Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most metastatic and deadly cancers. Despite the clinical significance of metastatic spread, our understanding of molecular mechanisms that drive PDAC metastatic ability remains limited. Using a novel genetically engineered mouse model of human PDAC, we uncover a transient subpopulation of cancer cells with exceptionally high metastatic ability. Global gene expression profiling and functional analyses uncovered the transcription factor Blimp1 as a key driver of PDAC metastasis. The highly metastatic PDAC subpopulation is enriched for hypoxia-induced genes and hypoxia-mediated induction of Blimp1 contributes to the regulation of a subset of hypoxia-associated gene expression programs. These findings support a model in which up-regulation of Blimp1 links microenvironmental cues to a metastatic stem cell character.
Keywords: Blimp1, PDAC, hypoxia, cancer metastasis, tumor heterogeneity
Introduction
PDAC is an almost uniformly lethal cancer that is projected to become the second leading cause of cancer-related deaths in the United States by 2030 (1). Most PDAC patients die from metastatic disease, underscoring the need to better understand the molecular mechanisms that drive disease progression and metastasis (2). Genomic analyses of PDAC have uncovered oncogenic KRAS and loss of function mutations in the CDKN2A, SMAD4, and TP53 tumor suppressors as key recurrent drivers of pancreatic cancer development (3-6). While these studies have also offered clues about metastatic progression, they have not uncovered consistent genetic alterations that explain the progression to a highly metastatic state (7-10).
While genomic alterations create stable changes that increase cancer growth, transient alterations in the metastatic state of cancer cells can be induced by interactions with stromal cells, diverse physical cues, as well as by changes in the local tumor microenvironment. For example, the epithelial-to-mesenchymal transition (EMT) is a well characterized transcriptional program that endows cancer cells with a transient high metastatic ability (11). However, EMT might not be critical for PDAC dissemination or metastasis (12,13). Subpopulations of PDAC cells with cancer stem cell-like properties have also been described but it is unclear whether these cells are the major source of metastases (14,15).
In many cancer types, metastasis is thought to be driven by diverse extracellular cues that increased stem-like behavior as well as invasion and metastasis (16). PDAC in particular has an extensive desmoplastic stromal response that generates unique physical properties, including increased extracellular matrix stiffness and areas with limited oxygen and nutrient availability (17). However, whether or not PDAC metastasis is driven by features of the tumor microenvironment is unclear. Identification of key environmental factors could provide insights into the process of metastasis as well as aid in the development of novel therapeutic strategies.
Genetically engineered mouse models of PDAC recapitulate key genetic events of the human disease. Cre-mediated expression of oncogenic KrasG12D in pancreatic cells of loxP-Stop-loxP KrasG12D knock-in mice (KrasLSL-G12D/+) leads to the development of early stage pancreatic intraepithelial neoplasms (PanINs) (18). Concomitant expression of point mutant p53 or deletion of p53, Cdkn2a, and/or Smad4 allows for the development of PDAC that can progress to gain multi-organ metastatic ability (19-23). Importantly, tumors arise in vivo from genetically defined lesions and evolve in their native context, providing the opportunity to identify the cancer cell intrinsic and extrinsic processes that contribute to tumor progression.
Here, we developed a novel mouse model of human PDAC, which enabled the isolation and molecular characterization of a highly metastatic subpopulation of pancreatic cancer cells. We demonstrate that these highly metastatic cancer cells exists within hypoxic tumor areas and that the transcription factor Blimp1 drives their high metastatic potential. Gene expression signatures of the metastatic state, as well as of hypoxia-induced Blimp1-dependant genes predicts PDAC patient outcome. These findings highlight microenvironment-induced heterogeneity as a driver of pancreatic cancer progression toward its deadly metastatic phase.
Results
Generation of a system to identify and isolate a highly metastatic population of PDAC cells
The chromatin-associated protein HMGA2 is a marker of increased malignancy in many tumor types, and high HMGA2 expression predicts poor prognosis in several major human cancer types, including PDAC (24-30). To determine whether neoplastic cells in genetically engineered mouse models of human PDAC also express Hmga2, we performed immunohistochemistry (IHC) on tumors at different stages of development. Hmga2 was not expressed in cells in the normal adult pancreas or PanINs in KrasLSL-G12D;p53LSL-R172H/+;Pdx1-Cre (KPC) mice, but was expressed in a subset of PDAC cells (Supplementary Fig. S1A and data not shown). In human PDAC, HMGA2 expression correlates with metastasis to lymph nodes and poor prognosis, and we confirmed that high HMGA2 expression in PDAC patients predicts shorter survival (Supplementary Fig. S1B-S1D) (28,31). Together, these results document the expression of Hmga2 in a subset of cancer cells in mouse models of PDAC and confirm the correlation of the presence of cancer cells in the HMGA2pos state with poor PDAC patient outcome.
To uncover the cellular and molecular features of Hmga2positive and Hmga2negative cancer cells, we generated a mouse model that would allow the isolation of these PDAC cell sub-populations. We incorporated two additional alleles into the KPC mouse model: a Cre-reporter allele (R26LSL-Tomato) to fluorescently mark all neoplastic cells, and an Hmga2 knock-in allele, which is converted by Cre from its wild-type conformation (Hmga2CK) into a GFP reporter (Hmga2GFP; Fig. 1A) (20,32). In the heterozygous state (Hmga2CK/+), the potential for GFP expression is restricted to cells in which Cre has inverted a lox-flanked region and GFP expression remains under control of all endogenous Hmga2 regulatory elements (20). In KPC;R26LSL-Tomato/+;Hmga2CK/+ mice (referred to as KPCcolors mice), all cancer cells were Tomato positive and Hmga2-expressing cancer cells were both Tomato and GFP positive (Supplementary Fig. S1A and S1E).
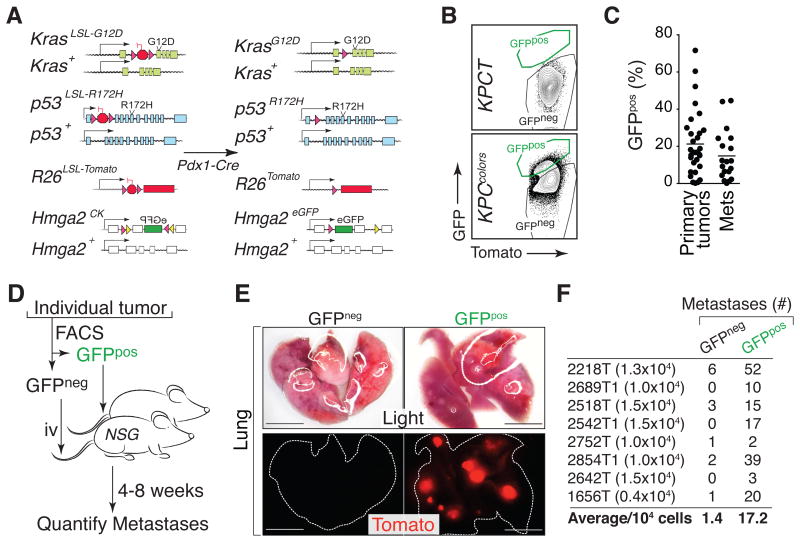
Figure 1. Identification of a subpopulation of highly metastatic pancreatic cancer cells.
(A) Alleles in the KPCcolors model (KrasLSL-G12D/+;p53LSL-R172H/+;Hmga2CK/+;R26LSL-Tom;Pdx1-Cre) before and after Cre-mediated recombination.
(B) Representative FACS plots of dissociated pancreatic cancer cells from KrasLSL-G12D/+;p53LSL-R172H/+;Pdx1-Cre;R26LSL-Tom (KPCT) and KPCcolors mice. FSC/SSC-gated lineageneg (CD45negCD31negF4/80negTer119neg) viable (DAPIneg) Tomatopos cells are shown.
(C) Individual primary tumors and metastases (Mets) have variable proportions of GFPpos cells. Each dot is a tumor and the bar is the mean.
(D) Metastatic ability of GFPneg and GFPpos subpopulations from individual tumors was assessed by intravenous (i.v.) transplantation into recipient mice.
(E) Light and fluorescent dissecting scope images of lungs from recipient mice after i.v. transplantation of GFPneg or GFPpos PDAC cells from an individual tumor from a KPCcolors mouse. Scale bars = 0.5 cm.
(F) Number of cells injected and the number of metastases is indicated for each matched pair. The average number of metastases per 104 GFPneg and GFPpos PDAC cells is shown. p-value < 0.008 by Wilcoxon matched-pair signed rank test.
The dual fluorescent marking of cancer cells in KPCcolors mice provided us with the ability to isolate TomposGFPneg and TomposGFPpos cancer cells by fluorescence-activated cell sorting (FACS) (Supplementary Fig. S1F-S1G). Consistent with Hmga2 expression observed by IHC, variable percentages of cancer cells in individual tumors were GFPpos (Fig. 1B-1C). In the KPC model, progression from PanINs to adenocarcinoma is driven by loss of the wild-type p53 allele (19). TomposGFPneg and TomposGFPpos samples contained less than 10% remaining p53wt allele and loss of the p53wt allele led to the stabilization of mutant p53 protein in both GFPneg and GFPpos cells (Supplementary Fig. S1H-S1J). Thus, TomposGFPneg and TomposGFPpos cells represent two distinct sub-populations of pancreatic cancer cells.
We next performed cell culture and transplantation-based in vivo metastasis assays on GFPneg and GFPpos PDAC cells. GFPpos cells consistently formed more spheres when plated into ultralow-attachment plates and formed more colonies when plated at low density under standard tissue culture conditions (Supplementary Fig. S1K and data not shown). Most importantly, for eight out of eight tumors from KPCcolors mice, the GFPpos PDAC cells formed more metastases than their GFPneg counterparts when transplanted intravenously into recipient mice (Fig. 1D-1F). On average, GFPpos cells were more than 10 times more metastatic than GFPneg cells (p-value < 0.008; Fig. 1F). Interestingly, the tumors that arose from GFPpos cells almost always had heterogeneous GFP expression, suggesting that GFPpos cells may be in a transient state with the potential to give rise to both GFPpos and GFPneg cells (Supplementary Fig. S1L-S1M).
Gene expression profiling reveals a dynamic metastatic state
To uncover pro-metastatic programs within the highly metastatic GFPpos PDAC cell state, we performed RNA-Seq-based gene expression profiling on six pairs of GFPneg and GFPpos cells (Fig. 2A and Supplementary Fig. S2A-S2B). Global clustering of all samples did not clearly separate GFPpos from GFPneg samples (Fig. 2B). However, direct pairwise comparison of GFPneg and GFPpos cells uncovered hundreds of genes with consistent and significant differences (Fig. 2C-2D). Neither canonical epithelial markers nor genes related to EMT were consistently different between GFPneg and GFPpos cells (data not shown). We also did not observe enrichment for previously described gene expression signatures of PDAC metastasis or putative cancer stem cells in GFPpos cells (13,14) (data not shown). Using flow cytometry, we confirmed that both GFPneg and GFPpos cancer cells had heterogeneous expression of the ductal/CSC marker CD133 and the epithelial marker EpCAM (Supplementary Fig. S2C-S2D) (33-35). Histologic features and IHC for differentiation markers confirmed that Hmga2 expression is largely independent from differentiated state (Fig. S2E).
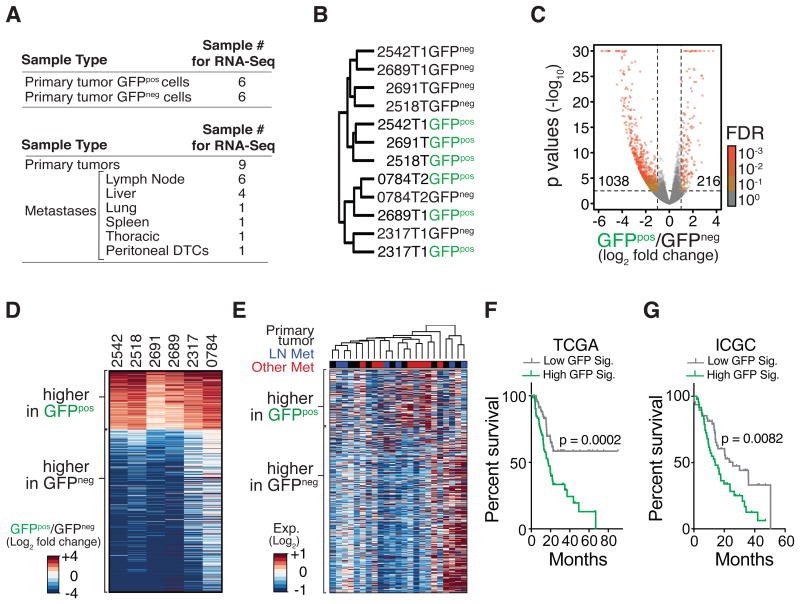
Figure 2. Highly metastatic PDAC cells have a unique gene signature, which is not preserved in metastases but predicts poor patient outcome.
(A) Samples used for gene expression profiling.
(B) Consensus clustering of GFPneg and GFPpos PDAC cell populations using Spearman correlation.
(C) Comparison of the gene expression in GFPneg and GFPpos cells. Number of genes with absolute log2 fold change > 1 and adjusted p-value < 0.05 (adjusted with maximum false discovery rate (FDR) of 0.1) is shown.
(D) Heatmap of genes differentially expressed between GFPneg and GFPpos PDAC cells, defined by paired comparison between GFPneg and GFPpos cells with a p-value < 0.05, false discovery rate < or = 0.001, and absolute log2 fold change > 1.
(E) Heatmap of the expression of differentially expressed genes between GFPneg and GFPpos PDAC cells in bulk cancer cells from primary tumors and metastases from KPCT mice.
(F, G) A gene expression signature based on genes that are more highly expressed in GFPpos cells (GFP Sig.) predicts shorter PDAC patient survival. PDAC patients from The Cancer Genome Atlas (F) and the International Cancer Genome Consortium (G) were split into top and bottom 50% (High GFP Sig. and Low GFP Sig., respectively) based on the single sample GSEA scores for GFP signature genes. p-values were calculated by log-rank test.
In addition to the paired GFPpos and GFPneg PDAC samples, we performed RNA-Seq analyses on FACS-sorted, bulk Tompos cancer cells from primary tumors and metastases (Fig. 2A and Supplementary Fig. S2F). If metastases had stable gene expression differences from primary tumors, this approach could identify gene expression alterations that contribute to metastatic ability or growth at secondary sites. Interestingly, comparison of primary tumors to all metastases identified very few significant differentially expressed genes (Supplementary Fig. S2G). Comparisons of primary tumors to liver metastases, but not to lymph node metastases, uncover several genes that were significantly differentially expressed in the liver metastases (Supplementary Fig. S2H and S2I). Consistent with a recent report on human PDAC metastasis (36), gene set analysis uncovered a trend towards enrichment for programs related to glucose metabolism in liver metastases (Supplementary Fig. S2J). Importantly, genes that were differentially expressed between GFPpos and GFPneg PDAC cells were not consistently different between primary tumors and metastases, consistent with the transient nature of the GFPpos cell state (Fig. 2E). Finally, high expression of a gene signature composed of genes that were more highly expressed in metastatic GFPpos cancer cells predicted worse PDAC patient outcome (Fig. 2F-2G).
Identification of the transcription factor Blimp1 as a driver of metastasis
To gain further insight into the metastatic process and identify potentially pro-metastatic factors, we focused on several genes that were among the most significantly and dramatically upregulated in GFPpos cells (fold change >2; p-value <10-6, Supplementary Fig. S3A). We stably knocked down five top candidate genes (Ero1l, Slc16a3, Glut1, Hilpda, and Blimp1) in a PDAC cell line (688M) derived from a liver metastasis from a KPCT mouse (Supplementary Fig. S3B-S3F). We assessed the importance of these genes in metastasis by quantifying the number of metastases that formed from subcutaneously and orthotopically transplanted tumors. These experiments suggested that the transcription factor Blimp1/Prdm1 could have pro-metastatic functions in PDAC (Supplementary Fig. S3G-S3L). Blimp1 is a transcription factor that was of particular interest due to its well-established role as a master regulator of cell fate determination during plasma B cell differentiation and primordial germ cell development (37,38). Blimp1 was one of the most highly upregulated genes in GFPpos cells, being 4- to 27-fold higher in GFPpos cells (p-value < 0.05; Fig. 3A). We confirmed increased Blimp1 protein expression in sorted GFPpos cells relative to GFPneg cells (Fig. 3B). Blimp1 expression was not consistently different between bulk cancer cells from primary tumors and metastases from the KPC mice, consistent with the unstable nature of the metastatic state (Supplementary Fig. S3M).
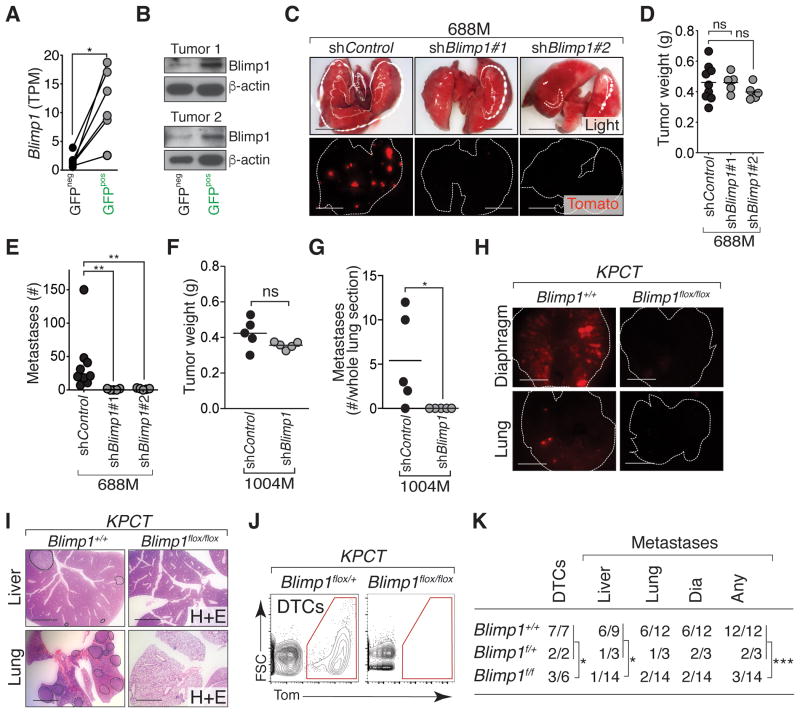
Figure 3. Highly metastatic PDAC cells express Blimp1, which is required for PDAC metastatic ability.
(A) Expression of Blimp1 (transcripts per million, TPM) in GFPpos and GFPneg cells (*, p-value < 0.05 by paired t test).
(B) Blimp1 protein expression in GFPpos and GFPneg PDAC cells from two KPCcolors tumors. Blimp1 size is estimated ∼60kD.
(C) Representative dissecting scope images of lung metastases in mice with subcutaneous tumors from 688M cells with Control or Blimp1 knockdown. Scale bars = 0.5 cm.
(D) Subcutaneous tumor growth of shControl and shBlimp1 PDAC cell line derivatives (688M). Each dot represents the average weight (g) of all tumors from a mouse and the bar is the average. In these experiments, mice were purposefully sacrificed when the subcutaneous tumors reached a designated size (Supplementary Methods). ns, not statistically significant by Student's t test.
(E) Quantification of lung metastases in mice with subcutaneous tumors. Each dot represents a mouse and the bar is the mean. Data represent pooled results from 2 experiments. **, p-value < 0.005 by Student's t test.
(F) Subcutaneous tumor growth of shControl and shBlimp1 PDAC cell line derivatives (1004M). Each dot represents the average weight (g) of all tumors from a mouse and the bar is the average. In these experiments, mice were purposefully sacrificed when the subcutaneous tumors reached a designated size (Supplementary Methods). ns, not statistically significant by Student's t test.
(G) Quantification of lung metastases in mice with subcutaneous tumors. Each dot represents a mouse and the bar is the mean. Data represent pooled results from 2 experiments. *, p-value < 0.05 by Student's t test.
(H) Representative images of lung and diaphragm metastases in KPTC;Blimp1+/+ and KPCT;Blimp1flox/flox mice. Scale bars = 0.5 cm. Lung and diaphragm are outlined with dotted white line.
(I) KPCT;Blimp1flox/flox mice have fewer metastases. Representative H&E stained sections of lung and liver metastases in KPTC;Blimp1+/+ and KPCT;Blimp1flox/flox mice. Scale bars = 0.5 cm. Organs with Tomatopos metastases are outlined.
(J) KPCT;Blimp1flox/flox mice have fewer disseminated tumor cells (DTCs). Representative FACS plots of viable, lineagenegTomatopos cancer cells in the peritoneal cavity of control KPCT;Blimp1flox/+ and KPCT;Blimp1flox/flox mice are shown. gated on lineageneg single cells.
(K) Number of mice with DTCs and metastases. *, p-value < 0.05; ***, p-value < 0.0005 by Fisher's exact test. Dia, diaphragm. p-values for other comparisons between control (KPTC;Blimp1+/+ and KPCT;Blimp1flox/+) mice and KPCT;Blimp1flox/flox mice are: lung, 0.1086 and Dia, 0.0502.
To further assess whether Blimp1 contributes to metastatic ability, we knocked down Blimp1 using two independent shRNAs (688M; Supplementary Fig. S3N). Blimp1 knockdown reduced the number of metastases seeded from subcutaneous tumors by >50-fold (p-value <0.005; Fig. 3C-3E). Blimp1 knockdown in a second metastasis-derived PDAC cell line (1004M) also significantly reduced metastasis (Fig. 3F-3G and Supplementary Fig. S3O). Interestingly, while Blimp1 appeared to be required for metastatic ability, overexpression of Blimp1 in multiple PDAC cell lines did not consistently enhance metastatic ability (Supplementary Fig. S3P-S3S), suggesting that expression of Blimp1 is required, but not sufficient, to drive PDAC metastasis.
Blimp1 contributes to the metastatic ability of PDAC cells in KPC mice
We next used a Blimp1 conditional knockout allele to further investigate Blimp1 function in autochthonous PDAC (37). Blimp1f/f;Pdx1-Cre mice were viable and their pancreata did not show obvious histological changes, suggesting that Blimp1 is not required for pancreas development or homeostasis (data not shown). KPCT;Blimp1f/f mice had similar overall pancreatic tumor burden but shorter survival compared to control KPCT;Blimp1+/+ mice (Supplementary Fig. S3T-S3U). Pancreata from KPCT;Blimp1f/f mice contained PanINs as well as adenocarcinomas that were similar to PDAC in control KPCT;Blimp1+/+ mice (Supplementary Fig. S3V-S3W and data not shown). To assess the effect of Blimp1 deficiency on metastatic progression in vivo, we carefully quantified the number of Tompos disseminated tumor cells (DTCs) in the peritoneal cavity as well as metastases in KPCT;Blimp1f/f and control mice. 14 out of 15 control mice (KPCT;Blimp1+/+ and KPCT;Blimp1f/+) developed metastases, which were often numerous and widespread in many different sites including the lymph nodes, diaphragm, lungs, and liver (Fig. 3H-3K). Conversely, only 3 out of 14 KPCT;Blimp1f/f mice developed metastases (Fig. 3K). Additionally, peritoneal DTCs could be detected in only half of KPCT;Blimp1f/f mice, but were present in all control mice (Fig. 3IJ-3K). Together with our observations from cell lines, these data suggest that Blimp1 promotes metastatic proclivity of PDAC.
The highly metastatic state of PDAC is associated with a strong hypoxia signature
To place Blimp1 in a pathway involved in metastasis, we next used gene set enrichment analysis (GSEA) and gene ontology (GO) enrichment analysis to identify pathways altered in the more metastatic GFPpos cells. These analyses uncovered an overwhelming enrichment for hypoxia-induced genes in GFPpos cells (Fig. 4A-4D and Supplementary Table S1). Genes expressed more highly in GFPpos cells were also enriched for Hif1-binding motifs near their transcription start sites and our analyses identified significant enrichments for both Hif1 and Hif2 regulated genes in GFPpos cells. (Fig. 4B, Supplementary Fig. S4A-S4B, and Supplementary Table S1). Conversely, genes downregulated in GFPpos cells were enriched for cell cycle processes, consistent with hypoxia-induced cell cycle arrest (Supplementary Table S1) (39). We confirmed the up-regulation of the canonical Hif1 target gene Ero1l at the protein level in sorted GFPpos PDAC cells (Supplementary Fig. S4C).
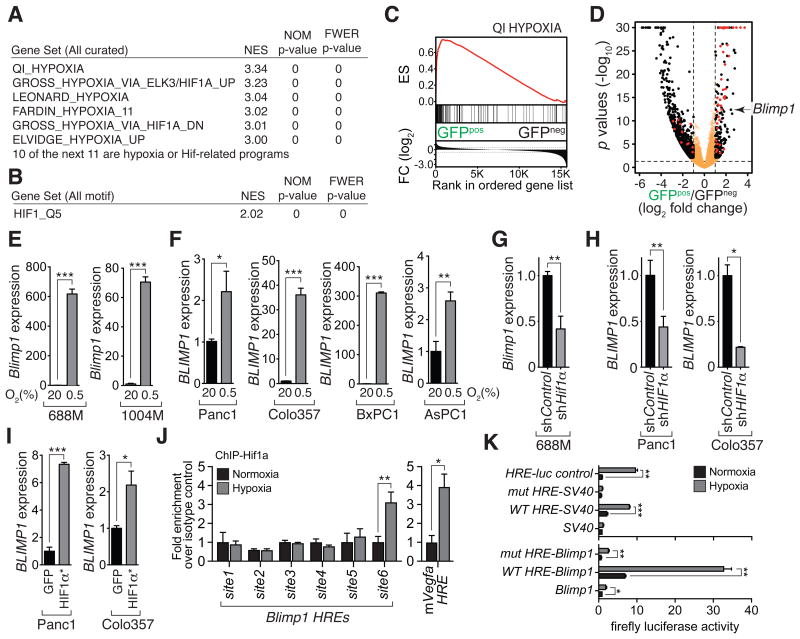
Figure 4. Blimp1 is regulated by hypoxia/Hif1 in murine and human pancreatic cancer cells.
(A) The GFPpos PDAC cell state is enriched for hypoxia-induced genes and Hif-targets.
(B) Enrichment for the Hif1-binding site in the promoters of genes up-regulated in the GFPpos cell state.
(C) Significant enrichment for hypoxia-regulated genes in GFPpos cells. ES, enrichment score; FC, fold change.
(D) Volcano plot of the gene expression differences between GFPpos and GFPneg cancer cell populations. Fold change and adjusted p-values were generated by taking paired samples into consideration. Red dots denote annotated Hif1 target genes.
(E,F) Hypoxia (0.5% O2 for 24 hours) induces Blimp1 expression in two mouse PDAC cell lines (E) and four human PDAC cell lines (F). *, p-value < 0.05; **, p-value < 0.005; ***, p-value < 0.0005 by Student's t test. Mean +/- SD of triplicate wells in shown.
(G,H) Knockdown of Hif1a in the murine 688M cells (G) and HIF1a in two human PDAC cell lines (H) reduces hypoxia-induced Blimp1 and BLIMP1 upregulation, respectively. Mean +/- SD of triplicate wells in shown. *, p-value < 0.05; **, p-value < 0.005 by Student's t test.
(I) Expression of stabilized HIF1α increases BLIMP1 expression in two human PDAC cell lines. *, p-value < 0.05; **, p-value < 0.0005 by Student's t test. Mean +/- SD of triplicate wells in shown.
(J) Chromatin immunoprecipitation (ChIP) qPCR-quantified Hif1α binding at an HRE-containing region 240 kb upstream of the Blimp1 TSS (site6). 688M cells were cultured under normoxia or hypoxia before ChIP. Six HRE were quantified for enrichment of Hif1a binding by qPCR. An HRE region in the Vegfa locus is a positive control for hypoxia-induced Hif1α binding. *, p-value < 0.05; **, p-value < 0.005 by Student's t test.
(K) The wildtype site6 (WT-HRE), but not site6 with all three HREs mutated (mut-HRE), conferred hypoxia responsiveness on a SV40 promoter or a 1.6kb Blimp1 promoter. Representative results of 688M cells transfected with indicated reporters cultured under normoxia or hypoxia. Means +/- SD of triplicated ratios of firefly luciferase normalized to co-transfected Renilla luciferase reporter are shown.
Pimonidazole-defined hypoxic areas were significantly enriched for Hmga2pos cells (Supplementary Fig. S4D-S4E). We also employed multicolor sequential immunoflourescence staining to show that Hmga2pos areas are enriched for the expression of the canonical hypoxic target protein Glut1 (Supplementary Fig. S4F-S4G) (40).
Based on the striking enrichment of Hif targets in GFPpos PDAC cells from KPCcolors mice, we also determined whether Hmga2 expression is regulated by hypoxia. Under hypoxia, we noted only a slight increase in Hmga2 protein levels in PDAC cell lines (Supplementary Fig. S4H). While Hif target genes were enriched in Hmga2-expressing PDAC cells, Hmga2 knockdown had no effect on the hypoxia-induced expression of canonical Hif1 target genes (Supplementary Fig. S4I). Thus, it remains unclear why Hmga2 marks this highly metastatic PDAC subpopulation but these data suggest that other aspects of the in vivo microenvironment either in conjunction with, or independent from, hypoxia induce Hmga2 expression in these cells.
Blimp1 is a novel hypoxia/Hif-regulated gene in human and murine PDAC
To determine whether Blimp1 expression is regulated by hypoxia in human and murine PDAC, we assessed Blimp1 mRNA and protein expression in PDAC cell lines exposed to hypoxia (0.5% O2 for 24 hours). Hypoxia led to the induction of multiple canonical Hif target genes, Hif1α stabilization, and the prominent and consistent induction of Blimp1 in two mouse and four human PDAC cell lines (Supplementary Fig. S4J-S4K and Fig. 4E-4F). Hypoxia-mediated induction of Blimp1 in mouse and human PDAC cells was attenuated by Hif1α knockdown, suggesting that Hif1α is at least partially required for Blimp1 induction under these conditions (Fig. 4G-4H). BLIMP1 induction in human PDAC cell lines was also partially HIF2 dependent (Supplementary Fig. S4B). Expression of stable HIF1α was sufficient to increase BLIMP1 expression in PDAC cells (Fig. 4I). Finally, human PDACs with the highest BLIMP1 expression are enriched for hypoxia signatures relative to those with the lowest BLIMP expression (Supplementary Fig. S4L and data not shown).
We next investigated how hypoxia and Hif induce Blimp1 expression. To determine whether Blimp1 can be induced indirectly by secreted factors, we measured Blimp1 levels in PDAC cells cultured with conditioned media from hypoxia-treated cells or recombinant VEGF-A which has been shown to induce Blimp1 in endothelial cells (41). In both cases, we did not observe robust Blimp1 induction (Supplementary Fig. S5A-S5C). Blimp1 was induced rapidly after exposure to hypoxia, paralleling the kinetics of canonical Hif target genes, suggesting that Blimp1 might be induced directly by Hif (Supplementary Fig. S5D). Analysis of chromatin accessibility around the Blimp1 locus (see below) enabled the prioritization of multiple putative distal regulatory regions that contained hypoxia-response elements (HREs) (Supplementary Fig. S5E). Hif1a ChIP qPCR identified a cluster of three adjacent HREs upstream of Blimp1 that were bound by endogenous Hif1α in PDAC cells under hypoxia (Fig. 4J). This HRE-containing putative distal regulatory region conferred hypoxia responsiveness in a heterologous reporter system, which was abolished by mutation of its HRE motifs (Fig. 4K and Supplementary Fig. S5F-S5G). Furthermore, Blimp1 knockdown significantly reduced the ability PDAC cells cultured under hypoxia to form spheres and had a variable effect of migratory ability in cell culture. These results suggest a role for Blimp1 in cellular behaviors related to metastatic ability (Supplementary Fig. S6A-S6I).
Blimp1 regulates a subset of hypoxic-mediated gene expression changes in PDAC
To characterize Blimp1's function in hypoxic cells, we profiled the gene expression and genome-wide chromatin accessibility of shControl and shBlimp1 PDAC cells cultured under normoxic and hypoxic conditions (Fig. 5A). Hypoxia can induce changes in chromatin state, and Blimp1 has been implicated in both plasma cell precursors and primordial germ cells as a regulator of chromatin structure (42-44). We uncovered widespread hypoxia-induced changes in chromatin accessibility, with differentially accessible regions being enriched for Hif-binding elements (Fig. 5B and Supplementary Fig. S6J-S6K). In addition, hypoxia induced genes associated with newly open chromatin regions more than those with constitutively open or close regions, suggesting that hypoxia likely regulate target gene induction in part through chromatin accessibility changes (Supplementary Fig. S6L). Interestingly, Blimp1 knockdown had minimal impact on hypoxia-induced changes in chromatin accessibility, indicating that the function of Blimp1 is largely independent of its ability to recruit factors that lead to changes in chromatin state (Supplementary Fig. S6M-S6N).
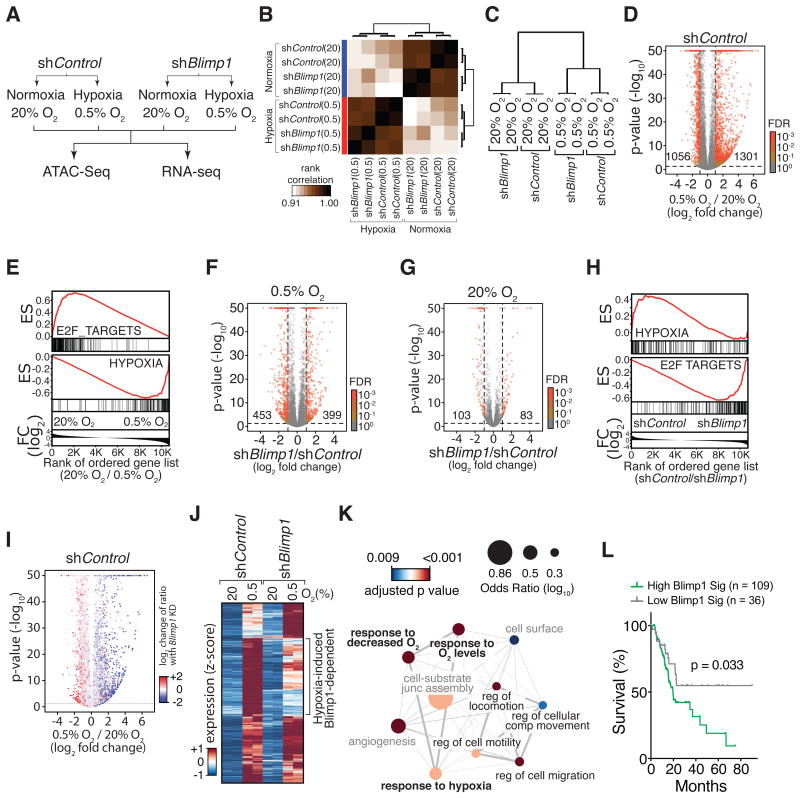
Figure 5. Blimp1 regulates the expression of a subset of hypoxia-induced genes.
(A) The PDAC cell line 688M was cultured for 24 hours under normoxia/hypoxia in vitro for RNA-Seq and ATAC-Seq analyses.
(B) Pearson correlation of all samples based on global chromatin accessibility determined from ATAC-Seq analysis.
(C) Clustering of correlation of samples based on the global gene expression derived from RNA-Seq analysis.
(D) Hypoxia-induced changes in gene expression. Numbers of differentially expressed genes that are significant with FDR < 0.001 are shown.
(E) Genes suppressed by hypoxia are significantly enriched for cell cycle-related programs (E2F_TARGETS). Genes induced by hypoxia are significantly enriched for a hypoxia signature. ES, enrichment score; FC, fold change.
(F,G) Gene expression differences between shBlimp1 and shControl cells cultured under hypoxia (F) and normoxia (G). Numbers of differentially expressed genes that are significant with FDR < 0.001 are shown.
(H) Blimp1-repressed genes under hypoxia are significantly enriched for cell cycle-related programs (bottom panel). GSEA was conducted by comparing the transcriptomes of shControl and shBlimp1 cells cultured under hypoxia. ES, enrichment score; FC, fold change.
(I) Blimp1 knockdown reduces the induction of hypoxia-induced genes (blue) while de-represses hypoxia-suppressed genes (red). Change of ratio under Blimp1 knockdown is defined as the ratio of a, “fold change of gene expression induced by hypoxia in shBlimp1 cells” over b, “fold change of gene expression induced by hypoxia in shControl cells”. p-values are adjusted with a FDR = 0.1.
(J) About 35% of hypoxia-induced genes required Blimp1 for their full induction and were less induced by hypoxia in shBlimp1 cells (log2 fold change < -0.5).
(K) GO term analysis of hypoxia-induced, Blimp1-dependent genes defined in (J). Significantly enriched biological processes are shown. Node size represents odds ratio (log10) and color shows adjusted p-values. Thickness of lines connecting nodes represents percent of shared genes between connected processes. Hypoxia (bold) and cell mobility (black font) related processes are highlighted.
(L) Hypoxia-induced, Blimp1-dependent genes (BLIMP1 Sig.) predict PDAC patient outcome. Single sample GSEA (ssGSEA) scores for the BLIMP1 Sig. were used to separate the top three from the bottom quartile of TCGA patients.
Our parallel RNA-Seq analysis identified many genes that were dramatically and significantly altered by hypoxia (Fig. 5C-5D). As expected, canonical genes related to hypoxia were induced while cell cycle-related programs were suppressed (Fig. 5E). Consistent with the induction of Blimp1 by hypoxia, Blimp1 knockdown affected the expression of more genes when the cells were cultured under hypoxic conditions (Fig. 5C, Supplementary Fig. S6O, and comparison between Fig. 5F and 5G). Blimp1 was required for both the induction and repression of subsets of hypoxia-regulated genes (Supplementary Table S2). Under hypoxia, cell cycle-related programs were enriched in shBlimp1 cells compared to shControl cells, suggesting that Blimp1 might play a role in hypoxia-induced cell cycle arrest (Fig. 5H). Approximately 12% of hypoxia-repressed genes required Blimp1 for their full suppression (N=95 of 825 hypoxia-repressed genes; Fig. 5I and Supplementary Fig. S6P-S6Q). The majority of these hypoxia-repressed, Blimp1-dependent genes were related to cell cycle processes, consistent with the role of Blimp1 in suppressing proliferation during plasma B cell differentiation (Supplementary Fig. S6R) (45,46).
Additionally, approximately 35% of hypoxia-induced genes required Blimp1 for their full induction and were less induced under hypoxia in shBlimp1 cells (N=833 of 2342 hypoxia-induced genes; Fig. 5J). Genes encoding proteins involved in hypoxic responses and cell mobility were reduced in shBlimp1 cells compared to shControl cells (Fig. 5K, Supplementary Fig. S7A-S7B, and Supplementary Table S3). We found that accessible distal regulatory regions within 500 kb of the transcription start sites of Blimp1-dependent, hypoxia-induced genes were enriched for transcription factor-binding motifs that closely resemble the Blimp1 motif (47) (IRF1/IRF2, Supplementary Fig. S7C-S7D). Although the regulation of these Blimp1-dependant genes is likely to be multifaceted, the enrichment of these motifs suggests that at least a subset of these genes may be regulated directly by Blimp1. Finally, high expression of a gene expression signature composed of hypoxia-induced, BLIMP1-dependent genes predicted worse PDAC patient outcome (Fig. 5L). These results suggest that Blimp1 is a hypoxia-regulated gene that regulates a defined subset of hypoxia-controlled genes in PDAC cells.
Blimp1 is required for hypoxia-induced cell cycle repression and the induction of pro-metastatic genes
To gain additional insight into the function of Blimp1 in PDAC, we integrated our ex vivo RNA-Seq data from GFPneg and GFPpos PDAC cells with our in vitro RNA-Seq data from shControl and shBlimp1 cells cultured under normoxia and hypoxia. As anticipated, a vast majority of genes that are more highly expressed in GFPpos cells were also up regulated by hypoxia in PDAC cells in cell culture (Fig. 6A and Supplementary Fig. S8A-S8B). Furthermore, many hypoxia-induced genes that were more highly expressed in GFPpos cells in vivo required Blimp1 for optimal induction under hypoxic conditions in vitro (Supplementary Fig. S8A and S8C). These results underscore the strong hypoxia signature in the GFPpos cells and highlight the contribution of Blimp1 to the expression of these genes.
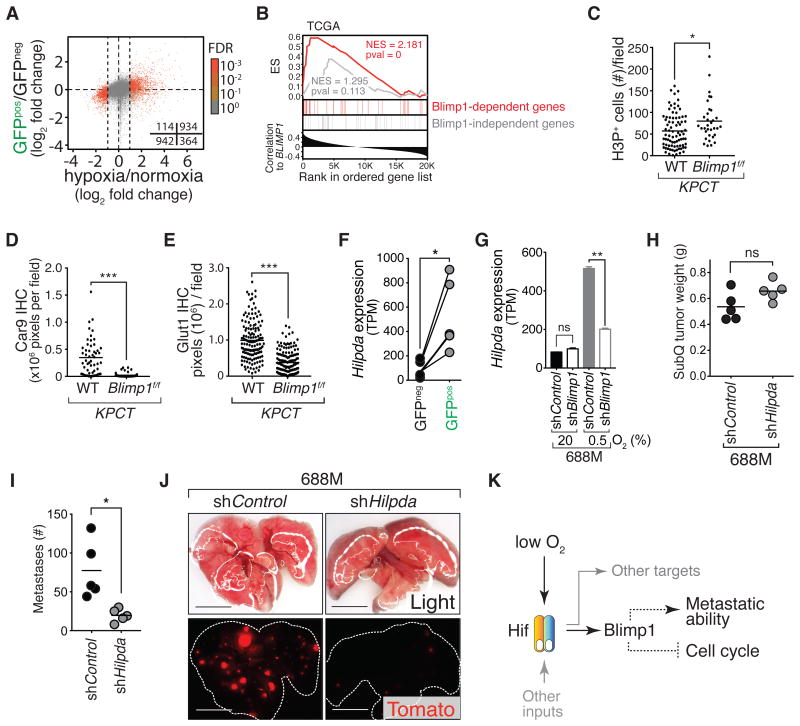
Figure 6. Blimp1 regulates the expression of pro-metastatic, hypoxia-induced target genes.
(A) Genes that are significantly differentially expressed under hypoxia (absolute log2 fold change > 1 and FDR < 0.001) are differentially expressed between GFPpos and GFPneg cells (Fisher's exact test, quadrant counts shown, p-values < 0.0001).
(B) BLIMP1 expression correlates with a subset of hypoxia-induced genes in human PDAC. All genes from the TCGA PDAC dataset are ranked by their correlation with BLIMP1 expression (Pearson r) and enrichments of 36 BLIMP1-dependent (red) and 36 BLIMP1-independent (grey), hypoxia-induced genes are shown. NS, enrichment score; NES, normalized enrichment score; pval, nominal p-value.
(C-E) Blimp1 deficiency in KPCT mice significantly increases PDAC cell proliferation (C) while reduces Car9 (D) and Glut1 (E) expression (N = 3). Proliferation was measured by IHC for phospho-histone 3 (H3P). H3P positive nuclei were quantified. (C) Each dot is the number of H3Ppos cells in a field and the bar is the mean. (D,E) Each dot is the sum of all pixels (brown color) above the cutoff in a field and the bar is the mean. *, p-value < 0.05; ***, p-value < 0.0005 by Student's t test.
(F) Expression (TPM) of Hilpda in GFPpos and GFPneg PDAC cells from KPCcolors mice. Paired samples are connected with a line. *, p-value < 0.05 by paired t test.
(G) Blimp1 knockdown significantly reduced hypoxia-induced Hilpda expression. **, p-value < 0.005. ns, not statistically significant.
(H-J) Hilpda is required for PDAC metastasis from subcutaneous tumors. (H) Each dot represents the mean of multiple subcutaneous tumor weight in a mouse and the bar is the mean of all mice in each cohort. (I) Each dot represents the number of lung metastases in a mouse and the bar is the mean. (J) Representative fluorescent images of lung metastases from the subcutaneous tumors are shown. Lung lobes are outlined with dotted lines (J). ns, not statistically significant; *, p-value < 0.05 by Student's t test. Scale bars = 0.5 cm.
(L) Proposed model. Dotted lines indicate potentially indirect mechanisms.
To further relate these gene expression programs with BLIMP1 expression in human PDAC, we used these integrated datasets to define a 36-gene signature of Blimp1-dependent, hypoxia-induced genes that are also higher in the GFPpos state. Across multiple human PDAC gene expression datasets, this Blimp1 signature correlated with BLIMP1 expression, suggesting conserved mechanism of BLIMP1 function in human PDAC in vivo (Fig. 6B and Supplementary Fig. S8D-S8E).
Our gene expression profiling suggested that Blimp1 might be required for hypoxia-induced cell cycle arrest. To directly test this, we cultured shControl and shBlimp1 cells at 0.5% and 20% O2 and assessed proliferation by short term BrdU labeling. While shControl cells almost completely arrested under hypoxia, shBlimp1 cells continued to proliferate (Supplementary Fig. S8F-S8G). To determine whether Blimp1 reduces the proliferation of PDAC cells in tumors in vivo, we assessed the proliferation of cancer cells in pancreatic tumors in KPCT;Blimp1f/f and control KPCT;Blimp1+/+ mice. Cancer cells in autochthonous Blimp1-deficient tumors had a higher mitotic index (Fig. 6C and Supplementary Fig. S8H). The higher proliferation of cancer cells in tumors from KPCT;Blimp1f/f mice is also consistent with the shorter survival of KPCT;Blimp1f/f mice (Supplementary Fig. S3U).
Many of the genes that were hypoxia-induced, Blimp1-dependent, and expressed at higher levels in the more metastatic GFPpos PDAC cells have been previously implicated as pro-metastatic factors in other cancer types. These genes included Pgf, Dusp1, Hmox1, Car9, Glut1, and Hilpda (48-53). Consistent with our RNA-Seq data, we observed reduced Glut1 and Car9 protein expression in PDACs in KPCT;Blimp1f/f mice compared to KPCT mice (Fig. 6D-6E and Supplementary Fig. S8H-S8M). High expression of the lipid droplet-associated protein Hilpda in other cancer types correlates with disease progression and metastasis (53,54). Hildpa expression was higher in GFPpos PDAC cells, induced by hypoxia in murine and human PDAC cells, and its induction was partially Blimp1-dependent (Fig. 6F-6G and Supplementary Fig. S4J). Hilpda knockdown reduced metastasis in our initial analysis and we further confirmed that Hilpda knockdown in PDAC cells significantly reduced their metastatic ability (Fig. 6H-6J, Supplementary Fig. S8N, and Supplementary Fig. S5G-S5L). These data suggest that Hilpda is a Blimp1-regulated pro-metastatic factor in PDAC.
Discussion
To uncover the molecular mechanisms that drive the metastatic ability of PDAC, we initially took two unbiased gene expression-profiling approaches: analysis of Hmga2-GFPpos and Hmga2-GFPneg PDAC subpopulation as well as analysis of bulk cancer cells from large primary tumors and macro-metastases. In both cases, we specifically isolated cancer cells at high purity by FACS to avoid confounding our analyses with contaminating stromal cell populations. Analysis of bulk cancer cells from primary tumors and metastases uncovered few significant gene expression changes, implying that cancer cells in the largest primary tumors possess most of the molecular features required for metastatic spread.
These findings are in stark contrast to the extensive gene expression differences between large primary tumors and metastases that we uncovered in a parallel study on a KrasG12D-driven, p53-deficient mouse model of lung adenocarcinoma (55). In the lung cancer model, large primary tumors often existed in an earlier non-metastatic state that had profound gene expression differences from metastases. In the lung, oncogenic KrasG12D alone can drive extensive tumor growth and even tumors in KrasLSL-G12D;p53flox/flox mice do not immediate receive benefit from being p53 deficient (55-58). Thus, pancreatic tumors may be forced into a potentially metastatic state by the selective pressures of primary tumor growth, thereby explaining the high likelihood of metastatic spread even in patients with relatively small pancreatic tumors (59).
Despite these observations, multiple lines of evidence suggest that the metastatic ability of PDAC is still an acquired phenotype. We previously noted mice with widespread PanIN lesions that lack any disseminated tumor cells in the peritoneal cavity (60). Additionally, we and others have generated mice with clonally marked pancreatic tumors and documented that not all tumors give rise to metastases (60,61). Although we did not observe gene expression differences between large primary tumors and metastases, we have documented microenvironment-driven metastatic heterogeneity. Our results support a model in which the development of hypoxic regions generates cells with increased metastatic ability (62). Consistent with results from autochthonous mouse models, human PDAC is a highly hypoxic cancer type (63) and the metastatic ability of orthotopically-grown, patient-derived PDAC xenografts is predicted by their level of hypoxia (64).
Hypoxia has been shown to induce metastasis in multiple cancer types through various mechanisms (reviewed in 64, 67, and 68). For example, hypoxia is related to alterations in EMT/MET, angiogenesis, local invasion and intravasation, extravasation, as well as the formation of the pre-metastatic niche (65). While some consequences of hypoxia may be relatively generalizable across cancer types, some outputs of hypoxia may also be cancer type specific, thus the importance of Blimp1 in these different steps of the metastatic cascades as well as in different cancer types remain to be determined.
Hypoxia also has a tremendous impact on the self-renewal and differentiation of the progenitor/stem cell lineages. For example, hypoxia potentiates the engraftment of human hematopoietic stem cells in recipient mice (66,67) and also helps maintain the stemness of embryonic stem and iPS cells in culture (68,69). In several cancer types, hypoxia has also been shown to play important roles in maintaining cancer stem cells (CSCs). In brain tumors, hypoxia promotes and/or maintains cancer cell stemness similar to the effect of hypoxia on bona fide stem cells (70,71). Several studies have identified subpopulations of murine and human PDAC cells with CSC characteristics based on their ability to generate new tumors upon transplantation (14,72). Importantly, the highly metastatic PDAC state that we identified is not directly related to previously reported CSC populations, the differentiation state of the cancer cells, or EMT. Thus, whether the highly metastatic PDAC cell state and these CSC states represent parallel or partially overlapping programs will be an important area for future study.
We initially anticipated that the high metastatic ability of Hmga2-expressing cells would be driven by Hmga2 itself. Surprisingly, this is not the case, as Hmga2 deficiency has no impact on the metastatic ability of tumors in the KPC PDAC mouse model nor does it influence the induction of canonical hypoxia target genes (BMG, S-HC, MMW; manuscript in preparation and Supplementary Fig. S4I). Hmga2 could play a subtle role in the later stages of metastatic outgrowth or may simply be a marker of the metastatic state.
Mechanistically, our results uncover hypoxia-mediated induction of the transcription factor Blimp1 as one molecular link between the tumor microenvironment and transient induction of pro-metastatic gene expression programs in PDAC. Although our data show that Blimp1 can be induced through hypoxia-mediated stabilization of Hif, other factors within the tumor microenvironment may also impact Hif activity (Fig. 6L). In PDAC, Blimp1 functions as a molecular switch that promotes metastatic ability while suppressing cell division under hypoxia (Fig. 6L). Our results are consistent with the link between Blimp1 and migratory ability of human lung and breast cancer cell lines in vitro (73,74). Blimp1 has not been described as a hypoxia/Hif target gene in normal cell types but hypoxia may also influence Blimp1 expression in those settings. In early embryos, where oxygen levels are low prior to the formation of major blood vessels (75), Blimp1 is expressed in primordial germ cells where it represses somatic programs and helps maintain pluripotency (38,76). Blimp1 is also critical for the differentiation of plasma cells that are generated in secondary lymphoid organs and maintained in bone marrow, both of which have hypoxic regions (37,77,78).
In summary, our findings support the concept of microenvironmental, rather than mutational, evolution being a critical factor that fosters PDAC metastatic ability. We found that intratumoral hypoxia, which is an inevitable feature of advanced human PDAC, induces the expression of the pro-metastatic transcription factor Blimp1. The co-option of this master regulatory transcription factor is required for metastatic ability, and the molecular output of Blimp1 expression is the modulation of discrete hypoxia-induced gene expression programs. A greater understanding of the origins and molecular features of cancer cells with transient high metastatic ability could provide therapeutic opportunities to reduce metastatic spread and further our appreciation of the obligate plasticity of these cells during the metastatic process.
Methods
Mice
KrasLSL-G12D, p53LSL-R172R, Blimp1flox, Pdx1-Cre, Rosa26LSL-tdTomato, and Hmga2CK mice have been described (18,20,32,37,79,80). Mice with the KrasLSL-G12D and the R26LSL-tdTomato alleles in cis on chromosome 6 were used to maximize retention of the R26LSL-tdTomato allele even in genomically unstable tumors. 6-10-week-old NOD/Scid/γc (NSG) mice (The Jackson Laboratory, Stock number: 005557) were used for transplantation experiments. The Stanford Institutional Animal Care and Use Committees approved all animal studies and procedures.
Histology and quantification of IHC
All histological staining were performed on paraffin-embedded, formalin-fixed sections as described previously (60). Briefly, 4 μm sections were re-hydrated and subjected to antigen retrieval before IHC was performed using Vector Lab ABC Vectastain kit. We used custom FIJI macro scripts for the quantification of IHC. See Supplemental Experimental Procedures for the detail of staining procedures and IHC quantification.
RNA-Seq data analyses
RNA and genomic DNA samples were extracted from 104 to 5×104 sorted cancer cells using Qiagen AllPrep DNA/RNA Micro Kit. RNA from ex vivo FACS-purified cells (15 ng/sample) were converted to cDNA and amplified with the NuGEN Ovation RNA-Seq system. Subsequently, amplified cDNA was sonicated and subjected to library preparation using the Illumina TruSeq DNA sample preparation kit. Total RNA from shControl or shBlimp1 688M cells cultured in 0.5% or 20% O2 were used for the preparation of RNA-Seq libraries with Illumina's TruSeq RNA Library Prep Kit v2 according to manufacturer's protocol. Sequencing was performed on Illumina HiSeq 2000 for 100-bp paired-end (ex vivo samples) and non-paired-end (in vitro samples) reads. See Supplemental Experimental Procedures for RNA-Seq analysis details.
ATAC-Seq data analysis
Murine PDAC 688M cells cultured in 0.5% or 20% O2 were also used for ATAC-Seq library preparation. Briefly, nuclei were extracted before incubation with TDE1 Tn5 transposase (Illumina). The fragmented genomic DNA was PCR amplified and ATAC-Seq libraries were sequenced on an Illumina NextSeq with paired-end 76bp reads using an Illumina High Output kit. ATAC-Seq data were processed as previously described with some modifications (81). See Supplemental Experimental Procedures for ATAC-Seq analysis details.
Western blotting
Cell lysates were prepared with RIPA buffer supplemented with protease inhibitors. Proteins were separated by PAGE before being transferred onto a Bio-Rad PVDF membrane. Primary antibodies were incubated in the presence of 5% skimmed milk at 4°C overnight, followed by stringent washes (3×) before staining with horseradish-peroxidase-conjugated secondary antibodies. After additional washes, enhanced chemiluminescence was performed to visualize the proteins of interest. See Supplemental Experimental Procedures for more detail of western blot analyses.
Hypoxia induction and qRT-PCR
To induce hypoxia in vitro, cancer cells were seeded at sub-confluency and cultured in a hypoxia chamber (Invivo2-400, Ruskin Technologies) with 0.5% O2 for 24 hours before harvest. Cells were subsequently lysed with TRIzol (Thermo Fisher Scientific, 15596-018) directly on tissue culture dishes for RNA extraction. RNA concentration was quantified on a NanoDrop spectrophotometer (Thermo Fisher Scientific, NanoDrop 2000 UV-Vis Spectrophotometer) and converted to cDNA according to manufacturer's protocol (Thermo Fisher Scientific, 4368814). For the quantification of transcripts, SYBR green (Sigma-Aldrich, S9194) was used with specific primer pairs. β-actin was used as internal control. See Supplementary Data for more detailed information.
Cell lines
None of the cell lines used in this study were authenticated. The years when the PDAC cell lines were obtained are as follow: 688M, 2014; 1004M, 2014; 887M, 2017; 1814, 2015; 1810, 2015; human PDAC cell Panc1, Colo357, BxPC1, AsPC1, and Capan1 were all obtained in 2014. All PDAC cell lines that were used in experiments reported in this study were expanded early in the passage and aliquots were stored in liquid nitrogen. Thawed cells were passaged within 1-2 months for any experimental usage.
Subcutaneous transplantation of cell lines into NSG mice
688M and 1004M PDAC cells were cultured at sub-confluency shortly before harvest for transplantation. All cells used in the transplantation experiments were validated for knockdown efficacies of designated genes. Briefly, cells were trypsinized and washed 3× in cold PBS before subcutaneous injection. 2.5 × 105 cells were used per injection on the dorsal flank. The numbers of Tompos metastases in the lung were quantified by direct counting using a fluorescence dissecting scope. Alternatively, H&E sections were used to estimate lung metastases seeded by the Tomato-negative 1004M cells. Metastases in the lung were validated by histology.
Pancreatic orthotopic transplantation
688M PDAC cell derivatives validated for efficient knockdown were washed 3× in cold PBS before resuspension in 100% Matrigel (Corning, 356231). Subsequently, a surgical procedure was performed with direct injection of the cells/Matrigel mixture into the pancreas of NSG mice. See Supplemental Experimental Procedures for more detail of the orthotopic transplantation.
Statistical analysis
For comparison between two quantitative variables, we used Student's t test when samples were not paired and paired t test for paired samples. When more than two variables were compared, both one-way ANOVA and Kruskall-Wallis test were used. For comparison of survival in Kaplan-Meier analyses, we used log-rank test for univariate survival analyses. Fisher's exact test was used in the analysis of contingency tables. Analyses were performed using Prism 6.0 (Graphpad Software Inc.).
Supplementary Material
Significance.
PDAC is an almost uniformly lethal cancer, largely due to its tendency for metastasis. We define a highly metastatic subpopulation of cancer cells, uncover a key transcriptional regulator of metastatic ability, and define hypoxia as an important factor within the tumor microenvironment that increases the metastatic proclivity.
Acknowledgments
We thank the Stanford Shared FACS Facility and the Protein and Nucleic Acid Facility for expert assistance; Carolyn Sinow, Santiago Naranjo, and Shashank Cheemalavugu for technical assistance; Chen-Hua Chuang and Nicholas Denko for experimental advice; Teri A Longacre for the analysis of human PDAC IHC samples; Justin A. Kenkel for help with pancreatic orthotopic transplantation procedure; Stephano Mello, Edward LaGory, and Julia Arand for reagents; Louis Leung, Chris Probert, Peyton Greenside, Andrew Seung-Hyun Koh, and Xun Lan for bioinformatics advice; Laura Attardi, Julien Sage, Jennifer Brady, Kenneth Olive, David Feldser, the Winslow laboratory, the Greenleaf laboratory, and the Stanford pancreatic cancer research community for helpful comments; and Sean Dolan and Alexandra Orantes for administrative support.
Financial Support: M.M. Winslow received a Pancreatic Cancer Action Network-AACR Award in memory of Skip Vinagh (13-20-25-WINS), Stanford Cancer Institute support grant (P30-CA124435) from the National Cancer Institute, and the award (NIH-R00CA151968) from the National Institute of Health (NIH). S. Chiou and B.M. Grüner received the Stanford Dean's Fellowship. V.I. Risca received the Walter V. and Idun Berry Fellowship. B.M. Grüner received a Pancreatic Cancer Action Network-AACR Fellowship in memory of Samuel Stroum (14-40-25-GRUE). E.E. Graves received a NIH R01 grant (CA197136). G.X. Wang received the John Merck Fund and National Institute of Mental Health R01 grant (MH099647). P. Mourrain received a NIH grant (MH09964) and the Brightfocus foundation and John Merck Fund. A.J. Giaccia received the NIH R01 grants CA67166 and CA198291.
Footnotes
Additional information: The authors declare no competing financial interests.
Accession number: The accession number for all the next generation sequencing data is included in the following superseries: GSE90825.
Supplementary Data: Supplementary Data includes Supplementary Methods, thirteen Supplementary Figures, and three Supplementary Tables.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M. Pancreatic cancer. The New England journal of medicine. 2010;362(17):1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer research. 1997;57(11):2140–3. [PubMed] [Google Scholar]
- 4.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: Evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer research. 2000;60(7):2002–6. [PubMed] [Google Scholar]
- 6.Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, et al. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. American Journal of Pathology. 2001;158(5):1677–83. doi: 10.1016/S0002-9440(10)64123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Notta F, Chan-Seng-Yue M, Lemire M, Li YL, Wilson GW, Connor AA, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538(7625):378. doi: 10.1038/nature19823. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nature genetics. 2017;49(3):358–66. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–30. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell. 2015;161(6):1345–60. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox RG, Lytle NK, Jaquish DV, Park FD, Ito T, Bajaj J, et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature. 2016;534(7607):407–11. doi: 10.1038/nature17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell stem cell. 2007;1(3):313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine. 2013;19(11):1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The Pancreas Cancer Microenvironment. Clinical Cancer Research. 2012;18(16):4266–76. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4(6):437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 19.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell. 2005;7(5):469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Chiou SH, Kim-Kiselak C, Risca VI, Heimann MK, Chuang CH, Burds AA, et al. A conditional system to specifically link disruption of protein-coding function with reporter expression in mice. Cell reports. 2014;7(6):2078–86. doi: 10.1016/j.celrep.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes & development. 2006;20(22):3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes & development. 2003;17(24):3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(15):5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raskin L, Fullen DR, Giordano TJ, Thomas DG, Frohm ML, Cha KB, et al. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. The Journal of investigative dermatology. 2013;133(11):2585–92. doi: 10.1038/jid.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hristov AC, Cope L, Reyes MD, Singh M, Iacobuzio-Donahue C, Maitra A, et al. HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22(1):43–9. doi: 10.1038/modpathol.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46(7):503–11. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 27.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(8):2334–40. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 28.Piscuoglio S, Zlobec I, Pallante P, Sepe R, Esposito F, Zimmermann A, et al. HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology. 2012;60(3):397–404. doi: 10.1111/j.1365-2559.2011.04121.x. [DOI] [PubMed] [Google Scholar]
- 29.Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(24):9920–5. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(8):2570–80. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, et al. An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. Br J Cancer. 2003;89(11):2104–9. doi: 10.1038/sj.bjc.6601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee S, Nomura A, Sangwan V, Chugh R, Dudeja V, Vickers SM, et al. CD133+ tumor initiating cells in a syngenic murine model of pancreatic cancer respond to Minnelide. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(9):2388–99. doi: 10.1158/1078-0432.CCR-13-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(17):2806–12. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 35.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nature genetics. 2017;49(3):367–76. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–20. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 38.Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–13. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 39.Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. The Journal of biological chemistry. 2001;276(11):7919–26. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 40.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55(1):25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arulanandam R, Batenchuk C, Angarita FA, Ottolino-Perry K, Cousineau S, Mottashed A, et al. VEGF-Mediated Induction of PRD1-BF1/Blimp1 Expression Sensitizes Tumor Vasculature to Oncolytic Virus Infection. Cancer cell. 2015;28(2):210–24. doi: 10.1016/j.ccell.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Melvin A, Rocha S. Chromatin as an oxygen sensor and active player in the hypoxia response. Cell Signal. 2012;24(1):35–43. doi: 10.1016/j.cellsig.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nature cell biology. 2006;8(6):623–30. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 44.Minnich M, Tagoh H, Bonelt P, Axelsson E, Fischer M, Cebolla B, et al. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nature immunology. 2016;17(3):331–43. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276(5312):596–9. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 46.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 47.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. Journal of immunology. 2004;173(9):5556–63. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 48.Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A, et al. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer research. 2011;71(9):3364–76. doi: 10.1158/0008-5472.CAN-10-4261. [DOI] [PubMed] [Google Scholar]
- 49.Kawamura T, Kusakabe T, Sugino T, Watanabe K, Fukuda T, Nashimoto A, et al. Expression of glucose transporter-1 in human gastric carcinoma: association with tumor aggressiveness, metastasis, and patient survival. Cancer. 2001;92(3):634–41. doi: 10.1002/1097-0142(20010801)92:3<634::aid-cncr1364>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 50.Chaturvedi P, Gilkes DM, Wong CC, Kshitiz, Luo W, Zhang H, et al. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. The Journal of clinical investigation. 2013;123(1):189–205. doi: 10.1172/JCI64993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moncho-Amor V, Ibanez de Caceres I, Bandres E, Martinez-Poveda B, Orgaz JL, Sanchez-Perez I, et al. DUSP1/MKP1 promotes angiogenesis, invasion and metastasis in non-small-cell lung cancer. Oncogene. 2011;30(6):668–78. doi: 10.1038/onc.2010.449. [DOI] [PubMed] [Google Scholar]
- 52.Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. The Journal of clinical investigation. 2015;125(7):2592–608. doi: 10.1172/JCI78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo T, Konda R, Sugimura J, Iwasaki K, Nakamura Y, Fujioka T. Expression of hypoxia-inducible protein 2 in renal cell carcinoma: A promising candidate for molecular targeting therapy. Oncology letters. 2010;1(4):697–701. doi: 10.3892/ol_00000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim SH, Wang D, Park YY, Katoh H, Margalit O, Sheffer M, et al. HIG2 promotes colorectal cancer progression via hypoxia-dependent and independent pathways. Cancer letters. 2013;341(2):159–65. doi: 10.1016/j.canlet.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chuang CH, Greenside P, Rogers Z, Brady J, Yang D, Caswell D, et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109/Jak/Stat axis. Nature medicine. 2017 doi: 10.1038/nm.4285. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–5. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muzumdar MD, Dorans KJ, Chung KM, Robbins R, Tammela T, Gocheva V, et al. Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers. Nature communications. 2016;7:12685. doi: 10.1038/ncomms12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers ZN, McFarland CD, Winters IP, Naranjo S, Chuang CH, Petrov D, et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nature methods. 2017 doi: 10.1038/nmeth.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fortner JG, Klimstra DS, Senie RT, Maclean BJ. Tumor size is the primary prognosticator for pancreatic cancer after regional pancreatectomy. Annals of surgery. 1996;223(2):147–53. doi: 10.1097/00000658-199602000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes & development. 2015;29(14):1576–85. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maddipati R, Stanger BZ. Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer discovery. 2015;5(10):1086–97. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nature reviews Cancer. 2008;8(12):967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koong AC, Mehta VK, Le QT, Fisher GA, Terris DJ, Brown JM, et al. Pancreatic tumors show high levels of hypoxia. International journal of radiation oncology, biology, physics. 2000;48(4):919–22. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 64.Chang Q, Jurisica I, Do T, Hedley DW. Hypoxia predicts aggressive growth and spontaneous metastasis formation from orthotopically grown primary xenografts of human pancreatic cancer. Cancer research. 2011;71(8):3110–20. doi: 10.1158/0008-5472.CAN-10-4049. [DOI] [PubMed] [Google Scholar]
- 65.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future oncology. 2013;9(11):1623–36. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danet GH, Pan Y, Luongo JL, Bonnet DA, Simon MC. Expansion of human SCID-repopulating cells under hypoxic conditions. The Journal of clinical investigation. 2003;112(1):126–35. doi: 10.1172/JCI17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cipolleschi MG, Dello Sbarba P, Olivotto M. The role of hypoxia in the maintenance of hematopoietic stem cells. Blood. 1993;82(7):2031–7. [PubMed] [Google Scholar]
- 68.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(13):4783–8. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell stem cell. 2009;5(3):237–41. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 70.Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28(45):3949–59. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 71.Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell cycle. 2009;8(20):3274–84. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishizawa K, Rasheed ZA, Karisch R, Wang QJ, Kowalski J, Susky E, et al. Tumor-Initiating Cells Are Rare in Many Human Tumors. Cell stem cell. 2010;7(3):279–82. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romagnoli M, Belguise K, Yu Z, Wang X, Landesman-Bollag E, Seldin DC, et al. Epithelial-to-mesenchymal transition induced by TGF-beta1 is mediated by Blimp-1-dependent repression of BMP-5. Cancer research. 2012;72(23):6268–78. doi: 10.1158/0008-5472.CAN-12-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu Z, Sato S, Trackman PC, Kirsch KH, Sonenshein GE. Blimp1 activation by AP-1 in human lung cancer cells promotes a migratory phenotype and is inhibited by the lysyl oxidase propeptide. PloS one. 2012;7(3):e33287. doi: 10.1371/journal.pone.0033287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walls JR, Coultas L, Rossant J, Henkelman RM. Three-dimensional analysis of vascular development in the mouse embryo. PloS one. 2008;3(8):e2853. doi: 10.1371/journal.pone.0002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132(6):1315–25. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 77.Caldwell CC, Kojima H, Lukashev D, Armstrong J, Farber M, Apasov SG, et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. Journal of immunology. 2001;167(11):6140–9. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 78.Cho SH, Raybuck AL, Stengel K, Wei M, Beck TC, Volanakis E, et al. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537(7619):234–8. doi: 10.1038/nature19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15(24):3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 81.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10(12):1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.