Abstract
Emerging evidence links exosomes to cancer progression by the trafficking of oncogenic factors and neoplastic reprogramming of stem cells. This necessitates identification and integration of functionally validated exosome-targeting therapeutics into current cancer management regimens. We employed quantitative high throughput screen on two libraries to identify exosome-targeting drugs; a commercially available collection of 1280 pharmacologically active compounds and a collection of 3300 clinically approved compounds. Manumycin-A (MA), a natural microbial metabolite, was identified as an inhibitor of exosome biogenesis and secretion by castration-resistant prostate cancer (CRPC) C4-2B, but not the normal RWPE-1, cells. While no effect was observed on cell growth, MA attenuated ESCRT-0 proteins Hrs, ALIX and Rab27a and exosome biogenesis and secretion by CRPC cells. The MA inhibitory effect is primarily mediated via targeted inhibition of the Ras/Raf/ERK1/2 signaling. The Ras-dependent MA suppression of exosome biogenesis and secretion is partly mediated by ERK-dependent inhibition of the oncogenic splicing factor hnRNP H1. Our findings suggest that MA is a potential drug candidate to suppress exosome biogenesis and secretion by CRPC cells.
Keywords: Manumycin A, exosome biogenesis and secretion, Ras signaling, hnRNP H1, prostate cancer
1. Introduction
Prostate cancer (PC) is the most common and the second leading cause of cancer-related deaths among American males. Despite the initial response to various treatment regimens [1], some PC patients inevitably progress to castration-resistant PC (CRPC). Thus, there is a need to develop new, highly effective therapeutic agents to circumvent advanced disease.
Recently, the breadth of knowledge about biogenesis, ‘cargo’ contents and intercellular communication of diverse types of cancer-derived extracellular vesicles (EVs) in a variety of physiologic contexts [2] has expanded considerably. EVs encompass a broad range of secreted vesicles, including exosomes, microvesicles (MVs), and apoptotic blebs [3]. In addition to their own selective markers, the membranous nano-sized exosomes harbor surface markers indicative of their cellular origin. The exosome ‘cargo’ contains a wide variety of RNAs (including mRNAs, non-coding RNAs), proteins, DNA and lipids [4]. The endosomal sorting complex required for transport (ESCRT) machinery and their associated proteins, such as tumor susceptibility gene 101 (TSG101), hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and ALG-2 interacting protein X (Alix), are pivotal to biogenesis, cargo sorting and secretion of exosomes [5].
Exosomes are implicated in cell-cell communication and modulation of the biology of recipient cells [6]. The PC-derived exosomes are detected in the prostatic secretions, seminal fluid, tissue, urine, and blood [7], implicating their clinical utility as ‘liquid biopsies’ in the diagnosis and prognosis of PC. The tumor-derived exosomes promote growth, angiogenesis, and metastasis of recipient cells [8]. We recently demonstrated that trafficking of oncogenic factors by PC cell-derived exosomes subvert tumor microenvironment and prime oncogenic reprogramming of tumor-tropic PC patients’ derived adipose stem cells, leading to tumor clonal expansion in vivo [9]. However, currently, there are no drugs that selectively target pathways involved in exosome biogenesis and secretion by cancer cells and their uptake by recipient cells. Such drugs may prove to be clinically effective for the prevention and/or treatment of advanced PC.
The family of heterogeneous nuclear ribonucleoproteins (hnRNPs) regulates pre-mRNA biogenesis, metabolism, and transport [10]. As a bona fide component of the nuclear matrix, the hnRNP H/F subfamily (hnRNP H1, hnRNP H2, hnRNP F, and hnRNP 2H9) are characterized by the possession of the quasi-RNA binding recognition motif (qRRM). Notably, we recently demonstrated the selective expression and growth promotion ability of the oncogenic slicing factor hnRNP H1 in CRPC cells via transcriptional upregulation of androgen receptor (AR) and it is spliced variant AR-V7 [11]. However, the functional role of hnRNPs, including hnRNP H1, in the biogenesis and/or section of exosomes remains elusive.
Ras proteins are small GTPases that function as molecular switches by alternating between inactive GDP-bound to active GTP-bound states. Active Ras (GTP bound Ras) binds to and activates downstream effectors, such as PI3K, RAF/MEK/ERK pathway, and initiates a cascade of cellular events related to tumor and non-tumor pathologies [12, 13]. Although Ras mutations in prostate cancer are infrequent, they play a pivotal role in multiple pathways that have been implicated in prostate cancer growth, transformation, differentiation, stress responses to androgen independence [14].
The present study provides evidence for the first time that Manumycin (MA) suppresses exosome biogenesis and secretion in CRPC cells via inhibition of Ras signaling pathway and hnRNP H1 expression. The shRNA silencing of hnRNP H1 attenuated the endogenous levels of Alix, Rab27a, and Ras, suggesting that hnRNP H1 is pivotal to MA-mediated inhibition exosome biogenesis and secretion in CRPC cells.
2. Materials and Methods
2.1 Materials
RPMI 1640, K-SFM, penicillin/streptomycin solution, fetal bovine serum (FBS) were from Invitrogen (Camarillo, CA). Manumycin A (MA) and GW4869 were purchased from Cayman Chemical Company (Denver, CO). The NCGC Pharmaceutical Collection (NPC) was custom assembled at National Center for Advancing Translational Sciences (NCATS, NIH, Bethesda, MD). The Ras activation assay kit (17–218) was purchased from Millipore (Darmstadt, Germany). U0126 and SB203580 were from Promega (Madison, WI) and SP600125 and the LOPAC collection were purchased from Sigma (Sigma-Aldrich, St. Louis, Missouri). Unless otherwise indicated, all other drugs were purchased from Sigma (St. Louis, MO, USA).
2.2 Cell culture and plasmids
The human normal prostate epithelial RWPE1 cells and CRPC cells (PC-3 and 22Rv1) were purchased from ATCC (Manassas, VA). The C4-2B cells were a kind gift of Dr. LW Chung (Cedars-Sinai, LA). The PC cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2mM L-glutamine and 1% penicillin/streptomycin (P/S). The RWPE-1 cells were maintained in K-SFM media with supplements (ATCC). For routine maintenance, each cell line was cultured as a monolayer at 37°C in a 5% CO2, 95% air incubator. Two C4-2B cell lines stably expressing a control shRNA or hnRNP H1 shRNA were generated as we described [11]. All cell lines were authenticated by short tandem repeat (STR) profiling at Genetica DNA Laboratories (LabCorp, Burlington, NC).
2.3 Cell proliferation
The experiments were performed as we described [15]. Briefly, a panel of control (RWPE-1) and CRPC (C4-2B, PC-3, and 22Rv1) cells were seeded in a 96-well plate and treated with MA at a concentration range of 0 (DMSO vehicle) to 250 nM in triplicates for 48 h. Fresh media containing 0.5 mg/ml MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide] was then added for 4 h. The supernatants were removed and the resulting formazan crystals were solubilized in DMSO and measured at 570 nm using a BioTek microplate reader (BioTek, Winooski, VT).
2.4 Immunoblot and PCR analyses
Oligonucleotide primers for both convention and Real-Time PCR were synthesized by Integrated DNA technologies (Coralville, Iowa) and are listed in Supplementary Table S1. Total RNA and proteins were isolated from various using standard protocols as we described [11]. Total RNA was subjected to conventional PCR and/or Real-Time PCR analyses using a master mix from Bio-Rad Laboratories (Hercules, CA). Protein extracts were subjected to immunoblot analysis using with antibodies against Alix, Hrs, NA+-K2+ ATPase, Rab5, Ras, p-Raf, Raf, p-ERK, and ERK (Cell Signaling Technology, Danvers, MA), TSG101, CD9, CD81, CD63, and GAPDH (Santa Cruz Biotechnology, Dallas, TX), Rab27a (Proteintech, Chicago, IL). Immune complexes were detected with appropriate secondary antibodies from Santa Cruz Biotechnology (Dallas, TX) and chemiluminescence reagents (Pierce, Rockford, IL, USA). Immunoblot signals were captured using the Image Quant Las 300 (GE Healthcare, Piscataway, NJ). Densitometric analysis was performed using ImageJ (NIH, Bethesda, MD, USA, http://imagej.nih.gov/ij/)
2.5 Ras activity assay
The Ras Activity assay kit (Catalog # 17–218) was purchased from Millipore and Ras affinity precipitation assay was performed as described in the manufacturer’s protocol [16]. Briefly, C4-2B cells were stimulated for with MA or DMSO for 30 min or 48 hours and rinsed twice with PBS, following which MLB lysis buffer was added and the cell lysates were collected by centrifugation. For the negative and positive loading controls, 5μL of GDP, (1mM final concentration) and 5μL of GTPγS (100μM final concentration) were added, respectively, to cell lysate extracts for 30 minutes at 30°C with agitation. Active Ras was pulled down with 10 μg purified Raf-1 RBD-agarose beads by incubating 1 mg of cell extracts for 45 min at 4°C with gentle agitation. The beads were washed, collected and reduced in Laemmli sample buffer. Active Ras (Ras-GTP) bound to the GST–RBD was dissociated by addition of SDS/PAGE loading buffer, and fractionation onto a 4–20% Mini-PROTEIN TGX™ polyacrylamide gel (Bio-Rad, Hercules, CA, USA). After transfer, the membranes were blotted with an anti-Ras Ab, clone RAS10 (EMD Millipore, Billerica, MA).
2.6 Sucrose gradient and exosome biomarker analyses
To examine the effect of MA on exosome biogenesis, C4-2B cells were incubated in RPMI media and treated with vehicle or MA for 48 h. Cells were washed, pelleted, and re-suspended in 0.5 ml of PBS. The cells were then lysed by drawing through a 21-gauge needle 20 times, centrifuged at 14,000 rpm for 15 min at 4°C to pellet the insoluble fraction. The supernatant (1 mg protein) was then applied to the top of a tube containing discontinuous (8%/15%/20%/30%/35%) sucrose gradient and centrifuged for 16 h at 100,000 × g at 4°C. Fractions of 500 μL each were collected by a needle and then separated by SDS-polyacrylamide gradient gel electrophoresis and analyzed by Western blotting for exosome markers Alix (exosome biogenesis marker), Hrs (multivesicular bodies marker), Rab5 (early endosome marker), Rab27a (exosome secretion marker), Rab7 (late endosome marker), GAPDH and Na+-K+ ATPase (plasma membrane Marker). Immunoblot signals were captured and densitometric analysis was performed as described above.
2.7 Exosome isolation
Exosomes from conditioned media (CM) of C4-2B cells cultured in exosome-free media containing MA or control vehicle were purified by differential ultracentrifugation (DU) as we described [9], with minor modifications (Supplementary Fig. S1) or by an exoEasy Maxi Kit per manufacturer’s instructions (QIAGEN GmbH, Hilden, Germany).
2.8 Analysis of exosomes by the qNano-IZON system and NanoSight 300
The impact of MA on exosome secretion was examined by the qNano-IZON and NanoSight 300 systems by measuring the concentrations, particle diameter, and size-distribution of exosomes harvested from the CM of C4-2B cells cultured in exosome-free media containing MA or vehicle (DMSO). The qNano system (Izon, Cambridge, MA, USA) allows the detection of exosomes using Tunable Resistive Pulse Sensing (TRPS) technology by driving vesicles through a pore using a combination of electrophoretic and convective flow induced by the applied voltage and an external pressure across the pore, respectively [17]. We first calibrated the voltage, stretch, pressure, and baseline current by using two standard beads: CPC100B (mode diameter of 110 nm (Izon) and a concentration of 1.1E13/mL) and CPC70D (mode diameter of 70nm (Izon) and a concentration of 2.8E13/mL). Finally, we optimized the system at a stretch of 48.50 mm, with a voltage of 0.30 V and 5.0 mbar pressure level. For analyses, 40 μL of the diluted sample was placed in the upper fluid cell under identical conditions. To prevent cross-contamination between samples, the upper fluid cell was washed 3 times with 100 μL PBS to remove residual particles after each application. We employed two pore types applicable to the intended measurement size range for extracellular vesicles; NP100 for 50–200nm size range, and NP400 for 200–800nm size range. For analysis with NP100, the samples were filtered by 0.22-micron Millex-HV Syringe Filter (EMD Millipore, Billerica, MA, USA). The data analysis was performed with qNano-IZON software. Size distribution within exosome preparations was also validated by measuring the rate of Brownian motion using the NanoSight 300 system, which is equipped with particle-tracking software (NanoSight, Amesbury, U.K.).
2.9 Analysis of exosomes by the NanoFACS system
The assay was performed as previously described (18–20). The NanoFACS system consists of Beckman Coulter Astrios-EQ MoFlo Sorter, configured with Side Scatter (SSC) detectors on each laser path, including the 405, 488, 561, and 640 nm lasers. With this configuration, a 561-nm SSC threshold was set, to allow a stable rate of background scattered light detection from the fluidics stream at the instrument noise floor. The 488 nm SSC and Forward Scatter (FSC) were used to detect scattered light from the exosomes and reference beads, with a resolution range of ~50–500 nm, as defined by control particles. Reference particles with known concentrations, validated by NanoSight NTA (LM-10, 405nm), were used as counting reference beads for quantification of nanoFACS-analyzed exosomes. Exosomes derived from the CM of CD63-GFP-expressing C4-2B cells maintained in exosome-free DMEM media and treated with DMSO, MA (250 nM) and/or GW4869 (0.25 or 10μM) were analyzed. The approximate size and concentration of the particles using the FSC and SSC parameters were measured relative to reference material controls. The FSC parameter along the X-axis depicts measurement of the amount of the laser beam that passes around the particle. The SSC parameter along the Y-axis detects the light scattered in the perpendicular direction, and, due to a larger collection angle, is more sensitive than FCS for particles with ~100 nm in size. Additionally, the GFP signal of the secreted CD63-GFP particles was also measured at 523 nm.
2.10 Statistical analysis
Data are presented as Means ± S.E.M. of more than three independent experiments performed in triplicate. For Western blots, a case representative experiment is depicted in the figures section. Comparisons between multiple groups were performed with ANOVA with Bonferroni’s test using GraphPad Prism. Statistical significance was considered at P< 0.05.
3. Results
3.1 High throughput screen (HTS) for CD63-GFP biogenesis
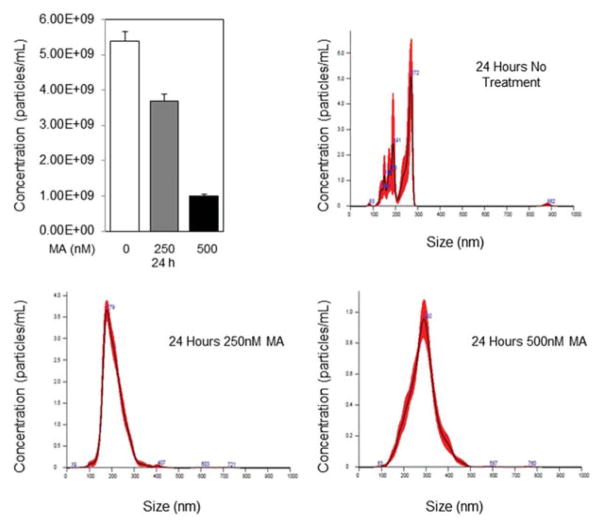
Two libraries were screened to examine a variety of compounds on exosome biogenesis; the LOPAC, a commercially available collection of 1280 pharmacologically active compounds, and the NPC (NCATS Pharmaceutical Collection, NIH), a collection of 3300 clinically approved compounds were screened for this assay. CD63-GFP-expressing C4-2B cells were generated and the screen was implemented in a dose response mode and the compounds were selected on the basis of GFP signal. One hundred and twenty-eight biogenesis activators and inhibitors were selected and re-tested in the same assay using 11-point dose responses to confirm activity and generate more accurate IC50 for these compounds. Analysis of secreted exosome in the CM of CD63-GFP-expressing C4-2B cells treated with MA or DMSO for 24 h was performed by NanoSight 300. Manumycin A produced a robust and reproducible dose response (0, 250 and 500 nM) in attenuating exosome biogenesis (Fig. 1).
Fig. 1.
Measurement of exosome secretion by NanoSight 300 Nanoparticle Tracking Analysis (Malvern, U.K.). Distribution curves of CD63-GFP-expressing C4-2B cell-derived exosomes after treatment with DMSO, 250nM Manumycin A for 24 or 72 h. Video of exosomes was captured in five replicates of one minute each by the NanoSight 300 using camera level 11, slider shutter 890, and slider gain 146. Videos were analyzed using detection threshold of 11. * denotes significance at P<0.05.
3.2 MA inhibits exosome biogenesis and secretion by CRPC cells
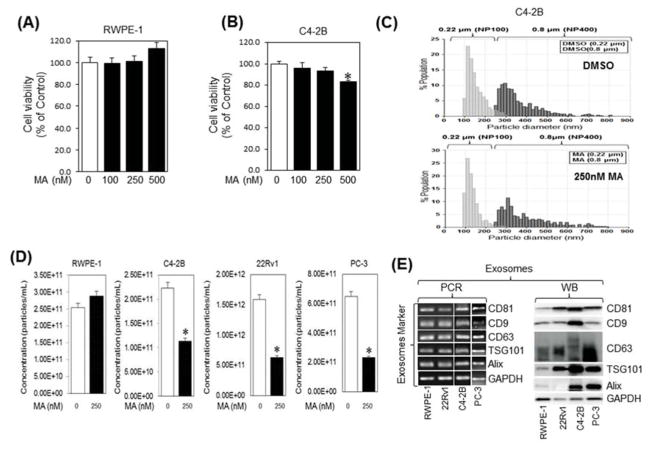
Dose response cell viability studies were performed to determine the non-cytotoxic dose (sub-IC50 concentrations) of MA to evaluate its effect on exosome biogenesis and secretion. While no effect was observed on RWPE-1 (Fig. 2A) and PC-3 cells (Supplementary Fig. S1), cellular exposure to MA (250 nM) for 48 hr caused ~8% and 10% cell death in C4-2B (Fig. 2B) and 22Rv1 (Supplementary Fig. S2), respectively, in comparison to controls. Since the pathways involved in the biogenesis and secretion of exosomes (3–150 nm) and microvesicles (MVs; >200 nm) differ, particle measurements were carried out to determine if their size-distribution and concentrations are modulated by the MA treatment in CRPC cells. Supplementary Fig. S2 shows the workflow of exosome and MV isolation by DU and sequential ultrafiltration. Analysis of secreted exosome and MVs in the CM of C4-2B, 22Rv1, PC-3 or RWPE-1 cells treated with MA or DMSO for 48 h was performed by the TRPS system using NP100 (50–200) and NP400 (200–800 nm) nanopores, respectively. The analysis showed no difference in the particle size-distribution, particle diameter or diameter mode (average size) of exosomes and MVs secreted by C4-2B (Fig. 2C; upper and lower panels, respectively), 22Rv1, and PC-3 cells (Supplementary Fig. S3A–C) treated with MA in comparison with DMSO. However, exosome quantification using NP100 nanopore (Izon) indicated that MA (250 nM) significantly suppressed exosome secretion in C4-2B, 22Rv1, and PC-3 cells by 50%, 65%, and 60%, respectively, as compared to the controls, but not in the normal RWPE-1 cells (Fig. 2D). While no effect was observed on particle diameter, similar pattern, but to a lower extent, of exosome biogenesis inhibition was observed by GW4869 in 22RV1 and PC-3 cells (Supplementary Fig. S4). Notably, a significant exosome biogenesis inhibition was detected upon combination of MA and GW4869 as opposed to either treatment alone (Supplementary Fig. S4). The secreted exosomes were validated by the expression of tetraspanins (CD9, CD63, and CD81) and exosome biogenesis markers Alix and TSG101 by Immunoblot and PCR analyses (Fig. 2E).
Fig. 2.
Manumycin A (MA) inhibits the exosome release by C4-2B cells. RWPE-1 (A) and C4-2B (B) cells were incubated overnight in 96-well cell culture plates with the indicated concentrations of Manumycin A or vehicle (DMSO) and analyzed for cell viability for 48 h, respectively. The IC50 concentration values for C4-2B cells were extrapolated from the sigmoid dose response curve fitting (GraphPad Prism software). The error bars represent the 95% confidence intervals of three independent experiments. (C) Following differential ultracentrifugation, extracellular vesicles and exosomes populations in the conditioned medium (CM) of DMSO (upper panel) or MA (250 nM; lower panel) were prepared by filtration through 0.8 μM and 0.22 μM filters, respectively, and their size distribution was analyzed by qNano-IZON system using NP400 and NP-100 nanopores, respectively. (D) qNano-IZON particle quantitative analysis (NP-100 nanopore) depicting a significant decrease in exosome concentrations (50–200 nm size) in the CM of RWPE-1, C4-2B, 22Rv1 and PC-3 cells treated with MA compared to vehicle treated controls. (E) Expression of exosome markers tetraspanins (CD9, CD63, and CD81) and exosome biogenesis markers Alix and TSG101 in the exosomes of RWPE-1, C4-2B, 22Rv1, and PC-3 cells harvested by UC and filtration (0.22 μm). GAPDH was used a control. *denotes significance at p<0.05 compared to control.
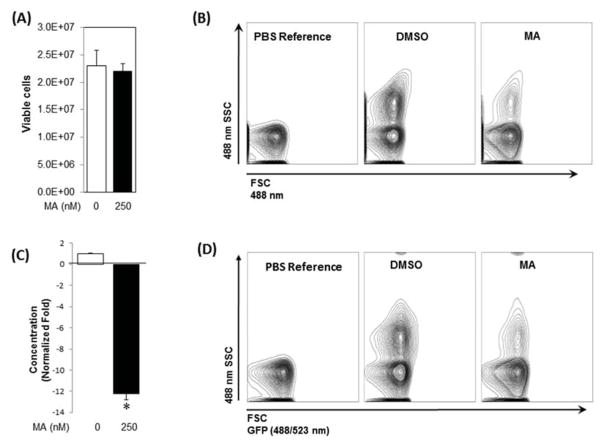
Next, using pCMV-CD63-GFP expression plasmid, we generated a stable C4-2B cell clone expressing the exosomal marker CD63 fused with GFP. Treatment of these cells with MA (250 nM) or DMSO for 48 h had no significant effect on cell viability (Fig. 3A). In order to independently corroborate the results of the qNano-IZON experiments, the exosome secretion by MA-, and DMSO-treated CD63-GFP-expressing cells was analyzed by a NanoFACS system. This is a high-resolution and high-throughput flow cytometry for quantification and multiparameter characterization of nano-sized cell-derived vesicles. The purified exosomes were analyzed and quantified in tandem with 200-nm reference beads, as a numeric counting control. As shown in Fig. 3B and C, the secretion of exosomes in MA-treated C4-2B-CD63-GFP was significantly reduced (12.25 fold) compared with controls. The data was further validated by measuring GFP signal of the secreted CD63-GFP-labeled exosomes as an indirect quantitation of exosome (Fig. 3D). Taken together, data from qNano-IZON and NanoFACS analyses indicate that MA inhibits exosome secretion in CRPC cells.
Fig. 3.
Analysis of exosome release by CD-63-GFP-expressing C4-2B cells. A stable C4-2B cell clone expressing CD63-GFP was generated as described in “Materials and Methods”. (A) Cell viability of CD-63-GFP-expressing C4-2B cells cultured in presence of MA or vehicle (DMSO) for 48 h. (B) The exosomes in the conditioned media (48 h) derived from CD-63-GFP-expressing C4-2B cells exposed for MA or DMSO were isolated and analyzed by the NanoFACS system. The size and particle concentrations were determined using the Forward SCatter (FSC) and Side SCatter (SSC) parameters. The FSC (488 nm) is depicted along the X-axis, whereas SSC (488 nm) on the Y-axis. PBS without EVs was used as a reference control. (C) MA significantly suppressed (12.5-fold) exosome secretion by the cells compared to controls. (D) The GFP signal in the tagged exosomes measured by the NanoFACS corroborated attenuation of exosomes secretion by MA in comparison to controls. Mean values and standard errors were derived from four independent experiments. *denotes significance at p<0.05 compared to control
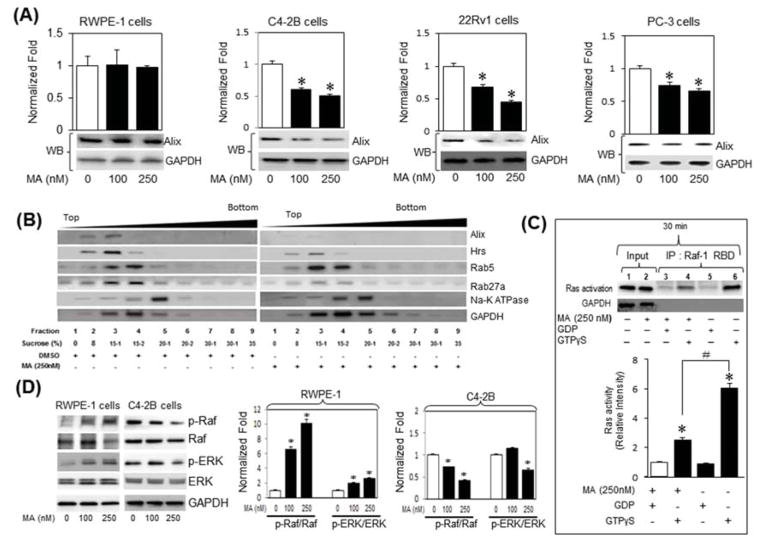
We examined key regulatory cellular targets involved in biogenesis and secretion in MA treated CRPC cells. Dose response experiments demonstrated that MA decreases Alix expression at a concentration range of 100 to 250 nM in C4-2B cells, but not in the RWPE-1 cells, as evidenced by qRT-PCR (bar graphs) and immunoblot (bottom panels) analyses (Fig. 4A). Subsequent time course experiments demonstrated a robust decrease in Alix mRNA transcripts and protein levels (Supplementary Fig. S5 A, B) in C4-2B cells when treated with 250nm of MA for 12 to 48 h period. Dose response experiments demonstrated that MA decreases Alix expression at a concentration range of 100 to 250 nM in C4-2B cells, but not in RWPE-1 cells as evidenced by qRT-PCR (bar graph) and immunoblot (bottom panels) analyses (Fig. 4A). Next, the marker expression of exosome biogenesis and secretion was examined in discontinuous sucrose gradient centrifugation fractions (8%/15%/20%/30%/35%) procured from C4-2B cells treated with vehicle (DMSO) or MA (250 nM) for 48 h. The sucrose fractions were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting for the presence of exosome biogenesis markers, Alix; the multivesicular body marker, Hrs; early endosome marker, Rab 5, and late endosome/secretion marker Rab27a. Whereas no change observed in Rab 5 levels, MA decreased the levels of Alix, Hrs, and Rab27a in C4-2B cells compared with vehicle-treated cells (Fig. 4B), suggesting a direct and selective role for MA in inhibiting exosome biogenesis in C4-2B cells. Notably, none of the exosome markers examined was detected in the membrane-enriched (Na+-K+ ATPase) fraction.
Fig. 4.
Manumycin A (MA) attenuates exosomes biogenesis and secretion through inhibition of the Ras signaling pathway in C4-2B cells. (A) RWPE-1, C4-2B, PC-3 or 22Rv1 cells were treated with various concentrations of MA or control vehicle (DMSO) for 48 h. The expression of Alix in cell lysates was examined by qRT-PCR (bar graph) and immunoblot (bottom panels) analyses using primers and antibodies specific to Alix and GAPDH. (B) Analysis of exosome biogenesis and secretion markers in fractions prepared by discontinuous sucrose gradient centrifugation (8%/15%/20%/30%/35%) of lysates (1 mg) procured from C4-2B cells treated with vehicle or MA (250 nM) for 48 h. Sucrose fractions were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting for the presence of exosome biogenesis marker (Alix), a multivesicular body marker, (Hrs), an early endosome marker (Rab 5), and late endosome secretion marker (Rab27a). (C) Cell lysates of C4-2B treated with vehicle or MA (250 nM) for 30 min and subjected to analysis by a Ras activation assay kit. The samples were pulled down with Raf-1 RBD-agarose beads, analyzed by SDS PAGE and blotted with an anti-Ras antibody (upper panel). Lane 1 and 2 are inputs. Lanes 5 (GDP alone) and 6 (GTPγS alone) are negative and positive controls, respectively. Lanes 3 and 4 show the levels of inactive and active Ras, respectively, upon treatment with MA. The immunoblots were normalized to inactive Ras (GDP-negative control) and quantified by ImageJ (NIH, Bethesda, MD, USA, http://imagej.nih.gov/ij/) (lower panel). (D) Lysates of RWPE-1 (left panel) or C4-2B (right panel) cells treated with vehicle or MA (100 or 250 nM) were analyzed by immunoblotting with antibodies against the phosphorylated form of Raf (p-Raf) and ERK1/2 (p-ERK) and total ERK. Immunoblots for p-Raf and p-ERK in RWPE1 (left bar graph) and C4-2B (right bar graph) were quantified using ImageJ (right panel) and data are expressed as the fold-change from control. *denotes significance at p<0.05 compared to controls
3.3 Manumycin A attenuates exosomes biogenesis and secretion via inhibition of Ras/RAF/ERK signaling pathway in C4-2B cells
Ras farnesylation is important for its activation and involvement in key cellular processes associated with prostate cancer progression [12]. Manumycin is a potent farnesyltranferase inhibitor (FTI) with tumoricidal activity against several cancers, especially the ones with constitutively active Ras [21]. Thus, we sought to examine if Ras signaling mediates the MA-induced suppression of exosome biogenesis and secretion in CRPC cells. We employed a Ras activity assay kit, which detects binding of activated Ras (Ras-GTP) to the Ras-binding domain (RBD) of Raf-1. The active GTP-bound Ras was then pulled down from C4-2B cell lysates with GST–Raf-RBD coupled to glutathione–agarose. The activated Ras fraction was determined by immunoblotting with an anti-Ras antibody. As shown in Fig. 4C (30 min; upper panel; lane 4) and Supplementary Fig. S6 (48 hr), MA inhibited GTPγS activated Ras in C4-2B cells. Quantitative band density blots indicated that Ras activation by GTPγS was significantly decreased (~ 50%) by MA (250 nM) in C4-2B cells (Fig. 4C; lower panel).
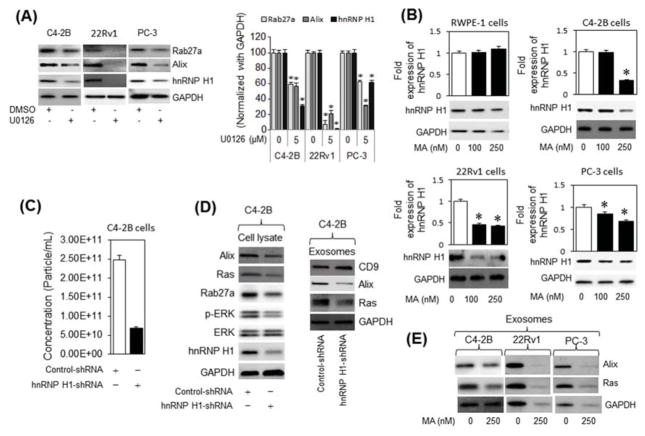
We examined if inhibition of Ras activation by MA reduces the activation of its downstream effectors using antibodies specific to the phosphorylated forms of c-Raf (p-Raf), ERK1/2 (p-ERK) and total ERK. MA (250 nM) significantly decreased the endogenous p-Raf and p-ERK protein levels relative to the total Raf and ERK in C4-2B, but not in RWPE-1, cells, as evidenced by quantitative immunoblot analysis (Fig 4D). Next, we examined whether Ras pathway selective inhibitors modulate the expression of exosome biogenesis (Alix) and secretion (Rab27a) markers. The selective inhibitory efficacies of U0126, SB203580 and SP600125 on ERK, p38 and JNK activation, respectively, were initially validated in PC-3 cells treated with vehicle or TNF-α (Supplementary Fig S7A). The selective inhibition of Alix and Rab27a expression by the ERK inhibitor (U0126), but not by p38 (SB203580) or JNK (SP600125) inhibitors, suggests a direct role for Ras signaling in mediating MA’s inhibition of exosome biogenesis and secretion in CRPC cells (Fig. 5A and Supplementary Fig. S7B, C). Taken together, MA attenuates exosome biogenesis and secretion via inhibition of Ras/Raf/ERK1/2 pathway in CRPC cells.
Fig. 5.
MA suppresses exosomes biogenesis and secretion via ERK-dependent inhibition of hnRNP H1 in C4-2B cells. (A) Immunoblot analysis of hnRNP H1 and the exosome markers Alix and Rab27a in C4-2B, 22Rv1 and PC-3 cells treated with an ERK inhibitor (U0126), or DMSO (vehicle) (left panel). The protein expression levels are quantified relative to GAPDH by ImageJ (right panel). (B) RWPE-1, C4-2B, 22Rv1 and PC-3 cells were treated with vehicle (DMSO) or various concentrations (up to 250 nM) of MA for 48 h. Cell protein lysates were subjected to immunoblot analysis with antibodies specific to hnRNP H1 or GAPDH. (C) Quantitative qNano-IZON particle analysis (NP-100 nanopore) depicting a significant decrease (~ 70%) in exosome concentrations (50–200 nm size) in conditioned media (CM) of C4-2B cells transfected with control short-hairpin RNA (Control-shRNA) or hnRHP H1-shRNA expression plasmid with MA compared to vehicle (DMSO) treated controls. (D) Total and exosome protein lysates were prepared from C4-2B cells transfected with control or hnRNP H1 shnRNA expression plasmid. Proteins were subjected to immunoblot analysis with antibodies against Alix, Ras, Rab27a, CD9, pERK, ERK, hnRNP H1, and GAPDH. (E) Immunoblot analysis of Alix and Ras in the exosomes derived from C4-2B, 22Rv1, and PC-3 cells. * denotes significance at p<0.05 compared to controls.
3.4 MA suppresses exosomes biogenesis and secretion through ERK-dependent inhibition of hnRNP H1 in PC cells
We examined the role of the nuclear matrix protein hnRNP H1 on Ras activation and exosome biogenesis and secretion by CRPC cells. The decreased levels of Alix and Rab27a following inhibition of ERK activation, but not p38 or JNK, was associated with down-regulation of hnRNP H1 protein levels in C4-2B, 22Rv1 and PC-3 cells (Fig 5A and Supplementary Fig S7C). We also found that MA inhibits the hnRNP H1 expression in a dose-dependent manner in C4-2B, 22Rv1, and PC-3 cells, but not in the RWPE-1 cells (Fig. 5B). Quantitative particle analysis (qNano-IZON; NP-100 nanopore) revealed that hnRNP H1 shRNA-silencing significantly decreased (~70%) the exosome release by C4-2B cells as opposed to control-transfected cells (Fig. 5C), suggesting that this nuclear matrix protein is involved in the MA-mediated inhibition of exosome biogenesis and/or secretion in CRPC cells. While no difference was observed in the particle size-distribution and particle diameter of secreted exosomes (Supplementary Fig. S8), a decrease in cellular Ras, pERK, Alix, Rab27a and in exosomal CD9, Ras and Alix transcript levels was detected in the shRNP H1 shRNA-silenced C4-2B cells compared with the control transfected cells (Fig. 5D, left and right panels, respectively). Similar to hnRNP H1 shRNA-silenced C4-2B cells, MA inhibited Ras and Alix protein sorting in the exosome “cargos” of C4-2B, 22Rv1 and PC-3 cells (Fig. 5E). Taken together, the results suggest that MA targeted inhibition of Ras/Raf/ERK signaling reduces exosome biogenesis and secretion in an hnRNP H1-dependent manner, as evidenced by a reduction in cellular expression and exosomal sorting of oncogenic Ras and biogenesis markers in the CRPC cells.
5. Discussion
Exosomes and other microvesicles are released into the extracellular space under both physiological and disease conditions by virtually all cells [2]. Exosomes are essentially involved in cellular communications through the trafficking of cytosolic components and membrane proteins, from donor cells into recipient cells [5]. The exosome-mediated transfer of such factors is implicated in cancer development, progression, and clonal expansion by provoking immune suppression, angiogenesis, metastasis, development of drug resistance and neoplastic reprogramming of tumor-recruited stem cells [3, 9]. Thus the discovery of targeted strategies aimed at inhibiting exosome biogenesis and secretion by tumor cells and/or their uptake by recipient cells may have important therapeutic implications.
The underlying mechanisms that govern EV formation, packaging of their ‘cargo’ and secretion by cancer cells remain largely elusive. Such a gap in knowledge clearly hampers efforts to unravel their pathophysiologic relevance and therapeutic targeting in cancer initiation and progression. In this study, our two-library screens identified MA, a natural, well-tolerated microbial metabolite and a potent experimental tumoricide [22], as one of the lead compounds that effectively target and inhibit exosome biogenesis and secretion by CRPC cells, but not normal prostate cells. The EV analysis by TRPS, Nanosight300 and NanoFACS analyses revealed that MA selectively inhibits biogenesis and secretion of exosomes (<200 nm in size) in a number of CRPC cells. This was evident by an appreciable decrease in secreted exosomes and their biogenesis markers, Alix, Hrs and Rab27a in these cells. The functional role of the ESCRT-0 member Hrs in exosome formation and secretion has been documented in various cell types, including Hela cells and head and neck squamous cell carcinoma [23]. Hrs recruits TSG101 of the ESCRT-I complex, which then recruits ESCRT-III via the ESCRT-accessory protein Alix [23]. Thus, targeting exosome biogenesis and secretion by MA may be triggered by diverse cell-type specific mechanisms. On the other hand, Trajkovic et al. [24] reported that exosomes are released independently from this ESCRT machinery via neutral sphingomyelinase 2 (nSMase2).
Ras, a member of the protein superfamily of a small GTPases, promotes survival pathways in cancer cells. Ras activation is reported in many cancers, including PC [25]. Farnesylation of Ras is required for its activation, translocation, and stimulation of mitogenic signals of the downstream tyrosine kinase receptors [26, 27]. Farnesyltransferase inhibitors suppress farnesylation of Ras and prevent its plasma membrane translocation and inhibit downstream signal transduction pathways, such as Raf-MEK-ERK (28). We documented herein that MA inhibits Ras activity (Ras-GTP) in CRPC cell. MA targeted inhibition of Ras in CRPC cells was coupled with a decrease in phosphorylation of its downstream effectors Raf and ERK, suggesting that MA-induced inhibition of Ras activity may be attributed to farnesyltransferase inactivation. The attenuation of exosome biogenesis by ERK inhibitors further attests to the functional significance of Ras/Raf/ERK signaling in exosome biogenesis and secretion and its crucial role in mediating MA effects in CRPC cells.
While MA (250 nM) or GW4869 alone reduced exosome release by CRPC cells, a robust inhibition was induced by their combination. However, MA inhibition of nSMase2 was reported at higher concentrations (10 μM) of MA [29], suggesting that lower concentrations (250 nM) of MA may inhibit exosome biogenesis primarily through inhibition Ras pathway in CRPC cells. The fact that low concentration of MA exerted no effect on exosome biogenesis and secretion in normal RWPE1 cells may be attributed to its inability to nSMase2 inhibition of low concentrations of MA and the paucity of constitutively active Ras in these cells. Thus unlike GW4869, MA potentially attenuates exosome biogenesis (and secretion) by targeting both Ras and nSMase2 pathways in a dose-dependent manner in CRPC cells.
Members of the hnRNP family are implicated in activation of Ras/Raf/ERK pathway [30]. We recently documented selective expression of the oncogenic and splicing factor hnRNP H1 in the promotion of PC progression [11]. The present study demonstrated a potential role for hnRNP H1 in exosome biogenesis and secretion and sorting of Ras in exosomal ‘cargo’ in CRPC cells. The decrease in hnRNP H1 transcripts by the ERK inhibitor, but not p38 MAPK or JNK inhibitors, suggests that hnRNP H1 is a Ras downstream effector involved in regulation of exosome biogenesis and secretion in CRPC cells. We also demonstrated a decrease in cellular Ras, pERK, Alix, Rab27a in the shRNP H1 shRNA-silenced C4-2B cells. This finding was in agreement with a previous study demonstrating that inhibition of Raf expression levels with siRNA hnRNP H1 results in inhibition of the ERK signaling and cellular transformation by binding and blocking activated Ras [31]. A positive feedback loop between Ras and hnRNP H1 was also evident; the latter is potentially critical to transcriptional upregulation of Ras in the CRPC cells. Our recent demonstration of the pivotal role of CRPC cell-derived exosomal Ras in the neoplastic reprogramming of tumor-tropic adipose stem cells [9], suggests that MA may potentially reduce cancer progression by suppressing Ras exosomal sorting in hnRNP H1-dependent manner in the CRPC cells. Taken together, hnRNP H1 facilitates MA’s suppression of exosome biogenesis and secretion either directly through targeted inhibition of Alix and Rab27a or indirectly via targeted inhibition of Ras expression in CRPC cells (Fig. 6).
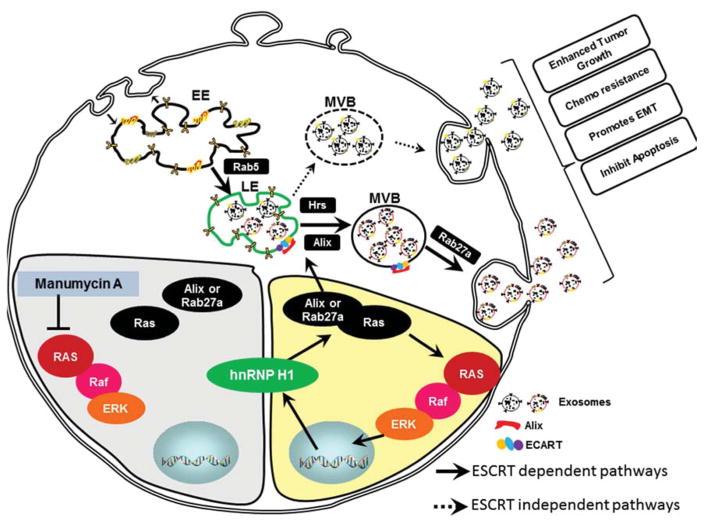
Fig. 6.
Schematic representation of inhibition of exosome biogenesis and secretion by Manumycin A in CRPC cells. The inhibition of Ras/Raf/ERK1/2 pathway by MA leads to transcriptional down-regulation of hnRNP H1. A decrease in hnRNP H1 transcripts leads to inhibition of exosome biogenesis and secretion by suppressing ALIX and Rab27a. (EE) early endosome; LE, late endosome; MVB, multivesicular bodies; ESCRT, endosomal sorting complex required for transport.
Overall, the present study provides evidence that MA suppresses ESCRT-dependent exosome biogenesis and secretion via inhibition of the Ras signaling pathway. The hnRNP H1 is a pivotal downstream effector required for MA-Ras mediated inhibition of exosome biogenesis and secretion and exosomal Ras sorting in the CRPC cells. Given their critical role in cell-to-cell communications and neoplastic reprogramming of stem cells, our findings suggest that MA is a potential exosome-targeting drug to curb PC progression and exosome-mediated cell-cell communications other disease types.
Supplementary Material
Highlights for Review.
There are no known approved drugs targeting exosome biogenesis and secretion by castration-resistant prostate cancer (CRPC) cells.
MA suppresses exosome biogenesis and secretion via inhibition of Ras/Raf/MEK/ERK1/2 signaling in CRPC cells.
The Ras-dependent MA suppression of exosome biogenesis and secretion is partly mediated via an ERK-dependent inhibition of hnRNP H1 in CRPC cells
MA is a potential adjuvant therapeutic drug in patients presenting with CRPC.
Acknowledgments
We thank Jessica A Daigle for editing the manuscript
Grant Support
This work was supported by grants from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (1UH2TR000928-01; 1UH3TR000928-04), to A.B.A.
Abbreviations
- Alix
ALG-2 interacting protein X
- CRPC
castration-resistant prostate cancer
- ESCRT
endosomal sorting complex required for transport
- EVs
extracellular vesicles
- hnRNP H1
heterogeneous nuclear ribonucleoprotein H1
- Hrs
hepatocyte growth factor-regulated tyrosine kinase substrate
- MA
Manumycin-A
- MVB
microvesicular bodies
- MVs
microvesicles
- PC
prostate cancer
- qRRM
quasi-RNA binding recognition motif
- TRPS
tunable resistive pulse sensing
- TSG101
tumor susceptibility gene 101
Footnotes
Disclosure of Potential Conflict of Interest
The authors declare no conflict of interest.
Authors’ Contributions
Conception and design: A. B. Abdel-Mageed, D. Mondal and M. Ferrer
Development of Methodology: A. Datta, H. Kim, Madhu Lal, Lauren McGee, Adedoyin Johnson, Ahmed Moustafa, Jennifer C. Jones
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fu W, Madan E, Yee M, Zhang H. Progress of molecular targeted therapies for prostate cancers. Biochimica et biophysica acta. 2012;1825:140–152. doi: 10.1016/j.bbcan.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saleem SN, Abdel-Mageed AB. Tumor-derived exosomes in oncogenic reprogramming and cancer progression. Cellular and molecular life sciences. Cell Mol Life Sci. 2015;72:1–10. doi: 10.1007/s00018-014-1710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoorvogel W. Resolving sorting mechanisms into exosomes. Cell Res. 2015;25:531–532. doi: 10.1038/cr.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma MM, Lam TK, Hebert E, Divi RL. Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin Pathol. 2015;15:6. doi: 10.1186/s12907-015-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake RR, Kislinger T. The proteomics of prostate cancer exosomes. Expert Rev Proteomics. 2014;11:167–177. doi: 10.1586/14789450.2014.890894. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury R, Webber JP, Gurney M, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger mesenchymal stem cell differentiation into pro-angiogenic and pro-invasive myofibroblasts. Oncotarget. 2015;6:715–731. doi: 10.18632/oncotarget.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32:983–997. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Jia D, Kim H, Abd Elmageed ZY, Datta A, Davis R, et al. Dysregulation of miR-212 Promotes Castration Resistance through hnRNPH1-Mediated Regulation of AR and AR-V7: Implications for Racial Disparity of Prostate Cancer. Clin Cancer Res. 2016;22:1744–1756. doi: 10.1158/1078-0432.CCR-15-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu S, Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R, et al. Ras Conformational ensembles, allostery, and signaling. Chem Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitaker HC, Neal DE. RAS pathways in prostate cancer-mediators of hormone resistance? Curr Cancer Drug Targets. 2010;10:834–839. doi: 10.2174/156800910793358005. [DOI] [PubMed] [Google Scholar]
- 15.Datta A, Bhasin N, Kim H, Ranjan M, Rider B, Abd Elmageed ZY, et al. Selective targeting of FAK-Pyk2 axis by alpha-naphthoflavone abrogates doxorubicin resistance in breast cancer cells. Cancer Lett. 2015;363:25–35. doi: 10.1016/j.canlet.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Cho JH, Yoon MS, Koo JB, Kim YS, Lee KS, Lee JH, et al. The progesterone receptor as a transcription factor regulates phospholipase D1 expression through independent activation of protein kinase A and Ras during 8-Br-cAMP-induced decidualization in human endometrial stromal cells. Biochem J. 2011;436:181–191. doi: 10.1042/BJ20101614. [DOI] [PubMed] [Google Scholar]
- 17.Coumans FA, van der Pol E, Böing AN, Hajji N, Sturk G, van Leeuwen TG, et al. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J Extracell Vesicles. 2014;3:25922. doi: 10.3402/jev.v3.25922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danielson KM, Estanislau J, Tigges J, Toxavidis V, Camacho V, Felton EJEJ, et al. Diurnal variations of circulating extracellular vesicles measured by Nano Flow Cytometry. PLoS One. 2016;11:e0144678. doi: 10.1371/journal.pone.0144678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Vlist EJ, Nolte-‘t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7:1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 20.Zhu S, Ma L, Wang S, Chen C, Zhang W, Yang L, et al. Light-scattering detection below the level of single fluorescent molecules for high-resolution characterization of functional nanoparticles. ACS Nano. 2014;8:10998–11006. doi: 10.1021/nn505162u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 22.Li JG, She MR, Lu CY, Wei SS, Xia PF, Lu ZS. Manumycin induces apoptosis in prostate cancer cells. Onco Targets Ther. 2014;7:771–777. doi: 10.2147/OTT.S60253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399:384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 24.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ce ramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 25.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy LA, Moore T, Nesnow S. Propiconazole-enhanced hepatic cell proliferation is associated with dysregulation of the cholesterol biosynthesis pathway leading to activation of Erk1/2 through Ras farnesylation. Toxicol Appl Pharmacol. 2012;260:146–154. doi: 10.1016/j.taap.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Lerner EC, Qian Y, Blaskovich MA, Fossum RD, Vogt A, Sun J, et al. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic ras signaling by inducing cytoplasmic accumulation of inactive ras-raf complexes. J Biol Chem. 1995;270:26802–26806. doi: 10.1074/jbc.270.45.26802. [DOI] [PubMed] [Google Scholar]
- 29.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282–291. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, Rauch J, et al. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA. 2014;20:505–515. doi: 10.1261/rna.042259.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauch J, Moran-Jones K, Albrecht V, Schwarzl T, Hunter K, Gires O, et al. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res. 2011;71:4664–4674. doi: 10.1158/0008-5472.CAN-10-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.