep.2016.06.051. [DOI] [PubMed] [Google Scholar]
- 10.Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–8. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 11.Thompson E, Taube JM, Elwood H, Sharma R, Meeker A, Warzecha HN, et al. The immune microenvironment of breast ductal carcinoma in situ. Mod Pathol. 2016;29:249–58. doi: 10.1038/modpathol.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikandar B, Qureshi MA, Mirza T, Khan S, Avesi L. Differential immune cell densities in ductal carcinoma In-Situ and invasive breast cancer: Possible role of leukocytes in early stages of carcinogenesis. Pak J Med Sci. 2015;31:274–9. doi: 10.12669/pjms.312.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lal A, Chan L, Devries S, Chin K, Scott GK, Benz CC, et al. FOXP3-positive regulatory T lymphocytes and epithelial FOXP3 expression in synchronous normal, ductal carcinoma in situ, and invasive cancer of the breast. Breast Cancer Res Treat. 2013;139:381–90. doi: 10.1007/s10549-013-2556-4. [DOI] [PubMed] [Google Scholar]
- 14.Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Breast Cancer Special Feature: Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24:5373–80. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- 16.Semeraro M, Adam J, Stoll G, Louvet E, Chaba K, Poirier-Colame V, et al. The ratio of CD8+/FOXP3 T lymphocytes infiltrating breast tissues predicts the relapse of ductal carcinoma in situ. Oncoimmunology. 2016;5:e1218106. doi: 10.1080/2162402X.2016.1218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–8. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma C, Zhang Q, Ye J, Wang F, Zhang Y, Wevers E, et al. Tumor-infiltrating gammadelta T lymphocytes predict clinical outcome in human breast cancer. J Immunol. 2012;189:5029–36. doi: 10.4049/jimmunol.1201892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sautes-Fridman C, Lawand M, Giraldo NA, Kaplon H, Germain C, Fridman WH, et al. Tertiary Lymphoid Structures in Cancers: Prognostic Value, Regulation, and Manipulation for Therapeutic Intervention. Front Immunol. 2016;7:407. doi: 10.3389/fimmu.2016.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, et al. Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. and 623-56. [Google Scholar]
- 25.Srivastava SK, Robins HS. Palindromic nucleotide analysis in human T cell receptor rearrangements. PLoS ONE. 2012;7:e52250. doi: 10.1371/journal.pone.0052250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koning D, Costa AI, Hoof I, Miles JJ, Nanlohy NM, Ladell K, et al. CD8+ TCR repertoire formation is guided primarily by the peptide component of the antigenic complex. J Immunol. 2013;190:931–9. doi: 10.4049/jimmunol.1202466. [DOI] [PubMed] [Google Scholar]
- 27.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166:1500–11. doi: 10.1016/j.cell.2016.08.052. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chauvin JM, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, et al. TIGIT and PD-1 impair tumor antigen-specific CD8(+) T cells in melanoma patients. J Clin Invest. 2015;125:2046–58. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V, et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 2008;68:9532–40. doi: 10.1158/0008-5472.CAN-08-3082. [DOI] [PubMed] [Google Scholar]
- 30.Barrett MT, Anderson KS, Lenkiewicz E, Andreozzi M, Cunliffe HE, Klassen CL, et al. Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget. 2015;6:26483–93. doi: 10.18632/oncotarget.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory Pathways in Immunotherapy for Cancer. Annu Rev Immunol. 2016;34:539–73. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 32.Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur J Immunol. 2015;45:17–31. doi: 10.1002/eji.201444972. [DOI] [PubMed] [Google Scholar]
- 33.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372:311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell. 2017;168:487–502. doi: 10.1016/j.cell.2016.12.022. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Blake SJ, Yong MC, Harjunpaa H, Ngiow SF, Takeda K, et al. Improved Efficacy of Neoadjuvant Compared to Adjuvant Immunotherapy to Eradicate Metastatic Disease. Cancer Discov. 2016;6:1382–99. doi: 10.1158/2159-8290.CD-16-0577. [DOI] [PubMed] [Google Scholar]
- 36.Kristensen VN, Vaske CJ, Ursini-Siegel J, Van Loo P, Nordgard SH, Sachidanandam R, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A. 2012;109:2802–7. doi: 10.1073/pnas.1108781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward JP, Gubin MM, Schreiber RD. The Role of Neoantigens in Naturally Occurring and Therapeutically Induced Immune Responses to Cancer. Adv Immunol. 2016;130:25–74. doi: 10.1016/bs.ai.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–27. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan X, Durrett RE, Zhu H, Tanaka Y, Li Y, Zi X, et al. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc Natl Acad Sci U S A. 2013;110:594–9. doi: 10.1073/pnas.1217322109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–9. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 48.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Terfve C, Rose JC, Markowetz F. HTSanalyzeR: an R/Bioconductor package for integrated network analysis of high-throughput screens. Bioinformatics. 2011;27:879–80. doi: 10.1093/bioinformatics/btr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–1. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 52.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCulloch CE, Neuhaus JM. Generalized linear mixed models. Wiley Online Library. 2001 [Google Scholar]
- 57.Akaike H. A new look at the statistical model identification. IEEE transactions on automatic control. 1974;19:716–23. [Google Scholar]
- 58.Baddeley A, Turner R. Spatstat: an R package for analyzing spatial point patterns. Journal of statistical software. 2005;12:1–42. [Google Scholar]
- 59.Lotwick H, Silverman B. Methods for analysing spatial processes of several types of points. Journal of the Royal Statistical Society Series B (Methodological) 1982:406–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.