Abstract
Phytochemical investigation of Premna odorata Blanco, Lamiaceae, leaves afforded three new acylated iridoid glycosides 1–3, and two new acylated rhamnopyranoses 9 and 10, in addition to ten known compounds. The structures of the new compounds were confirmed using extensive 1D and 2D NMR analysis. Molecular modeling study suggested the potential of the acylated rhamnopyranoses to bind at the c-Met kinase domain. Cell free Z’-LYTE assay testing revealed the good c-Met phosphorylation inhibitory activity of 9, followed by 8, and 10, with IC50 values of 2.5, 6.9 and 12.7 µM, respectively. The MTT cell proliferation assay testing against the human c-Met-expressing highly invasive MDA-MB-23 suggested compound 9 as the most active with IC50 value of 13.3 µM. Testing of compound 9 against multiple phenotypic breast cancer cell lines including MCF-7, BT-474 cells, and MDA-MB-468 proved enhanced activity against the highly c-Met expressing triple-negative breast cancer cell lines. Acylated rhmnopyranoses are potential novel c-Met inhibitors appropriate for future optimizations to control c-Met-dependent breast malignancies.
Keywords: Premna odorata, Iridoid glycosides, Acylated rhamnopyranoses, Molecular modeling, c-Met inhibitors
INTRODUCTION
Premna odorata Blanco, Lamiaceae, popularly known as “fragrant Premna” is a small tree native to temperate and tropical Asia, including the Philippines, and cultivated in different global locations (Lirio et al., 2014). In the Philippines; a decoction of the leaves was considered as diuretic, carminative and febrifuge; and used for vaginal irrigation; coughs; beri-beri; abdominal pains and dysentery (Lirio et al., 2014, Pinzon et al., 2011). The saturated linear fatty acid ester 1-heneicosyl formate was identified as the most potent antituberculosis ingredient of P. odorata leaves, with MIC of 8 µg/mL (Lirio et al., 2014). Generally; P. odorata is considered among the top plant sources of acylated iridoid glycosides and rhmnopyranoses (Zhang et al., 2016, Cui, 2013, Otsuka et al., 1992, Otsuka et al., 1991, Otsuka et al., 1989). P. odorata iridoids exhibited a wide range of pharmacological activities as antimutagenic, antitumor, antiviral and anti-inflammatory (Lirio et al., 2014, Viljoen et al., 2012, Pinzon et al., 2011,).
Breast cancer is the most commonly diagnosed cancer in women, with >40,000 expected women deaths in 2017 (Siegel et al., 2015). The triple negative breast cancer (TNBC) lacks positive staining for the estrogen receptor, progesterone receptor (PR) and human epidermal growth factor 2, therefore, TNBC is resistant to the selective effective regimens targeting these receptors like HER2-directed and endocrine therapies (Lehmann et al., 2011).
c-Met is an RTK proto-oncogene, which triggers activation of several critical downstream signaling pathways (Zhang et al., 2016, Furlan et al., 2014, Cui, 2013). c-Met dysregulation correlates with aggressive proliferation, invasive character, and pathological motility, especially in TNBC (Zhang et al., 2016, Furlan et al., 2014, Cui, 2013). Met amplification also correlates with escape from the anticancer effects of EGFR inhibitors and cetuximab (Zhang et al., 2016, Cui, 2013). More than 240 small-molecule c-Met inhibitors are currently in different clinical trial phases, with potential to delay the progression of several Met-dependent malignancies (Zhang et al., 2016, Furlan et al., 2014, Cui, 2013). Crizotinib (Xalkori) is a dual Met and ALK inhibitor approved by FDA for ALK-driven lung cancer (Zhang et al., 2016, Furlan et al., 2014, Cui, 2013). Cabozantinib (Cometriq) was approved by FDA in 2012 for medullary thyroid cancer (Furlan et al., 2014, Cui, 2013). Validated efficacies of these marketed agents, as well as of promising ones still undergoing clinical trials, validates c-Met as a molecular target in cancer therapy.
Few c-Met inhibitory natural products were identified (Akl et al., 2014). The current study describes the five new natural products from P. odorata and the discovery of its acylated rhmnopyranoses as a novel c-Met inhibitory class with activities against a panel of c-Met-dependent human breast cancer cell lines.
MATERIALS AND METHODS
General experimental procedures
The 1H, 13C and 2D NMR spectra were recorded at 400 and 100 MHz, respectively, using TMS as internal standard in methanol-d4, using the residual solvent peak (δH= 3.34 and δC= 49.9) as references, on Bruker Avance III 400 MHz with BBFO Smart Probe and Bruker 400 MHz AEON Nitrogen-Free Magnet (Bruker AG, Switzerland). Carbon multiplicities were determined using DEPT-Q experiments. The optical rotation values were determined using a Jasco P-1020 polarimeter (Easton, MD, USA). The UV spectrum in methanol was obtained using a Shimadzu UV 2401PC spectrophotometer. The IR spectra were obtained using a Jasco, FT IR 300E infrared spectrophotometer. ESI-MS data were obtained using a Thermo Scientific LTQ/XL Orbitrap (Waltham, MA USA). Column chromatography was performed using silica gel 60 (63–200 µm, Fluka, Sigma-Alderich, Germany); Polyamide-6 (50–160 µm, Sigma-Aldrich, Germany) and Sephadex LH-20 (Sigma-Aldrich, Germany). HPLC separations were conducted using an Agilent 1260 Infinity preparative pump (G1361A), Agilent 1260 Diode array detector VL (G1315 D), Agilent 1260 Infinity Thermostand column compartment (G1361 A) and Agilent 1260 Infinity preparative Autosampler (G2260A). YMC-Pack ODS-A A-324 column (i.d. 10 × 300 mm, YMC, Kyoto, Japan).
Plant material
P. odorata leaves were collected between the third week of March and the second week of May 2016 from the Giza Zoo garden, Egypt. P. odorata was kindly identified by Dr. Abd El-Halim A. Mohammed, Horticultural Research Institute, Department of Flora and Phytotaxonomy Researches, Dokki, Cairo, Egypt. A voucher specimen (2016-BuPD 45) was deposited at Department of Pharmacognosy, Faculty of Pharmacy, Beni Suef University, Egypt.
Extraction and isolations
Air-dried and finely powdered P. odorata leaves (2 kg) were exhaustively extracted with 70% ethanol (5L, 3X) at room temperature and concentrated under vacuum to afford 350 g crude extract. Dry extract was suspended in 400 mL distillated water and successively portioned with solvents of different polarities (n-hexane, CH2Cl2, EtOAc, and n-butanol). A portion of EtOAc extract (8 g) was fractionated on polyamide using gradient elution starting with H2O and ending with MeOH to obtain six fractions F1-F6. Fractions F2 and F3 eluted with H2O-MeOH, 8:2, were separately chromatographed on Sephadex LH20, eluted with MeOH. This was followed by chromatography on Si gel 60, eluting with CH2Cl2-MeOH (gradient elution) to afford compounds 9 (20 mg) and 10 (7mg). Fraction F3 eluted with H2O-MeOH, 6:4, was subjected to Sephadex LH20 chromatography, eluted isocratically with MeOH, affording compound 15 (123 mg). Fraction F5 eluted with H2O-MeOH, 2:8, was fractionated on Si gel 60 followed by Sephadex LH20 chromatography, affording compounds 11–13 (16 mg, 8 mg and 3 mg, respectively).
A portion of n-butanol extract (25g) was fractionated on polyamide using H2O-MeOH gradient to obtain fractions F1-F5. Fraction F2 eluted with H2O-MeOH; 8:2 was sub-fractionated on Si gel 60 followed by Sephadex LH20 to obtain two sub-fractions. One of these subfrations was further subjected to C-18 RP-HPLC using H2O-CH3CN (10–60%, 30 min, 5 mL/min) followed by preparative TLC on Si gel using EtOAc-AcOH- HCOOH-H2O, 10:1:1:1, to obtained compounds 1 (10 mg), 2 (8 mg), 4 (13 mg), 5 (9 mg), and 8 (7 mg). The other sub-fraction was subjected to Sephadex LH20 chromatography followed by preparative TLC on Si gel to obtain compounds 3 (10 mg) and 6 (7 mg). Fraction F4 eluted with H2O-MeOH; 5:5 was repeatedly sub-fractionated on Si gel 60 and polyamide to obtain compound 7 (18 mg). Fraction F5 eluted with H2O-MeOH; 3:7 was sub-fractionated on Si gel 60 followed by Sephadex LH20 to obtain compound 14 (8 mg).
Premnoside F (1)
Pale yellow amorphous solid; (c 0.05, MeOH); UV (MeOH) λmax (log ε) 225 (5.9), 310 (4.2), 318 (4.8), 341(4.4) nm; IR υmax (KBr) 3422, 2924, 1699, 1632, 1604, 1570, 1267, 1015, 835 cm−1; NMR data: see Table 1; HRESIMS m/z 801.2621 [M+H]+ (calc. for C39H44O18, 801.2606).
Table 1.
13C and 1H NMR Data of Compounds 1–3a
| Position | 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|---|
| Aglycone | 1, CH | 93.8 | 5.08, d (9.6) | 93.8 | 5.05, d (9.6) | 93.9 | 5.05, d (9.6) |
| 3, CH | 140.9 | 6.39, d (6.5) | 140.9 | 6.40, d (.56) | 140.9 | 6.39, d (6.5) | |
| 4, CH | 102.1 | 5.08, dd (6.5, 4.7) | 102.1 | 5.13, dd (6.5, 4.7) | 102.3 | 5.13, dd (6.5, 4.7) | |
| 5, CH | 35.9 | 2.46, m | 35.9 | 2.46, m | 35.9 | 2.46, m | |
| 6, CH | 82.4 | 4.02, dd (8, 1.6) | 82.4 | 4.02, dd (8, 1.6) | 82.4 | 4.02, dd (8, 1.6) | |
| 7, CH | 58.0 | 3.68, m | 58.1 | 3.68, m | 58.0 | 3.68, m | |
| 8, qC | 65.3 | 65.2 | 65.3 | ||||
| 9, CH | 41.8 | 2.59, dd (9.6, 7.5) | 41.8 | 2.59, dd (9.6, 7.5) | 41.9 | 2.59, dd (9.6, 7.5) | |
| 10, CH2 | 60.1 | 3.85, d (12) | 60.1 | 3.85, d (12) | 60.0 | 3.85, d (12) | |
| 4.17, d (12) | 4.17, d (12) | 4.17, d (12) | |||||
| β-D-Gluc. | 1′, CH | 98.3 | 4.80, d (8) | 98.3 | 4.79, d (8) | 98.3 | 4.79, d (8) |
| 2′, CH | 73.4 | 3.31, dd (8, 9.3) | 73.4 | 3.31, dd (8, 9.3) | 73.4 | 3.31, dd (8, 9.3) | |
| 3′, CH | 76.2 | 3.46, t (9.3) | 76.2 | 3.46, t (9.3) | 76.2 | 3.46, t (9.3) | |
| 4′, CH | 70.3 | 3.32, t (9.3) | 70.4 | 3.32, t (9.3) | 70.3 | 3.32, t (9.3) | |
| 5′, CH | 77.2 | 3.34, m | 77.2 | 3.34, m | 77.2 | 3.34, m | |
| 6′, CH2 | 61.5 | 3.67, dd (12, 6.6) | 61.5 | 3.67, dd (12, 6.6) | 61.5 | 3.67, dd (12, 6.6) | |
| 3.93, dd (12, 2) | 3.93, dd (12, 2) | 3.93, dd (12, 2) | |||||
| α-L-Rhamn. | 1″, CH | 98.8 | 5.00, d (1.6) | 98.8 | 5.00, d (1.6) | 98.9 | 5.00, d (1.6) |
| 2″, CH | 69.1 | 3.98, dd (1.6, 3.6) | 69.1 | 3.96, dd (1.6, 3.6) | 69.1 | 3.96, dd (1.6, 3.6) | |
| 3″, CH | 72.8 | 5.18, dd (3.6, 9.4) | 72.8 | 5.18, dd (3.6, 9.4) | 72.9 | 5.18, dd (3.6, 9.4) | |
| 4″, CH | 74.0 | 5.11, t (9.4) | 74.0 | 5.11, t (9.4) | 73.9 | 5.11, t (9.4) | |
| 5″, CH | 69.0 | 3.83, m | 68.9 | 3.83, m | 69.0 | 3.83, m | |
| 6″, CH3 | 16.7 | 1.33, d (6) | 16.6 | 1.33, d (6) | 16.6 | 1.28, d (6) | |
| trans-p-Coum. | 1‴, qC | 126.0 | 127.0 | 125.7 | |||
| 2‴/6‴,CH | 130.0 | 7.49, d (8) | 129.8 | 7.59, d (8) | 129.8 | 7.49, d (8) | |
| 3‴/5‴ CH | 115.5 | 6.84, d (8) | 114.1 | 6.98, d (8) | 115.6 | 6.84, d (8) | |
| 4‴, qC | 160.0 | 161.9 | 160.0 | ||||
| 7‴, CH | 146.0 | 7.67, d (16) | 145.5 | 7.71, d (16) | 145.5 | 7.61, d (16) | |
| 8‴, CH | 114.0 | 6.40, d (16) | 115.0 | 6.46, d (16) | 113.8 | 6.40, d (16) | |
| 9‴, qC | 167.6 | 167.3 | 167.5 | ||||
| 10‴, OCH3 | 54.5 | 3.84, s | |||||
| trans-p-Coum. | 1″″, qC | 125.8 | 126.9 | ||||
| 2″″/6″″ CH | 129.9 | 7.49, d (8) | 129.6 | 7.59, d (8) | |||
| 3″″/5″″CH | 115.5 | 6.84, d (8) | 114.1 | 6.98, d (8) | |||
| 4″″, qC | 159.8 | 161.8 | |||||
| 7″″, CH | 145.5 | 7.71, d (16) | 145.0 | 7.79, d (16) | |||
| 8″″, CH | 113.5 | 6.45, d (16) | 114.4 | 6.50, d (16) | |||
| 9″″, qC | 167.4 | 167.1 | |||||
| 10″″, OCH3 | 54.5 | 3.84, s | |||||
| trans-Caff. | 1″″, qC | 125.8 | |||||
| 2″″, CH | 114.1 | 7.10, d (2.1) | |||||
| 3″″, C | 148.2 | ||||||
| 4″″, C | 145.3 | ||||||
| 5″″, CH | 115.2 | 6.83, d (8) | |||||
| 6″″, CH | 121.7 | 6.98, dd (2.1, 8) | |||||
| 7″″, CH | 145.9 | 7.68, d (16) | |||||
| 8″″, CH | 113.9 | 6.46, d (16) | |||||
| 9″″, C | 167.7 | ||||||
In CD3OD, 400 MHz for 1H, 100 MHz for 13C NMR, J in Hz, carbon multiplicities were determined by DEPT-Q experiments, qC = quaternary, CH = methine, CH2 = methylene, CH3 = methyl carbons.
Premnoside H (2)
Pale yellow amorphous solid; (c 0.08, MeOH); UV (MeOH) λmax (log ε) 230 (5.4), 308 (3.0), 315 (3.3), 330 (2.8) nm; IR υmax (KBr) 3429, 2927, 1712, 1670, 1630, 1354, 1073, 864 cm−1; NMR data: see Table 1; HRESIMS m/z 851.2758 [M+Na]+(calc. for C41H48O18, 851.2738).
Premnoside I (3)
Pale yellow amorphous solid; (c 0.1, MeOH); UV (MeOH) λmax (log ε) 222 (5.6), 275 (2.0), 309 (3.4), 321(4.0), 335 (3.8) nm; IR υmax (KBr) 3431, 2922, 1666, 1603, 1070, 850 cm−1; NMR data: see Table 1; HRESIMS m/z 817.2553 [M+H]+ (calc. for C39H44O19, 817.2555).
1-O-trans-p-coumaroyl-2-O-trans-caffeoyl-α-L-rhamnopyranose (9)
Pale yellow amorphous solid; (c 0.04, MeOH); UV (MeOH) λmax (log ε) 218 (sh), 225 (6.0), 310 (3.0), 322 (2.8), 352 (3.2) nm; IR υmax (KBr) 3421, 2930, 1692, 1604, 1270, 1167, 1056, 850 cm−1; NMR data: see Table 2; HRESIMS m/z 473.1426 [M+H]+ (calc. for C24H24O10, 473.1448).
Table 2.
13C and 1H NMR Data of Compounds 9 and 10a
| Position | 9 | 10 | |||
|---|---|---|---|---|---|
| α-L-Rhamn. | 1, CH | 91.9 | 5.19, d (1.6) | 94.4 | 5.14, d (1.6) |
| 2, CH | 73.7 | 5.19, dd (2, 3.6) | 69.7 | 4.09, dd (2, 3.6) | |
| 3, CH | 69.1 | 4.11, dd (3.6, 9.4) | 74.0 | 5.24, dd (3.6, 9.4) | |
| 4, CH | 73.2 | 3.62, t (9.4) | 70.4 | 3.72, t (9.4) | |
| 5, CH | 68.3 | 3.99, m | 68.1 | 3.94, m | |
| 6, CH3 | 17.0 | 1.34, d (6) | 17.0 | 1.33, d (6) | |
| trans-p-Coum. | 1′, qC | 125.9 | 126.0 | ||
| 2′/6′, CH | 130.1 | 7.39, d (8) | 130.0 | 7.39, d (8) | |
| 3′/5′, CH | 115.6 | 6.81, d (8) | 115.6 | 6.81, d (8) | |
| 4′, qC | 159.6 | 159.5 | |||
| 7′, CH | 146.0 | 7.66, d (16) | 146.0 | 7.66, d (16) | |
| 8′, CH | 113.6 | 6.38, d (16) | 113.6 | 6.38, d (16) | |
| 9′, qC | 168.0 | 167.9 | |||
| trans-Caff. | 1″, qC | 126.5 | 126.7 | ||
| 2″, CH | 114.2 | 7.08, d (2.4) | 114.2 | 7.08, d (2.4) | |
| 3″, qC | 148.0 | 148.1 | |||
| 4″, qC | 145.1 | 145.1 | |||
| 5″, CH | 115.4 | 6.81, d (8) | 115.4 | 6.81, d (8) | |
| 6″, CH | 122.1 | 6.91, dd (2.4, 8) | 122.0 | 6.91, dd (2.4, 8) | |
| 7″, CH | 145.7 | 7.57, d (16) | 146.4 | 7.57, d (16) | |
| 8″, CH | 114.0 | 6.31, d (16) | 114.0 | 6.31, d (16) | |
| 9″, qC | 168.0 | 167.8 | |||
In CD3OD, 400 MHz for 1H, 100 MHz for 13C NMR, J in Hz, carbon multiplicities were determined by DEPT-Q experiments, qC = quaternary, CH = methine, CH2 = methylene, CH3 = methyl carbons.
1-O-trans-p-coumaroyl-3-O-trans-caffeoyl-α-L-rhamnopyranose (10)
Pale yellow amorphous solid; (c 0.04, MeOH); UV (MeOH) λmax (log ε) 214 (sh), 225 (5.8), 315 (2.8), 330 (3.0), 355 (3.0) nm; IR υmax (KBr) 3421, 2933, 1692, 1604, 1275, 1167, 1056, 850 cm−1; NMR data: see Table 2; HRESIMS m/z 473.1426 [M+H]+ (calc. for C24H24O10, 473.1448).
Molecular modeling studies
The in silico experiments were carried out using the Schrödinger molecular modeling software package installed on an iMac 27-inch Z0PG workstation with a 3.5 GHz Quad-core Intel Core i7, Turbo Boost up to 3.9 GHz, processor, and 16 GB RAM (Apple, Cupertino, CA, USA).
The X-ray crystal structures of the c-Met tyrosine kinase domain PDB codes 2WD1, 4XYF, and 3CD8 were retrieved from the Protein Data Bank. The Protein Preparation Wizard was implemented to prepare the kinase domain of c-Met protein (Olsson et al., 2011, Schiering et al., 2003). Each protein was reprocessed by assigning bond orders, adding hydrogens, creating disulfide bonds, and optimizing H-bonding networks using PROPKA (Jensen Research Group, Denmark). Finally, energy minimization with RMSD value of 0.3°A was applied using an Optimized Potentials for Liquid Simulation (OPLS_2005, Schrödinger, New York, USA) force field.
The chemical structure of each compound was sketched on the Maestro 9.3 panel interface (Maestro, version 9.3, 2012, Schrödinger, USA). The Lig Prep 2.3 module (Lig Prep, version 2.3, 2012, Schrödinger, USA) was implemented to generate the 3D structure and to search for different conformers. The OPLS (OPLS_2005, Schrödinger, USA) force field was applied to geometrically optimize each ligand structure and to compute partial atomic charges. Finally, 32 poses per ligand were generated with different steric features for subsequent docking studies.
The prepared X-ray crystal structure of c-Met was used to generate receptor energy grids applying the default value of the protein atomic scale (1.0°A) within the cubic box centered on the co-crystallized ligand of c-Met crystal structure. Compounds were then docked using the Glide 5.8 module (Glide, version 5.8, 2012, Schrödinger, USA) in extra-precision (XP) mode (Friesner et al., 2006). Modeling scores were generated using the Glide-Dock program’s empirical scoring functions.
Biochemical cell free kinase assay
The Z’-LYTE™ Kinase Assay-Tyr6 Peptide kit (Life Sciences) was used to assess the ability of in silico hits to inhibit the catalytic activity of c-Met kinase (Product# PV3143) (Mohyeldin et al., 2016). Briefly, 20 µL/well reactions were set up in 96-well plates containing kinase buffer, 200 µM ATP, 4 µM Z’-LYTE™ Tyr6 Peptide substrate, 2500 ng mL−1 c-Met kinase and tested compound as an inhibitor. After 1 h of incubation at rt, 10 µL development solution containing site-specific protease was added to each well. Incubation was continued for 1 h. The reaction was then stopped, and the fluorescent signal ratio of 445 nm (coumarin)/520 nm (fluorescein) was determined on a plate reader (BioTek FLx800™), which reflects the peptide substrate cleavage and kinase inhibitory activity in the reaction (Mohyeldin et al., 2016).
Cell lines, culture conditions and cell viability assessments
The human breast cancer cell lines were purchased from the ATCC (Manassas, VA). All cancer cell lines were maintained in RPMI-1640 (Gibco® by Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products); 100 U/mL penicillin G, 100 µg/mL streptomycin and 2 mmol/L glutamine. Cells were subcultured upon attaining 80% confluency. All cells were maintained at 37°C in a humidified incubator under 5% CO2. A stock solution was prepared by dissolving each tested compound in sterilized DMSO at a concentration of 10 mM for all assays. Working solutions at their final concentrations for each assay were prepared in appropriate culture medium immediately prior to use. The vehicle (DMSO) control was prepared by adding the maximum volume of DMSO, used in preparing test compounds, to the appropriate media type such that the final DMSO concentration was maintained as the same in all treatment groups within a given experiment and never exceeded 0.1%. (−)-Oleocanthal was used in all experiments as a positive control based on earlier studies (Akl et al., 2014, Mohyeldin et al., 2016).
Viable cell count was determined using the MTT colorimetric assay. The optical density of each sample was measured at 570 nm on a Synergy 2 microplate reader (BioTek, VT, USA). The number of cells per well was calculated against a standard curve prepared at the start of each experiment by plating various concentrations of cells (1,000–60,000 cells per well), as determined using a hemocytometer.
Cell proliferation assay
Breast cancer cells, in exponential growth, were seeded at a density of 1×104 cells per well (6 wells/group) in 96-well culture plates and maintained in RPMI-1640 media supplemented with 10% FBS and allowed to adhere overnight at 37°C under 5% CO2 in a humidified incubator (Holliday and Speirs, 2011). The next day, cells were washed with phosphate buffer saline (PBS), divided into different treatment groups and then fed serum-free defined RPMI-1640 media containing HGF (40 ng/mL as a mitogen which induced maximum growth in the two cell lines after 48 h) and experimental treatments (containing designated concentrations of the tested compounds) or vehicle-treated control media and incubation resumed at 37°C under 5% CO2 for 48 h. Control and treatment media were then removed, replaced with fresh media, and 50 µL fresh MTT solution (1 mg mL−1) was added to each well and plates were re-incubated for 4 h at 37 °C. The color reaction was stopped by removing the media and adding 100 µL DMSO in each well to dissolve the formed formazan crystals. Incubation at 37°C was resumed for up to 20 minutes to ensure complete dissolution of crystals. Absorbance was determined at λ 570 nm using an ELISA plate microreader (BioTek, Winooski, VT, USA) (Mohyeldin et al., 2016). The % cell survival was calculated as follows: % cell survival = (Cell No.treatment/Cell No.DMSO) × 100.
Cell migration assay
MDA-MB-231 cells were plated in sterile 24-well plates and allowed to form a confluent monolayer per well overnight (Mohyeldin et al., 2016, Akl et al., 2014). Wounds were then inflicted in each cell monolayer using a sterile 200 µL pipette tip. The media were removed and cells were washed twice with PBS and once with fresh RPMI medium to remove cell debris. Test compound concentrations were prepared in fresh serum-free defined media, containing 40 ng/mL HGF as a mitogen, and were added to wells in triplicate. Cells were incubated for 24 h after which, the medium was removed and cells were washed, fixed, and stained using Giemsa stain. Wound healing was visualized at 0 and 24 h by a Nikon ECLIPSE TE200-U microscope and digital images were captured using Nikon NIS Elements software (Nikon Instruments Inc., Melville, NY). The distance traveled by the cells was determined by measuring the wound width after 24 h. Percentages cell migration were calculated using the following formula:
Where, T0 is wound thickness at zero time, Tdmso is wound thickness in DMSO-treated control wells and Tt is wound thickness in treated wells.
Statistical analysis
All in vitro experiments were performed in triplicates. Pooled data are presented as the means ± standard error of mean (SEM) of at least three independent experiments. Differences among various treatment groups were determined by ANOVA followed by Dunnett’s test using PASW statistics® version 18 (Quarry Bay, Hong Kong). A difference of p<0.05 was considered statistically significant compared to the vehicle-treated control group and indicated by * symbol. A difference of p<0.05 was considered statistically significant compared to the oleocanthal-treated control group and indicated by # symbol. The IC50 values were determined using a non-linear regression curve fitting analysis using GraphPad Prism software version 6 (La Jolla, CA, USA).
RESULTS AND DISCUSSION
Phytochemical investigation of P. odorata
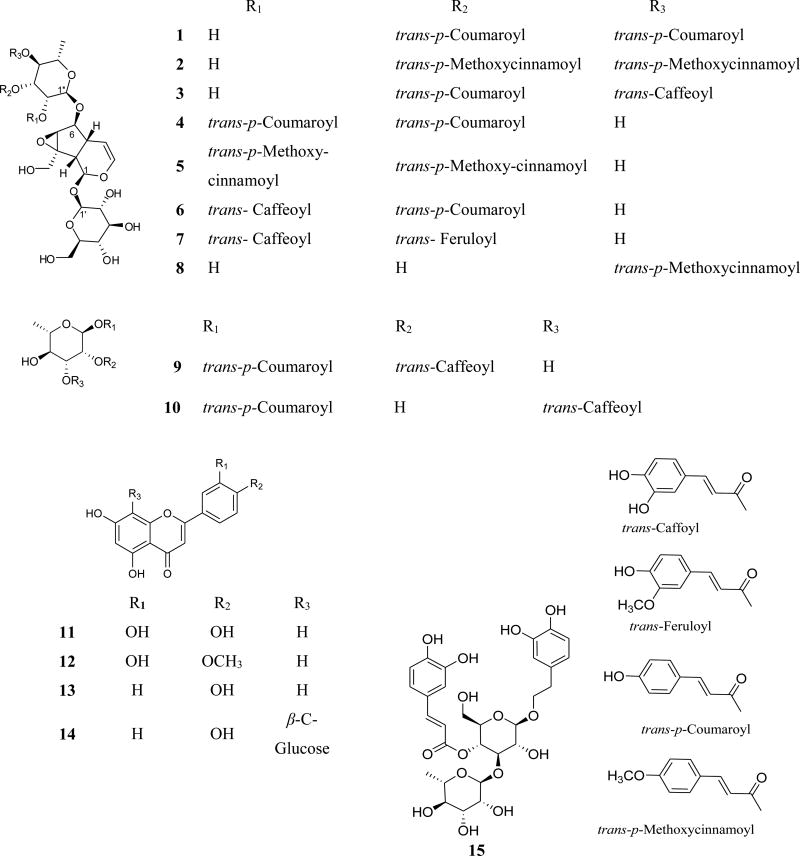
The ethanolic P. odorata leaf extract afforded the new acylated iridoid glycosides 1–3 and the acylated rhamnopyranoses 9 and 10, along with the known premnosides E (4) (Gu et al., 2013), G (5), B (6), and C (7) (Otsuka et al., 1989). In addition, 6-O-α-L-(4”-O-p-methoxycinnamoyl) rhamnopyranosylcatalpol (8) (Otsuka et al., 1991), luteolin (11) (Chaturvedula and Prakash, 2013), diosmetin (12) (Pinzon et al., 2011), apigenin (13) (Chaturvedula and Prakash, 2013), vitexin (14) (El-Sayed et al., 1991), and verbascoside (15) (Yalcin et al., 2003) were also identified.
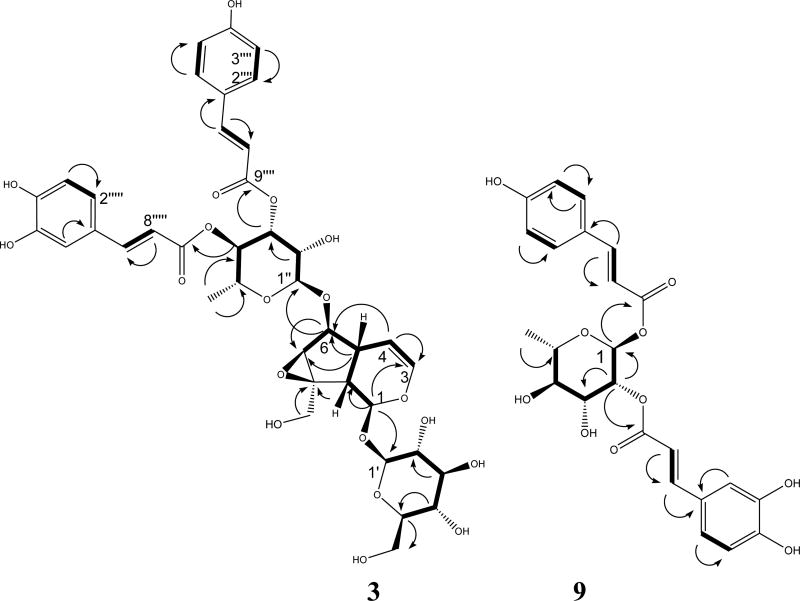
The HRESIMS data for compound 1 showed an adduct pseudo-molecular ion peak at m/z 801.2621 [M+H]+, consistent with the molecular formula C39H44O18 and suggesting eighteen degrees of unsaturation. The 1H and 13C NMR data (Table 1), along with the HSQC experiment suggested six characteristic resonances for β-D-glucopyranose, six resonances for disubstituated α-L-rhamnopyranose and eighteen resonances attributed to two trans-p-coumaric acid moieties. The remaining nine resonances perfectly matched a catalpol iridoid core with C-1-O-β-D-glucopyranose and 6-O-α-L-rhamnopyranose substitution (Otsuka et al., 1989). The 3J-HMBC correlations of the proton H-3″ (δH 5.18) with the quaternary carbonyl carbon C-9″″ (δC 167.6) and the correlation of the proton H-4″ (δH 5.11) with the carbonyl carbon C-9″″ (δC 167.4) confrmed the location of the two trans-p-coumaric moieties at carbons C-3″ and C-4″ of the 6-O-α-L-rhamnopyranosyl moiety. Compound 1 was compared to the known related positional isomer premnoside E (4) (Gu et al., 2013) in which the trans-p-coumaric acid moieties were attached at C-2″ and C-3″. The 13C NMR chemical shift of carbons C-1″ and C-4″ were significantly downfield shifted, ΔδC +2.5 and +5.1, respectively, while carbons C-2″, C-5″ and C-6 were upfield shifted by ΔδC −4.9, −1.0 and −0.5, respectively, in compound 1 compared to compound 4, confirming its C-4″ trans-p-coumaroyl substitution. Accordingly, compound 1 was identified as 6-O-α-L-(3″,4″-di-O-trans-p-coumaroyl) rhamnopyranosylcatalpol, which was given the trivial name premnoside F.
The HRESIMS analysis of compound 2 showed a pseudo-molecular ion peak at m/z 851.2758 [M+Na]+, suggesting a molecular formula of C41H48O18 and 28 mass units larger than premnoside F (1). The 1H NMR spectrum for 2 was quite similar to 1, with an additional oxygenated singlet (δH 3.85, 6H) assigned as C-10‴ and C-10″″ methoxy groups. The NMR data of compound 2 (Table 1) confirmed the C-3″ and C-4″ p-methoxycinnamoyl moieties, instead of the trans-p-coumaroyl moieties of 1. Similar to the comparison of premnosides E and F, the NMR data of 2 confirmed its distinct substitution relative to the related known positional isomer premnoside G (5) (Otsuka et al., 1989). Thus, compound 2 identified as 6-O-α-L-(3″,4″-di-O-trans-p-methoxycinnamoyl)-rhamnopyranosylcatalpol; to which it was given the trivial name premnoside H.
The HRESIMS data of compound 3 proved a molecular formula of C39H44O19 by showing a pseudo-molecular ion peak at m/z 817.2553, [M+H]+. The mass of 3 was 16 Daltons higher than the mass of 1. The 1H NMR data of 3 (Table 1) showed characteristic resonances for a trans-caffeoyl unit in addition to a trans-p-coumaroyl moiety, differing from compound 1. The 1H-1H COSY and HMBC data of 3 (Figure 1) suggested the location of the trans-p-coumaric and trans-caffeoyl moieties at C-3″ (δC 72.9) and C-4″ (δC 73.9), respectively. The proton H-3″ (δH 5.18) showed a 3J-HMBC correlation with the quaternary carbonyl carbon C-9‴ while the proton H-4″ (δC 5.11) showed a 3J-HMBC correlation with the carbonyl carbon C-9″″. The 13C NMR data of 3 clearly distinguished its identity as it compared to the related positional isomer premnoside B (6) (Otsuka et al., 1989). Hence, compound 3 was identified as 6-O-α-L-(3″-O-trans-p-coumaroyl-4″-O-trans-caffeoyl)-rhamnopyrano-sylcatalpol, which was named premnoside I.
Figure 1.
Selected HMBC (
 ) and 1H-1H COSY (
) and 1H-1H COSY (
 ) correlations of compounds 3 and 9.
) correlations of compounds 3 and 9.
Analysis of the HRESIMS, 1D and 2D NMR data of compounds 9 and 10 indicated their acylated rhamanopyranosyl identity, with trans-p-coumaroyl and trans-caffeoyl disubstitution (Gu et al., 2013). The HRESIMS data of 9 showed an adduct molecular ions at m/z 473.1426 [M+H]+, which suggested the molecular formula of C24H24O10 and thirteen degrees of unsaturation. Analysis of the 1H and 13C NMR data (Table 2), HSQC, HMBC, and 1H-1H COSY results (Figure 1) aided the assignment of the trans-p-coumaroyl moiety at C-1 (δC 91.9) and the trans-caffoyl moiety at C-2 (δC 73.7). The anomeric proton H-1 (δH 5.19) showed a 3J-HMBC correlation with the trans-p-coumaroyl carbonyl carbon C-9′, confirming the C-1 trans-p-coumaroyl esterification. Similarly, proton H-2 (δH 5.19) showed a 3J-HMBC correlation with the trans-caffeoyl carbonyl carbon, confirming the C-2 hydroxy trans-caffeoyl esterification. Accordingly compound 9 was identified as 1-O-trans-p-hydroxycinnamoyl-2-O-trans-caffeoyl-α-L-rhamnopyranose.
The HRESIMS and molecular formula of compound 10 was identical to that of compound 9. Their 1H and 13C NMR data were also closely similar (Table 2). The HSQC, HMBC and 1H-1H COSY experiments confirmed C-1 location of the trans-p-coumaroyl moiety while the trans-caffeoyl moiety was assigned at C-3. This was achieved through the 3J-HMBC correlation of the proton H-3 (δH 5.24) with the trans-caffeoyl carbonyl carbon C-9″. Meanwhile, carbons C-1 and C-3 were downfield shifted (ΔδC +2.5 and +4.9) while carbons C-2 and C-4 were upfield shifted (ΔδC +4.0 and 2.8) compared to those of compound 9. Accordingly, compound 10 was identified as 1-O-trans-p-coumaroyl-3-O-trans-caffeoyl-α-L-rhamnopyranose.
Primary screening of virtual binding affinity at the c-MET kinase domain and cell free c-MET inhibitory activity confirmation
The structures 1–10 were docked at the ATP-binding site of the highly resolved c-Met kinase domain crystal structure PDB 2WD1 using Schrödinger molecular modeling software package in extra-precision (XP) mode. The docking scores indicated the ability of only three compounds to fit within the ATP-binding site of c-Met kinase (Table 3). Compounds are ranked in decreasing order are: 9, 8, and 10 according to their docking scores.
Table 3.
Virtual Binding Affinity at c-Met Kinase Domain and Cell-Free Percentage Z’-LYTE Assay c-Met Phosphorylation Inhibition at A Single 5 µM Dose of Compounds 1–10a
| Compound | Docking Score | % c-Met Inhibition using Cell-Free assay (5 µM dose) |
|---|---|---|
| 1 | N/A | 0 % |
| 2 | N/A | 0 % |
| 3 | N/A | 10.9 % |
| 4 | N/A | 0 % |
| 5 | N/A | 0 % |
| 6 | N/A | 0 % |
| 7 | N/A | 0 % |
| 8 | −8.4 | 56.7 % |
| 9 | −8.8 | 79.8 % |
| 10 | −8.1 | 38.8 % |
Modeling scores were also generated for all compounds using the Glide-Dock program’s empirical scoring functions. c-Met crystal structure PDB code: 2WD1. N/A: Structure had no ability to fit within the ATP-binding site of c-Met kinase.
To avoid the possibility of hit attrition that may arise due to scoring functions inaccuracy, all compounds (1–10) were screened in a cell-free Z’-LYTE™ enzymatic assay using a single 5 µM dose of each compound (Table 3). This assay directly measures the in vitro ability of tested compounds to inhibit c-Met phosphorylation (activation) of the purified kinase domain of c-Met, that was in vitro phosphorylated to achieve the highest level of intrinsic kinase activity. The Z’-LYTE assay results typically matched the docking data and confirmed the in silico activity of the three hits 8–10 (Table 3). Compound 9 was the most active in the primary screening stage showing around 80% c-Met phosphorylation inhibition, followed by 8 with 56.7% inhibition, and finally compound 10 with 38.8 % inhibition at 5 µM dose (Table 3).
Secondary screening and assessment of structural determinants of the c-MET inhibitory activity
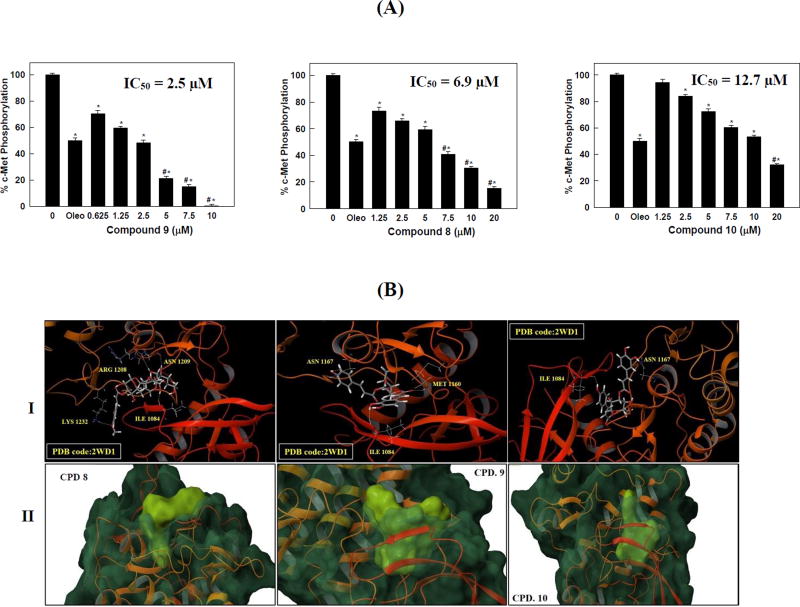
To validate the promising activity obtained in the primary screening, the inhibitory effects of various doses of hits 8–10 against the purified c-Met kinase domain have been re-investigated using the cell-free Z’-LYTE™ enzymatic assay (Figure 2.1). The extra-virgin olive oil phenolic secoiridoid S (−)-oleocanthal, with defined c-Met inhibitory activity (Akl et al., 2014), was used as a standard positive control at a 5 µM dose. As expected, compound 9 was the most active; with c-Met phosphorylation inhibitory IC50 value of 2.5 µM, followed by 8 (IC50 6.9 µM), and finally 10 (IC50 12.7 µM, Figure 2.1).
Figure 2.
(A) c-Met phosphorylation inhibition by various doses of the most active hits 8–10 using the Z’-LYTE assay kit. Error bars indicate the SEM of N=3/dose. Oleocanthal was used as a positive control at 5 µM (Akl et al., 2014). *P < 0.05 as compared with vehicle treated control group. #P < 0.05 as compared to oleocanthal- treated control group. The IC50 values were included for activity comparison. (B). In silico binding mode of 8–10 at the ATP binding site of the c-Met kinase domain (PDB code: 2WD1). I. Important hydrogen bonding interactions shown in dotted yellow lines along with the corresponding amino acids. II. Surface fitting of 8–10 at the c-Met kinase domain. The transparent protein surface, in aquamarine color, and the solid ligand surface, in yellow green color, are shown to emphasize the degree of shape fitting of each compound within the target pocket.
To identify the structural basis for the c-Met binding affinity difference among 8–10, docking simulation studies have been carried out at the c-Met crystal structure PDB 2WD1 (Figure 2.2). Interestingly, this docking study justified the potency of 9 over compounds 8 and 10 in the cell-free assay. The docking study showed a dramatic difference in the binding mode of 8 compared to 9 and 10 (Figure 2.2). Due to its bulkiness, compound 8 showed a partial fitting with its trans-p-methoxycinnamate moiety protruding outside the c-Met kinase pocket and forming a hydrogen bond via its p-methoxy with Lys1232 at the tip of the activation loop (Figure 2.2). In addition, compound 8 showed three additional hydrogen bonding interactions with Ile1084, Arg1208, and Asn1209 at the c-Met kinase domain. It can be clearly seen that the buried part of 8 within the pocket adopted an extended conformation compared to the U-shaped conformation observed in compounds 9 and 10 (Figure 2.2). This observation could suggest that compound 8 might act as a class-II inhibitor, unlike compounds 9 and 10, which typically represent class-I c-Met inhibitors (Zhang et al., 2016, Furlan et al., 2014, Cui, 2013). Despite showing more hydrogen bonding interactions than 9, compound 8 lacked the critical interactions with Met1160 and Pro1158 at the hinge region of the c-Met kinase. Such interactions are well-known to confer potency for any c-Met inhibitor hit (Zhang et al., 2016, Cui, 2013). The lack of these interactions at the hinge region, along with the incomplete fitting of 8 within the c-Met kinase pocket could justify why 9 was more potent than 8 in the cell-free assay. On the other hand, compounds 9 and 10 showed complete fitting within the c-Met kinase pocket compared to compound 8 (Figure 2.2). Both compounds showed two hydrogen bonding interactions with Ile1084 at one of the hydrophobic subpockets and Asn1167 near the activation loop of the c-Met kinase. However; only compound 9 was able to satisfy the critical hydrogen bonding interaction with Met1160 at the hinge region via the carbonyl group of its trans-p-coumaroyl moiety.
Compound 9 binding pose at the c-Met kinase domain
The c-Met′s ATP binding site includes: (1) Hinge region: Met1160 and Pro1158 (Zhang et al., 2016, Cui, 2013). Interaction at this site is highly characteristic for all compounds targeting the ATP binding site in kinase domains; (2) Central hydrophobic region; (3) Two smaller hydrophobic subpockets; and (4) c-Met activation loop (Asp1222-Lys1245) (Zhang et al., 2016, Cui, 2013). The good binding affinity for any c-Met inhibitor hit is hypothesized to be through interactions with at least one of Asp1222, Phe1223, or Tyr1230 at the activation loop and either Pro1158 or Met1160 at the hinge region, along with a good fitting within one or more of the hydrophobic regions (Zhang et al., 2016, Furlan et al., 2014).
The binding mode of 9 at the c-Met′s ATP binding pocket was studied via docking studies using different c-Met crystal structures, in addition to the previously mentioned 2WD1, to minimize false positive results due to conformational variations. Two highly resolved c-Met kinase domain crystal structures PDB codes 3CD8 and 4XYF were used to investigate the possible binding modes of 9 within the catalytic domain of the non-phosphorylated c-Met using Schrödinger software (Figure 3.1).
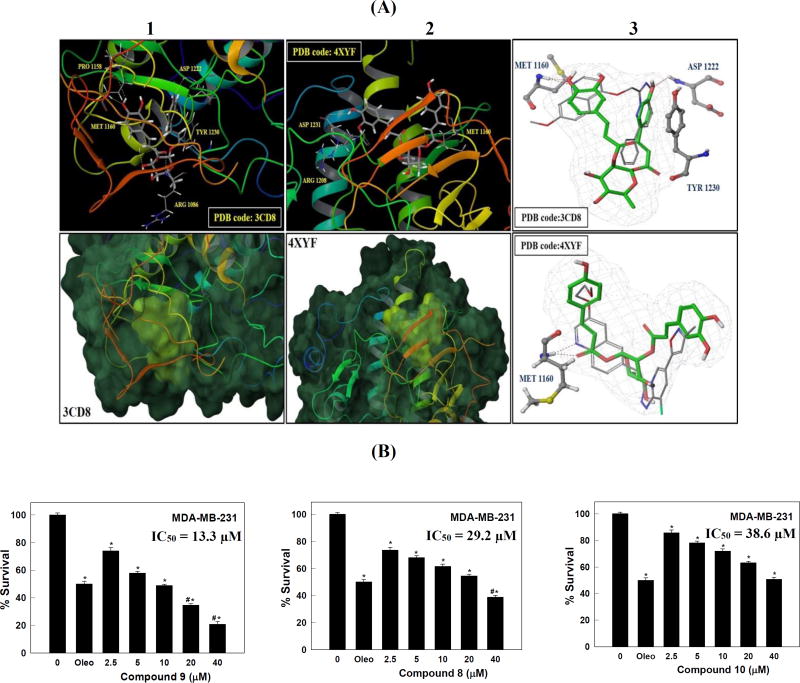
Figure 3.
(A) In silico binding mode of 9 at the ATP binding site of the c-Met kinase domain of crystal structure 3CD8 and 4XYF. Upper panel (1 and 2); Important hydrogen bonding interactions shown in dotted yellow lines along with the corresponding amino acids at each c-Met crystal structure. Lower panel (1 and 2); the transparent protein surface, in aquamarine color, and the solid surface of 9, in yellow-green color, emphasize the shape fitting of 9 within the target pocket of the crystal structures 3CD8 and 4XYF, respectively. (3) The structure overlay for 9 (green tube) with the original ligand conformation (grey thin tube) obtained from c-Met crystal structure 3CD8 (upper panel) and 4XYF (lower panel) using docking simulations; important hydrogen bonding interactions are shown in dotted red lines along with the corresponding amino acids at each c-Met crystal structure. (B) Effect of various doses of compounds 8–10 on the viability of the highly metastatic MDA-MB-231 breast cancer cells, after HGF stimulation, compared to DMSO as a vehicle control. Viable cell count was determined using the MTT assay. Error bars indicate the SEM of N = 3/dose. *P < 0.05 as compared with vehicle treated control groups. Oleocanthal was used as a positive control at 10 µM (Akl et al., 2014). #P < 0.05 as compared to oleocanthal-treated control group. The IC50 values were included for activity comparison.
In all crystal structures, 9 assumed a perfect U-shaped binding mode at the c-Met kinase domain with ideal wrapping around Met1211, which was the typical conformation of class-I c-Met inhibitors. Consistent with the results obtained in 2WD1 crystal structure, the visualization of the docked pose of 9 in 3CD8 and 4XYF emphasized its complete shape fitting at the ATP binding pocket of c-Met kinase domain. Two possible binding poses were noticed for 9, mainly due to the structural symmetry of the acylating ester parts around the sugar moiety. In crystal structure 3CD8, the p-phenolic hydroxyl group of the caffeoyl moiety is forming a pair of bidentate HB interactions with the backbone of both Met1160 and Pro1158 in the hinge region. In contrast, the same hydroxyl group interacted with the side chain of Asp1231 in the activation loop’s DFG motif of crystal structure 4XYF. The m-phenolic hydroxyl interacted with Arg1208 in crystal structure 4XYF, whilst lacking any interactions in crystal structure 3CD8. Instead, the carbonyl group of the trans-p-caffeoyl moiety engaged in a hydrogen bonding interaction with the backbone NH of Arg1086 in crystal structure 3CD8. The trans-p-coumaroyl moiety of 9 interacted via its carbonyl group with the backbone NH of Met1160 and extended to form hydrophobic interactions via its phenyl ring with Ile1084, Tyr1159, and Ala1221 at one of the hydrophobic subpockets in crystal structure 4XYF. In the lipophilic back pocket of the 3CD8’s activation loop, the same aromatic ring is sandwiched between Met1211 and Tyr1230, forming a strong π-π stacking interaction with Tyr1230, thus hindering its autophosphorylation necessary for c-Met activation and stabilizing the inhibitory conformation of the activation loop. The sugar part tethered the two cinnamate moieties in 9 appeared not to contribute any binding role, yet it provided the proper U-shaped conformation necessary for proper alignment of 9 binding pharmacophores at the c-Met kinase domain and thus, satisfied the critical HB interactions with both the hinge region and the activation loop.
To validate the docking results, the original ligand of each of the c-Met crystal structures 4XYF and 3CD8 was docked into it’s the ATP binding pocket by using the same parameters which have been used for docking 9 (Figure 3.1.3). The bound conformation of the co-crystal ligand was generated with a good root mean square displacement (RMSD) of 0.2 Å, indicating the robustness of the docking protocols. Compound 9 almost overlaid the original co-crystallized ligand of each crystal structure, demonstrating the same critical interaction with the hinge region's Met1160. In addition, the original ligand engaged in a HB interaction with the backbone amide hydrogen of Asp1222 in addition to a π-π stacking interaction with Tyr1230, which were also satisfied by 9 in crystal structure 3CD8 (Figure 3.1.3, upper panel). Despite the fact that the sugar moiety in 9 adopted a slight different conformation compared to the linker present in each of the two original ligands, it was still able to provide the proper U-shaped conformation and properly align the 9 binding pharmacophores at the c-Met kinase domain in order to satisfy the critical HB interactions with both the hinge region and the activation loop (Figure 3.1.3). Together, the docking studies showed the good fitting of 9 in the c-Met kinase. The binding mode of 9 within the catalytic c-Met domain is consistent with the kinase assay data, implying the potential of 9 as a promising c-Met inhibitory hit.
Antiproliferative activity against various human breast cancer cell lines
Compounds 1–10 were screened using the MTT cell proliferation assay for ability to inhibit hepatocyte growth factor (HGF)-induced proliferation of the c-Met-expressing highly invasive MDA-MB-231 breast cancer cells at 10 and 40 µM of each compound (Table 4). HGF, scatter factor, is the natural ligand for the activation of the c-Met receptor tyrosine kinase (Zhang et al., 2016, Furlan et al., 2014, Cui, 2013). Interestingly, the results obtained in the MTT assay typically matched the docking and the cell-free findings. Compound 9 was the most active, at 10 and 40 µM, it inhibited 50% and 80% of the HGF-mediated cell proliferation of MDA-MB-231 cells, followed by 8 with 32% and 61% cytotoxicity and finally compound 10 with 28% and 49% cytotoxicity, respectively (Table 4).
Table 4.
Antiproliferative Activity of Compounds 1–10 against The Human TNBC Cells MDA-MB-231 at 10 and 40 µM Screening Doses Using The MTT Assay.a
| Compound | % Survival Inhibition | |
|---|---|---|
| 10 µM | 40 µM | |
| 1 | ND | 26 |
| 2 | ND | 17 |
| 3 | 27 | 30 |
| 4 | ND | 7 |
| 5 | ND | 20 |
| 6 | 10 | 13 |
| 7 | 13 | 18 |
| 8* | 32 | 61.3 |
| 9* | 51.3 | 79.3 |
| 10* | 28.2 | 49.2 |
ND: Not determined
Hits selected for secondary screening.
In an attempt to follow up this two-doses screening assay, the three active compounds 8–10 were tested at different doses against the same breast cancer MDA-MB-231 cells to calculate an IC50 value for each compound using the MTT assay (Figure 3.2). (−)-Oleocanthal with defined antiproliferative activity was used as a standard positive control at 10 µM dose (Akl et al., 2014). Compound 9 was able to inhibit the HGF-mediated cell proliferation in a dose-dependent manner, with an IC50 value of 13.3 µM, followed by compound 8, with an IC50 value of 29.2 µM, and finally compound 10, with an IC50 value of 38.6 µM (Figure 3.2).
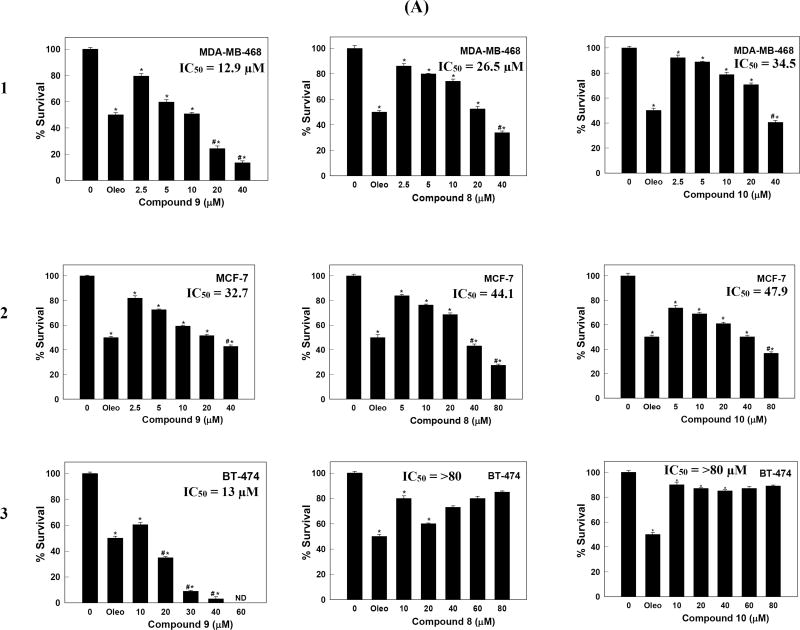
The antiproliferative effects of 8–10 were analyzed against additional key human breast cancer cell lines using the MTT assay. The cell lines were chosen to represent a wide range of breast cancer phenotypes. ERα, one of the most important targets in human breast cancer therapy, is expressed in MCF-7 and BT-474 cells, whereas MDA-MB-231 and MDA-MB-468 cells lack the expression of ERα. On the other hand, c-Met is expressed at higher levels in TNBC MDA-MB-231 and MDA-MB-468 cells, and expressed at relatively lower levels in MCF-7 and BT-474 cells. Accordingly, the human breast cancer cell lines expressing c-Met, namely MDA-MB-468, MCF-7, and BT-474, in addition to the previously tested MBA-MD-231, were chosen to determine the antiproliferative activities of 8–10 (Figure 4.1). HGF was added to the media to induce cell proliferation via c-Met activation. (−)-Oleocanthal with defined antiproliferative activity was used as a standard positive control at 20 µM dose.
Figure 4.
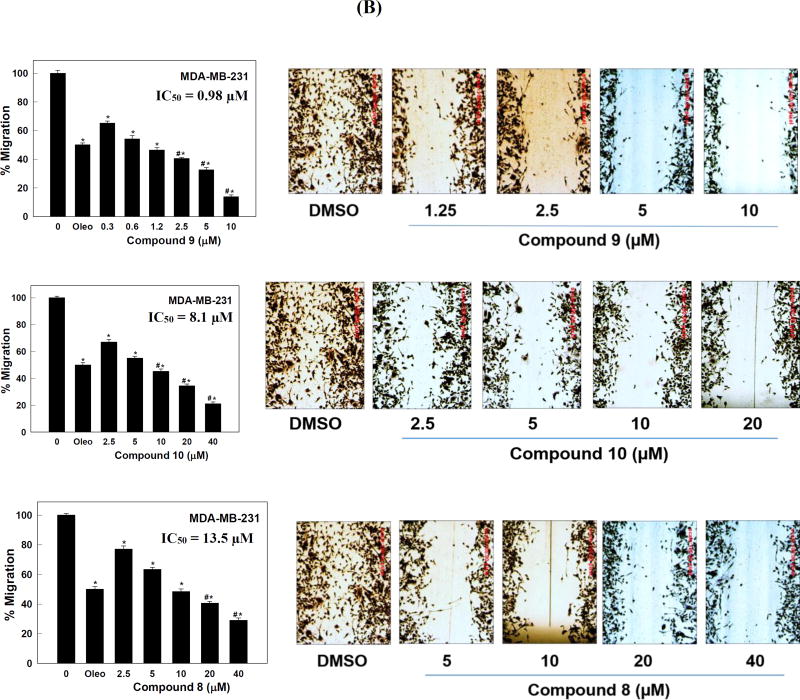
(A) Effect of various doses of compounds 8–10 on the viability of (1) MDA-MB-468, (2) MCF7, and (3) BT-474 breast cancer cells after HGF stimulation, compared to DMSO as a vehicle control. Viable cell count was determined using the MTT assay. Error bars indicate the SEM of N = 3/dose. *P < 0.05 as compared with vehicle treated control groups. Oleocanthal was used as a positive control at 20 µM (Akl et al., 2014). #P < 0.05 as compared to oleocanthal-treated control group. The IC50 values were included for activity comparison. (B) Antimigratory activity of compounds 8–10 against the human TNBC MDA-MB-231 cells. Left panel; effects of various doses of 8–10 on the HGF-induced MDA-MB-231 cells migration in wound healing assay (WHA), compared to DMSO as a vehicle control. Error bars indicate the SEM of N = 3/dose. *P < 0.05 as compared with vehicle treated control group. Oleocanthal was used as a positive control at 10 µM (Akl et al., 2014). #P < 0.05 as compared to oleocanthal-treated control group. The IC50 values were included for activity comparison. Right panel; photomicrographs showing the migration inhibitory activity of each compound against the MDA-MB-231 cells in WHA at different concentrations, compared to DMSO as a vehicle control. Scale bars are provided in red to ensure that the results are comparable.
Treatment with 9 caused a dose-dependent suppression of HGF-induced proliferation of the three cell lines (Figure 4.1). Consistent with biochemical data, compound 9 was more potent than 8 and 10 in all tested breast cancer cell lines. In addition, compound 9 was more active against the highly c-Met-expressing TNBC MDA-MB-231 and MDA-MB-468 cells, compared to the MCF-7 cell line, which has lower c-Met expression level. Compound 9 maintained antiproliferative potency against the HER2-c-Met expressing hormone-dependent luminal B BT-474 breast cancer cells. These results indicated that compound 9 significantly inhibited the HGF-dependent growth across multiple c-Met-expressing breast cancer cell lines in a dose-responsive manner as compared to their vehicle-treated control groups, supporting the hypothesis that c-Met inhibition can be the main molecular target of this unique natural product in breast cancer (Figure 2.1 and 2.2).
Antimigratory activity against the TNBC MDA-MB-231 cells
In addition to cell proliferation, the c-Met/HGF signaling axis plays an important role in cell motility and migration. Therefore, the hits 8–10 active in the primary and secondary screening were further evaluated at different doses in the wound-healing scratch assay (WHA) using the highly metastatic TNBC MDA-MB-231 cells. HGF was added to the media as activating mitogen to induce cell migration via c-Met activation. S (−)-Oleocanthal with defined antimigratory activity was used as a standard positive control at 10 µM dose (Akl et al., 2014). Compound 9 showed impressive cell migration inhibitory activity and potently inhibited the HGF-induced cell migration in a dose-dependent manner with an IC50 value of 0.98 µM (Figure 4.2). Surprisingly, compound 10 was more active than compound 8 in the cell migration assay despite being less active than 8 in all previous assays, which may suggest another molecular target, in addition to the c-Met kinase, for its cell migration inhibitory capacity. The IC50 values for compounds 10 and 8 in WHA were 8.1 and 13.5 µM, respectively (Figure 4.2).
In conclusion, this study shows the potential of the acylated iridoids and rhmnopyranoses represented by hits 8–10 as novel classes of c-Met kinase inhibitors appropriate for future optimizations to control c-Met-dependent breast cancer.
Supplementary Material
Acknowledgments
Research reported in this manuscript was supported in-part by the National Cancer Institute of the National Institutes of Health under Award Number R15CA167475. Authors declare no conflict of interest.
Footnotes
ASSOCIATED CONTENT
The associated Supporting Information including the 1H, DEPT-Q, HSQC, and HMBC NMR spectra along with the HRESIMS spectra of 1, 9, and 10, in addition to the 1H and DEPT-Q NMR spectra and HRESIMS spectra of 2 and 3 (Figures S1–S21) are available at DOI:
References
- Akl MR, Ayoub NM, Mohyeldin MM, Busnena BA, Foudah AI, Liu YY, El Sayed KA. Olive phenolics as c-Met inhibitors:(−)-Oleocanthal attenuates cell proliferation, invasiveness, and tumor growth in breast cancer models. PloS one. 2014;9:e97622. doi: 10.1371/journal.pone.0097622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedula V, Prakash I. Flavonoids from Astragalus propinquus. J Chem Pharm Res. 2013;5:261–5. [Google Scholar]
- Cui JJ. Targeting receptor tyrosine kinase MET in cancer: Small molecule inhibitors and clinical progress. J Med Chem. 2013;57:4427–4453. doi: 10.1021/jm401427c. [DOI] [PubMed] [Google Scholar]
- El-Sayed NH, Ahmed AA, Ishak MS, Kandil FE. Luteolin 7, 4′-dimethyl ether 6-C-glucoside from Parkinsonia aculeata. Phytochem. 1991;30:2442. [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Furlan A, Kherrouche Z, Montagne R, Copin MC, Tulasne D. Thirty years of research on Met receptor to move a biomarker from bench to bedside. Cancer Res. 2014;74:6737–6744. doi: 10.1158/0008-5472.CAN-14-1932. [DOI] [PubMed] [Google Scholar]
- Gu W, Hao XJ, Liu HX, Wang YH, Long CL. Acylated iridoid glycosides and acylated rhamnopyranoses from Gmelina arborea flowers. Phytochem Lett. 2013;6:681–685. [Google Scholar]
- Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lirio SB, Macabeo APG, Paragas EM, Knorn M, Kohls P, Franzblau SG, Wang Y, Aguinaldo MAM. Antitubercular constituents from Premna odorata Blanco. J Ethnopharmacol. 2014;154:471–474. doi: 10.1016/j.jep.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Mohyeldin MM, Akl MR, Ebrahim HY, Dragoi AM, Dykes S, Cardelli JA, El Sayed KA. The oleocanthal-based homovanillyl sinapate as a novel c-Met inhibitor. Oncotarget. 2016;7:32247. doi: 10.18632/oncotarget.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson MH, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: consistent treatment of internal and surface residues in empirical p K a predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Otsuka H, Kashima N, Hayashi T, Kubo N, Yamasaki K, Padolina WG. Premnaodorosides A, B and C, iridoid glucoside diesters of an acyclic monoterpenediol from leaves of Premna odorata. Phytochem. 1992;31:3129–3133. [Google Scholar]
- Otsuka H, Kubo N, Sasaki Y, Yamasaki K, Takeda Y, Seki T. Iridoid diglycoside monoacyl esters from stems of Premna japonica. Phytochem. 1991;30:1917–1920. [Google Scholar]
- Otsuka H, Kubo N, Yamasaki K, Padolina WG. Premnosides A-D: Diacyl 6-O-α-l-rhamnopyranosylcatalpols from Premna odorata. Phytochem. 1989;28:3063–3067. [Google Scholar]
- Pinzon LC, Uy MM, Sze KH, Wang M, Chu IK. Isolation and characterization of antimicrobial, anti-inflammatory and chemopreventive flavones from Premna odorata Blanco. J Med Plants Res. 2011;5:2729–2735. [Google Scholar]
- Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci. 2003;100:12654–12659. doi: 10.1073/pnas.1734128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA cancer J Clinic. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Viljoen A, Mncwangi N, Vermaak I. Anti-inflammatory iridoids of botanical origin. Curr Med Chem. 2012;19:2104–2127. doi: 10.2174/092986712800229005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin FN, Ersoez T, Akbay P, Calis I, Donmez AA, Sticher O. Iridoid and Phenylpropanoid Glycosides from Phlomis samia, P. monocephala and P. carica. Turk J Chem. 2003;27:295–306. [Google Scholar]
- Zhang J, Jiang X, Jiang Y, Guo M, Zhang S, Li J, He J, Liu J, Wang J, Ouyang L. Recent advances in the development of dual VEGFR and c-Met small molecule inhibitors as anticancer drugs. Eur J Med Chem. 2016;108:495–504. doi: 10.1016/j.ejmech.2015.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.