Abstract
Tumor necrosis factor-inducible gene 6 protein (TSG-6) has recently been shown to protect the liver from acute damage. However, the mechanism underlying the effect of TSG-6 on the liver remains unclear. Autophagy is a catabolic process that targets cell components to lysosomes for degradation, and its functions are reported to be dysregulated in liver diseases. Here we investigate whether TSG-6 promotes liver regeneration by inducing autophagic clearance in damaged livers. Mice fed a methionine choline-deficient diet supplemented with 0.1% ethionine (MCDE) for 2 weeks were injected with TSG-6 (the M+TSG-6 group) or saline (the M+V group) and fed with MCDE for 2 additional weeks. Histomorphological evidence of injury and increased levels of liver enzymes were evident in MCDE-treated mice, whereas these symptoms were ameliorated in the M+TSG-6 group. Livers from this group contained less active caspase-3 and more Ki67-positive hepatocytic cells than the M+V group. The autophagy markers ATG3, ATG7, LC3-II, LAMP2A and RAB7 were elevated in the M+TSG-6 group compared with those in the M+V group. Immunostaining for LC3 and RAB7 and electron microscopy analysis showed the accumulation of autophagy structures in the M+TSG-6 group. TSG-6 also blocked both tunicamycin- and palmitate-induced apoptosis of hepatocytes and increased their viability by inducing autophagy formation in these cells. An autophagy inhibitor suppressed TSG-6-mediated autophagy in the injured hepatocytes and livers of MCDE-treated mice. These results therefore demonstrate that TSG-6 protects hepatocytes from damage by enhancing autophagy influx and contributes to liver regeneration, suggesting that TSG-6 has therapeutic potential for the treatment of liver diseases.
Introduction
The liver is the second-largest organ in the body and performs many roles, such as nutrient metabolism, detoxification and the production of bile, blood clotting proteins, albumin and so on.1, 2, 3 Most liver functions are carried out by hepatocytes, which are parenchymal cells that occupy more than 70% of liver tissues.4 Although hepatocytes are quiescent, these cells enter cell cycles, proliferate and repair damaged tissues in response to hepatic injury.5, 6 Under physiologic conditions, the rate of hepatocyte proliferation is higher than the rate of hepatocyte death, leading to successful regeneration of the liver.6 However, severe and/or chronic injuries lead to massive death of hepatocytes, where the death rate exceeds hepatocyte proliferation. This is followed by the compensatory proliferation of other types of cells, such as activated hepatic stellate cells (HSCs) and progenitor cells.7, 8 These cells start to occupy the parenchymal areas instead of hepatocytes and distort the liver structure by promoting fibrosis, eventually leading to the loss of liver function.9 Hence, the protection of hepatocytes from damage is a key strategy to prevent the initiation of progressive liver disease.
Tumor necrosis factor-inducible gene 6 protein (TSG-6) is one of the cytokines released from mesenchymal stem cells (MSC).10 TSG-6 has been identified as an inflammatory factor and is also involved in various cellular responses depending on the tissue per cell type.11 The administration of TSG-6 for treating corneal wounds, myocardial infarction or an injured central nervous system has been shown to decrease both inflammation and cell death and to increase cell proliferation, eventually alleviating the injury and improving the functions of these organs.12, 13, 14 It has recently been reported that TSG-6 reduces inflammation and fibrosis and promotes liver regeneration in mice with acute liver damage.15 These findings highlight the therapeutic potential of TSG-6 in the treatment of liver disease. However, the mechanism underlying the protective role of TSG-6 in the liver remains poorly understood.
Autophagy is a catabolic process that plays an essential role in degrading long-lived proteins and other abnormal cellular contents by targeting these cellular constituents for trafficking to lysosomes.16, 17 In the process of autophagy formation, the cleaved LC3-I of LC3 soluble proteins are conjugated to lipids through ATG7 and ATG3. This lipid conjugation generates the autophagic double membrane-associated LC3-II protein, which allows the closure of the autophagic vacuole and the formation of the autophagosome.18 The autophagosome subsequently fuses with endosomes and then lysosomes, and the internal material is degraded.17, 19 This process is tightly regulated and highly inducible.17, 20 Autophagy is activated in response to the deprivation of nutrients or growth factor to provide the energy required for cell survival, which indicates that autophagy is necessary for the maintenance of cellular homeostasis.19 In addition, autophagy protects cells from various kinds of damage by eliminating unnecessary or dysfunctional components and toxic substances.19, 21 Alteration of the autophagy process has been shown to increase chromosomal instability and induce dysfunction in the degradation of damaged cellular contents, contributing to the development of various diseases, including liver cancer.22, 23 These findings suggest that autophagy is involved in the pathogenesis of various human diseases. Recent studies have shown that autophagy reduces liver cell death caused by acetaminophen and is involved in caffeine-mediated hepatic fat clearance in a nonalcoholic steatohepatitis animal model.24, 25
In previous studies, we have demonstrated that TSG-6 promotes liver regeneration in mice with acute liver injury. However, it remains unclear whether and how TSG-6 influences the liver repair process in chronically damaged livers. In addition, the relationship between TSG-6 and autophagy has not yet been elucidated. We therefore investigated whether TSG-6 activates autophagy formation in injured hepatocytes and chronically damaged livers using mice fed a methionine choline-deficient (MCD) diet supplemented with ethionine (MCDE). Our results show that TSG-6 increases hepatocyte survival and attenuates liver injury by inducing autophagy influx both in vivo and in vitro. In addition, we found that pharmacological suppression of autophagy blocked TSG-6-mediated autophagy and led to extensive hepatic injury. These findings suggest that TSG-6-regulated autophagy may be a viable therapeutic target for developing pharmacologic agents that treat chronic liver disease.
Materials and methods
Animal studies
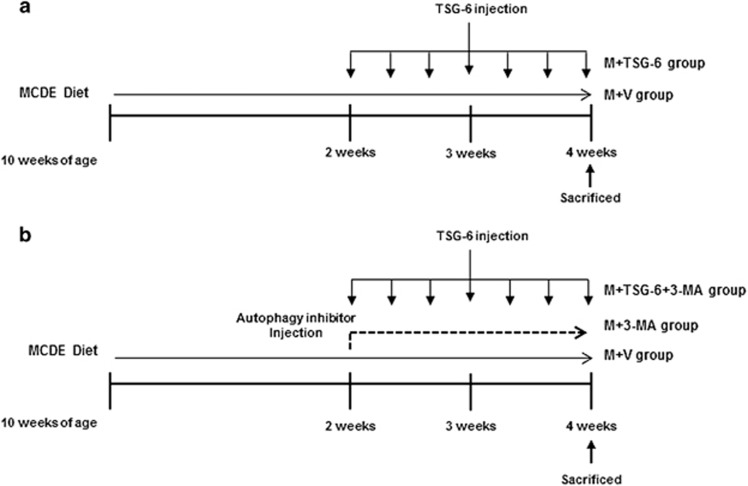
Eight-week-old male C57BL6 mice were purchased from Hyochang (Dae-gu, Korea), fed with a normal diet, watered and housed with a 12-h light–dark cycle. To examine the effects of TSG-6 in vivo, 10-week-old mice were fed an MCD diet supplemented with 0.1% ethionine (MCDE) for 2 weeks. The mice were then randomly divided into two groups, which were treated with MCDE for an additional 2 weeks along with intraperitoneal injections of saline (M+V, n=12) or 50 ng of recombinant TSG-6 (M+TSG-6, n=6) every other day (Figure 1a). As a control, chow-fed mice were injected with equal volumes of the vehicle (control (CON), n=4). The surviving mice were killed to obtain serum and liver samples 2 weeks after injection with TSG-6 or the vehicle (M+V group, n=6; M+TSG-6 group, n=6; CON group, n=4). To assess autophagy associated with TSG-6, mice that had been MCDE-treated for 2 weeks were randomly divided into two further groups, which were additionally treated with MCDE for another 2 weeks in parallel with daily intraperitoneal injections of 40 mg kg−1 of 3-MA (M9281; Sigma-Aldrich, St. Louis, MO, USA) either with TSG-6 (M+TSG-6+3-MA, n=10) or without (M+3-MA, n=8) (Figure 1b). As a CON, chow-fed mice were injected with an equal volume of vehicle (CON) or 3-MA (CON+3-MA, n=4) for 2 weeks. The surviving mice were killed to obtain serum and liver samples 2 weeks after injection with TSG-6 or 3-MA (M+TSG-6+3-MA group, n=5; M+3-MA group, n=4). The period of MCDE treatment was determined based on previous experiments. MCDE-fed mice started to die after 3 weeks, showing 50% and 0% survival at 4 and 5 weeks, respectively. To get serum and liver samples from live mice, we treated mice with MCDE for 4 weeks. Animal care and surgical procedures were approved by the Pusan National University Institutional Animal Care and Use Committee and carried out in accordance with the provisions of the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Figure 1.
Design of mouse experimental model. (a) After being fed MCDE for 2 weeks, mice were treated with MCDE for 2 additional weeks in parallel with i.p. injection of saline (M+V, n=6) or 50 ng of recombinant TSG-6 (M+TSG-6, n=6) every other day for 2 weeks. Chow-fed mice were injected with equal volume of vehicle (CON, n=4). (b) Mice fed MCDE for 2 weeks were additionally treated with MCDE for 2 weeks in parallel with everyday i.p. injections of 40 mg kg−1 of 3-MA with (M+TSG-6+3-MA, n=5) or without TSG-6 (M+3-MA, n=4) for 2 weeks. As a CON, chow-fed mice were injected with equal volumes of vehicle (CON) or 3-MA (CON+3-MA, n=4) for 2 weeks.
Cell experiments
The murine hepatocyte cell line AML12 (CRL-2254; ATCC) was cultured at a density of 2 × 106 in Dulbecco’s modified Eagle’s medium/F-12 (DMEM/F-12, Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco, Life Technologies) and 1 × antibiotics along with insulin, transferrin, selenium and dexamethasone, as per ATCC instructions, at 37 °C in a humidified atmosphere containing 5% CO2.26, 27 To perform biochemical analyses to assess growth and gene expression, AML12 cells at 70–80% confluence were serum-starved in medium containing no fetal bovine serum overnight. After starvation, cells were cultured with tunicamycin (TM) (5 μg ml−1) (ME-654380l; EMD Millipore, Billerica, MA, USA) or palmitic acid (400 μM) (P0500; Sigma-Aldrich)28 in order to induce cell damage for 24 h, and the cell medium was then changed after washing with phosphate-buffered saline (PBS) to withdraw TM. These cells were treated with 3-MA (10 nM) (M9281; Sigma-Aldrich) or PBS for 3 h, and they were then stimulated with TSG-6 (10 ng ml−1) for 24 and 48 h. These experiments were repeated at least three times.
lamp2a siRNA transfection
AML12 cells at 50–60% confluence were serum-starved overnight, cultured in antibiotic-free medium with 2% fetal bovine serum for 24 h and then transfected with 25 nM of lamp2a (L-059036, a mixture of four siRNAs, SMART-pools; Dharmacon, Lafayette CO, USA) or scramble siRNA (D-001206 non-targeting siRNA, siCONTROL; Dharmacon) using Lipofectamine RNAiMAX (13778150; Invitrogen, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. These cells were collected at 24, 48 and 72 h post transfection, and both RNA and protein were extracted for gene expression analysis. The efficiency of gene knockdown was evaluated by qRT-PCR and western blot assays. To examine the effects of TSG-6 in the LAMP2A-suppressed AML12 cells, either TM or palmitate acide (PA) was added to these transfected cells for 24 h. After washing with PBS, these cells were cultured with new medium containing TSG-6 (10 ng ml−1) for 24 and 48 h. As a CON group, scramble siRNA-transfected AML12 cells were treated with equal volumes of the vehicle during these experiments. These experiments were repeated at least three times.
EM assay
Cells and tissues were pre-fixed with 2.5% glutaraldehyde in PBS (pH 7.4) at 4 °C for 2 h and were post-fixed with 1% osmium tetroxide in PBS. The materials were dehydrated with a series of graded ethyl alcohol immersions and embedded in epoxy resin (Epon 812 mixture). Thick sections (1 μm) were stained with 1% toluidine blue for light microscopy. Thin sections, (50–60 nm) prepared using an ultramicrotome (EM UC7; Leica), were double-stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (JEM 1200EX-II; JEOL). To quantify the number of autophagic vacuoles in electron microscopy (EM) images, 10 different fields per mouse or cell group were randomly selected, and morphometric analysis on these selected fields was performed using ImageJ software as described previously.29
Cell viability assay
Cell viability was measured with a Cell Titer Proliferation Assay (MTS; Promega, Madison, WI, USA). In brief, 50 000 cells per well were plated in 96-well plates and treated with TM, PA, TSG-6 and/or 3-MA as described in the above cell experiments. After treatment, 10 μl of MTS reagent was added, and cell viability was measured by reading the absorbance at 490 nm using an ELISA plate reader.
Liver histology and immunohistochemistry
Liver specimens were fixed in 10% neutral buffered formalin, embedded in paraffin and cut into 4-μm-thick sections. Specimens were dewaxed, hydrated and stained per standard protocols with standard hematoxylin and eosin (H&E) staining to examine morphology. For immunohistochemistry (IHC), sections were incubated for 10 min in 3% hydrogen peroxide to block endogenous peroxidases. Antigen retrieval was performed by heating in 10 mm sodium citrate buffer (pH 6.0). After washing with TBS, sections were treated with Dako protein block (X9090; Dako Envision, Dako) for 30 min and incubated with primary antibodies targeting KI67 (NCL-Ki67, Novocastra, Leica Microsystems, Newcastle, Upon Tyne, UK), active caspase-3 (AF835; R&D Systems, Minneapolis, MN, USA) or LC3-II (12741s; Cell Signaling) at 4 °C overnight. Additional sections were also incubated at 4 °C overnight in non-immune sera to demonstrate staining specificity. Polymer horseradish peroxidase anti-rabbit (K4003; Dako Envision, Dako) or anti-mouse (K4001; Dako) secondary antibodies were used. 3,3′-diaminobenzidine (DAB, K3466; Dako) was employed for the detection procedure. For immunofluorescent staining, frozen liver sections or cells were employed. Samples were fixed and permeabilized with acetone and methanol, respectively. They were washed with TBS and incubated with blocking solution for 30 min. Sections or cells were incubated with primary antibody, anti-RAB7 (9367s; Cell Signaling, Danvers, MA, USA) or anti-LC3-II (12741s; Cell Signaling), at 4 °C overnight. Fluorescein-labeled anti-rabbit IgG (AlexaFluor568, Invitrogen, Carlsbad, CA, USA) was used as a secondary antibody for 30 min. 4′,6-diamidino-2-phenylinole (DAPI) was employed in the counter staining procedure.
Quantitative real-time PCR
Total RNA, which had been stored at −80 °C, was extracted with TRIZOL (Ambion by Life Technologies). After assuring sufficient RNA quality and concentration, gene expression was evaluated by qRT-PCR analysis. mRNAs were quantified by real-time RT-PCR per the manufacturer’s instructions (Eppendorf, Mastercycler Real-Time PCR). The sequences of primers for mice are listed in additional file 1: Supplementary Table 1. Samples were analyzed in duplicate according to the ΔΔCt method. All PCR products were directly sequenced for genetic confirmation by Macrogen Inc. (Seoul, Korea).
Western blot assay
Total protein was extracted from freeze-clamped liver tissue samples that had been stored at −80 °C. Whole tissues were homogenized in RIPA buffer (78510; Thermo) supplemented with protease inhibitors (Complete Mini 11 836 153 001; Roche, Indianapolis, IN, USA). Equal amounts of total protein (100 μg) were separated by polyacrylamide gel electrophoresis and transferred to PVDF (polyvinylidene difluoride) membranes. Primary antibodies against LC3 (12741s; Cell Signaling), LAMP2A (sc-19991; Santa Cruz, CA, USA) and RAB7 (9367s; Cell Signaling) were used in this experiment. Membranes were developed by chemiluminescence (ATTO Corporation). Blots obtained from three independent experiments were scanned, and a region of interest (ROI) around the band of interest was defined. Band intensities were calculated using the CS analyzer 2.0 program (ATTO Corporation, Tokyo, Japan).
Triglyceride measurement
Total liver triglyceride levels were measured using a Triglyceride Fluorometric Assay Kit from Cayman Chemical (Nashville, TN, USA) following the manufacturer’s specifications.
Measurement of AST/ALT
Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured using Chemi Lab GOT/GPT (IVD Lab Co., Uiwang, Korea) according to the manufacturer’s instructions.
Cell quantification
To quantify the number of RAB7-positive cells, 10 randomly chosen X40 fields per section were evaluated for each mouse. Cells showing ⩽5 μm of DAPI-stained nucleus in each field were excluded from consideration. The RAB7-positive cells were quantified by counting the total number of RAB7-positive cells per field and dividing by the total number of the cells per field for each mouse.
Statistical analysis
Results are expressed as the mean±s.d. and mean±s.e.m. Statistical differences were determined by Student’s t-test or a one-way ANOVA using SPSS statistics 20 software, followed by the Scheffe post hoc test. P-values <0.05 were considered statistically significant.
Results
TSG-6 attenuates liver damage
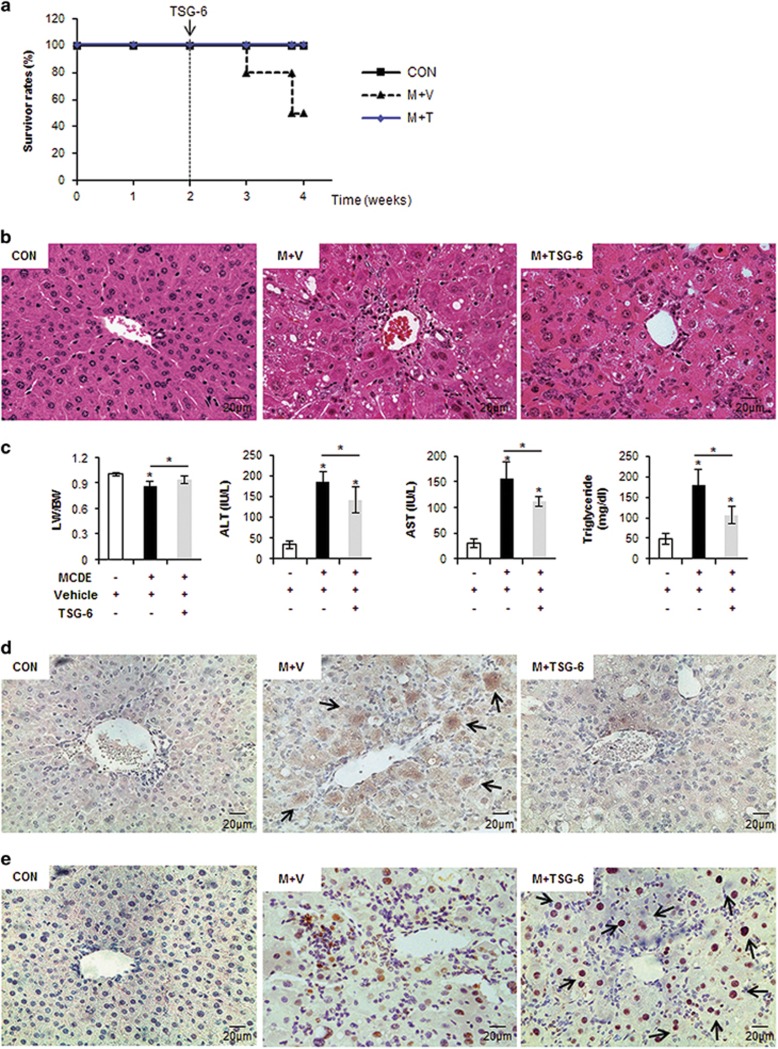
To investigate the functional effects of TSG-6 in chronically damaged livers, mice were fed MCDE (Figure 1). MCD induces lipid accumulation in the liver, and the addition of ethionine exacerbates liver injury by inhibiting replication and survival of mature hepatocytes in rodents.30, 31, 32, 33 Only 50% of mice survived during the MCDE treatment. However, TSG-6 treatment induced a 100% survival rate in MCDE-fed mice (Figure 2a). Severe hepatic injuries, such as cytoplasmic vacuolation, macro or microvesicular fatty changes, loss of cellular boundaries, congestion in the sinusoids and ballooned hepatocytes, were more frequently observed in the liver sections of MCDE-treated mice than in the livers of chow-fed mice (control), as examined by H&E staining. These abnormal histomorphological changes were improved to a significant degree in the M+TSG-6 group (Figure 2b). The ratio of liver weight to body weight (LW/BW) was similar between the CON and the M+TSG-6 group, whereas it was significantly lower in the M+V group (0.861±0.051 vs CON, *P<0.05). The MCDE diet elevated AST and ALT serum levels in the M+V group, whereas TSG-6 treatment reduced the increases in ALT and AST levels in the M+TSG-6 group (AST—CON: 29.61±8.62, M+V: 156.14±32.34, and M+TSG-6: 111.9±10.39; ALT—CON: 34.83±9.01, M+V: 184.36±26.02 and M+TSG-6: 140.91±31.33, *P<0.05). Triglyceride (TG) levels in the liver were lower in the M+TSG-6 group than in the M+V group (CON: 47.4±13.76, M+V: 178.43±39.7 and M+TSG-6: 105.73±21.13) (Figure 2c). In addition, the inflammatory markers tnf-α, il-1β and cxcl1 were downregulated in the M+TSG-6 group compared with those in the M+V group (Supplementary Figure 1).
Figure 2.
TSG-6 improves histomorphology and function of livers with chronic damage. (a) Survival rates of MCDE-fed mice treated with vehicle or TSG-6 were plotted (CON group: n=4, M+V group: n=12, M+TSG-6 groups: n=6). (b) Liver morphology was assessed by H&E staining of liver sections from representative CON and MCDE groups treated with saline (M+V) or TSG-6 (M+TSG-6) (X40). (c) The LW/BW, serum levels of ALT and AST and TG levels in the livers of mice treated with vehicle or MCDE with or without TSG-6 (50 ng). The results were plotted as the mean±s.d. (n⩾4 mice per group) (*P<0.05). (d, e) IHC for active CASPASE-3 (d) and KI67 (e) in liver sections from the representative CON, M+V and M+TSG-6 groups (X40).
To determine whether liver damage improvement in the mice treated with TSG-6 and MCDE was associated with hepatocyte survival, liver sections from MCDE-fed mice with or without TSG-6 treatment were stained for active caspase-3, an apoptosis marker,33 and Ki67, an S phase marker.15, 33 Hepatocytic cells expressing active caspase-3 accumulated in the M+V group, whereas such cells were rare or only occasionally observed in the CON and M+TSG-6 groups, respectively (Figure 2d). Ki67-positive cells were found to be mainly hepatocytic cells (indicated by arrows) in the livers of TSG-6-injected mice fed with MCDE, whereas these positive cells were mostly HSCs or progenitor-looking cells in the livers of MCDE-treated mice, with a smaller number of hepatocytic cells (Figure 2e). These results indicate that TSG-6 protects hepatocytes from chronic hepatic damage, contributing to the alleviation of liver injury.
TSG-6 enhances autophagy in chronically injured livers
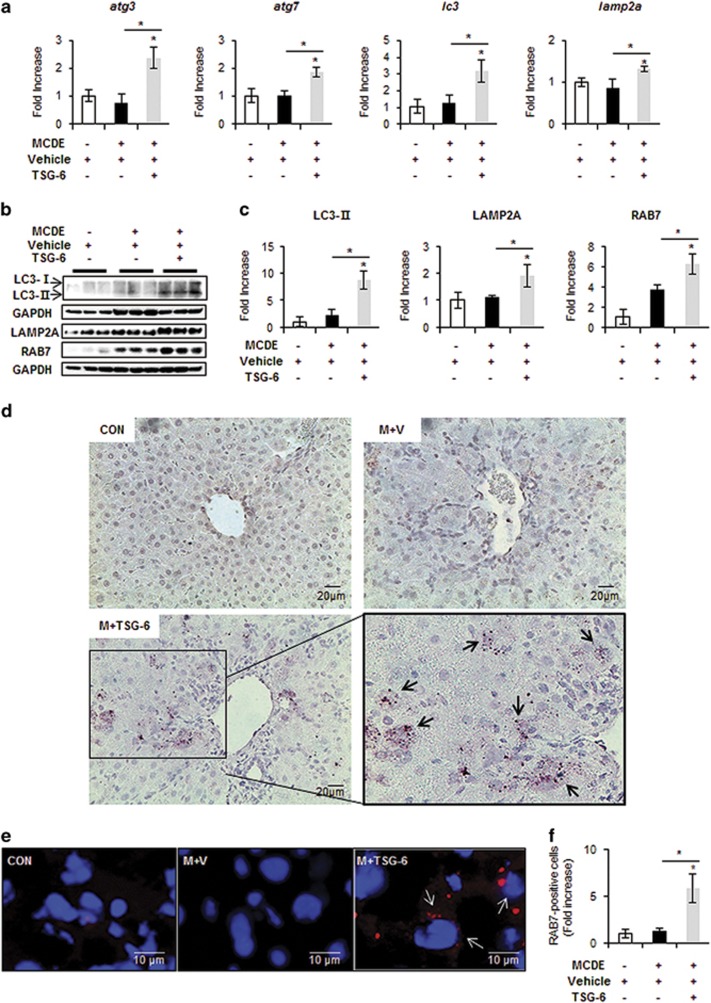
Autophagy is known to protect liver cells from a broad range of pathological stresses by selectively removing damaged organelles and providing a stable intracellular environment, contributing to liver regeneration.34, 35 As the protective effect of TSG-6 was shown in the livers of MCDE-treated mice, we investigated whether TSG-6 promoted autophagy formation and subsequent liver regeneration in MCDE-fed mice treated with TSG-6. The expression levels of autophagy markers, including atg3, atg7 and lc3,18 were significantly higher in the M+TSG-6 group than those in the CON and the M+V groups, as assessed by qRT-PCR (Figure 3a). In addition, the expression of lamp2a, the receptor for chaperone-mediated autophagy (CMA) and one of the lysosome-associated membrane glycoproteins,36 was upregulated in the M+TSG-6 group compared to the CON and the M+V groups (Figure 3a). The increased levels of LC3-II and LAMP2A proteins confirmed the qRT-PCR data in mice treated with TSG-6 and fed MCDE. The expression of RAB7, regulating autolysosome-mediated lipid degradation,37 was also significantly higher in the M+TSG-6 group than that in the CON and the M+V groups (Figure 3b and c). IHC staining for LC3 showed clearly that LC3-positive hepatocytic cells accumulated in the livers of the M+TSG-6 group to a significant extent, whereas these cells were rarely observed in the CON and the M+V groups (Figure 3d). The increased number of RAB7-positive cells in the M+TSG-6 group was also supported by increased expression of RAB7 at both the RNA and protein levels (Figure 3e and f). This elevated formation of autophagy structures in the livers of MCDE-fed mice treated with TSG-6 was evident in EM analysis (Figure 4). Therefore, these results suggest that TSG-6 promotes autophagy formation in damaged livers and attenuates hepatic injury.
Figure 3.
TSG-6 increases expression of autophagy markers in livers of MCDE-fed mice. (a) qRT-PCR analysis of atg3, atg7, lc3 and lamp2a in livers from CON, M+V, and M+TSG-6-treated mice results are plotted (mean±s.d. n⩾4 mice per group). (b) Western blot analysis and (c) cumulative densitometry analyses for LC3 (LC3-II: 14 kDa, processed form) (inducer of autophagosomes), LAMP2A (lysosome membrane protein) and RAB7 (autophagosome-lysosome docking marker) in livers of three representative mice from each group. GAPDH was used as an internal CON. Data shown represent one of three experiments with similar results. The results are displayed as the mean±s.d. (n⩾4 mice per group) (*P<0.05). (d) IHC for LC3-II in liver sections from representative chow- or MCDE-fed mice treated with vehicle or TSG-6 (X40). (e) Immunofluorescent staining for RAB7 in liver sections from each group. Representative images are shown (× 100, Scale bar: 10 μm). (f) Quantitative RAB7 immunofluorescent staining data from each group (n⩾4 mice per group). The number of RAB7-positive cells was counted in 10 fields per section. RAB7-positive cells were quantified by counting the total number of RAB7-positive cells with ⩾5 μm-sized DAPI-stained nuclei per field and dividing by the total number of cells with ⩾5 μm-sized DAPI-stained nuclei. Mean±s.d. results are plotted (*P<0.05).
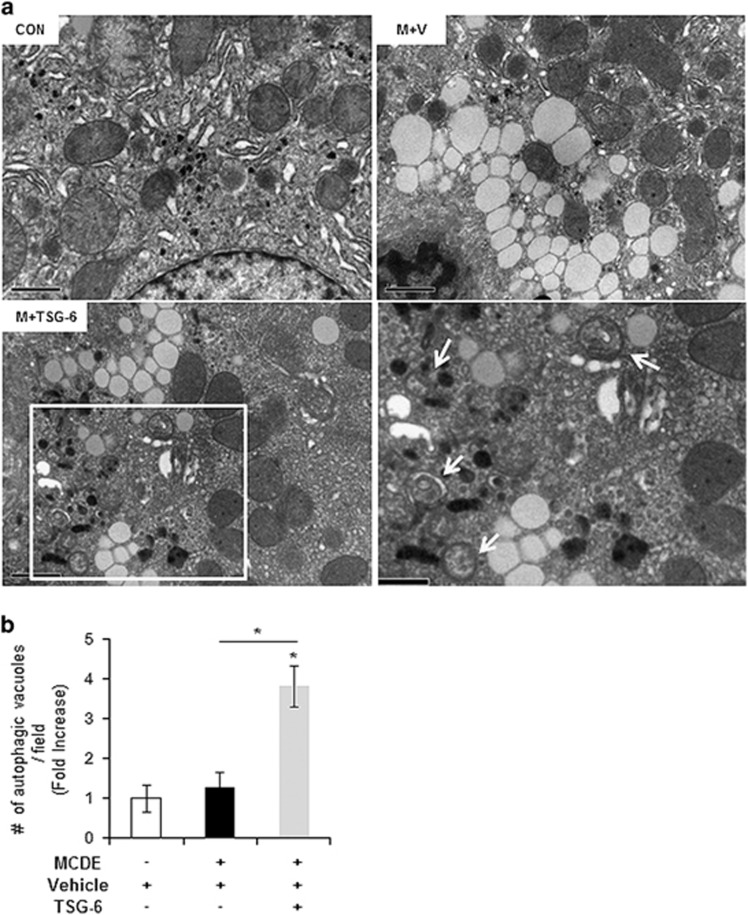
Figure 4.
Ultrastructural analysis shows increasing autophagy formation in damaged livers with TSG-6 treatment. (a) EM analysis showing the elevated autophagy formation (indicated by arrows) in livers of TSG-6-treated mice. These autophagy structures were rarely detected in livers of the CON and M+V groups. The inset box in the image was magnified in the lower-right image. Representative images are shown (Scale bar: 1 μm). (b) Morphometric data of the autophagic vacuoles from all mice were plotted as the mean±s.d. results (*P<0.05).
TSG-6 contributes to the survival of hepatocytes by improving autophagy formation
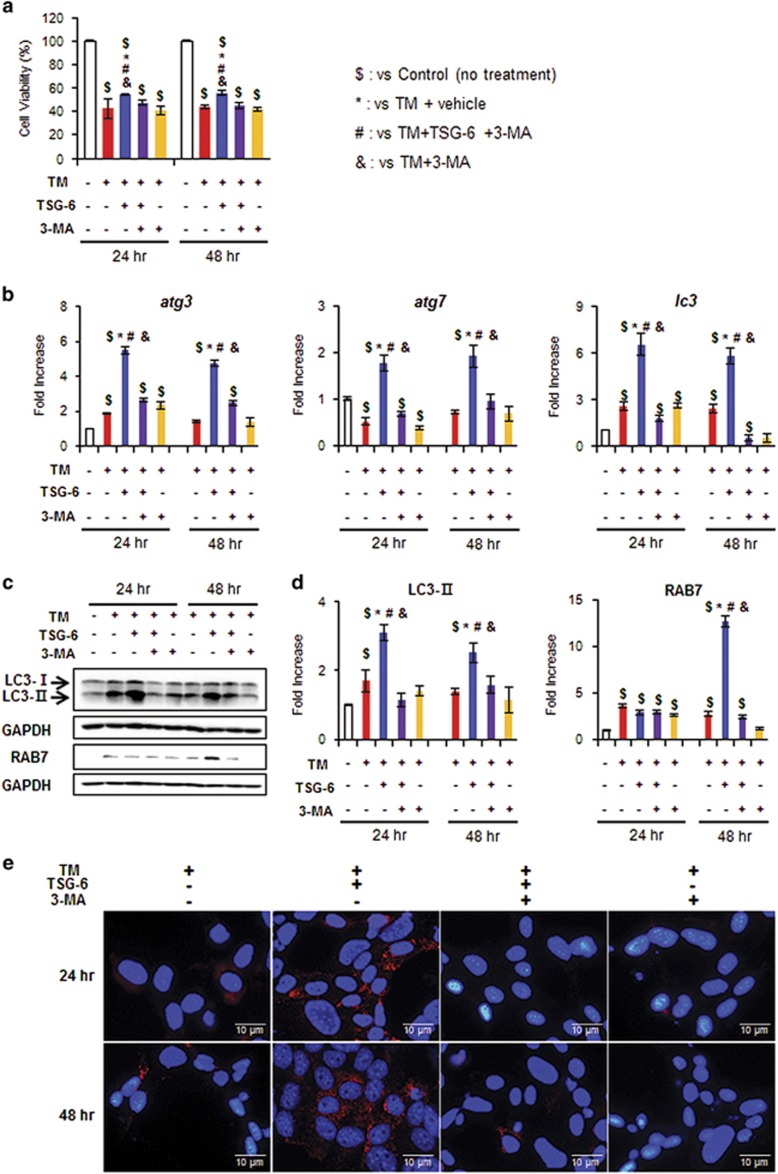
To determine whether TSG-6 directly influences the survival of hepatocytes by promoting autophagy formation, we treated AML12 cells, a normal mouse hepatocyte cell line, with TSG-6 for 2 days after inducing endoplasmic reticulum (ER) stress using tunicamycin TM.38 In these in vitro systems, we employed 3-methyladenine (3-MA), a well-known autophagy inhibitor,19, 37 to block autophagy formation. TM effectively induces cell death, with up to a 58% reduction of cell viability (42.5±8.41 at 24 h, 43.7±1.77 at 48 h post withdrawal of TM). However, TSG-6 suppressed this rapid decrease in cell viability (54.4±0.95 at 24 h, 55.8±1.91 at 48 h). This viability effect of TSG-6 in hepatocytes was blocked by 3-MA (TM+TSG-6+3-MA: 47.6±1.79 at 24 h, 45.2±2.8 at 48 h; TM+3-MA: 40.6±3.5 at 24 h, 42.1±1.88 at 48 h) (Figure 5a). On the basis of these data, we investigated whether TSG-6-induced autophagy formation to protect hepatocytes from ER stress. The RNA levels of the macroautophagy markers atg3, atg7 and lc3 were greatly elevated in TSG-6-treated hepatocytes treated with TM, whereas there was no significant change in the other groups (Figure 5b). The protein expression of LC3-II, which is known to function earlier in the autophagy process, increased at 24 and 48 h, and RAB7, which mediates later steps in the autophagy process, was upregulated 48 h after TM withdrawal in the hepatocytes of the TM+TSG-6 group compared with the other groups (Figure 5c and d). In addition, LC3-positive cells were observed in the TM+TSG-6 groups, whereas these cells were rarely observed in the other groups (Figure 5e). EM analysis showed the accumulation of autophagy structures in the TM+TSG-6-treated hepatocytes at both 24 and 48 h (Figure 6). In addition, we assessed whether TSG-6 protected hepatocytes from lipotoxicity-induced ER stress. To induce fatty acid toxicity in hepatocytes, AML12 cells were treated with PA, which has been reported to induce lipotoxicity,26, 27, 28 and TSG-6 or vehicle CON was added to these cells at 3 h post PA treatment. TSG-6 improved the number of viable cells in PA-treated cells, whereas 3-MA suppressed this TSG-6-mediated effect on cell viability (Supplementary Figure 2a). The protein level of LC3-II was also higher in the PA-treated AML12 cells treated with TSG-6 at 24 and 48 h than in the other groups (Supplementary Figure 2b and c). Therefore, these data support the finding that TSG-6 reduces hepatocyte damage by inducing autophagy flux.
Figure 5.
TSG-6 protects hepatocytes from tunicamycin-induced cell death by increasing autophagy influx. (a) Cell viability of AML12 cells, a normal murine hepatocyte cell line, treated with TSG-6 was analyzed using MTS assay. After being exposed to TM (5 ng ml−1) for 24 h, AML12 cells were treated with vehicle (TM), 3-MA (TM+3-MA) or TSG-6 with (TM+TSG-6+3-MA) or without 3-MA (TM+TSG-6) for 24 and 48 h. As a CON, AML12 cells were treated with equal volumes of vehicle without TM. The mean±s.e.m. results obtained from three identical experiments are plotted. (b) qRT-PCR analysis of atg3, atg7 and lc3 in these cells. Data represent the mean±s.e.m. of three independent experiments. (c) Western blot assay and (d) cumulative densitometry analyses for LC3-II and RAB7 in these cell groups. GAPDH was used as an internal CON. Data shown represent one of three experiments with similar results, and the mean±s.e.m. results obtained from three identical experiments are shown (*P<0.05 vs TM, #P<0.05 vs TM+TSG-6 with 3-MA, &P<0.05 vs TM+3-MA, $P<0.05 vs CON). (e) Representative images of LC3 immunofluorescence-stained AML12 cells (Scale bar: 1 μm) (X80).
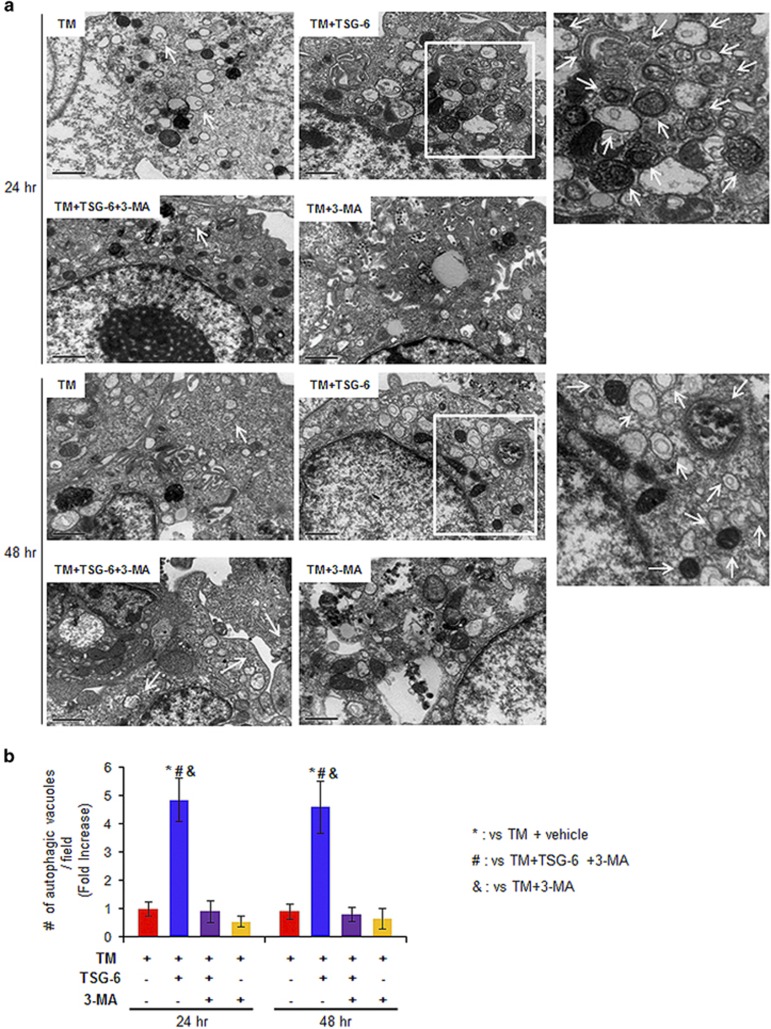
Figure 6.
Accumulation of autophagy in TM-treated hepatocytes with TSG-6. (a) EM analysis shows increased autophagy formation (indicated by arrows) in TSG-6-treated AML12 cells at 24 (top panel) and 48 h (bottom panel) after TM withdrawal. Inset boxes in images are magnified in the images in the right panel. Representative images are shown (Scale bar: 1 μm). (b) Morphometric data for the autophagic vacuoles from all groups were plotted as the mean±s.d. (*P<0.05 vs TM, #P<0.05 vs TM+TSG-6 with 3-MA, &P<0.05 vs TM+3-MA).
To investigate whether TSG-6 also promoted CMA to enhance hepatocyte viability, lamp2a siRNA was transfected into AML12 cells to block CMA, and TSG-6 was subsequently added to these cells after TM or PA treatment. Scramble siRNA (scr) was transfected into AML12 cells as a negative CON. lamp2a siRNA efficiently reduced the RNA and protein levels of LAMP2A in the AML12 cells compared with scr-treated cells (Supplementary Figure 3a and b). TSG-6 elevated the viability of the scr-transfected cells, which were injured by either TM or PA, whereas it rarely increased the number of viable cells in the lamp2a siRNA-transfected cells (Supplementary Figure 3c). The deletion of lamp2a also suppressed the expression of atg3, lc3, atg7 and lamp2a in the TSG-6-treated cells that were also treated with either TM or PA compared to the scr-treated cells with TSG-6 (Supplementary Figure 3d and e). Therefore, these results indicated that TSG-6 was involved in activating CMA to protect hepatocytes from ER stress-induced damage.
TSG-6 reduces liver damage by enhancing autophagy formation in mice
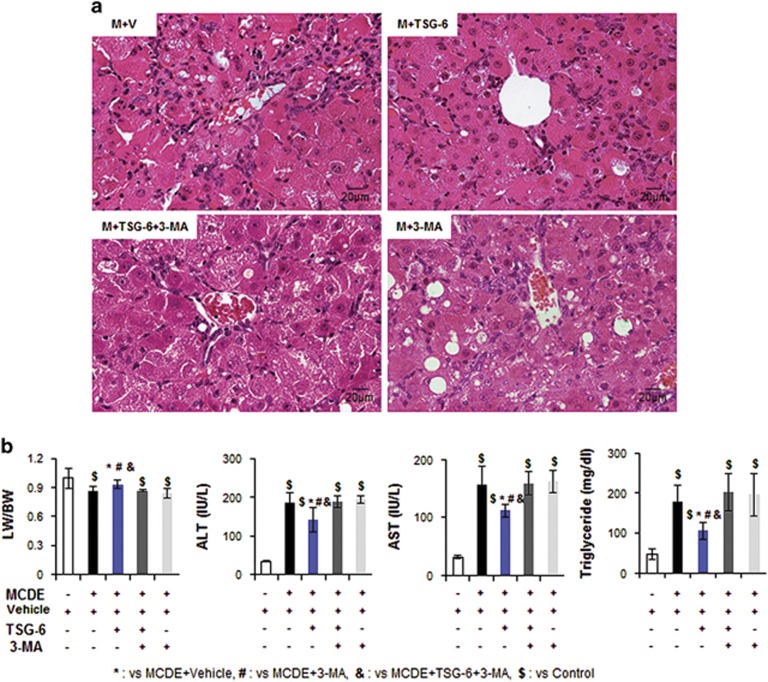
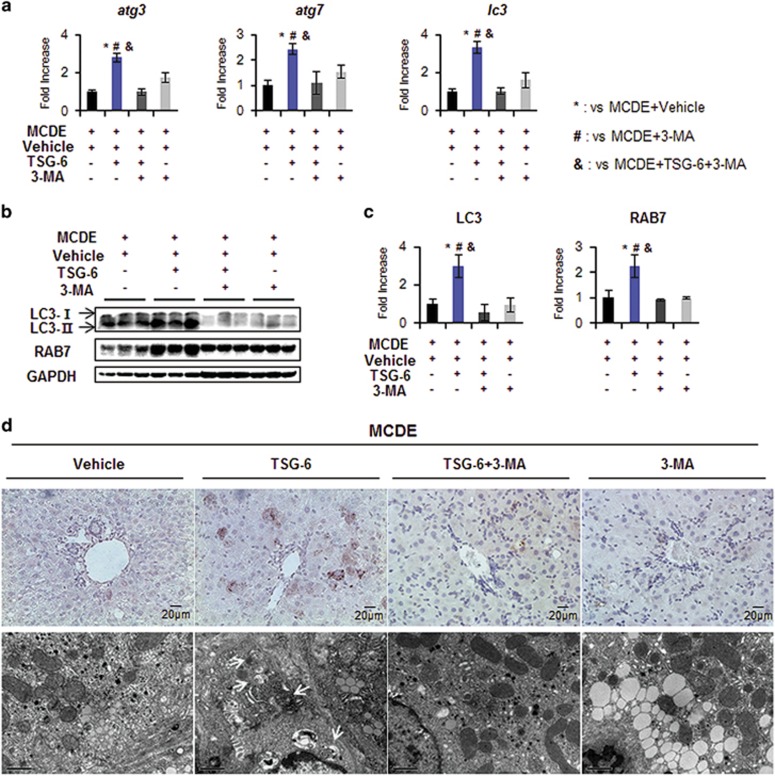
To investigate whether TSG-6 reduced hepatic damage by mediating autophagy influx in an experimental animal model, we suppressed autophagy formation in the livers of TSG-6-injected mice fed with MCDE using 3-MA, an autophagy inhibitor. When the livers of chow-fed mice, with or without 3-MA, were examined, liver morphology and LW/BW were similar between the two groups (Supplementary Figure 4a and b). There was no significant change in the expression levels of atg3, atg7 and lc3 RNA or LC3-II protein in the livers of mice treated with 3-MA compared to the normal livers of mice treated with vehicle (Supplementary Figure 4c and d). After confirming that 3-MA rarely influenced autophagy flux in healthy murine livers, 3-MA or vehicle was administered via intraperitoneal injection into MCDE-fed mice with or without TSG-6 treatment. Mice were killed 2 weeks after 3-MA treatment (n⩾4 per group) (Figure 1b). H&E staining showed intense hepatic damage in MCDE-fed mice treated with 3-MA (M+3-MA group) or vehicle (M+V group), whereas the hepatic distortion in the M+TSG-6 group was lower. Interestingly, these improved morphological changes were not seen in the livers of MCDE-fed mice that had been injected with TSG-6 and 3-MA (Figure 7a). The LW/BW ratios were lower (M+V group: 0.86±0.051, M+TSG-6 group: 0.93±0.047, M+TSG-6+3-MA group: 0.86±0.01, M+3-MA group: 0.84±0.048-fold increase, *,#,$,&P<0.05) and serum ALT and AST levels were higher in the M+V, M+TSG-6+3-MA and M+3-MA groups than those in the CON and M+TSG-6 groups (ALT, M+V: 184.36±26.1, M+TSG-6: 140.91±31.33, M+TSG-6+3-MA: 187.72±14.72, M+3-MA: 193.88±10.78; AST, M+V: 153.14±32.35, M+TSG-6: 119.91±10.38, M+TSG-6+3-MA: 158.88±20.04, M+3-MA: 162.08±19.4, *,#,$,&P<0.05). TG levels in the liver were significantly reduced in the M+TSG-6 group compared with the other groups damaged by MCDE (M+V: 178.43±39.7, M+TSG-6: 105.73±21.13, M+TSG-6+3-MA: 195.91±47.41 and M+3-MA: 202.25±52.69) (Figure 7b). These data show that the protective effects of TSG-6 in the liver were abrogated by 3-MA. Compared with the M+TSG-6 groups, the expression levels of atg3, atg7 and lc3 were reduced in the 3-MA-injected groups of mice fed with the MCDE diet, as assessed by qRT-PCR (Figure 8a). In addition, these 3-MA-treated groups contained lower levels of LC3 and RAB7 proteins than TSG-6-treated mice that were not injected with 3-MA, indicating that 3-MA effectively suppresses autophagy formation (Figure 8b and c). In the LC3-stained liver sections, LC3-puncta were remarkably reduced in all MCDE-treated groups, with the exception of the M+TSG-6 group (Figure 8d, upper panel). EM analysis also revealed that the distinct autophagy structures in the livers of the M+TSG-6 groups were rarely observed in the other groups (Figure 8d, bottom panel). Taken together, these findings suggest that TSG-6 attenuates liver damage by enhanced autophagy formation in chronically injured livers of mice.
Figure 7.
Ablation of TSG-6-induced autophagy prevents amelioration of liver damage in mice. (a) H&E staining of liver sections from the M+V, M+TSG-6, M+TSG-6+3-MA and M+3-MA groups (X40). (b) The LW/BW, serum levels of ALT and AST, and TG levels in the livers of these mice groups. The results were plotted as the mean±s.d. (n⩾4 mice per group) (*P<0.05 vs M+V, #P<0.05 vs M+3-MA, &P<0.05 vs M+TSG-6+3-MA, $P<0.05 vs CON).
Figure 8.
3-MA suppresses TSG-6-mediated autophagy in livers of MCDE-fed mice. (a) qRT-PCR analysis of atg3, atg7 and lc3 in livers from the M+V, M+TSG-6, M+TSG-6+3-MA and M+3-MA groups. Mean±s.d. results are plotted (n⩾4 mice per group) (*P<0.05 vs M+V, #P<0.05 vs M+3-MA, &P<0.05 vs M+TSG-6+3-MA). (b) Western blot analysis and (c) cumulative densitometry analyses for LC3-II and RAB7 in livers of three representative mice from these groups. GAPDH was used as an internal CON. Data shown represent one of three experiments with similar results. The results are displayed as the mean±s.d. (n⩾4 mice per group) (*P<0.05 vs M+V, #P<0.05 vs M+3-MA, &P<0.05 vs M+TSG-6+3-MA). (d) Representative images of IHC for LC3-II (upper panel, × 40) and EM in liver sections from these groups (arrows indicate autophagy structures. Scale bar: 1 μm).
Discussion
Chronic liver diseases, such as cirrhosis and liver cancer, are accompanied by extensive hepatocyte mortality, which induces the activation and proliferation of HSCs, ultimately leading to a distorted fibrotic liver structure with loss of liver function.8, 39, 40 Hence, it is important to protect hepatocytes from damage to prevent the development of end-stage liver disease. Recently, the MSC secretome, the broad range of secreted tropic and immunomodulatory cytokines produced by MSCs, has been shown to have therapeutic potential for repairing damaged tissues, providing a new and promising treatment strategy for the treatment of liver diseases.41 As TSG-6 has been reported to be a component of the MSC secretome and to have anti-inflammatory effects in the cornea, heart and brain,12, 13, 14 we previously investigated the effects of TSG-6 in the liver and showed that TSG-6-containing conditioned medium from MSCs improved repair processes in mice with acute damage by modulating the immune and fibrotic responses.15 However, the mechanism underlying the effects of TSG-6 on hepatocytes during the repair process needs further explanation. In this study, we have demonstrated the direct function of TSG-6 on the liver repair process in mice with chronic hepatic injury. We employed the MCDE diet for 4 weeks to induce chronic liver damage in mice. An MCDE diet causes defects in hepatic lipid metabolism and hepatocyte replication, injures hepatic parenchymal cells and stimulates the compensatory proliferation of activated HSCs and progenitor cells. This results in severe steatosis and apoptosis of hepatocytes with inflammation and fibrosis, such as the nonalcoholic steatosis that accompanies hepatic fibrosis.30, 31, 32, 33 Although only 50% of mice survived during the MCDE treatment, MCDE-fed mice with TSG-6 showed 100% survival rates. In addition, the improved histomorphology and serum levels of liver enzymes and the similar ratio of LW/BW in the M+TSG-6 group compared with the CON group supports the idea that TSG-6 directly regulates the repair process in damaged livers, contributing to the increased survival rate of mice (Figures 2 and 7). The expansion of Ki67-positive hepatocytic cells and fewer apoptotic hepatocytes in the liver sections of the M+TSG-6 group clearly revealed that TSG-6 protected hepatocytes from damage and promoted their proliferation (Figure 2). In addition, enhanced autophagy influx in the hepatocytes and livers of mice treated with TSG-6 strongly supported the idea that TSG-6 exerts a protective effect in hepatocytes. Therefore, our findings suggest that autophagy mediated by TSG-6 helps repair liver damage and has significant potential as a therapeutic strategy for treating liver disease.
Autophagy has diverse physiological effects on the liver through cellular recycling and degradation processes. There are three types of autophagic pathways: macroautophagy, microautophagy and CMA.17, 19 Macroautophagy is initiated with the formation of the limiting membrane of the autophagosome using lipids and proteins from different organelles. Cargo sequestration occurs in bulk or is specifically mediated by soluble protein receptors. After engulfment of cargo, the sealed vesicles (autophagosomes) travel along microtubules and deliver cargo to lysosomes through membrane fusion, forming an autolysosome where cargo is degraded by lysosomal hydrolases.17 In CMA, substrates carrying a KFERQ-like pentapeptide are recognized by the cytosolic chaperone HSC70, and the substrate–chaperone complex binds to the CMA receptor, LAMP2A, on the lysosomal membrane. A luminal HSC70 helps to translocate these substrates into lysosomes, where the substrates are finally degraded.17 Microautophagy is mediated by direct lysosomal engulfment of the cytoplasmic cargo.17 In the current study, we employed a nonalcoholic steatohepatitis-like experimental animal model using a MCDE diet. Growing evidence shows that reduction of macroautophagy and CMA is associated with nonalcoholic steatohepatitis development.19, 42, 43 Suppression of hepatic macroautophagy through ATG7 deletion has been shown to cause lipid drop accumulation, and inhibition of CMA has been shown to increase lipogenic enzymes.29, 36 In line with these findings, our results indicate that TSG-6 protects hepatocytes from MCDE-mediated lipotoxity. The macroautophagy-related makers, atg3, atg7 and lc3, were upregulated in TSG-6-treated hepatocytes and livers, as shown by qRT-PCR, western blot and IHC analysis. Expression of the CMA marker LAMP2A increased in these groups. In addition, TSG-6 failed to promote autophagy formation in CMA-suppressed cells, leading to the increased loss of hepatocytes (Supplementary Figure 3). These results suggest that TSG-6 induces both macroautophagy and CMA in MCDE-treated livers and contributes to liver regeneration. The microautophagy process was not identified in this research because the molecular mechanisms and physiological associations of microautophagy in the liver are poorly defined.20 In addition, various forms of microautophagy are known to exist, and proteins regulating microautophagy have not been clearly identified.17, 20, 44 However, we cannot exclude the potential action of microautophagy in removing hepatic lipids, although it is known to directly trap lipid droplets in yeast (but not in mammalian lysosomes). Further studies are required to provide evidence for the involvement of microautophagy in clearing hepatic lipid accumulation.
The autophagy-associated pathway has emerged as an attractive target for developing treatments for liver disease because of its hepatoprotective and anti-inflammatory effects.19, 45 In damaged liver tissues, apoptotic hepatocytes result in increased inflammation and activation or transition of HSCs into myofibroblasts.46 These cells are major contributors to hepatic fibrosis, which is a common characteristic of liver disease.47 Hence, suppression of the transition of quiescent HSCs into myofibroblasts is a key antifibrotic strategy for liver fibrogenesis. A growing body of evidence shows that autophagy is related to the activation of HSCs and promotes a profibrotic response in the liver.42, 48 However, several studies have reported that autophagy protects hepatocytes from apoptosis and reduces the inflammatory response, leading to a decrease in profibrogenic signals.49, 50 These findings are a challenge for the application and development of autophagy-based treatments for liver diseases with fibrosis because of the cell type-specific effects of autophagy on the liver. Because TSG-6 was administered by intraperitoneal injection into MCDE-fed mice and not specifically targeted into the hepatocytes, it seems possible that TSG-6 may influence HSCs, immune cells and other types of cell in the liver. Hence, it is possible that the repairing actions of TSG-6 on the injured liver are mediated by macrophage autophagy because macrophage and/or dendritic cell autophagy is known to prevent excessive release of inflammatory factors.51, 52 In addition, we need to consider the effect of TSG-6 on HSC activation. However, we have demonstrated that TSG-6 elevated autophagy influx and viability in TM-treated hepatocytes in in vitro systems. TSG-6 also led to decreased apoptosis and increased proliferation of hepatocytic cells in MCDE-treated livers. These results clearly show that TSG-6 protects hepatocytes from apoptosis, suggesting that hepatocytes protected by TSG-6 are able to decrease the profibrotic signals sent to HSCs with reduced inflammation, thus contributing to liver regeneration. In line with our findings, Amir et al. and Lodder et al.43, 53 have also reported that autophagy decreased profibrotic signals by protecting hepatocytes from apoptosis and macrophage autophagy. However, given that activated HSCs are the main cells producing collagen and that TSG-6-regulated autophagy needs a more targeted approach in treating liver disease, further study is required to clarify the cell-specific roles of TSG-6.
In conclusion, we have demonstrated that TSG-6 promotes liver regeneration in mice with chronically damaged livers by enhancing autophagy influx both in cell culture and in vivo. TSG-6 directly mediated the decreased apoptosis and increased viability of hepatocytes. A pharmacological inhibitor of autophagy blocked the protective effects of TSG-6 in the liver, suggesting that the therapeutic potential of TSG-6 in the damaged liver was associated with TSG-6-mediated autophagy. Therefore, these findings indicate that TSG-6 has great potential as a therapeutic agent for treating liver disease.
Acknowledgments
This work was supported by the National Research Foundation (NRF) of Korea funded by the Korean Government (MEST) (2016R1A2B2007922) to YJ.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Garcia-Martinez R, Caraceni P, Bernardi M, Gines P, Arroyo V, Jalan R. Albumin: pathophysiologic basis of its role in the treatment of cirrhosis and its complications. Hepatology 2013; 58: 1836–1846. [DOI] [PubMed] [Google Scholar]
- Grompe M, Strom S. Mice with human livers. Gastroenterology 2013; 145: 1209–1214. [DOI] [PubMed] [Google Scholar]
- Reinke H, Asher G. Circadian clock control of liver metabolic functions. Gastroenterology 2016; 150: 574–580. [DOI] [PubMed] [Google Scholar]
- Mohammed FF, Khokha R. Thinking outside the cell: proteases regulate hepatocyte division. Trends Cell Biol 2005; 15: 555–563. [DOI] [PubMed] [Google Scholar]
- Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology 2004; 39: 1477–1487. [DOI] [PubMed] [Google Scholar]
- Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol 2004; 5: 836–847. [DOI] [PubMed] [Google Scholar]
- Yang S, Koteish A, Lin H, Huang J, Roskams T, Dawson V et al. Oval cells compensate for damage and replicative senescence of mature hepatocytes in mice with fatty liver disease. Hepatology 2004; 39: 403–411. [DOI] [PubMed] [Google Scholar]
- Chung C, Iwakiri Y. Activated hepatic stellate cells: negative regulators of hepatocyte proliferation in liver diseases. Hepatology 2012; 56: 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K, Burt AD, Wright MC. Liver fibrosis. Biochem J 2008; 411: 1–18. [DOI] [PubMed] [Google Scholar]
- Tsukahara S, Ikeda R, Goto S, Yoshida K, Mitsumori R, Sakamoto Y et al. Tumour necrosis factor alpha-stimulated gene-6 inhibits osteoblastic differentiation of human mesenchymal stem cells induced by osteogenic differentiation medium and BMP-2. Biochem J 2006; 398: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner CM, Day AJ. TSG-6: a multifunctional protein associated with inflammation. J Cell Sci 2003; 116: 1863–1873. [DOI] [PubMed] [Google Scholar]
- Zhang R, Liu Y, Yan K, Chen L, Chen XR, Li P et al. Anti-inflammatory and immunomodulatory mechanisms of mesenchymal stem cell transplantation in experimental traumatic brain injury. J Neuroinflammation 2013; 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Kim DH, Ryu JS, Ko AY, Ko JH, Kim MK et al. Topical TSG-6 administration protects the ocular surface in two mouse models of inflammation-related dry eye. Invest Ophthalmol Vis Sci 2015; 56: 5175–5181. [DOI] [PubMed] [Google Scholar]
- Lee RH, Pulin AA, Seo MJ, Kota DJ, Ylostalo J, Larson BL et al. Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell 2009; 5: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lee JS, Hyun J, Kim J, Kim SU, Cha HJ et al. Tumor necrosis factor-inducible gene 6 promotes liver regeneration in mice with acute liver injury. Stem Cell Res Ther 2015; 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology 2008; 47: 1773–1785. [DOI] [PubMed] [Google Scholar]
- Madrigal-Matute J, Cuervo AM. Regulation of liver metabolism by autophagy. Gastroenterology 2016; 150: 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zois CE, Giatromanolaki A, Sivridis E, Papaiakovou M, Kainulainen H, Koukourakis MI. ‘Autophagic flux’ in normal mouse tissues: focus on endogenous LC3A processing. Autophagy 2011; 7: 1371–1378. [DOI] [PubMed] [Google Scholar]
- Czaja MJ, Ding WX, Donohue TM Jr, Friedman SL, Kim JS, Komatsu M et al. Functions of autophagy in normal and diseased liver. Autophagy 2013; 9: 1131–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 2011; 7: 673–682. [DOI] [PubMed] [Google Scholar]
- Shin JY, Park HJ, Kim HN, Oh SH, Bae JS, Ha HJ et al. Mesenchymal stem cells enhance autophagy and increase beta-amyloid clearance in Alzheimer disease models. Autophagy 2014; 10: 32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21: 1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Hou J, Wang X, Jiang R, Yin Y, Ji J et al. PTPRO-mediated autophagy prevents hepatosteatosis and tumorigenesis. Oncotarget 2015; 6: 9420–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, McGill MR, Chao X, Du K, Williams JA, Xie Y et al. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J Hepatol 2016; 65: 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y et al. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology 2014; 59: 1366–1380. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, McCullough RL, Sanz-Garcia C, Saikia P, Alkhouri N, Matloob A et al. Receptor interacting protein 3 protects mice from high-fat diet-induced liver injury. Hepatology 2016; 64: 1518–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MJ, Kim DI, Lim SK, Choi JH, Kim JC, Yoon KC et al. Thioredoxin-interacting protein mediates hepatic lipogenesis and inflammation via PRMT1 and PGC-1alpha regulation in vitro and in vivo. J Hepatol 2014; 61: 1151–1157. [DOI] [PubMed] [Google Scholar]
- Ning H, Sun Z, Liu Y, Liu L, Hao L, Ye Y et al. Insulin protects hepatic lipotoxicity by regulating ER stress through the PI3K/Akt/p53 Involved pathway independently of autophagy inhibition. Nutrients 2016; 8: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M et al. Autophagy regulates lipid metabolism. Nature 2009; 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams T, Yang SQ, Koteish A, Durnez A, DeVos R, Huang X et al. Oxidative stress and oval cell accumulation in mice and humans with alcoholic and nonalcoholic fatty liver disease. Am J Pathol 2003; 163: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhurst B, Croager EJ, Farley-Roche CA, Ong JK, Dumble ML, Knight B et al. A modified choline-deficient, ethionine-supplemented diet protocol effectively induces oval cells in mouse liver. Hepatology 2001; 34: 519–522. [DOI] [PubMed] [Google Scholar]
- Syn WK, Jung Y, Omenetti A, Abdelmalek M, Guy CD, Yang L et al. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology 2009; 137: e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Witek RP, Syn WK, Choi SS, Omenetti A, Premont R et al. Signals from dying hepatocytes trigger growth of liver progenitors. Gut 2010; 59: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Chen YS, Lin CC, Chen YJ, Lo GH, Lee PH et al. Amiodarone as an autophagy promoter reduces liver injury and enhances liver regeneration and survival in mice after partial hepatectomy. Sci Rep 2015; 5: 15807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima T, Shirabe K, Fukuhara T, Ikegami T, Yoshizumi T, Soejima Y et al. Suppression of autophagy during liver regeneration impairs energy charge and hepatocyte senescence in mice. Hepatology 2014; 60: 290–300. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol 2015; 17: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizaso A, Tan KT, Lee YH. Beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 2013; 9: 1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Hong IK, Kim BR, Shim SM, Sung Lee J, Lee HY et al. Activation of sphingosine kinase 2 by endoplasmic reticulum stress ameliorates hepatic steatosis and insulin resistance in mice. Hepatology 2015; 62: 135–146. [DOI] [PubMed] [Google Scholar]
- Moreira RK. Hepatic stellate cells and liver fibrosis. Arch Pathol Lab Med 2007; 131: 1728–1734. [DOI] [PubMed] [Google Scholar]
- Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest 2013; 123: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath SH, Levy O, Inamdar MS, Karp JM. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell 2012; 10: 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallat A, Lodder J, Teixeira-Clerc F, Moreau R, Codogno P, Lotersztajn S. Autophagy: a multifaceted partner in liver fibrosis. Biomed Res Int 2014; 2014: 869390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir M, Czaja MJ. Autophagy in nonalcoholic steatohepatitis. Expert Rev Gastroenterol Hepatol 2011; 5: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Elazar Z. Shedding light on mammalian microautophagy. Dev Cell 2011; 20: 1–2. [DOI] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 2010; 22: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology 2015; 61: 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I et al. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 2016; 7: 10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoen LF, Guimaraes EL, Grunsven LA. Autophagy: a new player in hepatic stellate cell activation. Autophagy 2012; 8: 126–128. [DOI] [PubMed] [Google Scholar]
- Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology 2012; 55: 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canbay A, Friedman S, Gores GJ. Apoptosis: the nexus of liver injury and fibrosis. Hepatology 2004; 39: 273–278. [DOI] [PubMed] [Google Scholar]
- Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab 2012; 15: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA 2012; 109: E3168–E3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder J, Denaes T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM et al. Macrophage autophagy protects against liver fibrosis in mice. Autophagy 2015; 11: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.